Abstract
The initial molecular events that lead to malignant transformation of the fimbria of the fallopian tube (FT) through high-grade serous ovarian carcinoma (HGSC) remain poorly understood. In this study, we report that increased expression of signal transducer and activator of transcription 3 (pSTAT3 Tyr705) and suppression or loss of protein inhibitor of activated STAT3 (PIAS3) in FT likely drive HGSC. We evaluated human tissues-benign normal FT, tubal peritoneal junction (TPJ), p53 signature fallopian tube tissue, tubal intraepithelial lesion in transition (TILT), serous tubal intraepithelial carcinoma (STIC) without ovarian cancer, and HGSC for expression of STAT3/PIAS3 (compared with their known TP53 signature) and their target proliferation genes. We observed constitutive activation of STAT3 and low levels or loss of PIAS3 in the TPJ, p53 signature, TILT, and STIC through advanced stage IV (HGSC) tissues. Elevated expression of pSTAT3 Tyr705 and decreased levels of PIAS3 appeared as early as TPJ and the trend continued until very advanced stage HGSC (compared to high PIAS3 and low pSTAT3 expression in normal benign FT). Exogenous expression of STAT3 in FT cells mediated translocation of pSTAT3 and c-Myc into the nucleus. In vivo experiments demonstrated that overexpression of STAT3 in fallopian tube secretory epithelial cells (FTSEC) promoted tumor progression and metastasis, mimicking the clinical disease observed in patients with HGSC. Thus we conclude that the STAT3 pathway plays a role in the development and progression of HGSC from its earliest pre-malignant states.
Keywords: Ovarian cancer, High-grade serous carcinoma, STAT3, p53 signature, STIC, fallopian tube, PIAS3
Introduction
High grade serous carcinoma (HGSC) of the ovary, peritoneum, or fallopian tube is the most lethal gynecologic malignancy in the United States. The majority of patients are diagnosed with advanced disease at presentation as symptoms are uncommon in early stage disease and there are no effective screening measures (1–4). Understanding the molecular processes involved in the initiation and progression of HGSC is a crucial step in improving our ability to detect disease at its earliest stages.
It is now widely believed that, in a majority of cases, cells in the fallopian tube give rise to HGSC (rather than the ovary itself) (5–9). It has been postulated that a proliferating intraepithelial neoplasm could exist in the distal fallopian tube for a period of time before spreading to nearby or distant structures. The tubal-peritoneal junction (TPJ) and secretory cell outgrowths (SCOUTS) are non-neoplastic and p53 negative. The p53 signature is characterized by strong p53 immunostaining and low proliferative index while tubal intraepithelial lesion (TILT) is positive for both p53 and Ki67. With further malignant transformation, serous tubal intraepithelial carcinoma (STIC) appears, which is considered the direct precursor to invasive HGSC (10–14)
Several critical molecular abnormalities, including mutations in tumor suppressor genes TP53 (15, 16), PTEN (17), and FOXO3A (18, 19) have been well documented in HGSC; however, the early molecular events remain elusive. There is emerging evidence for a role of the STAT3 signaling pathway in the development of HGSC. High STAT3 expression is present in more than 70% of human HGSC and has been associated with tumor progression and metastasis (20–22). PIAS3 is an E3-type small ubiquitin-like modifier (SUMO) ligase involved in regulating STAT3 signaling via inhibiting STAT3 DNA-binding and suppressing cell growth(23, 24), but the intracellular kinetics and trafficking of PIAS3 in the setting of solid tumors is still unknown. Negative regulation of STAT3 activity by PIAS3 is a well-documented pathway impacting cancer biology (25) and low PIAS3 expression has been linked to increased STAT3 activation in malignant mesothelioma coupled with poor patient survival (26). However, STAT3 activation and PIAS3 downregulation in the FT has never been addressed as a potential driver of the progression and development of ovarian cancer. There is no definitive evidence as to when and how the upregulation of STAT3 expression starts or if PIAS3 influences the activation of STAT3 directly or indirectly through upstream signaling pathways.
The primary goal of this study was to record the spectrum of molecular changes in the FT epithelium leading to the development of HGSC through STAT3 activation and low levels or loss of PIAS3. The secondary aim was to define and comprehend the role of STAT3 activation in carcinogenesis in the fallopian tube using STAT3/PIAS3 overexpression in FTSECs. The overarching hypothesis that we investigated was that activation of STAT3, under the influence of decreased levels of PIAS3 in FT epithelial cells, is an early event in the initiation of malignant transformation, and that these changes govern the transformation from STIC lesions to HGSC.
Materials & Methods
Cell-culture medium (RPMI 1640 and DMEM), fetal bovine serum (FBS), antibiotics, sodium pyruvate, trypsin, and phosphate-buffered saline (PBS) were purchased from Gibco (Grand Island, NY). Polyvinylidene fluoride (PVDF) membrane and molecular-weight markers were obtained from Bio-Rad (Hercules, CA). Antibodies, along with the seller information and dilutions, used in the current study have been listed in Sup. Table 1. Enhanced chemiluminescence (ECL) reagents were obtained from Amersham Pharmacia Biotech (GE Healthcare, Piscataway, NJ). All other reagents, of analytical grade or higher, were purchased from Sigma-Aldrich.
Patient Samples
Patient samples were obtained from biorepositories at both The Ohio State University Wexner Medical Center, Columbus, OH and Cedars Sinai Medical Center, LA, CA. Tissues collected in these biorepositories were from patients undergoing surgery at the respective institutions. In total we received 9 high grade serous ovarian cancers (HGSOC), 2 Tubal Peritoneal Junctions (TPJ), 3 p53 signatures, 3 STIC lesions, 1 TILT and 10 benign ovarian neoplasms. 9 HGSOC samples were homogenized in non-denaturing lysis buffer and subjected to western blot analysis as described earlier. The use of stored human tissues in this study was approved by the Institutional Review Board (IRB) of The Ohio State University Wexner Medical Center under Study Number: 2004C0124 and The Ohio State University’s OHRP Federalwide Assurance #00006378. No human subjects were directly consented for this study as the tissues were obtained from a biorepository.
Cell lines and culture
We have obtained immortalized FT33 cell lines from Dr. Ronny Drapkin (University of Pennsylvania) in early 2014. The FT33 cells are very well characterized and published (6, 27, 28). These are even available for purchase with companies like ABM and BioCat. We used these cells only for a short duration of about 4 months and we confirmed them for mycoplasma activity using ATCC® Universal Mycoplasma Detection Kit, every 2 months. Once the frozen cells were thawed, they were passaged for 5 times only and discarded thereafter and a fresh vial was thawed.
Immunocytochemistry (ICC)
Cells in RPMI medium were seeded onto sterile glass coverslips in 6-well plates with an average population of 50,000 cells/well. After 24 hours of culture the cells were washed, fixed, and incubated with primary antibody according to a previously described ICC protocol (29).
Immunoblot analysis
Cell lysates were prepared in non-denaturing lysis buffer as previously described and subjected to immunoblot analyses (30).
STAT3 overexpression
Non-cancerous FTSECs (FT33) as well as human ovarian surface epithelial cells (hOSE, used as control) were transfected using Lentivirus (Human, CMV) bearing STAT3 overexpression construct (pLenti-GIII-CMV) (abm Inc., Richmond BC, Canada, Cat. No. LVP323577, construct map and exact transfection procedure is described in Sup. Fig. 1). Changes in STAT3 target gene expression were measured using qPCR and Western blot along with migration and invasion in comparison to the normal, non-transfected FTSECs.
Ubiquitin assay
To trace the ubiquitinated proteins in the cell lysates, agarose beads (Thermo Fisher Scientific, Cat. No. 78601) coated with domains having affinity to ubiquitin were incubated in the lysates at 4°C for 2 hours. After washing the beads, the ubiquitinated proteins were subjected to immunoblot for PIAS3 and blotted by the ubiquitin antibody.
Immunohistochemistry (IHC)
Fresh-frozen tissue embedded in OCT was sectioned at 5 μm and fixed in phosphate-buffered 4% paraformaldehyde and washed in PBS. For studies on formalin-fixed paraffin-embedded tissues, 5 mm sections were deparaffinized and hydrated, and antigen epitope retrieval was performed by boiling slides in 10 mmol/L sodium citrate buffer (pH 8.5) at 80°C for 20 minutes. Endogenous peroxidase activity was blocked by incubation in 3% H2O2, followed by blocking of nonspecific sites with SuperBlock blocking buffer (Pierce). Sections were then incubated with primary antibody overnight at 4°C. After washing in PBS, antibody binding was localized with appropriate secondary antibodies. Nuclei were counter-stained with 6-diamidino-2-phenylindole dihydrochloride hydrate (DAPI). The number of cells staining positive was counted by a blinded observer in 5 random 40x fields and compared by the use of Student t-test. Images were obtained with an Olympus AX70 fluorescence microscope and Spot v2.2.2 (Diagnostic Instruments, Sterling Heights, MI) digital imaging system.
Strong expression of p53 in >75% of at least 12 epithelial cells (with or without intervening ciliated cells) was considered p53(+). Foci showing a Ki-67 labeling index ≥20% were considered “Ki-67 high” since normal tubal mucosa typically has a labeling index <2%, whereas a Ki-67 proliferation index of <10% was considered “low”.
Cell migration and invasion Assay
Cell migration assays were performed on both control FT33 and FT33 cells overexpressing STAT3 using a previously-described wound-healing method (31) and quantified using freehand selection option on Image J software.
Development of orthotopic tumor model
Cultured FTSECs (control FT33 or FT33 overexpressing STAT3; n=6 for each group) were injected into the ovarian bursa (3 × 106 cells in 100 μL of PBS) of 6-week-old BALB/c nude mice from the OSU Transgenic Mice Core Lab. In vivo MRI imaging was done periodically to check upon the tumor growth. After sacrifice, the tumors were weighed in order to get tumor weight and metastases sites were counted in the form of number of nodules. The tumor tissues were then subjected to immunoblot analysis and histopathology experiments. Some of the tumor tissues were snap frozen in liquid nitrogen and stored at −80°C for the Real time quantitative PCR.
RNA isolation and Reverse Transcription PCR (RT PCR)
Freshly excised ovarian tumor tissues from FT OE mice (injected with FT33 cells overexpressing STAT3) were snap frozen in liquid nitrogen and stored at −80°C in appropriately labelled vials. Since the control mice injected with normal FT33 cells did not develop any tumor(s), normal ovaries from 2 mice were combined for extracting RNA for a total of n=3 (6 mice). At the conclusion of the experimental protocol, total RNA was isolated using the RNeasy Mini Kit (Qiagen, Valencia, CA). RNA samples with an optical density A260/A280 ratio between 1.8 and 2.1 were used. RT-PCR was then performed using the Transcriptor First Strand Complementary DNA (cDNA) Synthesis Kit (Roche Applied Science) to synthesis cDNA. RT-PCR was performed with 1μg of RNA template. The reaction was carried out using the Veriti Thermal Cycler (Applied Biosystems, Carlsbad, CA) and random hexamer primers. The real time quantitative PCR was performed with gene specific primers designed for human GAPDH, STAT3, c-myc, cyc D1, PIAS3 and STIP1 (primer sequences used are listed in Sup. Table 2) and SYBR green mix; ordered from Sigma –Aldrich. Each sample was normalized to the control gene glyceraldehyde 3-phosphate dehydrogenase (GAPDH).
Statistical Analysis
Results were expressed as mean ± S.E. Comparisons between groups were made by the Student t-test for all the graphs. The significance level was set at p ≤ 0.05.
Results
Identification of pSTAT3 Tyr705 and PIAS3 expression in dysplastic fallopian tube epithelium and HGSC
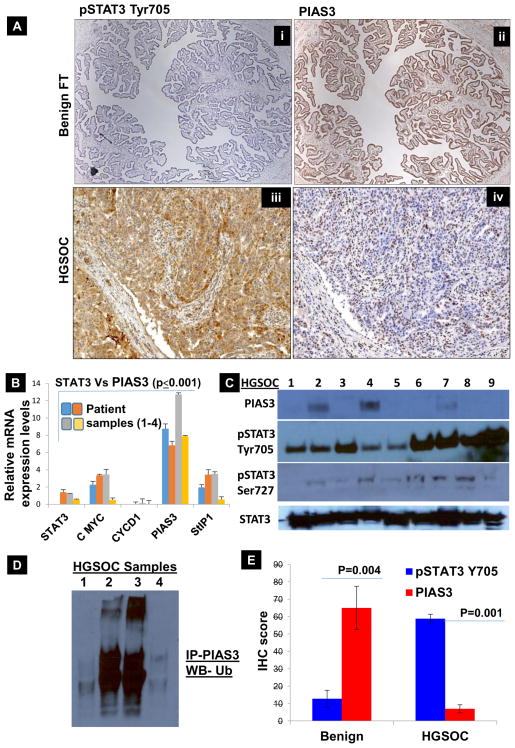
To determine the role of varied expression patterns of pSTAT3 Tyr705 and PIAS3 in the progression of HGSOC, we analyzed their expression patterns in consecutive sections from: A) normal, benign human fallopian tube and B) stage IV ovarian HGSC. We found that the normal FT tissue sample lacks pSTAT3 Tyr705 expression (absence of brown-colored stain, Fig. 1A–i, and higher magnification images in Sup. Fig. 2A&B) but has a high PIAS3 expression (as detected by the abundance of brown stain, Fig. 1A–ii). Stage IV HGSOC patient samples showed reverse expression patterns: high pSTAT3 Tyr705 (Fig. 1A–iii) and low PIAS3 (Fig. 1A–iv, higher magnification images in Sup. Fig. 3A&B). In order to confirm these results at the level of mRNA we proceeded with extracting RNA from FFPE ribbons of FT tissues, from 4 different patients, which was converted to cDNA and subjected to real-time PCR. Fig. 1B & Sup. Fig. 4 shows that the relative expression of PIAS3 is 2 to 4 fold higher in the normal FT tissue compared to STAT3, c-Myc, StIP1 and Cyclin D1, which are present at lower levels (p≤0.001). Thus, this confirms that increasing expression of pSTAT3 Tyr705 couples with diminishing levels of PIAS3 during the progression of HGSOC. Therefore, we can conclude that constitutive STAT3 activation and low PIAS3 expression might impart malignant characteristics to untransformed FTSECs through altering the STAT3 target genes such as c-Myc and Cyclin D1.
Fig. 1. Characterization of pSTAT3 Tyr705 and PIAS3 expression in benign fallopian tube tissue (Benign FT) and tissue from Grade IV HGSOC patient.
(A) pSTAT3 Tyr705/PIAS3 IHC scoring: benign fallopian tube tissues are characterized by the absence of pSTAT3 Tyr705 (A–i); HGSOC tissue samples are characterized by overexpression of pSTAT3 Tyr705 (A–iii). In contrast, benign fallopian tube tissues show marked expression of PIAS3 in 90–100% of cells (A–ii) whereas only <30% of HGSOC cells express PIAS3 (A–iv). This inverse relationship in pSTAT3 Tyr705 and PIAS3 holds true throughout our results. (B) In order to analyze the expression of PIAS3 along with pSTAT3 Tyr705 and its associated genes at the level of mRNA, we proceeded with extracting RNA from FFPE ribbons of benign FT tissues (4 different patients), which was converted to cDNA and subjected to real time PCR. Figure 1B displays the relative expression of PIAS3 is 2 to 4 folds higher in the benign FT tissue as compared to STAT3, c-Myc, StIP1 and Cyclin D1, which are present at lower levels (p≤0.005). (C) Protein lysates from 9 HGSOC tissues collected from consented patients was subjected to Western Blot analyses and probed with PIAS3, pSTAT3 Tyr705, pSTAT3 Ser727 and total STAT3. Only 3 patients show low expression of PIAS3 and it is entirely absent from the rest of the samples. There is very high pSTAT3 Tyr705 and total STAT3 expression in all the 9 samples, while pSTAT3 Ser727 is rather low. (D) To analyze if the proteasomal-mediated degradation is involved in lowering the levels of PIAS3 in HGSOC tissues, HGSOC tissue lysates were immunoprecipitated (IP) with PIAS3 antibody and immunoblotted (IB) using Ubiquitin. A clear increase in ubiquitination of PIAS3 was evident. (E) Additional scoring of PIAS3 and pSTAT3 Tyr705 expression was completed using IHC in 4 samples. PIAS3 scores were higher for all the benign FT tissues and low for the HGSOC tissues. On the contrary, pSTAT3 Tyr705 scored 5–15% for the benign FT tissues and >70% for the HGSOC tissues. It is noteworthy that there is an inverse relationship between pSTAT3 Tyr705 and PIAS3, which is shown in both mRNA and protein expression levels in both benign fallopian tube tissues and HGSOC. There was a trend of high PIAS3 levels and low pSTAT3 Tyr705 levels in benign tissues, while the inverse was true for HGSOC tissues.
Protein lysates from 9 HGSOC tissue samples were subjected to Western blot analyses and probed with PIAS3, pSTAT3 Tyr705, pSTAT3 Ser727, and total STAT3 (Fig. 1C). Only 2 samples show very low expression of PIAS3; otherwise, PIAS3 is absent from the remaining samples. There is very high pSTAT3 Tyr705 and total STAT3 expression in all 9 samples, but pSTAT3 Ser727 is rather low. This further shows that the expression profiles for pSTAT3 Tyr705 and PIAS3 observed in IHC are consistent with the expression seen at the RNA and protein levels.
It is well-known that many transcription factors are ubiquitinated and degraded by the proteasome. In fact, in many cases, transcriptional activation domains and signals for Ub conjugation directly overlap. PIAS3 is also known to be tightly regulated by the ubiquitin-dependent proteolytic pathway (12). Thus, we aimed to study if proteasome mediated degradation is involved in lowering the levels of PIAS3 in HGSC (Fig. 1D). When HGSC tissue lysates were immunoprecipitated with PIAS3 antibody and immunoblotted using ubiquitin, an increase in ubiquitination of PIAS3 was evident in at least half of the samples, suggesting that Ub may play a role in downregulating PIAS3 in HGSC. Additionally, we scored PIAS3 and pSTAT3 Tyr705 expression using IHC in 9 other samples of normal benign FT tissues as well as 9 HGSOC tissues. PIAS3 scores were higher (90–100%) for all the benign normal FT tissues and very low (<30%) for the HGSOC tissues (Fig. 1E). On the contrary, pSTAT3 Tyr705 scored 5–15% for the normal FT tissues and >70% for the HGSOC tissues.
Expression patterns of pSTAT3 Tyr705/PIAS3 from tubal peritoneal junction (TPJ) through STIC region
Non-neoplastic changes in the junctional epithelia
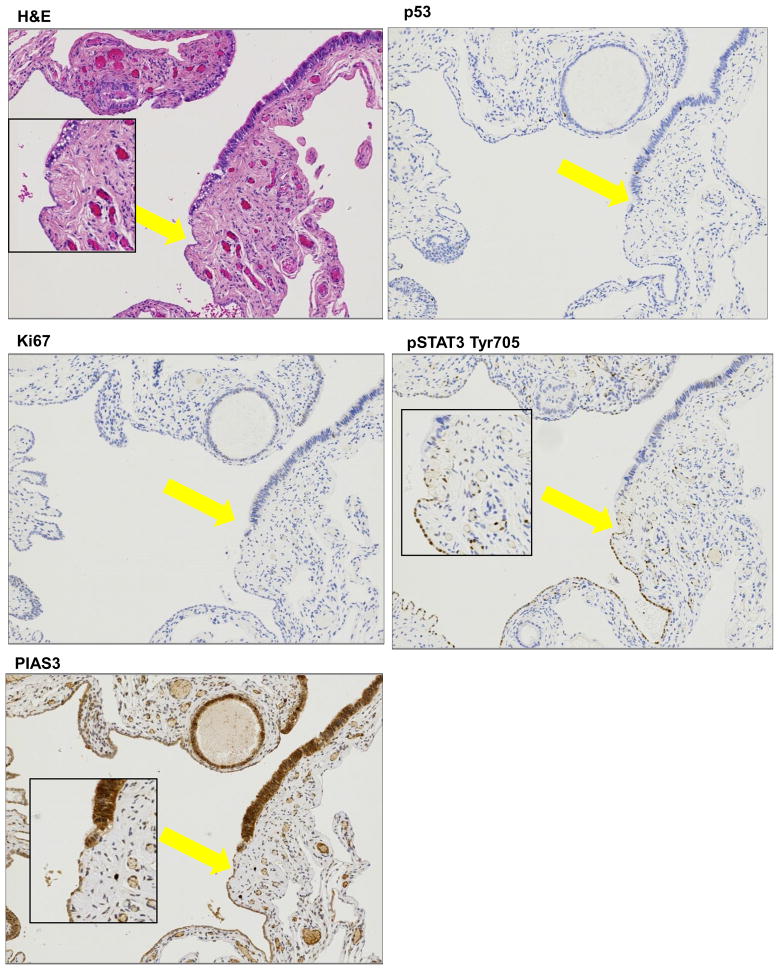
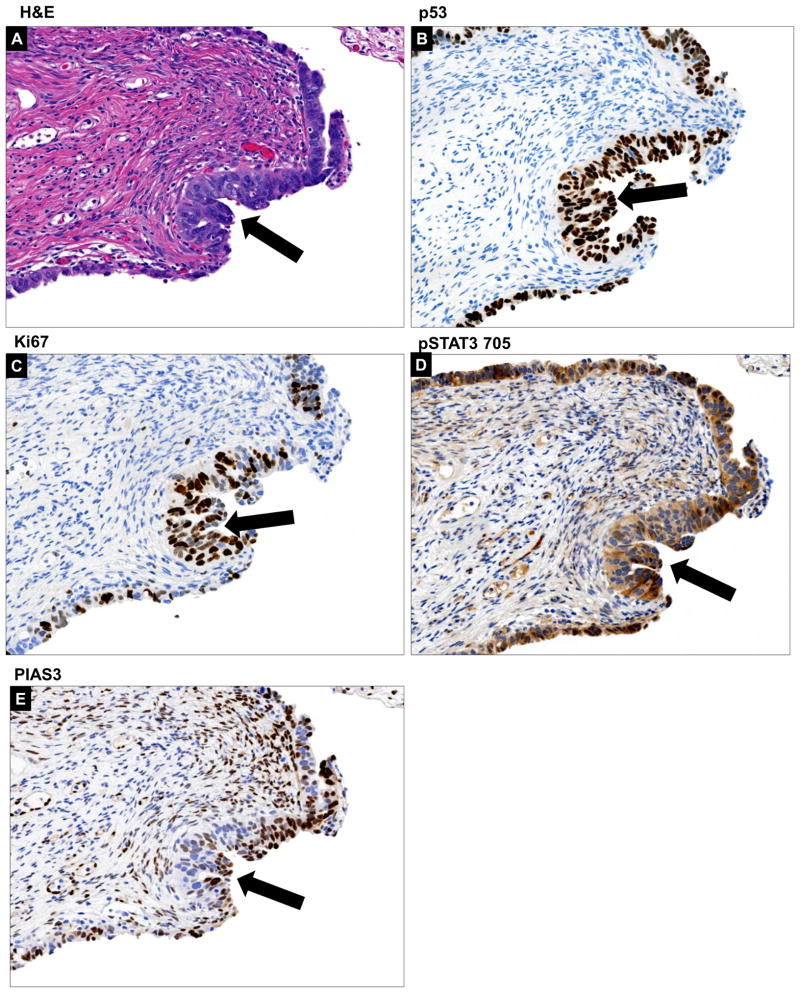
One of the most widely accepted theories regarding the evolution and spread of HGSOC considers that the junction of the fallopian tube epithelium with the mesothelium of the tubal serosa, termed the “tubal peritoneal junction” (TPJ), undergoes malignant transformation due its location, and can eventually metastasize to the nearby ovary and surrounding pelvic peritoneum. 2 separate TPJ sections were subjected to IHC with pSTAT3 Tyr705, PIAS3, Ki67 and p53 antibodies. As is evident from the H&E stain in Fig. 2A and Supp. Fig. 5 (additional TPJ from a different patient), TPJ is morphologically characterized by an abrupt transition from the tubal type serous epithelium to the flat mesothelium and physiologically is reported to have a novel stem cell niche prone to malignant transformation. We detected no p53 or Ki67 staining in any of the cells (Figs. 2B & C). Interestingly, pSTAT3 Tyr705 expression was also absent except for the TPJ where the flat mesothelial cells showed an abrupt positive signal for pSTAT3 Tyr705 (Fig. 2D). High PIAS3 staining was present in most of the cells but the cells which seemed to have gained pSTAT3 Tyr705 had less PIAS3 staining (Fig. 2E). It may be possible that even before the FT cells gain the p53 signature and Ki67 staining, they start losing PIAS3 coupled with a corresponding gain of pSTAT3 Tyr705 in the same region.
Figure 2. Histopathology of Distal fallopian tube showing Tubal Peritoneal Junction (TPJ).
(A) TPJ is morphologically characterized by an abrupt transition from the tubal type serous epithelium to the flat mesothelium. Normal surface tubal epithelium with ciliated cells is interrupted by patches of non –ciliated cells (H&E stain). (B) P53 staining is not markedly increased and proliferation (Ki67) is not significantly elevated (C). pSTAT3 Tyr705 staining is absent in most of the cells except an abrupt small patch (yellow arrow marks this area alonwith an inset picture) (D) PIAS3 staining is positive and intact but is again interrupted by the cells which seem to have lost PIAS3 (E) (marked by yellow arrow and inset picture).
Putative precursor lesions in earlier stages of serous carcinogenesis
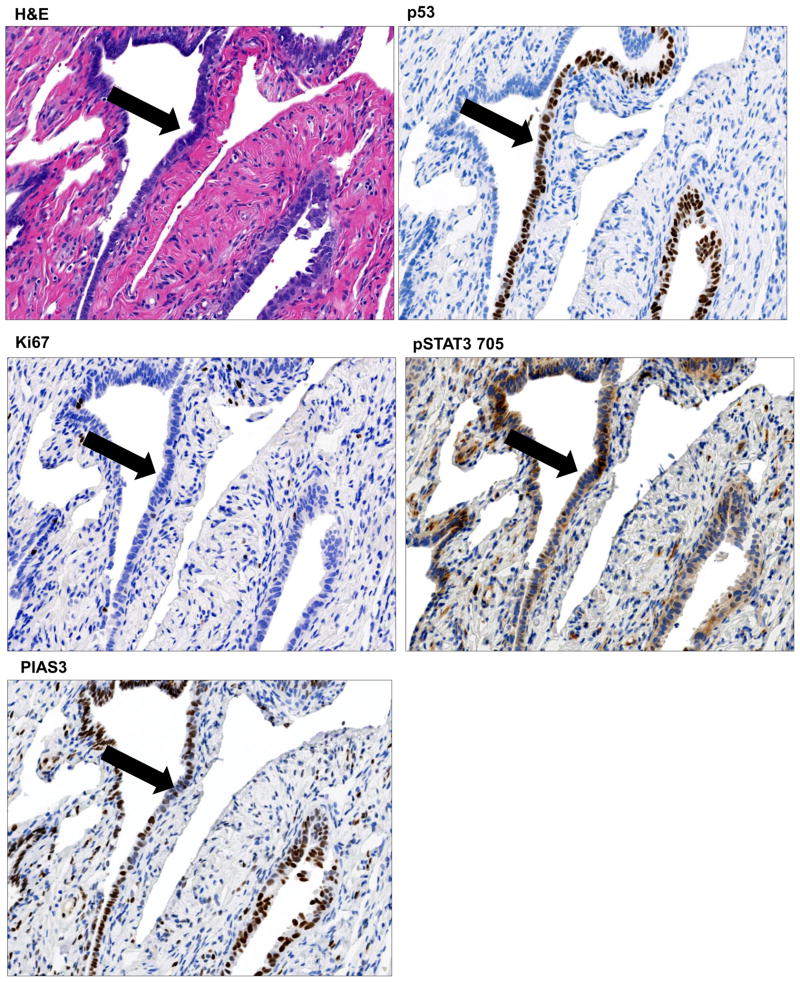
The biological and clinical significance of p53 signature and tubal intraepithelial lesions in transition (TILT) is yet to be well-delineated. We were able to identify a p53 signature in a FT section which also displayed a STIC lesion. P53 signature was classified as discrete cells which are morphologically indiscernible from the surrounding cells (Fig. 3A, additional p53 signature sample in Suppl. Fig. 6A & B) seen as a segment of linear p53-staining (Fig. 3B). The proliferative fraction (Ki67, Fig. 3C) was consistently lower in this section and the pSTAT3 Tyr705 expression was still moderate (Fig. 3D) coupled with low PIAS3 expression (Fig. 3E).
Figure 3. Histopathology of p53 signature.
We were able to identify a p53 signature in a discrete row of cells which are morphologically indiscernible from the surrounding cells as is evident from the H&E (A) and p53 staining (B). The proliferative fraction (Ki67) (C) was consistently lower. The pSTAT3 Tyr705 expression was still moderate (D) coupled with low PIAS3 expression (E). Histopathology of tubal intraepithelial lesions in transition (TILT). The H&E for TILT displays moderate nuclear atypia, intermediate N/C ratio, and distinct nucleoli, Tubal epithelia showed nuclear enlargement and molding, some stratification, mild nuclear enlargement, polarity loss, and prominent nucleoli (F). A short stretch of strong p53 expression (G) and <25% of Ki67 expression (Ki67 low) (H) was observed. Further there was an abundance of pSTAT3 Tyr705 expression (I) and relatively low but noticeable PIAS3 expression (J). All areas of interest are marked by a yellow arrow.
The term TILT has been used to describe a spectrum of epithelial changes ranging from normal-appearing tubal epithelium expressing p53, to lesions with increasing degrees of cytologic atypia that fall short of a STIC lesion. On the H&E stain, the tubal epithelial cells showed moderate nuclear atypia, intermediate N/C ratio, and prominent nucleoli (Fig. 3F); additionally, this area had strong p53 expression (Fig. 3G) and <25% of Ki67 expression (Fig. 3H). Further, there was abundance of pSTAT3 Tyr705 expression (Fig. 3I) and relatively low, but noticeable, PIAS3 expression (Fig. 3J, lower magnification image in Suppl. Fig. 7).
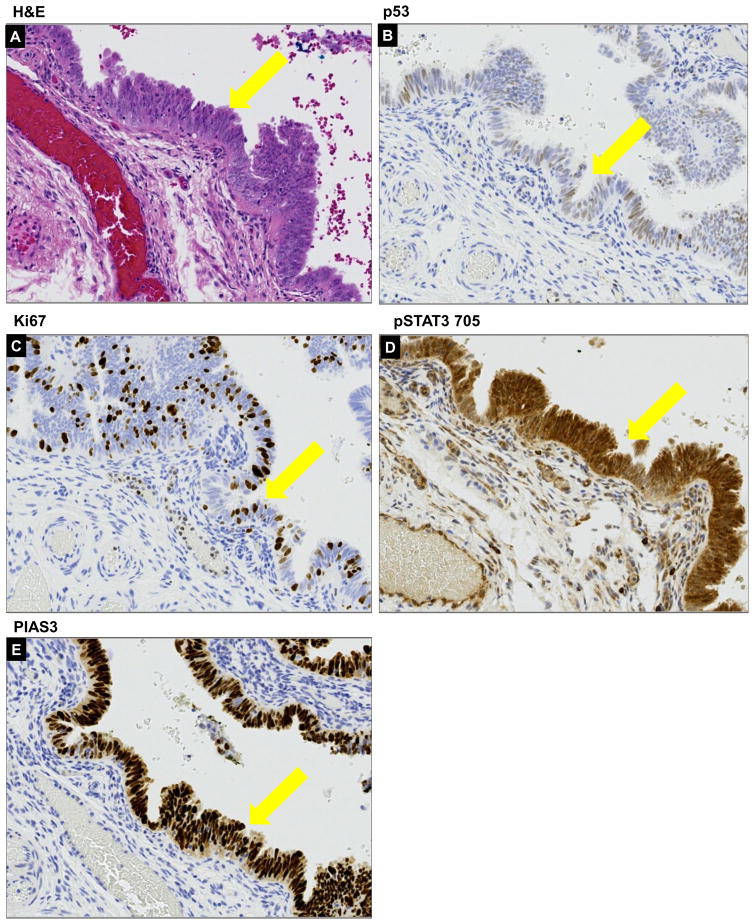
The STIC lesion is widely accepted as the immediate precursor of invasive HGSC. In our samples, the STIC lesion displayed morphological stratification (Fig. 4A) associated with diffuse moderate nuclear p53 expression in 25–30% of cells within the lesion (Fig. 4B) and moderate Ki-67 index (15–20%, Fig. 4C). pSTAT3 Tyr705 expression was high (Fig. 4D) and the PIAS3 expression was also still noticeable at this stage (Fig. 4E).
Figure 4. Histopathology of a STIC lesion.
The STIC lesion from our sample displayed morphological stratification manifested by H&E (A) associated with diffuse nuclear moderate to strong p53 expression in >75% of cells (B) within lesion and moderate Ki67 index (≥10%) (C). pSTAT3 Tyr705 expression was high (D) and the PIAS3 expression was also still noticeable at this stage (E). All areas of interest are marked by a yellow arrow.
The effects of perturbed STAT3/PIAS3 expression in secretory fallopian tube epithelial cells and roles in cell transformation leading to HGSC
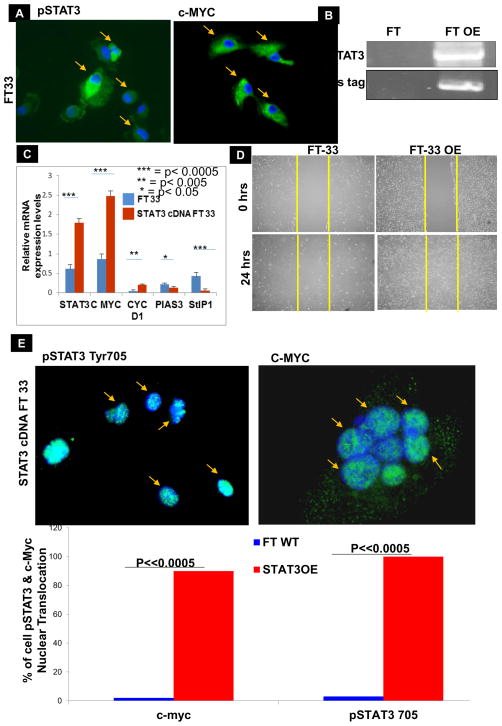
We then sought to determine whether the activation of STAT3 occurs in FTSECs through the depletion of PIAS3 and whether STAT3 activation is associated with the initiation of molecular changes that lead to the development of HGSC. We started by analyzing the expression of pSTAT3 Tyr705 and its target genes by immunocytochemistry (ICC) and found that pSTAT3 Tyr705 and c-Myc were present in normal FT cells (FT-33, Fig. 5A) as shown by the green fluorescence and a blue nucleus. We further performed in-depth in vitro studies using STAT3 overexpression strategy in FTSECs by transfecting with a mammalian STAT3 overexpression vector, which was confirmed by Western blot (Fig. 5B). RT-qPCR revealed that overexpression of STAT3 significantly increased the expression levels of c-Myc and Cyclin D1 and decreased the PIAS3 expression in FT33 cells at the mRNA level (Fig. 5C). Overexpression of STAT3 in FT-33 cells markedly increased migration as compared to the control un-transfected FT33 cells (Fig. 5D, Suppl. Fig. 8). On an intracellular level, FT cells overexpressing STAT3 displayed a more nuclear presentation for both pSTAT3 Tyr705 and c-Myc (green fluorescence for the genes against the blue nuclear background, Fig. 5E, 5F).
Figure 5. Effect of STAT3 overexpression in FT33 cells in vitro:
(A) ICC of normal FT 33 cells displayed pSTAT3 Tyr705 expressions in the cytoplasm of the cells as evident by the green fluorescence. c-MYC was also present in the cytoplasm of a few cells. All the cells of interest are marked by yellow arrows. (B) Confirmation of STAT3 overexpression in FT33 cells using a Western Blot where FT is control and FT OE is STAT3 overexpression. The bottom panel is his-tag expression which is tagged to STAT3 gene in the plasmid. In order to create STAT3 overexpression cells, FT33 cells were transfected with a vector harboring STAT3 gene or with the empty vector backbone for control. (C) Reverse Transcription qualitative PCR confirmed the relative expression of STAT3 and associated genes using RNA extracted from normal fallopian tube cells (FT) and fallopian tube cells overexpressing STAT3 (FTOE). The RNA was converted to cDNA and amplified with gene specific primers; relative expression of STAT3, c-Myc and cyclin D1 goes up while PIAS3 and StIP1 go down (values normalized to GAPDH; ***p<0.0005, ** p<0.005, *p<0.05). Immunocytochemistry (ICC) of control FT 33 and FT 33 STAT3 OE cells with pSTAT3 Tyr705 (left) and c-myc (right). (D) Quantification of scratch assay showing percent migration of control FT33 cells in comparison to FT33 cells overexpressing STAT3. (E & F) On an intracellular level, FT cells overexpressing STAT3 displayed a more nuclear presentation for both pSTAT3 Tyr705 and c-Myc (green fluorescence for the genes against the blue nuclear background).
Elucidating the role of STAT3/PIAS3 in vivo in the initiation and progression of ovarian HGSC
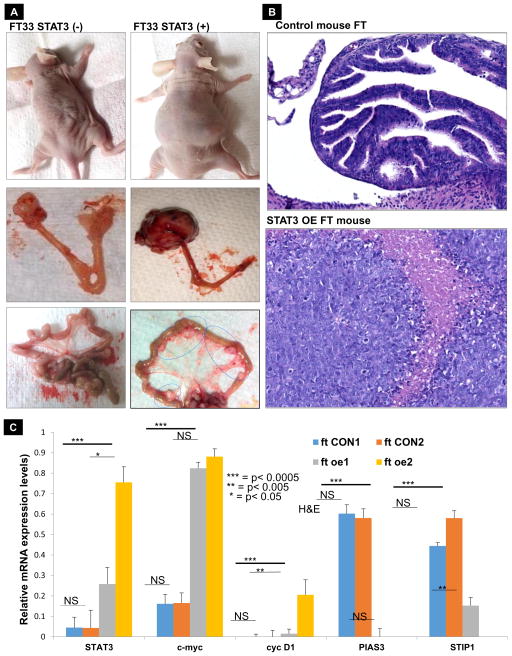
In order to validate our in vitro results at an in vivo level, we injected the ovarian bursa of mice with control FT cells and FT cells overexpressing STAT3; these mice were then sacrificed 4 weeks post inoculation (hOSE cells overexpressing STAT3 or normal hOSE cells were also injected in another set of mice as controls). The size of the primary tumor was significantly bigger in mice injected with FT cells overexpressing STAT3 as compared to the mice injected with normal FT cells which showed no tumor formation. The former also displayed widespread peritoneal metastasis (Fig. 6A) thereby confirming the in vitro results for STAT3 overexpression in normal FT cells. Tumor weight, as well as the number of metastatic nodules, was significantly higher in the mice injected with FT33OE cells (Suppl. Fig. 9A). Pathology of mouse ovarian tissue samples obtained from mice injected with control FT33 cells was observed using hematoxylin and eosin staining and showed normal cell morphology and structure (Fig. 6B top panel). Further, a higher PIAS3 expression and no pSTAT3 Tyr705 expression were seen for the control mice (Suppl. Fig. 9B). H&E staining of tumor tissue for mice injected with FT33 OE cells showed invasive cancer spanning the ovary and beyond (Fig. 6B, bottom panel). RT-qPCR of the organs collected from mice of both the groups also showed increased STAT3, c-Myc and cyclin-D1; decreased PIAS3 and STIP1 was seen for the mice injected with STAT3-overexpressing FT (Fig. 6C). In order to further demonstrate that the effects of STAT3 overexpression, which we observed, is lineage specific, we used transfected hOSE cells overexpressing STAT3 as well as control hOSE cells to inject in mice. In the hOSE cells overexpressing STAT3 did not show tumors or even any significant increase in ovary weight (Suppl. Fig. 10A&B). These results suggest that STAT3 overexpression in the FT plays a key role in the HGSC initiation and progression in mice. This will provide an understanding of the opportunities for disease prevention through targeting STAT3 activation in women at risk for HGSC.
Figure 6. STAT3 overexpression in FT-33 cells induces tumor growth and metastasis in orthotopic mouse.
(A) BALB/c nude mice were injected into the ovarian bursa with FT33 cells isolated and immortalized from FTSEC (left column) and FT33 cells overexpressing STAT3 (right column). As is evident, the STAT3 overexpression in FT33 cells causes the mice to develop bulky tumors in the ovary which metastasized to the omentum (indicated by arrow). Normal FT33 cells did not induce any tumor/ metastasis in the mice. (B) Pathology of mouse ovarian tissue samples obtained from ovary of mice injected with control FT cells (top panel) or ovarian tumor of mice injected with FT33 cells overexpressing STAT3 (bottom panel), observed using hematoxylin and eosin staining. (C) Tissues were collected from mice injected with FT33 control /FT33 STAT3 overexpressing cells 5 weeks post inoculation. The quantitative relative expression of STAT3 and regulatory genes in control, untransformed FT33 mice tissue (ft CON1 and ft CON2) and FT33 overexpressing mice tissues (ft oe1 and ft oe2) display an increase in STAT3, c-Myc and cyclin D1 and decrease in PIAS3 and StIP1 for both the mice replicates (p<0.005).
DISCUSSION
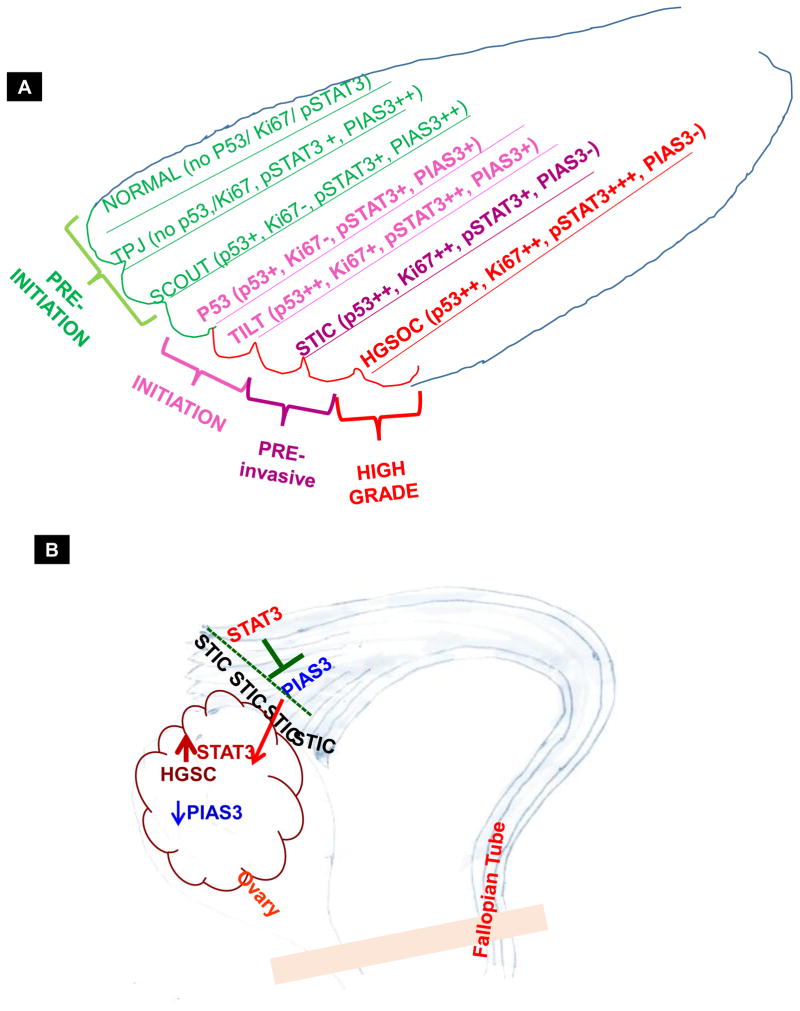
This study strongly supports the theory that HGSC originates in the fallopian tube and also suggests that the role of precursor lesions merits closer attention. Detection of STIC lesions may occur at the time of risk-reducing salpingo-oophorectomy or salpingectomy for other benign indications, however frequently it is only detected during pathologic examination when an invasive cancer has been diagnosed. Understanding the earliest gene expression pattern changes in HGSC may allow for early detection strategies to prevent progression to and/or death from this disease (32–34). For this reason, we aimed to reveal the expression pattern of pSTAT3 Tyr705 in human tissue samples ranging from benign fallopian tube samples through TPJ, TILT, p53 signature, STIC, and HGSC (Fig. 7, Sup. Fig. 11 & Sup. Table 3). In our earlier studies, we found a high expression of pSTAT3 Tyr 705 in various HGSOC tumor samples as well as cell lines (21, 35, 36). Therefore, with evidence that pSTAT3 Tyr705 is overexpressed in advanced HGSC, we considered it imperative to try to analyze this further in the pre-malignant tissues and determine the time point where pSTAT3 Tyr705 expression levels rise and PIAS3 diminishes. Using various pre-initiation, initiation, pre invasive and invasive stages of HGSC that were available to us, we found that the normal FT tissue lacks pSTAT3 Tyr705 expression and has very high PIAS3 expression. Interestingly, we observed the first signs of pSTAT3 Tyr705 expression, albeit low levels, in the TPJ which perfectly matches the diminished PIAS3 in the same area in consecutive sections. Therefore, we hypothesize that in the HGSC carcinogenesis process, expression of pSTAT3 Tyr705 may be one of the first molecular changes complemented by an abrupt disappearance of PIAS3. This may occur even before the area undergoes mutations in TP53 or begins the process of rapid division and display of strong Ki67 staining. This was reaffirmed in the TILT region where p53-positive patches of cells perfectly blended with a high pSTAT3 Tyr705 staining and low PIAS3 stain. This trend of increasing pSTAT3 Tyr705 and simultaneously declining PIAS3 continued in the p53 signature samples, STIC lesions, and the HGSC samples. It is likely that this disturbance in the STAT3-PIAS3 continuum initiates a cascade of molecular changes that allow cells to survive in the presence of DNA damage and oncogenic activation without undergoing senescence or apoptosis. Overexpression of PIAS3 suppresses cell growth and restores the drug sensitivity of human lung cancer cells in association with PI3-K/Akt inactivation (23). Loss of PIAS3 protein expression has been linked with an increase in STAT3 phosphorylation and activity in glioblastoma multiform (37). Dabir et al (38) also reported the role of phosphorylation at Tyr705 of STAT3 in the formation and intracellular shuffling of the PIAS3-STAT3 complex in a lung cancer model as well as the importance of STAT3 tyrosine phosphorylation site for this association. The association and nuclear translocation of the PIAS3-STAT3 complex is ligand and time dependent.
Figure 7.
Schematic showing the hypothesized pSTAT3 Tyr705/PIAS3 expression patterns at the latent precursor stages of HGSC within the fallopian tube.
It is well known that the ovarian surface epithelium, which is of mesothelial origin, displays gene expression profiles which are different from the fallopian tube epithelium, which is predominantly Mullerian-derived (39). Thus, it was vital to validate whether the overexpression of pSTAT3 Tyr705 and downregulation of PIAS3 genes observed in the IHC of tissues are indeed upregulated in the fallopian tube epithelium cells as well, rather than just the adjacent ovarian surface epithelium. Our STAT3 overexpression studies in FT cells in vitro as well as in vivo, further proves that our findings are tissue lineage specific rather than tumor specific (Suppl. Fig. 10). These findings further endorse the function of the pSTAT3 Tyr705-PIAS3 continuum in the pathogenesis of early HGSC.
Clinical studies have found that nuclear localization of activated STAT3 occurs in more than 70% of human HGSC and is associated with tumor progression and decreased survival (21, 40, 41). Another study compared the genetic profiles in the epithelium of the fallopian tubes from BRCA1 mutation carriers to those with a low-risk epithelium and showed nuclear accumulation of pSTAT3 Tyr705 in FT epithelium and HGSC tumor samples (15, 42). The mechanisms by which STAT3 activation in FTSECs leads to HGSC is not yet understood. Our current study strengthens the evidence for a connection between increased pSTAT3 Tyr705 expression and low levels of PIAS3 expression in the fimbria of fallopian tubes and ovarian HGSC. These data suggest a role for STAT3/PIAS3 in the development of pelvic HGSC from the fallopian tube. Other studies have also found that PIAS3 downregulation is closely correlated with STAT3 activation in adenocarcinomas of the uterine cervix, lung and glioblastoma (43–45).
Based on our results, we suggest that in addition to p53 mutations (and excessive accumulation of mutated p53) the precursor lesions to HGSC also overexpress STAT3 and lose PIAS3. These alterations may be clinically relevant as STAT3 activation and/PIAS3 suppression or loss may be utilized to identify women with premalignant changes or early stage HGSC. In order to map the wide gap between the molecular events associated with the earliest stages of serous carcinogenesis to its advanced ovarian cancer form, we need to contemplate other genes which could prove as biomarkers at an earlier stage. We acknowledge that our results are hypothesis generating and expansion of these findings in additional samples is critical. In addition, the lack of a precise pathogenetic sub-classification of this unique subset of serous carcinomas necessitates confirmation of our findings in future studies. Such investigations could aim to better understand whether activated STAT3 and low or absent PIAS3 play a role in early tubal secretory cell transformation and development of fallopian tube–derived HGSC, and to elucidate the role that STAT3 and PIAS3 play in the initiation and early tumorigenesis of HGSC in vivo. Arriving at a better understanding of the early events in the pathogenesis of HGSCs from the fallopian tube will improve our chances of detecting and intervening on such lesions before they become advanced and life-threatening.
Supplementary Material
Figure 8.
Acknowledgments
The authors are thankful to the GYN/Oncology Residency Fellow Dr. Brent Smith, MD, undergraduate students Roman Zingarelli, Riley Maria, John Fowler for the cell culture, basic assay help and Jason Shawn Scully for IHC pictures. Financial Support: This work was funded by NCI RO1-CA176078 (K. Selvendiran and D.E. Cohn), an internal Ohio State University Comprehensive Cancer Center grant and the Beucler Family Fund (D.E. Cohn).
Footnotes
Disclosure of Potential Conflicts of Interest: No potential conflicts of interest were disclosed.
References
- 1.Bartlett TE, Chindera K, McDermott J, Breeze CE, Cooke WR, Jones A, et al. Epigenetic reprogramming of fallopian tube fimbriae in BRCA mutation carriers defines early ovarian cancer evolution. Nature communications. 2016;7:11620. doi: 10.1038/ncomms11620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bast RC, Jr, Hennessy B, Mills GB. The biology of ovarian cancer: new opportunities for translation. Nature reviews Cancer. 2009;9:415–28. doi: 10.1038/nrc2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.SEER Stat Fact Sheets: Ovary Cancer. 2015 [Google Scholar]
- 4.Sarojini S, Tamir A, Lim H, Li S, Zhang S, Goy A, et al. Early detection biomarkers for ovarian cancer. Journal of oncology. 2012;2012:709049. doi: 10.1155/2012/709049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Karst AM, Levanon K, Drapkin R. Modeling high-grade serous ovarian carcinogenesis from the fallopian tube. Proc Natl Acad Sci U S A. 2011;108:7547–52. doi: 10.1073/pnas.1017300108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Perets R, Wyant GA, Muto KW, Bijron JG, Poole BB, Chin KT, et al. Transformation of the fallopian tube secretory epithelium leads to high-grade serous ovarian cancer in Brca;Tp53;Pten models. Cancer Cell. 2013;24:751–65. doi: 10.1016/j.ccr.2013.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eckert MA, Pan S, Hernandez KM, Loth RM, Andrade J, Volchenboum SL, et al. Genomics of Ovarian Cancer Progression Reveals Diverse Metastatic Trajectories Including Intraepithelial Metastasis to the Fallopian Tube. Cancer discovery. 2016;6:1342–51. doi: 10.1158/2159-8290.CD-16-0607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vang R, Shih Ie M, Kurman RJ. Fallopian tube precursors of ovarian low- and high-grade serous neoplasms. Histopathology. 2013;62:44–58. doi: 10.1111/his.12046. [DOI] [PubMed] [Google Scholar]
- 9.Ducie J, Dao F, Considine M, Olvera N, Shaw PA, Kurman RJ, et al. Molecular analysis of high-grade serous ovarian carcinoma with and without associated serous tubal intra-epithelial carcinoma. Nature communications. 2017;8:990. doi: 10.1038/s41467-017-01217-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Koshiyama M, Matsumura N, Konishi I. Recent concepts of ovarian carcinogenesis: type I and type II. BioMed research international. 2014;2014:934261. doi: 10.1155/2014/934261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ahmed AA, Etemadmoghadam D, Temple J, Lynch AG, Riad M, Sharma R, et al. Driver mutations in TP53 are ubiquitous in high grade serous carcinoma of the ovary. J Pathol. 2010;221:49–56. doi: 10.1002/path.2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jones PM, Drapkin R. Modeling High-Grade Serous Carcinoma: How Converging Insights into Pathogenesis and Genetics are Driving Better Experimental Platforms. Front Oncol. 2013;3:217. doi: 10.3389/fonc.2013.00217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Labidi-Galy SI, Papp E, Hallberg D, Niknafs N, Adleff V, Noe M, et al. High grade serous ovarian carcinomas originate in the fallopian tube. Nature communications. 2017;8:1093. doi: 10.1038/s41467-017-00962-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Perets R, Drapkin R. It’s Totally Tubular....Riding The New Wave of Ovarian Cancer Research. Cancer research. 2016;76:10–7. doi: 10.1158/0008-5472.CAN-15-1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.George SH, Greenaway J, Milea A, Clary V, Shaw S, Sharma M, et al. Identification of abrogated pathways in fallopian tube epithelium from BRCA1 mutation carriers. J Pathol. 2011;225:106–17. doi: 10.1002/path.2927. [DOI] [PubMed] [Google Scholar]
- 16.Quartuccio SM, Karthikeyan S, Eddie SL, Lantvit DD, Oh E, Modi DA, et al. Mutant p53 expression in fallopian tube epithelium drives cell migration. Int J Cancer. 2015 doi: 10.1002/ijc.29528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim J, Coffey DM, Creighton CJ, Yu Z, Hawkins SM, Matzuk MM. High-grade serous ovarian cancer arises from fallopian tube in a mouse model. Proc Natl Acad Sci U S A. 2012;109:3921–6. doi: 10.1073/pnas.1117135109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Levanon K, Sapoznik S, Bahar-Shany K, Brand H, Shapira-Frommer R, Korach J, et al. FOXO3a loss is a frequent early event in high-grade pelvic serous carcinogenesis. Oncogene. 2014;33:4424–32. doi: 10.1038/onc.2013.394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Karst AM, Jones PM, Vena N, Ligon AH, Liu JF, Hirsch MS, et al. Cyclin E1 deregulation occurs early in secretory cell transformation to promote formation of fallopian tube-derived high-grade serous ovarian cancers. Cancer research. 2014;74:1141–52. doi: 10.1158/0008-5472.CAN-13-2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rosen DG, Mercado-Uribe I, Yang G, Bast RC, Jr, Amin HM, Lai R, et al. The role of constitutively active signal transducer and activator of transcription 3 in ovarian tumorigenesis and prognosis. Cancer. 2006;107:2730–40. doi: 10.1002/cncr.22293. [DOI] [PubMed] [Google Scholar]
- 21.Saini U, Naidu S, ElNaggar AC, Bid HK, Wallbillich JJ, Bixel K, et al. Elevated STAT3 expression in ovarian cancer ascites promotes invasion and metastasis: a potential therapeutic target. Oncogene. 2016 doi: 10.1038/onc.2016.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yu H, Lee H, Herrmann A, Buettner R, Jove R. Revisiting STAT3 signalling in cancer: new and unexpected biological functions. Nature reviews Cancer. 2014;14:736–46. doi: 10.1038/nrc3818. [DOI] [PubMed] [Google Scholar]
- 23.Ogata Y, Osaki T, Naka T, Iwahori K, Furukawa M, Nagatomo I, et al. Overexpression of PIAS3 suppresses cell growth and restores the drug sensitivity of human lung cancer cells in association with PI3–K/Akt inactivation. Neoplasia. 2006;8:817–25. doi: 10.1593/neo.06409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sundvall M, Korhonen A, Vaparanta K, Anckar J, Halkilahti K, Salah Z, et al. Protein inhibitor of activated STAT3 (PIAS3) protein promotes SUMOylation and nuclear sequestration of the intracellular domain of ErbB4 protein. J Biol Chem. 2012;287:23216–26. doi: 10.1074/jbc.M111.335927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bromberg J. Stat proteins and oncogenesis. The Journal of clinical investigation. 2002;109:1139–42. doi: 10.1172/JCI15617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dabir S, Kluge A, Kresak A, Yang M, Fu P, Groner B, et al. Low PIAS3 expression in malignant mesothelioma is associated with increased STAT3 activation and poor patient survival. Clinical cancer research : an official journal of the American Association for Cancer Research. 2014;20:5124–32. doi: 10.1158/1078-0432.CCR-14-1233. [DOI] [PubMed] [Google Scholar]
- 27.Karst AM, Drapkin R. Primary culture and immortalization of human fallopian tube secretory epithelial cells. Nature protocols. 2012;7:1755–64. doi: 10.1038/nprot.2012.097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Levanon K, Ng V, Piao HY, Zhang Y, Chang MC, Roh MH, et al. Primary ex vivo cultures of human fallopian tube epithelium as a model for serous ovarian carcinogenesis. Oncogene. 2010;29:1103–13. doi: 10.1038/onc.2009.402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Selvendiran K, Koga H, Ueno T, Yoshida T, Maeyama M, Torimura T, et al. Luteolin promotes degradation in signal transducer and activator of transcription 3 in human hepatoma cells: an implication for the antitumor potential of flavonoids. Cancer research. 2006;66:4826–34. doi: 10.1158/0008-5472.CAN-05-4062. [DOI] [PubMed] [Google Scholar]
- 30.Selvendiran K, Tong L, Vishwanath S, Bratasz A, Trigg NJ, Kutala VK, et al. EF24 induces G2/M arrest and apoptosis in cisplatin-resistant human ovarian cancer cells by increasing PTEN expression. J Biol Chem. 2007;282:28609–18. doi: 10.1074/jbc.M703796200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Selvendiran K, Ahmed S, Dayton A, Ravi Y, Kuppusamy ML, Bratasz A, et al. HO-3867, a synthetic compound, inhibits the migration and invasion of ovarian carcinoma cells through downregulation of fatty acid synthase and focal adhesion kinase. Mol Cancer Res. 2010;8:1188–97. doi: 10.1158/1541-7786.MCR-10-0201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dutta DK, Dutta I. Origin of ovarian cancer: molecular profiling. J Obstet Gynaecol India. 2013;63:152–7. doi: 10.1007/s13224-013-0419-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kurman RJ. Origin and molecular pathogenesis of ovarian high-grade serous carcinoma. Ann Oncol. 2013;24(Suppl 10):x16–21. doi: 10.1093/annonc/mdt463. [DOI] [PubMed] [Google Scholar]
- 34.Coscia F, Watters KM, Curtis M, Eckert MA, Chiang CY, Tyanova S, et al. Integrative proteomic profiling of ovarian cancer cell lines reveals precursor cell associated proteins and functional status. Nature communications. 2016;7:12645. doi: 10.1038/ncomms12645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McCann GA, Naidu S, Rath KS, Bid HK, Tierney BJ, Suarez A, et al. Targeting constitutively-activated STAT3 in hypoxic ovarian cancer, using a novel STAT3 inhibitor. Oncoscience. 2014;1:216–28. doi: 10.18632/oncoscience.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rath KS, Naidu SK, Lata P, Bid HK, Rivera BK, McCann GA, et al. HO-3867, a safe STAT3 inhibitor, is selectively cytotoxic to ovarian cancer. Cancer research. 2014;74:2316–27. doi: 10.1158/0008-5472.CAN-13-2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brantley EC, Nabors LB, Gillespie GY, Choi YH, Palmer CA, Harrison K, et al. Loss of protein inhibitors of activated STAT-3 expression in glioblastoma multiforme tumors: implications for STAT-3 activation and gene expression. Clinical cancer research : an official journal of the American Association for Cancer Research. 2008;14:4694–704. doi: 10.1158/1078-0432.CCR-08-0618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dabir S, Kluge A, Dowlati A. The association and nuclear translocation of the PIAS3-STAT3 complex is ligand and time dependent. Mol Cancer Res. 2009;7:1854–60. doi: 10.1158/1541-7786.MCR-09-0313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sehdev AS, Kurman RJ, Kuhn E, Shih Ie M. Serous tubal intraepithelial carcinoma upregulates markers associated with high-grade serous carcinomas including Rsf-1 (HBXAP), cyclin E and fatty acid synthase. Modern pathology : an official journal of the United States and Canadian Academy of Pathology, Inc. 2010;23:844–55. doi: 10.1038/modpathol.2010.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chaluvally-Raghavan P, Jeong KJ, Pradeep S, Silva AM, Yu S, Liu W, et al. Direct Upregulation of STAT3 by MicroRNA-551b-3p Deregulates Growth and Metastasis of Ovarian Cancer. Cell reports. 2016;15:1493–504. doi: 10.1016/j.celrep.2016.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen MW, Yang ST, Chien MH, Hua KT, Wu CJ, Hsiao SM, et al. The STAT3-miRNA-92-Wnt Signaling Pathway Regulates Spheroid Formation and Malignant Progression in Ovarian Cancer. Cancer research. 2017;77:1955–67. doi: 10.1158/0008-5472.CAN-16-1115. [DOI] [PubMed] [Google Scholar]
- 42.Ruscito I, Dimitrova D, Vasconcelos I, Gellhaus K, Schwachula T, Bellati F, et al. BRCA1 gene promoter methylation status in high-grade serous ovarian cancer patients--a study of the tumour Bank ovarian cancer (TOC) and ovarian cancer diagnosis consortium (OVCAD) European journal of cancer. 2014;50:2090–8. doi: 10.1016/j.ejca.2014.05.001. [DOI] [PubMed] [Google Scholar]
- 43.Zhang P, Yang B, Yao YY, Zhong LX, Chen XY, Kong QY, et al. PIAS3, SHP2 and SOCS3 Expression patterns in Cervical Cancers: Relevance with activation and resveratrol-caused inactivation of STAT3 signaling. Gynecologic oncology. 2015;139:529–35. doi: 10.1016/j.ygyno.2015.09.087. [DOI] [PubMed] [Google Scholar]
- 44.Liu LH, Li H, Cheng XX, Kong QY, Chen XY, Wu ML, et al. Correlative analyses of the expression levels of PIAS3, p-SHP2, SOCS1 and SOCS3 with STAT3 activation in human astrocytomas. Molecular medicine reports. 2017;15:847–52. doi: 10.3892/mmr.2016.6079. [DOI] [PubMed] [Google Scholar]
- 45.Kluge A, Dabir S, Vlassenbroeck I, Eisenberg R, Dowlati A. Protein inhibitor of activated STAT3 expression in lung cancer. Mol Oncol. 2011;5:256–64. doi: 10.1016/j.molonc.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.