Abstract
Acquired thermotolerance (AT) is the ability of cells to survive a normally lethal temperature treatment as a consequence of pretreatment at an elevated but sublethal temperature. In yeast and cyanobacteria, the expression of the HSP100/ClpB protein is required for the AT response. To determine whether the HSP100/ClpB protein is associated with this response in lima bean (Phaseolus lunatus), we have cloned an HSP100/ClpB homolog and assessed expression of the two gene copies under heat stress conditions, which induce AT. Transcription of the cytoplasmically localized HSP100/ClpB protein genes is stringently controlled by heat stress in both of the laboratory and field heat stress conditions. From a heat-induced cDNA library, we identified a clone of a putative chloroplast-targeted (cp) HSP100/ClpB protein gene sequence. The cp HSP100/ClpB protein genes are constitutively expressed, but transcript levels increase post-heat stress in laboratory heat stress experiments. In field conditions the genes for the cp HSP100/ClpB are constitutively expressed. Although we were unable to correlate differences in the timing of AT response with the expression or genetic structure of the HSP100/ClpB genes in heat-tolerant or -sensitive varieties of lima bean, we clearly demonstrate the association of expression of HSP100/ClpB proteins with heat response in this species.
All plant, animal, insect, bacterial, and fungal species so far examined produce a new set of proteins (heat shock proteins [HSPs]) in response to temperatures above optimum. When plant species that are adapted to temperate environments, such as soybean, peas, maize, and wheat, are grown at elevated temperatures, expression of 20 to 40 different HSPs is induced (Vierling, 1991). These proteins function as molecular chaperones to prevent aggregation of denatured proteins, to assist in folding of nascent polypeptides, to aid in refolding of denatured proteins (Becker and Craig, 1994), or to resolubilize aggregated denatured proteins (Parsell et al., 1994).
Genes for HSPs of approximately 100 kD have been isolated and sequenced from various species. The gene products are termed Clp proteins because of their sequence similarity to Escherichia coli ClpA, which is thought to be involved in the regulation of proteolysis (Gottesman et al., 1990). Clp proteins are found in cytosolic/nuclear compartments and in organelles of eukaryotic and prokaryotic cells (Moore and Keegstra, 1993; Schirmer et al., 1994; Schmitt et al., 1995). Class-I Clp proteins contain two highly conserved nucleotide-binding sites, both of which appear to be essential for function (Parsell et al., 1991) and have two large blocks of sequence homology (approximately 200 amino acids) centered around these two ATP-binding sites. Subfamilies of the class-I Clp proteins are defined by the length of the spacer region between the two nucleotide-binding sites. ClpA proteins have a short (5 amino acids) spacer, ClpB proteins have the longest spacer (123–131 amino acids), whereas ClpC and ClpD protein spacers are intermediate in length (62–69 amino acids) (Schirmer et al., 1996). ClpB proteins are often associated with heat-inducible expression and are thus designated HSP100/ClpB. ClpC in plants is a constitutively expressed 92-kD protein that is targeted to the chloroplasts and may be involved in protein translocation.
Acquired thermotolerance (AT) is the ability of cells to adapt and survive a normally lethal-temperature treatment as a result of pretreatment at an elevated but sublethal temperature. Deleting the HSP104/ClpB protein gene from Saccharomyces cerevisiae decreases the ability of yeast cells to mount an AT response (Sanchez and Lindquist, 1990). Using photosynthesis as a metabolic indicator, Eriksson and Clarke (1996) have shown that deletion of the HSP100/ClpB protein gene in Synechococcus sp. prevents a full AT response and that accumulation of the protein is coordinated with AT. Combined, these results indicate that the HSP100/ClpB protein is a primary determinant in protection of or recovery of cells from extreme heat stress.
HSP100/ClpB gene orthologs have been cloned from Arabidopsis, soybean, corn, wheat, and tobacco (Lee et al., 1994; Schirmer et al., 1994; Wells et al., 1998, Nieto-Sotelo et al., 1999). Plant HSP100/ClpB proteins are thought to be soluble, cytoplasmically localized (cyt) proteins. The Arabidopsis and soybean cyt HSP100/ClpB proteins are 86% identical and both of these proteins and the tobacco HSP100/ClpB protein have been shown to complement the deletion of HSP104/ClpB in yeast and restore the AT response. Therefore, the plant cyt HSP100/ClpB proteins can function similarly to the yeast HSP104/ClpB protein. There has been no direct evidence that HSP100/ClpB is required for AT in plants or that the expression of HSP100/ClpB proteins is correlated with differences in AT observed among species or among cultivars of the same species in plants. However, the ability of HSP100/ClpB from plants to complement the yeast HSP104 deletion and restore the AT response strongly suggests that this protein may be essential to the mechanism of the AT response in plants.
There are two major gene pools in lima bean (Phaseolus lunatus) as described both by morphology and random-amplified polymorphic DNA marker analyses (Mackie, 1943; Nienhuis et al., 1995). The small seeded varieties, commonly called baby limas are in the Mesoamerican gene pool. The large seeded varieties, commonly called fordhooks belong to the Andean gene pool. It is generally thought that cultivars from the Mesoamerican gene pool have greater adaptability to heat stress than cultivars from the Andean gene pool (Mackie, 1943).
In this study we observed AT in several cultivars of lima bean from different heat response categories by measuring leakage of ions from heat-stressed leaf cells. A PCR-generated sequence encoding a cyt HSP100/ClpB gene and a cDNA encoding a putative chloroplast-localized (cp) HSP100/ClpB gene were identified. The expression of the HSP100/ClpB genes and the accumulation of HSP100/ClpB proteins were observed under laboratory heat stress conditions and in a field growth environment.
RESULTS
AT in Leaf Tissues
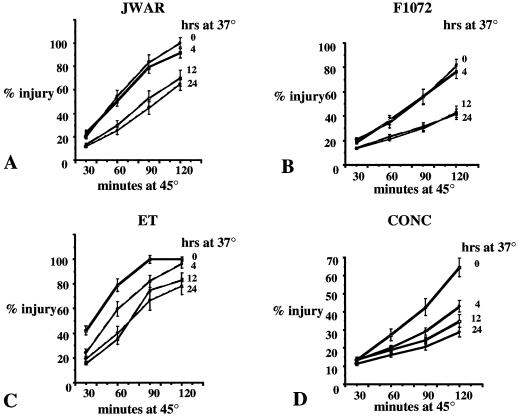
Four cultivars were assayed for tolerance to a 45°C test temperature using the electrical conductivity (EC) assay (Fig. 1). These cultivars were chosen as representatives of the Mesoamerican (generally more heat tolerant) and Andean (generally less heat tolerant) gene pools. The Mesoamerican variety cv Jackson WonderAR (JWAR) is a speckled seeded variety known to be heat tolerant in field and greenhouse yield trials. cv Early Thorogreen (ET), a Mesoamerican cultivar, and cv Fordhook 1072 (F1072), an Andean cultivar, are varieties commonly planted for production in Delaware. cv Concentrated Fordhook (CONC) was grouped in the average heat tolerance classification by Kleiner and Frett (1996) in greenhouse yield studies, but in actual field growth CONC is considered to yield poorly in above-average temperatures. From the data in Figure 1, the larger, thicker leaves of the Andean varieties (F1072 and CONC) had less total damage as measured by leakage of ions than did the thinner leaf Mesoamerican varieties, JWAR and ET. Our assay was used to observe the change in total injury rate at 45°C due to pretreatment at 37°C.
Figure 1.
Percent injury of leaf cells at 45°C after pretreatment of plants at 37°C. Injury of leaf cells was measured by assaying leakage of ions as an increase in conductivity. Eleven-day-old plants were incubated at 37°C for 0, 4, 12, or 24 h before leaf discs were assayed for leakage of ions at 45°C. The conductivity of the sample was measured after 30, 60, 90, or 120 min of incubation at 45°C (EC1). The tissues were frozen overnight and warmed to 45°C and a final conductivity measurement taken (EC2). Percent injury = EC1/EC2 × 100. Numbers are averages of three experiments ± se.
To observe AT in green leaf tissue, we measured leakage of ions from leaf discs as an indicator of heat injury as defined by March et al. (1982). Leaves of seedlings from JWAR reached 100% injury after 120 min of exposure to the 45°C test temperature when the plants had not been previously exposed to 37°C (Fig. 1A, 0 h). When plants were maintained at 37°C for 4 h before the 45°C test, no increase in thermotolerance was observed. However, if plants were exposed to 37°C for 12 or 24 h and then assayed at 45°C, the amount of injury measured was reduced significantly (approximately 60% injury at 120 min). This reduction in injury at 45°C reflects an AT to the normally lethal heat (45°C) as a result of previous exposure to less-extreme heat stress (37°C). F1072 showed similar results (Fig. 1B) in that 4 h at 37°C did not reduce the amount of injury at 45°C as compared to untreated plants, while 12 or 24 h of 37°C pretreatment did result in decreased injury at 45°C.
With CONC and ET, just 4 h of pretreatment at 37°C produced a significant decrease in injury at 45°C. These results suggest that pretreatment of lima bean plants with 4 to 12 h of 37°C is sufficient to increase AT as measured by this assay. The different patterns of AT observed in these four cultivars were not related to heat tolerance based on yield or to the different gene pools. ET, F1072, and CONC were all in category II (heat tolerance average) for yield but showed different AT response times. This result suggests that heat tolerance of leaf tissue is not the determining factor in yield under heat stress.
Identification of the cyt and cp HSP100/ClpB Genes of Lima Bean
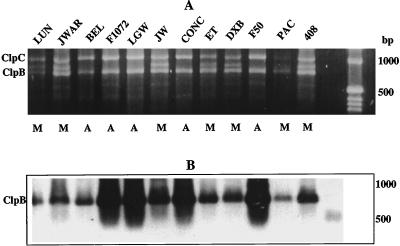
To determine if a relationship could be established between expression of the HSP100 protein and the timing of AT in leaf tissue of lima bean, we obtained a probe for the lima bean HSP100/ClpB ortholog by PCR amplification of the region between the conserved ATP-binding sites of class-I Clp proteins. From PCR amplification reactions with genomic DNA isolated from various cultivars of lima bean, either two or three PCR bands were observed (Fig. 2A). A 720-bp PCR-amplified fragment, seen with all of the lima bean genomic DNAs, was confirmed as being part of a cyt HSP100/ClpB protein gene by having high-sequence homology to the known soybean HSP101 sequence (GenBank accession no. L35272). In addition, all of the reactions produced an approximately 1,000-bp band that was homologous to the tomato ClpC CD4B sequence (GenBank accession no. M32604). A third PCR-amplified fragment of approximately 860 bp was apparent in reactions that used DNA from Mesoamerican-derived cultivars only. The sequence of this band did not match any known sequences in the National Center for Biotechnology Information (NCBI) database. At high stringency, a probe prepared from the cyt HSP100/ClpB fragment hybridized only to its fragment of origin (720 bp, HSP100/ClpB sequence, Fig. 2B) and not the other two PCR products.
Figure 2.
PCR-amplified Clp genes. A, PCR-generated fragments using lima bean genomic DNA as template and primers 1070 and 1790. Fragments were separated on a 1.2% (w/v) agarose gel and stained with ethidium bromide. Genomic DNAs from these cultivars: cv Luna (LUN), Jackson Wonder AR (JWAR), cv Belgreen (BEL), Fordhook 1072 (F1072), cv Large White (LGW), cv Jackson Wonder (JW), Concentrated Fordhook (CONC), Early thorogreen (ET), cv Dixie Butter Bean (DXB), cv Fordhook 5055 (F50), cv Packers (PAC), and cv ML408 (408). M, Mesoamerican gene pool; A, Andean gene pool. B, Southern blot of gel in A probed with antisense riboprobe derived from the lima bean cyt HSP100/ClpB.
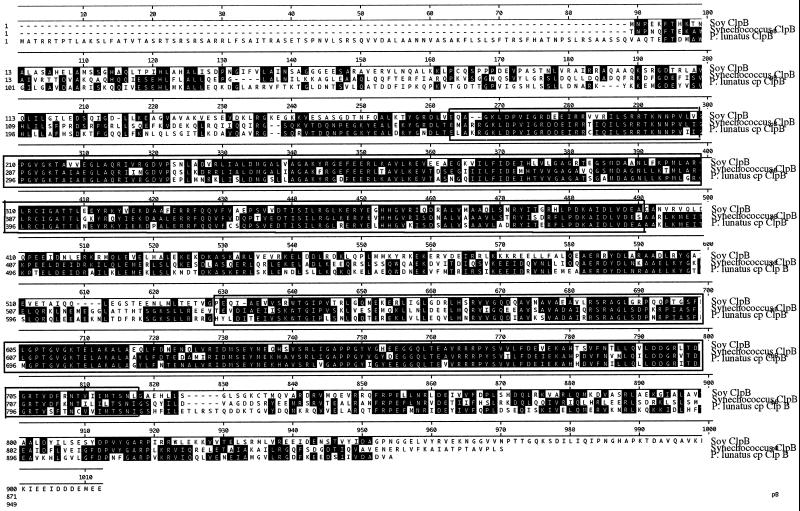
A cDNA library in Lambda ZAP XR (Stratagene, La Jolla, CA) was constructed from poly(A+) RNA isolated from leaf tissue of JWAR, which had been incubated at 40°C for 30-min, conditions under which HSP100/ClpB is strongly induced (see below). The first approximately 500 nucleotides from the 5′ end of 2,557 random clones from this library were sequenced, and the sequences were analyzed by comparison with the NCBI database to detect and evaluate the various populations of HSP mRNAs induced in heat-stressed tissues and to obtain the remainder of the HSP100/ClpB gene. As expected, this library contained a high proportion of HSP sequences (S. Keeler, unpublished data). Surprisingly, no cDNAs corresponding to the cyt HSP100/ClpB sequence were recovered. However, one cDNA insert (GenBank accession no. AF203700) was found to have extensive homology at the amino acid level to the Synechococcus sp. ClpB1 sequence (GenBank accession no. U20646). The complete amino acid sequence encoded by this cDNA is presented in Figure 3, aligned with the soybean cyt HSP100/ClpB and the Synechococcus sp. HSP100/ClpB1 amino acid sequences. Starting at amino acid 94 of this sequence, it is 50% identical to the soybean cyt HSP100/ClpB and 62% identical to the Synechococcus sp. ClpB1 sequence.
Figure 3.
Alignment of amino acid sequences. Sequences aligned were from soybean HSP101/ClpB (Soy ClpB) (GenBank accession no. L35272), Synechococcus sp. ClpB1 (GenBank accession no. U20646), and the cp HSP100/ClpB protein from lima bean (GenBank accession no. AF203700). Sequences were aligned using the MegAlign program (DNASTAR, Inc.).
The additional 94 amino acids on the amino terminus of the protein encoded by this clone have characteristics of a chloroplast transit sequence: a high percentage of Ser and Thr (23%) and a net positive charge (± = 12/2). The endosymbiotic theory of chloroplast evolution hypothesizes the absorption of photosynthesizing cyanobacterial cells by eukaryotic hosts. The homology of the lima bean cp HSP100/ClpB protein to the cyanobacterial HSP100/ClpB protein and the presence of a putative chloroplast transit sequence is strong evidence that this sequence represents the first identification of a cp form of HSP100/ClpB. Consequently, we have designated this gene cp HSP100/ClpB and the PCR-generated clone cyt HSP100/ClpB to denote their putative cellular locations.
By comparing the DNA sequences, we determined that the primers used to generate the cyt HSP100/ClpB fragment in Figure 2 would not have annealed to the cp HSP100/ClpB sequence. Therefore, a PCR fragment would not have been generated from the cp HSP100/ClpB gene using these primers. This was confirmed by using primers 1070 and 1790 in PCR reactions with plasmid DNAs containing cyt HSP100/ClpB, cp HSP100/ClpB, and ClpC (data not shown). In this reaction, the cyt HSP100/ClpB template generated the cloned 720-bp fragment, the cp HSP100/ClpB gave no PCR product, and the ClpC clone gave a predicted fragment.
Southern-Blot Analysis of HSP100/ClpB Gene Sequences
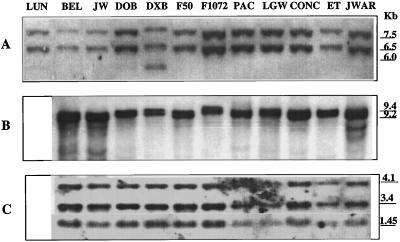
To determine the gene copy number of each of the ClpB genes in lima bean cultivars and to observe possible allelic variations among the heat-tolerant or -sensitive cultivars, Southern blots of restriction enzyme-digested genomic DNA isolated from 11 cultivars of lima bean were probed with cyt HSP100/ClpB or cp HSP100/ClpB. Five different restriction digests (BglII, EcoRI, BamHI, XhoI, and PstI) each produced only two bands, which hybridized with the cyt HSP100/ClpB probe, the EcoRI digest produced two bands (9.2 and 9.4 kb), appearing as a doublet (Fig. 4B). Only with the BglII digestion was there an observable RFLP and only in one cultivar (cv Dixie Butter Bean) in which a third band was visible (Fig. 4A). This indicates that there are at least two genes with considerable homology to the cyt HSP100/ClpB gene fragment in lima bean cultivars and little if any sequence polymorphism in these genes among cultivars suggesting that differences in heat tolerances are not related to allelic variations at these loci. Within the cpHSP100/ClpB gene there is an EcoRI site. Therefore, it is likely that the three bands that hybridized to the cp HSP100/ClpB probe in Figure 4C represent at least two genes with high homology to the cp HSP100/ClpB gene in the lima bean genome.
Figure 4.
Southern blots of restriction enzyme-digested genomic lima bean DNA. Sizes of hybridizing fragments are indicated on the right in kb. Cultivars: cv Luna (LUN), cv Belgreen (BEL), cv Jackson Wonder (JW), cv Dover Bush (DOB), cv Dixie Butter Bean (DIX), cv Fordhook 5055 (F50), F1072, cv Packers (PAC), cv Large White (LGW), CONC, ET, and JWAR. A, Probe = cyt HSP100/ClpB, digest = BglII; B, probe = cyt HSP100/ClpB, digest = EcoRI; C, probe = cp HSP100/ClpB, digest = EcoRI.
Expression of the HSP100/ClpB Gene Transcripts in Heat-Stressed Lima Bean Leaf Tissue
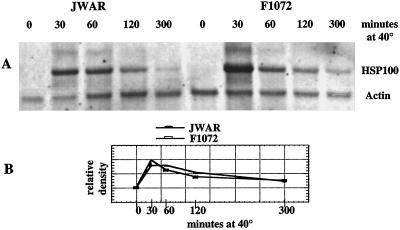
To compare the expression of the cyt HSP100/ClpB protein gene in lima bean leaf tissue from a heat-tolerant (Mesoamerican lineage) and average heat-tolerant (Andean lineage) cultivar, RNA was extracted from leaves of plants of JWAR and F1072, which had been incubated at 40°C for 0 to 300 min. Northern blots of these RNAs were hybridized simultaneously with the lima bean cyt HSP100/ClpB antisense RNA probe and an actin antisense RNA probe (Fig. 5A). The relative level of the cyt HSP100/ClpB signal was normalized using the level of the actin signal and relative densities plotted against time at 40°C (Fig. 5B). The cyt HSP100/ClpB probe gave a strong signal with RNA derived from leaf tissues exposed to 40°C for 30 min in both of the cultivars indicating no gross difference in expression of this gene in leaf cells from a Mesoamerican or Andean variety. The size of this message (approximately 3 kb) correlates well with the predicted size for the full-length cyt HSP100/ClpB coding region.
Figure 5.
Northern blot of total RNA. A, RNA was extracted from leaf tissues of 3- to 4-week-old lima bean JWAR and F1072. Three- to 4-week-old seedlings were incubated at 40°C for 0, 30, 60, 120, or 300 min. RNA was extracted from immature leaves, separated by formaldehyde, agarose gel electrophoresis, and blotted onto Hybond N+ (Amersham, Arlington Heights, IL) membrane. The northern blot was hybridized with antisense riboprobe derived from lima bean cyt HSP100/ClpB gene fragment and an actin gene fragment simultaneously. B, Quantification of hybridizing signals. An autoradiograph of the hybridized northern blot in A was converted to a digital image and the density of the bands measured using the NIH Image program. Relative densities were adjusted based on the density of the actin signal at that time point. The least actin signal was set at a value of 1.0, and all of the other actin signals adjusted to this signal. The actin ratio was then used to normalize the relative density of the signal from the HSP100 probe for the same sample.
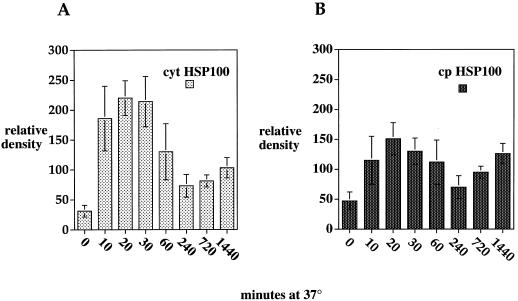
To compare the expression of both of the cp and cyt HSP100/ClpB genes at the 37°C temperature used in the AT assay, 3-week-old plants were incubated at 37°C for 0, 10, 20, 30, 60, 120, 240, 720, or 1,440 min (Fig. 6). Relative expression levels of HSP100/ClpB transcripts were measured at each time point. Although three cultivars (JWAR, F1072, and CONC), which differ in their heat responsiveness by yield, were used for this analysis; the relative amounts of HSP100/ClpB gene transcripts observed with samples from each cultivar and the patterns of gene expression under heat stress in each cultivar were not significantly different. Therefore, we used averaged data to compare general patterns of response of the cp and cyt HSP100/ClpB genes in lima bean.
Figure 6.
Steady-state level of HSP100 RNAs in lima bean leaf tissue. Autoradiographs of RNA slot blots hybridized to digoxigenin-labeled antisense RNA probes were converted to digital images and the density of the bands measured using the NIH Image program. Relative densities were adjusted based on the density of the actin signal at that time point as described in Figure 3. Densities are given as averages from three samples (one each of CONC, JWAR, and F1072) at each time point ± se. A, Probe = cyt HSP100/ClpB; B, probe = cp HSP100/ClpB.
The expression of the cyt HSP100/ClpB genes is strongly induced by heat stress with little or no transcript in unstressed leaf tissue (Fig. 6A). The amount of cyt HSP100/ClpB transcript was observable as early as 10 min after the start of the heat stress and reached maximum levels within the first 20 to 30 min of heat stress. The steady-state level of transcript then decreased over time from 60% of maximum at 60 min to a low of 32% of maximum level at 720 min. The timing of this response is substantially different from the timing of the AT response, which can be observed in these plants only after 4 to 12 h of continuous heat exposure. The temporal pattern of cp HSP100/ClpB transcription was similar to that observed for cyt HSP100/ClpB (Fig. 6B). However, the maximum accumulation of this transcript was relatively less than cyt HSP100/ClpB suggesting that cp HSP100/ClpB is less heat responsive and less well expressed in general than the cyt HSP100/ClpB genes.
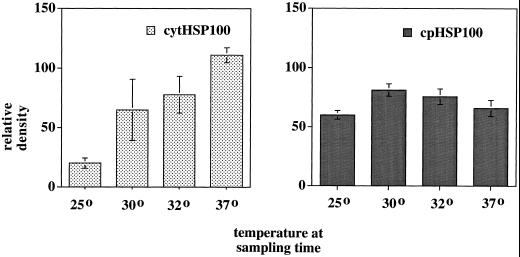
To observe the response to heat of the HSP100 genes in leaf tissues under field conditions, we set up a field experiment using five cultivars representing all of the three heat-response categories. When the plants were 3 weeks old, leaf samples were taken every 2 h starting just after sunrise on a day when the ambient temperature was predicted to rise above 36°C. Temperature measurements taken at plant height reached 37°C by 1 pm, a temperature sufficient to increase expression of these genes (Fig. 6). Using slot-blot transcript analysis, there was considerable variability in signals from samples from individual plants such that we could not discern substantial differences among cultivars. Therefore, we used the averaged data to get an overall observation of expression of the HSP100/ClpB genes under these conditions. In the samples presented in Figure 7, cyt HSP100/ClpB appeared to be strongly regulated by heat, increasing coincidentally with a gradual rise in temperature. The cp HSP100/ClpB transcripts appeared to have a higher background level of expression and did not appear to be heat responsive.
Figure 7.
Steady-state levels of HSP100 RNAs in lima bean leaf tissue from field samples. Slot blots of total RNA from leaf tissue samples taken from field grown plants on August 17, 1997, probed with antisense probes of cyt HSP100/ClpB (cyt100) or cp HSP100/ClpB (cp100). Relative densities of hybridizing signals were determined as described in the text and were ad-justed for the level of hybridizing signal to an antisense actin probe in the same sample. Densities were averaged from five separate samples at each time point ± se. Each sample was a pool of leaves from three plants each of CONC, JWAR, F1072, ET, or BEL. The air temperature at plant height in the field was: 7 am, 25°C; 9 am, 30°C; 11 am, 32°C; and 1 pm, 37°C.
Accumulation of ClpB and Related Proteins in Leaf Cells during Heat Stress
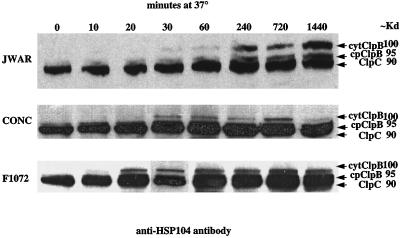
To observe HSP100/ClpB protein levels in leaf tissue during 37°C heat stress, we used an antibody directed against the conserved ATP1-binding site of the ClpB and ClpC proteins (Parsell et al., 1991). This antibody reacts with both the constitutively expressed ClpC protein and the heat-induced HSP100/ClpB gene products (Eriksson and Clarke, 1996). In Figure 8A, a strong approximately 90-kD band, corresponding to the constitutively expressed ClpC gene product (Schirmer et al., 1994), was detected in all of the samples. By 30 min of heat stress, an approximately 100-kD protein was detected in leaf tissue from all three cultivars. There does appear to be an increased accumulation of this protein in JWAR versus the other two cultivars, which may correlate with the increased thermotolerance of JWAR, however this difference was not observed consistently in repeated experiments (data not shown). The steady-state level of this heat-inducible protein remained constant from 60 min through the 1,440 min of heat stress. In Figure 8A, a third reactive band (approximately 95 kD) is visible between the ClpC and HSP100/ClpB-inducible band. This protein appears to be constitutively expressed (visible in lane 1, unstressed tissue) but increases with heat stress. Since the predicted cp HSP100/ClpB protein would have cross reactivity with antibody 3-2 , the additional band may represent the cp HSP100/ClpB protein.
Figure 8.
Detection of Clp proteins in leaf samples. Proteins were extracted from leaf tissue that had been incubated at 37°C. Proteins were separated on SDS-PAGE, blotted onto nitrocellulose, and reacted with antibody 3-2 specific for the ATP1-binding site of Clp proteins. Primary antibody was detected by reaction with secondary anti-rabbit HRP-conjugated antibody and reaction with CSPD (Amersham) chemiluminescent substrate as described in “Materials and Methods.” Approximate sizes of visible bands are indicated in kD. Probable identifications of reactive bands are indicated.
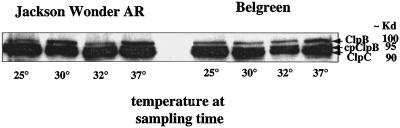
Western-blot analysis was also used to correlate the levels of cyt HSP100/ClpB with transcript levels observed in the field experiment. Although the cyt HSP100/ClpB transcript levels were low at the early time points in the leaf samples from field grown plants (Fig. 7), the cyt HSP100/ClpB protein was visible at approximately equal levels at every time point (Fig. 9). This result indicates that the protein may have accumulated and been maintained through the night following the heat stress of the previous day. We have observed a similar accumulation and maintenance of heat-induced HSP100 proteins through a cool overnight period in a greenhouse growth experiment (data not shown). There was no gross difference in accumulated levels of HSP100/ClpB proteins in heat-tolerant (JWAR) or heat-sensitive (BEL) varieties in this field sampling.
Figure 9.
Detection of Clp proteins in leaf tissues from field samples of lima bean. Total soluble proteins were extracted from leaf tissues of samples taken from field grown plants on August 17, 1997. These samples are the same as used for RNA extractions in Figure 7. The protein extracts for JWAR and BEL at each time point are shown. The proteins were separated on SDS-PAGE, blotted onto nitrocellulose, and reacted with antibody 3-2 specific for the ATP1-binding site of Clp proteins as described in “Materials and Methods.” Antibody 3-2 was detected by secondary anti-rabbit HRP-conjugated antibody and reaction with CSPD (Amersham) chemiluminescent substrate. Approximate sizes of visible bands are indicated in kD. 7 am, 25°C; 9 am, 30°C; 11 am, 32°C; and 1 pm, 37°C.
DISCUSSION
AT in Leaf Tissue
We determined that the length of 37°C pretreatment time needed to produce an observable decrease in leaf cell damage at 45°C in our assay was between 4 and 12 h. Lin et al. (1985), using a similar assay to observe leakage of solutes from heat-treated soybean seedlings, found that 2 h of pretreatment was sufficient to induce AT. CONC, ET, and F1072 all had an average heat tolerance in yield studies, yet their AT response measured as injury at 45°C after 4 h at 37°C was variable. This is contrary to the results of Li et al. (1991) who found a direct correlation between heat acclimation potential (ability of leaf cells to withstand injury at 50°C after exposure to 37°C for 24 h) and yield under heat stress in the field with cultivars of Phaseolus vulgaris. However, our designations as heat tolerant or heat sensitive in this study were derived from yield data from non-drought-stressed plants, whereas the field-grown plants used by Li et al. (1991) were likely to have been exposed to both heat and drought stress. In addition Li et al. (1991) assayed leaf tissues at 50°C, a more extreme temperature than was used in our assays. Yield under heat stress may be affected by various aspects of the heat response: the ability to detect temperatures as stressful; the ability to respond to temperature shifts quickly; the extent of the response; and the tissue specificity of the response. Whereas response of leaf tissue to heat may be important in maximizing the biomass of the plant, which can affect its ability to support sink material such as seeds, it is more likely that a direct effect on yield (no. of beans/plant) is the result of heat stress on reproductive tissues. It has been reported that pollen development is particularly sensitive to extreme heat exposure (Weaver and Timm, 1988).
Swan (1997) observed that thermotolerance in yeast is associated with increased membrane protein stability. Increased membrane protein stability may be the result of the protective actions of HSPs. Preserving membrane integrity during heat stress would provide an observable decrease in ion leakage during heat stress in our assay. HSP100/ClpB protein levels increase after exposure of plants to 37°C (Fig. 8). In the incubator heat stress experiments, HSP100/ClpB protein accumulated several hours before AT could be observed with this assay. This supports the theory that additional HSPs and other heat-induced factors may be necessary to achieve a maximized AT response in plants. Therefore, whatever the direct effect of HSP100/ClpB accumulation, it is not the sole causative agent of AT in plant leaf tissue as measured by solute leakage.
Conservation of cyt HSP100/ClpB Gene Sequences among Species and among Cultivars of Lima Bean
Unlike other HSPs that serve to aid in renaturation, refolding, or protection of unfolded or denatured proteins during heat stress, HSP100/ClpB appears to function in resolubilization of aggregated proteins (Parsell et al., 1994). This function is particularly important under extreme heat stress conditions or under combined multiple stress conditions in which the ability of HSP70 and other protecting chaperones to prevent aggregation of denatured proteins may be overwhelmed. In immobile organisms such as plants, avoidance of occasional extreme heat stress is not an option. Therefore, the ability to cope effectively with occasional extreme heat stress or multiple stress situations becomes critically important.
As with other HSPs, cyt HSP100/ClpB gene sequences are highly conserved among species, suggesting a critical and essential function for this HSP gene product. The Arabidopsis cyt HSP100/ClpB protein sequence is 43% identical to the yeast HSP104 sequence and 86% identical to the soybean HSP101/ClpB protein (Schirmer et al., 1994). The Synechococcus sp. ClpB sequence is 53% identical to the Arabidopsis and soybean cyt HSP100/ClpB sequences. Because of this high degree of sequence conservation, primers designed to anneal to the DNA sequence encoding the ClpB ATP-binding domains promoted the amplification of an internal portion of a lima bean nuclear cyt HSP100/ClpB gene sequence that was 90% identical to the soybean sequence at the amino acid level and 80% identical at the nucleotide level and clearly represented an ortholog of the soybean HSP101 gene.
Southern-blot analyses of soybean genomic DNA using the cyt HSP100/ClpB spacer region as probe detected a single band in soybean (Lee et al., 1994). Using a probe from the 3′ end of the Arabidopsis HSP101/ClpB gene on Southern blots of Arabidopsis DNA, Schirmer et al. (1994) also detected a single band. We determined that there are two copies of the cyt HSP100/ClpB gene in lima bean. The sequence of these genes appears to be highly conserved among cultivars of lima bean. Since lima bean is a true diploid, these results may suggest a duplication event or an altered genomic organization around the cyt HSP100/ClpB gene in this genus compared with Arabidopsis or soybean.
Characterization of a cp HSP100/ClpB Protein Gene in Lima Bean
A cDNA library constructed using mRNA isolated from heat-stressed lima bean leaves yielded a very high proportion of HSP-related sequences (S. Keeler, unpublished data). Among these, a sequence with homology at the amino acid level to the Synechococcus sp. ClpB1 gene sequence was identified and designated cp HSP100/ClpB. Southern blots indicate that there are two cpHSP100/ClpB genes in limabean that are distinct from the two cyt HSP100/ClpB genes. The homology of the lima bean cp HSP100/ClpB protein to the cyanobacterial HSP100/ClpB protein and the presence of a putative chloroplast transit sequence is strong evidence that this sequence represents the first identification of a cp form of HSP100/ClpB. Since the HSP100/ClpB protein in Synechococcus sp. is required for recovery of extreme heat stress, it follows that this cp protein may serve a similar purpose for the chloroplast.
The cp HSP100/ClpB protein sequence has all of the characteristics of other HSP100/ClpB sequences including the two conserved ATP-binding domains (amino acid 270–460 and 690–780 in Fig. 3), a spacer region of approximately 140 amino acids, and several other highly conserved regions outside the binding domains. PDZ domains (PSD-95/Discs large/Zo-1) are protein-protein interaction domains that often bind to COOH-terminal peptide sequences of partner proteins (Hanada et al., 1997). Some of the conserved regions in both of the cp and cyt HSP100/ClpB sequences are thought to function as PDZ domains controlling the recognition of and interaction with protein substrates (Levchenko et al., 1997).
Expression of HSP100/ClpB Gene Products in Heat-Stressed Leaf Tissue
As with Arabidopsis and soybean, the expression of the lima bean cyt HSP100/ClpB genes was strongly heat responsive in both the incubator and field heat stress experiments. Although the cp HSP100/ClpB genes were inducible by heat stress in incubator experiments, the response to heat was less stringent than the cyt HSP100/ClpB gene possibly due to differences in promoter elements. In the incubator heat experiments, even though the plants were maintained at 40°C for 300 min, the steady-state levels of the cyt HSP100/ClpB gene transcript decreased to approximately 25% of maximum by 300 min. According to a model proposed by Bharadwaj et al. (1999), heat shock transcription factors (HSTFs) are maintained in an inactive (monomeric) state by binding to constitutively present HSP90 chaperone complex. This complex includes HSP/c70 and several other chaperone-related proteins. HSP70 acts as a principal chaperone in refolding of proteins post-heat stress. The presence of excess denatured proteins post-heat stress may recruit away the HSP70 component allowing HSTF to polymerize into an active (trimeric) state. After the HSP genes have been transcribed and the level of HSP70 increases sufficiently to promote monomerization of HSTF, the expression of HSP genes should decrease accordingly. This method of auto regulation would serve to limit gene expression as long as sufficient levels of HSPs were present to deal with denatured or aggregated proteins. The decrease in the lima bean HSP100/ClpB signal over time under continuous heat stress is in agreement with this model. We observed lower but above non-stressed levels of signals for both the cyt HSP100/ClpB and cp HSP100/ClpB transcripts from 60 min throughout the 1,440 min under continuous heat stress. Assuming some level of turnover or inactivation of HSPs over time, a low level of transcription would need to be maintained for all of the HSP genes so that sufficient de novo-produced proteins are made to cope with continuous heat stress.
A heat-inducible 100-kD protein corresponding to cyt HSP100/ClpB was observed 20 to 30 min after the start of heat induction (Fig. 8). This correlates with an increase of cyt HSP100/ClpB transcript levels. As expected, the ClpC protein (90 kD) was constitutively expressed. A third band of approximately 95 kD is visible in unstressed samples but appears to increase after 240 min of heat stress. This 95-kD band may correspond to cp HSP100/ClpB. Schirmer et al. (1994) using the same anti-HSP104 antibody observed multiple approximately 100-kD molecular mass proteins induced by heat stress in Arabidopsis, tepary bean, wheat, maize, pea, and kidney bean leaf tissue. These bands may represent cyt HSP100/ClpB as well as cp HSP100/ClpB and possibly other species of heat-inducible Clp proteins.
The field experiments helped visualize how these genes responded and proteins accumulated in a real-life situation where many other environmental factors may have an influence. In the field samples in Figure 8, the cyt HSP100/ClpB protein is present regardless of transcript level. Even though the cyt HSP100/ClpB transcript level is low at 7 am, the cyt HSP100/ClpB protein is already present. It is likely that the HSP100/ClpB protein accumulated during the previous day's heat stress and was maintained through the night. We have observed a similar phenomenon in greenhouse heat stress experiments (S.L. Keeler, S.L. Kitto, and J.G. Haynes, unpublished data). Assuming some level of HSP protein turnover or inactivation, newly induced cyt HSP100/ClpB transcripts would be used to renew the levels of active cyt HSP100/ClpB for use during heat stress in the late afternoon of the same day or the following day.
In field samples, cp HSP100/ClpB transcript levels had a higher background level than cyt HSP100/ClpB and appeared to be much less heat responsive. Since photosynthesis is inactivated at temperatures that are several degrees below temperatures needed for inactivation of soluble enzymes (Santarius and Muller, 1979), additional protection of the photosystems from heat inactivation might be provided by constitutive production of the cp HSP100/ClpB protein. In Synechococcus sp., Erickson and Clarke (1996) have shown that HSP100/ClpB directly affects the ability of a Synechococcus sp. to acquire thermotolerance by reducing the susceptibility of photosystem II activity to heat. Alternatively, the cp HSP100/ClpB protein may have additional functions in chloroplasts along with heat or stress response. The cyt HSP100/ClpB protein of tobacco has been found to serve as a translational enhancer (Wells et al., 1998), whereas ClpC, which is a constitutively expressed cp protein, appears to be involved in protein translocation into chloroplasts (Nielsen et al., 1997). HSP100/ClpB proteins, in general, have been implicated in many processes in which they serve to regulate specific pathways by promoting the dissociation of key components (Patino et al., 1996). In higher plant chloroplasts, the cp HSP100/ClpB protein could be involved in turnover of photosystem components, regulation of chloroplast functions, translocation of proteins, or translational enhancement in addition to its heat protective capacity.
In the results presented here, we were unable to find variations in AT in leaf tissues or of HSP100/ClpB expression that could be correlated with cultivar-based differences in yield of lima bean under heat stress. However, we have identified a previously unknown chloroplast form of HSP100/ClpB protein. This novel HSP may be involved in mediating the response of the chloroplast to heat stress. Future investigation will focus on physiological measurements of heat responses in reproductive tissues of various cultivars or species of Phaseolus and in determining the activity of the cp HSP100/ClpB gene by complementation experiments in yeast or Synechococcus sp.
MATERIALS AND METHODS
Plant Sources and Treatments
Lima bean (Phaseolus lunatus) cultivars were chosen for these studies based on previous work by Kleiner and Frett (1996). Cultivars chosen for this study were used to represent three heat response categories: category I, (heat tolerant) yield was equivalent under heat stress conditions as under optimal temperatures (JWAR, cv Packers, cv Dixie Butter Bean, and cv Luna); category II, (heat average) yield was reduced 50% to 80% of optimal under heat stress conditions (F1072, CONC, ML408, and ET); category III, (heat sensitive) yield was <50% of optimal under heat stress conditions (BEL, cv Fordhook 5055, cv Dover Bush, and cv Large White).
Eleven-day-old seedlings of lima bean, which had been grown in 25°C day/15°C night were placed in a 37°C incubator for 4, 12, or 24 h in the dark for conductivity assays. For gene expression studies, 3- to 4-week-old plants that had first or second set of trifoliates were placed in an incubator at 37°C or 40°C. At set time intervals plants were removed, and the immature trifoliate leaves were harvested and frozen for RNA and protein extractions.
Samples representing field heat stress environments were taken from 3- to 4-week-old lima bean plants including five cultivars in a random-block design. Plants were irrigated and mulched with black plastic to prevent dehydration. The temperature at plant height adjacent to the sampled plants was recorded at the time of each sampling: 7 am, 25°C; 9 am, 30°C; 11 am, 32°C; 1 pm, 37°C.
Conductivity Assay for Release of Ions from Heat-Damaged Cells
Leaf discs (1.4 cm) were cut using cork borers, placed in 5.0 mL of distilled water in 15-mL polypropylene tubes, and shaken vigorously at 45°C. EC was measured using an EC bridge (Yellow Springs Instrutment, Yellow Springs, OH) at 30, 60, 90, and 120 min (EC1). After the final reading, tubes were frozen at −20°C overnight, warmed to 45°C, and a final conductivity reading taken (EC2). The ratio of EC1/EC2 × 100 was used to determine percent injury (March et al., 1982). Three replicates for each variable were done. The experiment was performed three times. Data were analyzed by ANOVA at each time point and for each cultivar.
Cloning and of an HSP100 Gene Fragment from Lima Bean
DNA was extracted from leaf tissue of the 12 cultivars listed above using an adaptation of the CTAB extraction method described by Rogers and Bendich (1988). All of the DNA manipulation and cloning procedures were as described in Sambrook et al. (1989). To obtain a genomic fragment of the cyt lima bean HSP100/ClpB gene, oligonucleotide primers were designed to anneal to regions of the HSP100/ClpB gene sequence encoding the two conserved ATP-binding sites. The primers were designed using the published sequences of the soybean (GenBank accession no. L35272) and Arabidopsis (GenBank accession no. U13949) cyt HSP100/ClpB genes. These primers were degenerate to reflect codon usage in both of these plant species and were also homologous to the ATP-binding regions of ClpC genes. Primer 1070 (5′-aagaattcaaaTGCAT- TGGTGCYACMACGCTTGA-3′) contained an additional 12 bases with an EcoRI restriction site on its 5′ end, and primer 1790 (5′-aaactcgagaaaGTMACTGGWATMCCKG- TCCA-3′) had an additional 12 bases containing an XhoI restriction site for the purpose of cloning. PCR reactions using these primers were done using standard conditions. The amplified cyt HSP100/ClpB fragment from F1072 was cloned into pBluescript SK+ (Stratagene).
PCR-amplified fragments were sequenced using dye terminator cycle sequencing and the 5′-degenerate primer 1070 as outlined in the Applied Biosystems (Foster City, CA) protocol. Sequence reactions were run on an ABI 371 sequencer. Chromatographic data were analyzed using Seqed 1.03 software (ABI, Sunnyvale, CA).
Southern Analyses
To analyze the number of homologous sequences and RFLP patterns in various cultivars, restriction digests were performed using appropriate conditions for each enzyme. Protocols for hybridizations, washes, and visualization using the Genius chemiluminescence detection system were as described by the manufacturer (Boehringer Mannheim, Indianapolis). High-stringency hybridization and wash conditions were 55°C hybridization, 68°C wash in 0.1× SSC.
RNA Extractions and Northern Analyses
RNA was isolated using an adaptation of a one-step guanidine thiocyanate separation (Chomczyuski and Sacchi, 1987). Poly(A+) RNA was isolated from the total RNA using the Promega Polyattract kit following the manufacturer's instructions (Promega, Madison, WI). Ten micrograms of total leaf RNA was separated on 1.0% (w/v) agarose/10% (v/v) formaldehyde in 1× MOPS buffer (0.2 m MOPS [3-(N-morpholino)propanesulfonic acid], 0.05 m sodium acetate, and 0.01 m EDTA) and transferred onto Hybond N+ membrane (Amersham) using 20× SSC. Northern blots were hybridized with antisense riboprobes from the cyt HSP100/ClpB gene and a soybean actin gene fragment at 65°C. Washes were performed with 0.5× SSC, 0.1% (w/v) SDS at 68°C. Hybridization to the actin riboprobe was used as a base line to control for loading assuming equivalent expression of this gene in all of the samples.
Slot blots were prepared by loading 10 μg of RNA per slot on a 72-well slot blotter (Schleicher & Schull, Keene, NH). The slots were rinsed with 10× SSC and baked at 80°C for 1 h. Hybridizations, washes, and development of signal were as described for northern blots. Digoxigenin-labeled probes were prepared as described below. Northern blots and slot blots were quantitated by converting the film images into digital form using an Alpha-Innotech Multi-Imager and analyzing the density of the spots on the images using the public domain NIH Image program (developed at the U.S. National Institutes of Health and available on the Internet at http://rsb.info.nih.gov/nih-image).
Hybridization with Digoxigenin-Labeled Probes
To obtain labeled riboprobe for Southern- and northern-blot hybridizations, linearized templates were used in in vitro transcription reactions with the Genius digoxigenin-labeling system (Boehringer Mannheim). Visualization of the bound digoxigenin-labeled probes was obtained by reaction with alkaline phosphate-conjugated anti-digoxigenin antibodies, followed by detection of phosphatase activity by addition of the chemiluminescent substrate, disodium 3-(4-methoxyspiro{1,2-dioxetane-3,2′-(5′-chloro)tricyclo[3.3.1.13,7]decan}4-yl) phenyl phosphate (CSPD).
Construction and Sequencing of a cDNA Library
Poly(A+) RNA was isolated from JWAR plants that had been heat stressed for 30 min at 40°C. cDNA clones were prepared using the ZAP-cDNA synthesis kit protocol (Stratagene) to give oriented inserts. A mass excision procedure was performed and the resultant ampicillin-resistant colonies were picked at random by an automated picker, grown in 96-well plates, and rapid extraction alkaline lysis miniplasmid preps (Qiagen, Valencia, CA) prepared. Sequencing reactions were done using Perkin Elmer reagents (PE Applied Biosystems, Foster City, CA), Amplitaq DNA polymerase, and a dye-conjugated M13 reverse sequencing primer. Reactions were run on ABI model 377 automated DNA sequencers (ABI). The first 500 to 600 bases from the 5′ end of each clone sequence was screened through the NCBI sequence database using BLASTN and BLASTX programs (Altschul et al., 1990).
A full-length cDNA for a putative cp HSP100/ClpB protein, was fully sequenced by sequential dye primer design allowing significant overlap of sequence fragments (GenBank accession no. AF203700). Comparisons to the sequences of other HSP100/ClpB genes was done using the MegAlign program (DNASTAR, Inc., Madison, WI).
Analysis of ClpB and ClpC Proteins
Two hundred milligrams of frozen leaf tissue was suspended and ground using a low-speed drill with mini-pestle in a total of 1.0 mL of lysis buffer (50 mm Tris [tris(hydroxymethyl)aminomethane], pH 8.0, 1 mm EDTA, and 100 mm NaCl) in a 1.5-mL eppendorf microcentrifuge tube. Cell debris was pelleted by centrifugation in a microcentrifuge at one-half speed for 15 min.
Twenty microliters (10 μg) of each leaf protein sample was loaded onto a 7.5% (w/v) acrylamide SDS-PAGE (acrylamide:bis, 36.5:1) minigel with a 5.0% (w/v) acrylamide stacking gel and electrophoresed at 50 V for 2 h, then at 26 mA for 3.5 h to achieve separation of high molecular mass proteins. Gels were electroblotted onto 0.45-μm nitrocellulose membrane in 25 mm Tris, 192 mm Gly, and 20% (v/v) methanol, pH 8.3, overnight at 30 V.
A polyclonal antibody to the yeast HSP104 protein was kindly provided by Dr. Susan Lindquist (University of Chicago). This rabbit antibody (no. 3-2) was raised against a peptide derived from a conserved region in the yeast HSP104 ATP1-binding site sequence (Parsell et al., 1991). This antibody reacts with both of the HSP100/ClpB and related ClpC proteins in plant extracts (Eriksson and Clarke, 1996). Western blots were developed using the enhanced chemiluminescence peroxidase system (Amersham).
Acknowledgments
Our sincere thanks to J. Frett, S. Antonins, and K. Kleiner for help with cultivar selection, to J. Pesek for assistance with statistical analyses, and to J. Odell for critical reading of the manuscript. Thanks to S. Lindquist for anti-HSP104 antibody and to E. Vierling for consultations on this project.
Footnotes
This work was supported in part by the U.S. Department of Agriculture-National Research Initiative (grant no. 95–37311–2458), by the University of Delaware, College of Agriculture and Natural Resources Competitive Grants Program, and by the State of Delaware Advanced Technology Fund.
LITERATURE CITED
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Becker J, Craig EA. Heat-shock proteins as molecular chaperones. FEBS Lett. 1994;219:11–23. doi: 10.1007/978-3-642-79502-2_2. [DOI] [PubMed] [Google Scholar]
- Bharadwaj S, Adnan A, Ovsenek N. Multiple components of the HSP90 chaperone complex function in regulation of HSF1 in vivo. Mol Cell Biol. 1999;19:8033–8041. doi: 10.1128/mcb.19.12.8033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- Eriksson M-J, Clarke AK. The heat shock protein ClpB mediates the development of thermotolerance in the cyanobacterium Synechococcus sp. strain PCC 7942. J Bacteriol. 1996;178:4839–4846. doi: 10.1128/jb.178.16.4839-4846.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesman S, Clark WP, Maurizi M. The ATP-dependent protease of Esherichia coli. J Biol Chem. 1990;265:7886–7893. [PubMed] [Google Scholar]
- Hanada T, Lin L, Chandy KG, Oh SS, Chishti AH. Human homologue of the Drosophila iscs large tumor suppressor binds to p56-lck tyrosine kinase and shaker type Kv1.3 potassium channel in T lymphocytes. J Biol Chem. 1997;272:26899–26904. doi: 10.1074/jbc.272.43.26899. [DOI] [PubMed] [Google Scholar]
- Kleiner KR, Frett JJ. The effects of high temperature on Phaseolus lunatus yield. Annu Rep Bean Improv Coop. 1996;39:65–66. [Google Scholar]
- Lee Y-RJ, Nagao RT, Key JL. A soybean 101-kD heat shock protein complements a yeast HSP104 deletion mutant in acquiring thermotolerance. Plant Cell. 1994;6:1889–1897. doi: 10.1105/tpc.6.12.1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levchenko I, Smith CK, Walsh NP, Sauer RT, Baker TA. PDZ-like domains mediate binding specificity in the Clp/HSP100 family of chaperones and protease regulatory subunits. Cell. 1997;91:939–947. doi: 10.1016/s0092-8674(00)80485-7. [DOI] [PubMed] [Google Scholar]
- Li PH, Davis DW, Shen Z-Y. High-temperature-acclimation potential of the common bean: can it be used as a selection criterion for improving crop performance in high temperature environments? Field Crops Res. 1991;27:241–256. [Google Scholar]
- Lin CY, Chen YM, Key JL. Solute leakage in soybean seedlings under various heat shock regimes. Plant Cell Physiol. 1985;26:1493–1498. [Google Scholar]
- Mackie WW. Origin, dispersal, and variability of the lima bean Phaseolus lunatus. Hilgardia. 1943;15:1–29. [Google Scholar]
- March L, Davis DW, Li PH, Silbernagel MJ. Two methods of evaluating genotypes of Phaseolus vulgaris under high temperature stress. Annu Rep Bean Improv Coop. 1982;25:55–56. [Google Scholar]
- Moore T, Keegstra K. Characterization of a cDNA clone encoding a chloroplast-targeted Clp homologue. Plant Mol Biol. 1993;21:525–537. doi: 10.1007/BF00028809. [DOI] [PubMed] [Google Scholar]
- Nielsen E, Akita M, Davila-Aponte J, Keegstra K. Stable association of chloroplastic precursors with protein translocation complexes that contain proteins from both envelope membranes and a stromal HSP100 molecular chaperone. EMBO J. 1997;16:935–946. doi: 10.1093/emboj/16.5.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nienhuis J, Tivang J, Skroch P. Genetic relationships among cultivars and landraces of lima bean (Phaseolus lunatus L.) as measured by RAPD markers. J Am Soc Hortic Sci. 1995;120:300–306. [Google Scholar]
- Nieto-Sotelo J, Kannan KB, Martinez LM, Segal C. Characterization of a maize heat shock protein 101 gene, HSP101, encoding a ClpB/Hsp100 protein homologue. Gene. 1999;230:187–195. doi: 10.1016/s0378-1119(99)00060-8. [DOI] [PubMed] [Google Scholar]
- Parsell DA, Kowal A, Singer MA, Lindquist S. Protein disaggregation mediated by heat-shock protein Hsp104. Nature. 1994;372:475–478. doi: 10.1038/372475a0. [DOI] [PubMed] [Google Scholar]
- Parsell DA, Sanchez Y, Stitzel JD, Lindquist S. HSP104 is a highly conserved protein with two essential nucleotide-binding sites. Nature. 1991;353:270–273. doi: 10.1038/353270a0. [DOI] [PubMed] [Google Scholar]
- Patino MM, Liu JJ, Glover JR, Lindquist S. Support for the prion hypothesis for inheritance of a phenotypic trait in yeast. Science. 1996;273:623–626. doi: 10.1126/science.273.5275.622. [DOI] [PubMed] [Google Scholar]
- Rogers SO, Bendich AJ. Extraction of DNA from plant tissues. Plant Mol Biol. 1988;A6:1–10. doi: 10.1007/BF00020088. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Ed 2. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Sanchez Y, Lindquist SL. HSP104 required for induced thermotolerance. Science. 1990;248:1112–1115. doi: 10.1126/science.2188365. [DOI] [PubMed] [Google Scholar]
- Santarius KA, Muller M. Investigations on heat resistance of spinach leaves. Planta. 1979;146:529–538. doi: 10.1007/BF00388828. [DOI] [PubMed] [Google Scholar]
- Schirmer EC, Glover JR, Singer MA, Lindquist S. HSP 100/Clp proteins: a common mechanism explains diverse functions. Trends Biochem Sci. 1996;21:289–296. [PubMed] [Google Scholar]
- Schirmer EC, Lindquist S, Vierling E. An Arabidopsis heat shock protein complements a thermotolerance defect in yeast. Plant Cell. 1994;6:1899–1909. doi: 10.1105/tpc.6.12.1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt M, Neupert W, Langer T. Hsp78, a Clp homologue within mitochondria, can substitute for chaperone functions of mt-hsp70. EMBO J. 1995;14:3434–3444. doi: 10.1002/j.1460-2075.1995.tb07349.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swan TM. Membrane fatty acid composition and membrane fluidity as parameters of stress tolerance in yeast. Can J Microbiol. 1997;43:70–77. doi: 10.1139/m97-010. [DOI] [PubMed] [Google Scholar]
- Vierling E. The roles of heat shock proteins in plants. Annu Rev Plant Physiol Plant Mol Biol. 1991;42:579–620. [Google Scholar]
- Weaver ML, Timm H. Influence of temperature and plant water status on pollen viability in beans. J Am Soc Hortic Sci. 1988;113:31–35. [Google Scholar]
- Wells DR, Tanguay RL, Le H, Gallie DR. HSP101 functions as a specific translational regulatory protein whose activity is regulated by nutrient status. Genes Dev. 1998;12:3236–3251. doi: 10.1101/gad.12.20.3236. [DOI] [PMC free article] [PubMed] [Google Scholar]