Abstract
Hydrogen-deuterium exchange mass spectrometry (HDX-MS) provides information about protein conformational mobility under native conditions. The area between exchange curves, Abec, a functional data analysis concept, was adapted to the interpretation of HDX-MS data and provides a useful measure of exchange curve dissimilarity for tests of significance. Importantly, for most globular proteins under native conditions, Abec values provide an estimate of the log ratio of exchange-competent fractions in the two states, and thus are related to differences in the free energy of microdomain unfolding.
Introduction
Hydrogen-deuterium exchange-mass spectrometry (HDX-MS) is a useful technique for characterizing the conformational mobility of proteins and protein complexes under native conditions [1–3]. Through the determination of deuterium incorporation into protein amide groups, HDX-MS provides an alternative method for obtaining structural information about proteins which, due to their size, limited solubility, or intrinsic conformational mobility, may not be suitable for other methods of structural biology. Individual aspects of the HDX-MS workflow, including sample handling, peptide separation, mass analysis, and post-processing analysis continue to be subjects for further development.
Hydrogen-deuterium exchange experiments provide information about the behavior of a complex system over time. In common with time course observations in fields such as gene expression, metabolomics, and toxicology studies, practical considerations generally limit the number of time points. Analytical tools that have been developed for extended or periodic time course data are unsuited to data with only a small number of time points. Functional data analysis (FDA) has been usefully applied to short time series data in several fields [4–6]. The dynamics of the specific system informs the choice of model curves. In the case of HDX-MS, although a theoretical framework for understanding the time dependence of amide hydrogen exchange in globular proteins has been appreciated for some time [1, 7–9], this framework has not been engaged as a basis for functional analysis of HDX-MS data. In this work, we present the area between the curves, Abec, as a measure of hydrogen-deuterium exchange curve dissimilarity and recall an interpretation that relates the area between exchange curves to differences in the fractional exposure of amide hydrogens [7]. We apply the method to published HDX-MS data [10] and show that Abec is useful for statistical analysis of conformational mobility differences.
Methods
We are interested in using the area between exchange curves to characterize differences in the conformational mobility of a protein between states a (e.g., free or apo) and b (e.g., ligand-bound). In the HDX-MS method, samples of the protein in the two conditions are exposed to high deuterium content buffers for times t1 … tn [1–3]. Samples are quenched and digested into peptides that are then chromatographically separated and analyzed by mass spectrometry. Data from nrep replicate experiments are usually reported as mean relative percent deuterium incorporated, D, with the associated standard deviations, and are often presented as plots of D vs. log time. We used a plot digitizer utility [11] to extract deuterium uptake values from published HDX data [10] and calculated the mean remaining hydrogen fractions ya,i = (100-Da,i)/100 and yb,i = (100-Db,i)/100, where i is the index over the timepoints. As depicted in Figure 1, we used four methods to estimate Abec. Two are geometric methods and two are parametric, curve-fitting methods. For additional details and statistical methods, see Supporting Information.
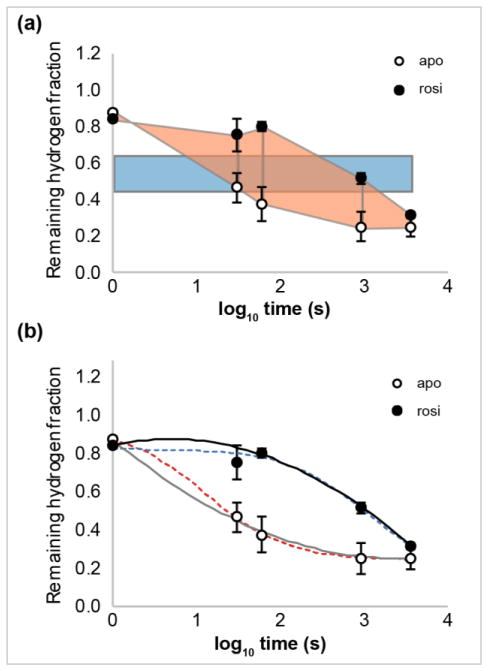
Figure 1.
Exchange curves and methods for estimating Abec, the area between exchange curves. Remaining hydrogen fractions for apo (○) and ligand-bound (●, rosiglitazone (rosi)) states of peptide aa 445–452 of Peroxisome proliferator-activated receptor gamma (PPARγ). HDX data from [10]. (a) Abec values are estimated by the difference of the averaged hydrogen fraction scaled by the span in log time (blue rectangle) or by trapezoidal approximation (peach trapezoids). (b) Time-dependent HDX data can be fitted by weighted second order polynomials (apo, solid grey; rosi, solid black) or logistical functions (apo, dotted red; rosi, dotted blue).
- Averaged incorporation. As this method is commonly implemented, deuterium uptake values are averaged over all timepoints [12]. Here, we average the remaining hydrogen fractions and scale the result by the span in log time:
-
Trapezoidal approximation. The area under the exchange curve for state a can be approximated as:The area between two exchange curves can be expressed as:
-
Weighted second order polynomial fit. The area under the exchange curve for state a is the integral of the fitted curve:The area between the curves is given by the difference:
-
Logistical function. The data are fitted byThe area between the curves is the difference between the areas under the fitted exchange curves:
where Aa,logi and Ab,logi are evaluated numerically.
Results and Discussion
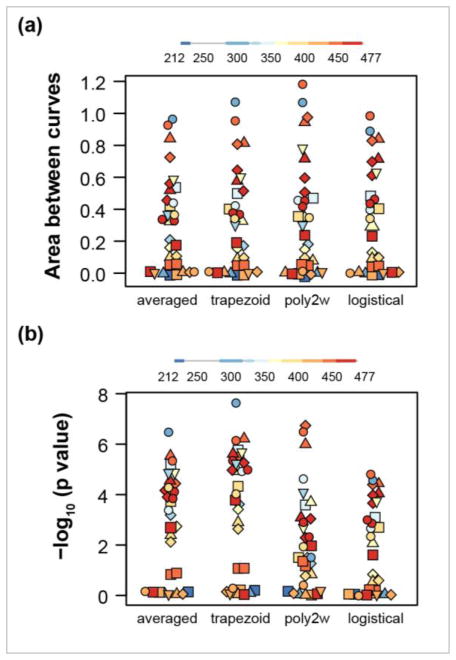
In a published account [10], HDX-MS was used to investigate conformational mobility in the ligand binding domain (LBD) of Peroxisome proliferator-activaed receptor gamma (PPARγ) in the ligand-free state (apo) compared with agonist- or partial agonist-bound states. We extracted deuterium uptake values from deuterium incorporation versus log time plots for 31 peptide/charge states for the apo and rosiglitazone-bound states using a digitizer tool [11]. We used two geometric methods and two parametric, curve-fitting methods to estimate the area between the exchange curves (see Methods and Supporting Information). We note that for these data, with only five exchange times, the parametric methods may be less appropriate. Specifically, for two peptides from the rosi-bound state, curve fitting to the three-parameter logistical function failed to converge; Abec values were calculated for only 29 peptides by this method. A visual representation of the distributions of the Abec values calculated by the four methods shows simlar patterns (Fig 2a). For the 29 peptides analyzed by all four methods, areas estimated by the two geometric methods were the most highly correlated (Pearson r = 0.995). Correlations of other pairs of methods ranged from 0.980 to 0.986. In addition, the numbers of peptides with substantially different exchange dynamics, as indicated by Abec values > 0.3, were similar by the four methods. The “averaged” method identified 16/31 substantially affected peptides, the ‘trapezoid’ and “poly2w” methods each identified 15/31 substantially affected peptides and the “logistical” method identified 14/29 substantially affected peptides. Values of Abec greater than 0.3 indicate at least a two-fold reduction in the average exchange-competent fraction of amide hydrogens in the stabilized state (see Supporting Information). Thus, the area between exchange curves is relatively insensitive to the method of area estimation, In addition, the magnitude of Abec provides molecular insight.
Figure 2.
Characterization of four methods for estimating Abec values and their statistical significance. Distributions of Abec values (a) and the negative log of the p value (b) are shown for 31 peptides comparing the exchange behavior of ligand-bound (rosiglitazone) and apo states of the LBD of PPARγ. The methods for area estimation are: averaged hydrogen fraction scaled by the span of log time (averaged), trapezoidal approximation (trapezoid), curve fitting to weighted second order polynomial (poly2w), curve fitting to a three parameter logistical function (logistical). Abec values from logistical functions were obtained for 29 out of 31 peptides. Symbol color indicates the approximate location of the peptide. Different charge states of a given peptide are represented by the same symbol.
The area between exchange curves provides a useful measure of dissimilarty in dynamic behavior for statistical tests (see Supporting Information). The estimated statistical significance varies among the four methods. Distributions of the negative of the log of the p values for the four methods show greater dispersion (Fig 2b). Visually, the distributions of significance values for the geometric methods appear the most similar and the “poly2w” distribution is the most divergent. For the 29 peptides analyzed by all four methods, negative log p values for the two geometric methods were the most highly correlated (Pearson r = 0.996). Correlation of “poly2w” negative log p values with those estimated by the other methods ranged from 0.775 to 0.818. The geometric methods identified 21/31 peptides to be significantly affected by ligand binding (p < 0.05), the “poly2w” method identified 19/31 significantly affected peptides, and the “logistical” method identified 16/29 significantly affected peptides.
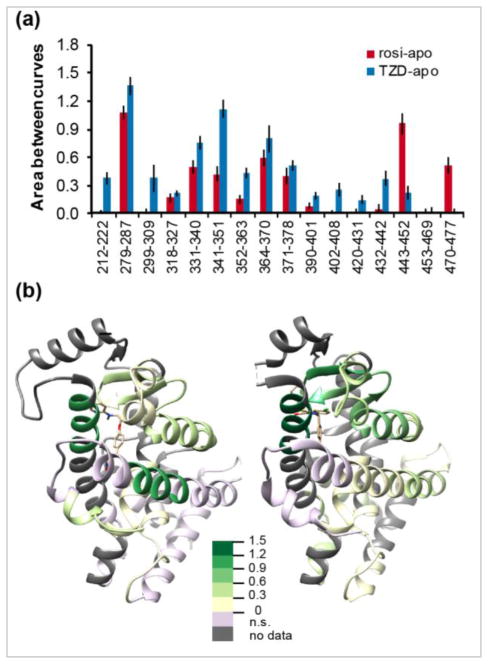
To illustrate molecular insights provided by Abec calculations, we used published HDX-MS data for the binding of a partial agonist, nTZDpa, compared with rosi, an agonist, on the conformational mobility of PPARγ LBD [10]. In particular, we used the trapezoid method for estimating the area between exchange curves as the number of timepoints in this study was relatively small, limiting the utility of parametric methods. Abec values for rosi-bound versus apo were significantly positive for 10 out of 16 LBD peptides whereas Abec values for nTZDpa-bound versus apo were significantly positive for 14 out of 16 LBD peptides (Fig 3a). Specifically, the binding of rosi has a strong stabilizing effect on the conformational mobility of peptides 279–287 and 443–452, resulting in an approximately tenfold decrease in the exposed fraction in the bound state. The effects of nTZDpa binding are more wide-spread. Peptide 279–287 shows a 20-fold decrease in exchange competency, whereas peptide 341–351 shows a 12-fold decrease, and peptides 331–341 and 364–370 each show a six-fold decrease. As shown in the spatial representation (Fig 3b), the conformational effects of rosi binding were smaller in magnitude and affected the binding pocket with additional stabilization of helix 12 (aa 470–477), whereas the effects of nTZDpa binding were larger in magnitude and were confined to the binding pocket without affecting the conformational mobility of helix 12.
Figure 3.
Effects of rosiglitazone (rosi, an agonist), and nTZDpa (TZD, a partial agonist) on hydrogen-deuterium exchange dynamics of PPARγ-LBD. HDX data are from [10]. (a) The area between exchange curves (Abec) for 16 peptides. (b) Abec values overlaid onto three-dimensional structures of PPARγ-LBD showing the effects of Rosi binding (left, PDB: 2PRG) and nTZDpa binding (right, PDB: 2Q5S). Color key: Abec values; n.s., not significant.
Conclusions
The time dependence of hydrogen-deuterium exchange of amide hydrogens in globular proteins is well understood to be described by a sum of exponential curves [1–3, 7]. In practice, recovery of individual exchange rate constants from HDX-MS data is rarely achieved due to the generally acknowledged ill-conditioned and under-determined characteristics of the problem, although many creative approaches toward extracting additional information have been developed [14–15].
The area between exchange curves is a practical and sensitive measure for statistical tests of significance for comparing the conformational dynamics of a protein under two conditions. Abec values also provide molecular insight into changes in the exposed fractions between the two states. By integration over the log of exchange times, differences in reaction progress are converted into information about changes in the solvent-exposed/exchange competent fractions in the two states.
Supplementary Material
Acknowledgments
In part, this research was supported by the Intramural Research Program of the Center for Cancer Research, National Cancer Institute, National Institutes of Health. The contributions of DPW were on his own time; Interclypse makes no claims on or regarding this work. The authors thank George Nelson and Maggie Cam of the CCR Collaborative Bioinformatics Resource for helpful discussions.
Footnotes
Supporting Information Available. Supporting information is available.
References
- 1.Englander SW. Hydrogen exchange and mass spectrometry: a historical perspective. J Am Soc Mass Spectrom. 2006;17:1481–1489. doi: 10.1016/j.jasms.2006.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Engen JR. Analysis of protein conformation and dynamics by hydrogen-deuterium exchange MS. Anal Chem. 2009;81:7870–7875. doi: 10.1021/ac901154s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gallagher ES, Hudgens JW. Mapping protein-ligand interactions with proteolytic fragmentation, hydrogen/deuterium exchange-mass spectrometry. Meth Enzym. 2016;566:357–404. doi: 10.1016/bs.mie.2015.08.010. [DOI] [PubMed] [Google Scholar]
- 4.Coffey N, Hinde J. Analyzing time-course microarray data using functional data analysis. Stat Appl Genom Mol Biol. 2011;10 Article 23. [Google Scholar]
- 5.Minas C, Waddell SJ, Montana G. Distance-based differential analysis of gene curves. Bioinformatics. 2011;27:3135–3141. doi: 10.1093/bioinformatics/btr528. [DOI] [PubMed] [Google Scholar]
- 6.Montana G, Berk M, Ebbels T. Modelling short time series in metabolomics: a functional data analysis approach. Adv Exp Med Biol. 2011;696:307–315. doi: 10.1007/978-1-4419-7046-6_31. [DOI] [PubMed] [Google Scholar]
- 7.Hvidt A, Wallevik K. Conformational changes in human serum albumen as revealed by hydrogen-deuterium exchange studies. J Biol Chem. 1972;247:1530–1535. [PubMed] [Google Scholar]
- 8.Engen JR, Wales TE. Analytical aspects of hydrogen exchange mass spectrometry. Ann Rev Anal Chem. 2015;8:127–148. doi: 10.1146/annurev-anchem-062011-143113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bai Y, Milne JS, Mayne L, Englander SW. Primary structure effects on peptide group hydrogen exchange. Proteins. 1993;17:75–86. doi: 10.1002/prot.340170110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bruning JB, Chalmers MJ, Prasad S, Busby SA, Kamemecka TM, He Y, Nettles KW, Griffin PR. Partial agonists activate PPARγ using a helix 12 independent mechanism. Structure. 2007;15:1258–1271. doi: 10.1016/j.str.2007.07.014. [DOI] [PubMed] [Google Scholar]
- 11.Rohatgi A. WebPlotDigitizer. 2016 arohatgi.info/WebPlotDigitizer.
- 12.Pascal BD, Chalmers MJ, Busby SA, Griffin PR. HD Desktop: an integrated platform for the analysis and visualization of H/D exchange data. J Am Soc Mass Spectrom. 2009;20:601–610. doi: 10.1016/j.jasms.2008.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.R Core Team. R A Language and Environment for Statistical Computing. R Foundation for Statistical Computing; Vienna, Austria: 2016. [Google Scholar]
- 14.Claesen J, Burzykowski T. Computational methods and challenges in hydrogen/deuterium exchange mass spectrometry. Mass Spectrom Rev. 2016 doi: 10.1002/mas.21519. [DOI] [PubMed] [Google Scholar]
- 15.Walters BJ, Jensen PF, Larraillet V, Lin K, Patapoff T, Schlothauer T, Rand KD, Zhang J. Conformational destabilization of immunoglobulin G increases the low pH binding affinity with the neonatal Fc receptor. J Biol Chem. 2016;291:1817–1825. doi: 10.1074/jbc.M115.691568. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.