Abstract
Natural rubber was extracted from the fig tree (Ficus carica) cultivated in Korea as part of a survey of rubber producing plants. Fourier transform infrared and 13C nuclear magnetic resonance analysis of samples prepared by successive extraction with acetone and benzene confirmed that the benzene-soluble residues are natural rubber, cis-1,4-polyisoprene. The rubber content in the latex of fig tree was about 4%, whereas the rubber content in the bark, leaf, and fruit was 0.3%, 0.1%, and 0.1%, respectively. Gel-permeation chromatography revealed that the molecular size of the natural rubber from fig tree is about 190 kD. Similar to rubber tree (Hevea brasiliensis) and guayule (Parthenium argentatum Gray), rubber biosynthesis in fig tree is tightly associated with rubber particles. The rubber transferase in rubber particles exhibited a higher affinity for farnesyl pyrophosphate than for isopentenyl pyrophosphate, with apparent Km values of 2.8 and 228 μm, respectively. Examination of latex serum from fig tree by sodium dodecyl sulfate-polyacrylamide gel electrophoresis revealed major proteins of 25 and 48 kD in size, and several proteins with molecular mass below 20 and above 100 kD. Partial N-terminal amino acid sequencing and immunochemical analyses revealed that the 25- and 48-kD proteins were novel and not related to any other suggested rubber transferases. The effect of EDTA and Mg2+ ion on in vitro rubber biosynthesis in fig tree and rubber tree suggested that divalent metal ion present in the latex serum is an important factor in determining the different rubber biosynthetic activities in fig tree and rubber tree.
Natural rubber is a polymer composed of 320 to 35,000 isoprene molecules. Among the natural rubber producing plant species, the Brazilian rubber tree (Hevea brasiliensis) is the only commercial source at present, due to its high rubber content and quality (Backhaus, 1985). However, diminishing acreage of rubber plantations and life-threatening allergies to the latex based products of rubber tree, coupled with increasing demand for high quality rubber, point to a need for alternative natural rubber resources. In recent years, guayule (Parthenium argentatum Gray), which accumulates rubber in parenchyma cells and contains high molecular mass-rubber comparable to rubber tree (Bowers, 1990), has attracted research interest for this reason. Despite some limitation as an alternative rubber crop due to its slow volume growth and low abundance of rubber particles, guayule has been proposed as a viable commercial alternative for hypoallergenic latex (Cornish, 1996, 1998).
Although more than 2,000 plant species are known to produce natural rubber, the molecular mechanism of rubber biosynthesis in most of these plants has not been studied in detail. Intense efforts to isolate and characterize key enzyme(s) involved in rubber biosynthesis in rubber tree, guayule, and Ficus elastica have been reported (Madhavan and Benedict, 1984; Dennis and Light, 1989; Light and Dennis, 1989; Cornish and Backhaus, 1990; Cornish, 1993; Siler and Cornish, 1993). However, the function and role of many additional proteins associated with the rubber biosynthetic machinery remain to be investigated. To better appreciate what contribution these proteins might make to the quality and size of rubber produced in plants, we have adapted a comparative approach of characterizing the enzymes and proteins related to the rubber biosynthetic machinery in different plant species.
Fig tree (Ficus carica) is cultivated for its fruit in southern parts of temperate zones. It has been extensively investigated for its proteolytic enzymes (Oner and Akar, 1993), amino acids, minerals and sugars (Kim et al., 1992), triterpenes (Ahmed et al., 1988), and organic acids (Shiraishi et al., 1996). Development of fig tree as an alternative rubber crop is promising because it generates a large latex volume, has a fast growth habit and long life expectancy, and is suitable for vegetative propagation, a means for amplifying genetically engineered trees. However, there have been no reports on rubber biosynthesis in fig tree. Evaluating the quantity and quality of the natural rubber produced in fig tree, and characterizing the rubber biosynthetic activity in rubber particles and latex serum remain important objectives in the development of fig tree as an alternative rubber producing plant.
In this report we compare the content and molecular size of cis-1,4-polyisoprene in fig tree to that in rubber tree, and we investigate the rubber biosynthetic activity of the latex serum and proteins associated with the rubber particles in fig tree. Our results show that the fig tree contains rubber biosynthetic activity distinct from rubber tree, including the divalent metal ion requirement necessary for optimal rubber biosynthetic activity in the latex serum of fig tree. Partial N-terminal amino acid sequencing and immunochemical analyses of the proteins tightly associated with the rubber particles revealed a novel 25-kD protein not related to any other previously characterized rubber transferases.
RESULTS
Fourier Transform Infrared (FTIR) and NMR Reveal That the Benzene Extract Is a Natural Rubber
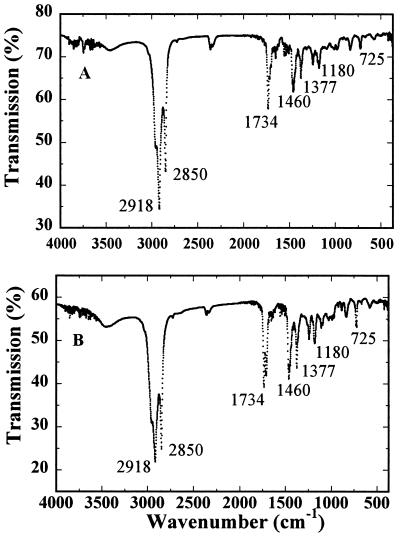
Natural rubber was obtained by successive extraction with acetone and benzene, and the benzene extracts were analyzed using FTIR and 13C NMR. Identical FTIR spectra were observed for the benzene extracts of fig tree and rubber tree (Fig. 1). The infrared (IR) spectra are also identical to cis-1,4-polyisoprene references, confirming that the benzene extract is a natural rubber.
Figure 1.
FTIR spectra of the rubber extracted from fig tree (A) and rubber tree (B) in potassium bromide disc. Four scans were co-added in the spectral range of 4,000 to 370 cm−1 at a resolution of 2 cm−1. Characteristic bands for cis-1,4-polyisoprene are indicated with their wave numbers.
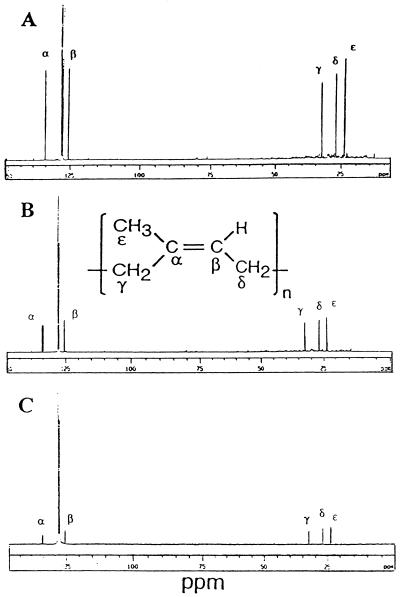
To further investigate the chemical nature of the benzene-soluble fractions, we obtained 13C NMR spectra for fig tree, F. elastica, and rubber tree. The 13C NMR spectra were referenced relative to the central peak of C6D6 at 128.5 ppm. As shown in Figure 2, five characteristic peaks with chemical shifts of 135.6, 124.8, 32.6, 27.0, and 23.0 ppm were observed. These peaks arise from two ethylenic, two methylene, and the methyl carbon atoms of the cis-1,4-polyisoprene (Duch and Grant, 1970). A peak at 29.6 ppm typical for high-molecular mass straight-chain hydrocarbons was not observed in the 13C NMR spectrum. These NMR spectra are consistent with the benzene-soluble fraction containing natural rubber.
Figure 2.
13C NMR spectra of the rubber extracted from fig tree (A), F. elastica (B), and rubber tree (C) in C6D6. About 5,000 scans were collected at a spectral width of 20,000 Hz. Representative peaks for cis-1,4-polyisoprene are indicated as α through ε.
Content and Mr of the Natural Rubber in Fig Tree
Table I summarizes the rubber contents in different parts of fig tree, and in the latex of F. elastica and rubber tree for comparison. The reported data represent averages of four replicate analyses. The latex of fig tree contained about 4% (w/v) natural rubber. However, other parts of fig tree like bark, leaf, and fruit contained much lower amounts of rubber (0.1%–0.3% [w/w]). In comparison, rubber tree and F. elastica latex contain about 32% (w/v) and 6% (w/v) natural rubber, respectively. To estimate the molecular size of the natural rubbers in fig tree, F. elastica, and rubber tree, the benzene-soluble residues were subjected to a gel-permeation chromatography. The average molecular mass of the rubber in the latex of fig tree was 190 kD, in comparison to 1,500 and 590 kD for the rubber in rubber tree and F. elastica, respectively.
Table I.
Content and size of the natural rubber in fig tree, F. elastica, and rubber tree
| Sample | Rubbera | Molecular Mass |
|---|---|---|
| % | kD | |
| Fig tree | ||
| Latex | 4.10 ± 0.35 | 190 |
| Bark | 0.35 ± 0.10 | |
| Leaf | 0.15 ± 0.05 | |
| Fruit | 0.10 ± 0.04 | |
| F. elastica | ||
| Latex | 6.08 ± 2.60 | 590 |
| Rubber tree | ||
| Latex | 32.10 ± 0.65 | 1,500 |
Rubbers were extracted by successive acetone-benzene extraction method, and molecular mass of the rubber was determined by a gel-permeation chromatography analysis.
All values are averages based on four determinations. Percentage in the latex is weight rubber in volume of latex, and percentage in bark, leaf, and fruit is per dry wt sample.
Comparison of in Vitro Rubber Biosynthetic Activities of the Latex from Fig Tree and Rubber Tree
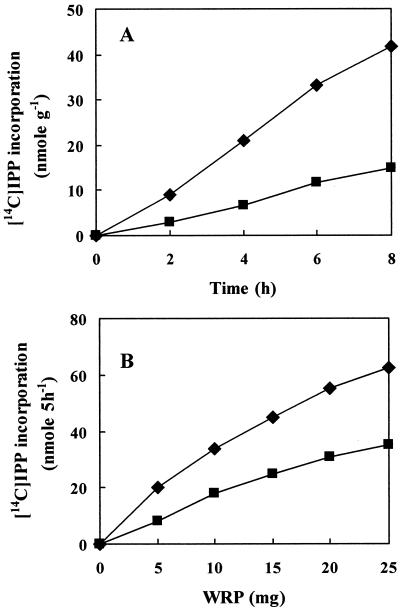
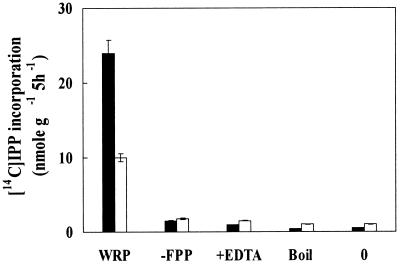
To investigate the rubber biosynthetic activity in fig tree, washed rubber particles (WRPs) were prepared and their in vitro activities tested. Initial attempts to measure the rubber transferase activity in rubber particles of fig tree were obscured by a large amounts of radiolabeled isoprenyl pyrophosphate (IPP) being physically trapped into WRP, as observed in F. elastica rubber particles (Cornish and Siler, 1996). We therefore carefully examined the background levels of IPP trapped by WRP for each analysis and used this background value to correct for each measurement. In time course experiments using the WRP as an enzyme source, [14C]IPP incorporation into rubber increased with incubation time (Fig. 3A). Rubber biosynthesis also increased with the amount of WRP added in the reaction mixture (Fig. 3B). To confirm that the increase of IPP incorporation resulted from the action of rubber transferase associated with WRP, a series of incubations containing WRP, farnesyl pyrophosphate (FPP), and EDTA were performed. Although it is difficult to define absolute units of rubber transferase activity because the observed enzymatic activities depend on the samples of WRP that contain varying amount of proteins, the overall rate of [14C]IPP incorporation into rubber by fig tree WRP was lower than that by rubber tree WRP (Fig. 4).
Figure 3.
Time course- (A) and WRP- (B) dependent incorporation of IPP into rubber. Reactions were carried out in 50 μL of 100 mm Tris-HCl, pH 7.5, containing 1 mm MgSO4, 1 mm DTT, 20 μm FPP, 0.1 mm [14C]IPP (55 mCi mmol−1), and 5 mg WRP in A and the indicated amount of WRP in B. Reactions in B were performed at 25°C for 5 h, the rubbers were extracted with benzene as described in text, and the resulting radioactivities of the 14C-labeled rubber were measured by a liquid scintillation counter. ♦, Rubber tree; ▪, fig tree.
Figure 4.
Rubber transferase activity of the WRP of fig tree (white bars) and rubber tree (black bars). Reactions were carried out in 50 μL of reaction mixture containing 5 mg of WRP as described in Figure 3. −FPP, Without FPP; +EDTA, with 25 mm EDTA; Boil, WRP-boiled for 5 min; 0, reaction at 0°C.
Incubations of fig tree WRP with EDTA still contained fairly high amount of radioactivity, indicating that the fig tree rubber particle exhibited relatively high levels of non-specific IPP trapping or incorporation. This distinct feature of fig tree rubber particle is similar to F. elastica rubber particle (Cornish and Siler, 1996). Boiling the rubber particles for 5 min (or heating at 60°C for 10 min) to destroy rubber transferase or incubating the reaction mixture at 0°C, inhibits IPP incorporation to a basal level (Fig. 4). Removing FPP, an efficient initiating molecule for rubber biosynthesis, from the reaction mixture resulted in a marked decrease in [14C]IPP incorporation (Fig. 4). These analyses using fig tree WRP as well as rubber tree WRP are consistent with the previously reported rubber biosynthetic activities in rubber tree, guayule, and F. elastica. Together, the results suggest that the enzyme(s) associated with the fig tree rubber particles catalyzes the condensation of IPP into rubber and that the [14C]IPP incorporated is into newly synthesized rubber chains.
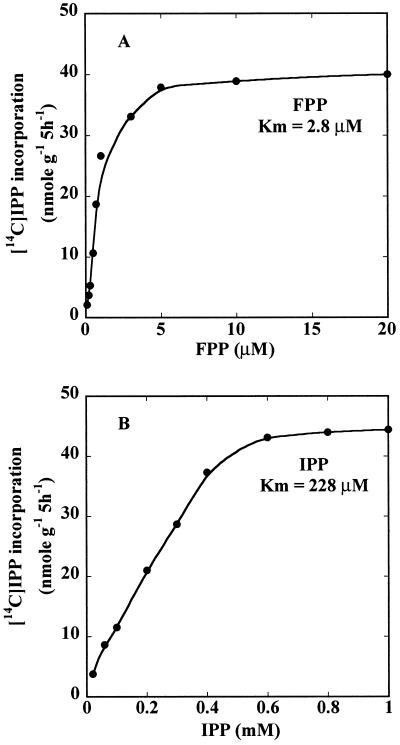
Kinetic analysis of rubber transferase activity in WRPs was performed by following IPP incorporation at varying substrate concentrations. At saturating IPP concentrations (1 mm), rubber biosynthesis increased up to 5 μm FPP. A Lineweaver-Burk analysis (1/rate plotted against 1/[substrate]) of the FPP concentration dependence (Fig. 5A) indicated an apparent Km for FPP of 2.8 μm. Similar kinetic analysis for IPP concentration dependence at saturating FPP concentration (20 μm) indicated an apparent Km for IPP of 228 μm (Fig. 5B).
Figure 5.
Substrate dependence of rubber transferase activity in fig tree WRP. Five milligrams of WRP was incubated with increasing amounts of FPP at saturating IPP concentration (1 mm; A), and increasing amounts of IPP at saturating FPP concentration (20 μm; B) at 25°C for 5 h. Product formation was plotted against substrate concentration, and the apparent Km values were calculated by a Lineweaver-Burk analysis.
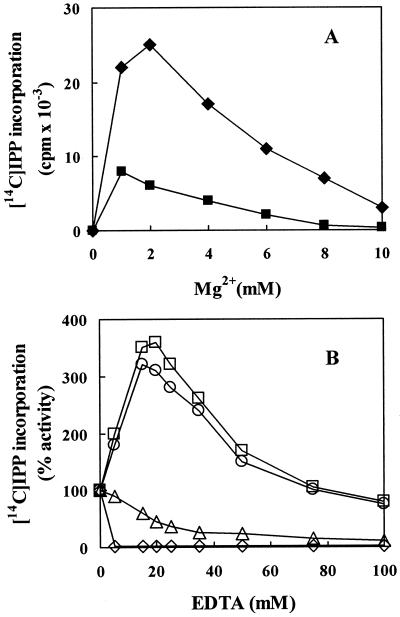
Rubber transferase activity was Mg2+ dependent, as shown in Figure 6A. Optimum Mg2+ concentrations for activities with fig tree WRP and rubber tree WRP were 1 and 2 mm, respectively. IPP incorporation was completely inhibited at Mg2+ concentrations higher than 10 mm. In vitro rubber biosynthesis activity with latex from fig tree was stimulated upon addition of EDTA to the reaction mixture (Fig. 6B). We, therefore, determined the effect of EDTA on rubber biosynthetic activity in the latex of fig tree and rubber tree. An addition of 5 to 10 mm EDTA to WRP of fig tree resulted in a marked decrease in IPP incorporation (data not shown). However, addition of EDTA up to 15 to 20 mm markedly increased [14C]IPP incorporation into rubber by fig tree latex. A gradual decrease in the [14C]IPP incorporation was measured at EDTA concentrations greater than 25 mm, but even at 50 to 100 mm EDTA, incorporation was higher than the non-treated control. This positive effect of EDTA on [14C]IPP incorporation was not observed in the fig tree latex subjected to washing on a membrane (3,000 Mr cutoff) to remove divalent ions. In contrast, a reduction in [14C]IPP incorporation was observed for the latex of rubber tree at concentrations of EDTA equal to or higher than 5 mm. These results suggested that different concentration of divalent ions in the latex of fig tree and rubber tree differentially affected the rubber biosynthesis in these two plant species. This was partially confirmed. Analysis of the latex of fig tree and rubber tree by inductively coupled plasma atomic emission spectrometer revealed that fig tree latex contained seven times more Mg2+ ion than rubber tree latex (120 versus 16 mm, data not shown). To confirm whether the increasing effect of EDTA on rubber biosynthesis resulted from the removal of excess magnesium ion in the latex, 120 mm EDTA was added to the fig tree latex to completely inhibit IPP incorporation, and then exogenous magnesium ion was added to the reaction mixture. IPP incorporation again increased proportional to the exogenous Mg2+ ion added (data not shown).
Figure 6.
Effect of Mg2+ ion on rubber transferase activity of fig tree and rubber tree WRPs (A), and dependence of IPP incorporation by the latex of fig tree and rubber tree on EDTA additions (B). Reactions were carried out as described in Figure 3 with 50-μL reaction mixtures containing 5 mg of WRP (black symbol) to determine the effects of Mg2+ (A), or 5-μL aliquots of latex serum (white symbol) were evaluated for effects of EDTA (B). ♦ and ⋄, Rubber tree; ▪, fig tree WRP; □ and ○, fig tree latex collected at different time of the year; ▵, fig tree latex filtered to 3-k membrane centricon (Amicon, Beverly, MA) to remove smaller molecules, including Mg2+ ions.
Major Proteins in the Latex of Fig Tree Involved in Rubber Biosynthesis in Vitro
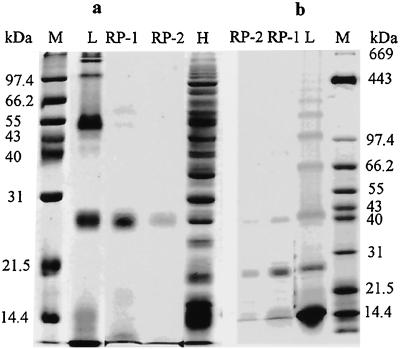
The latex of fig tree contains several distinct proteins in which 25- and 48-kD proteins are the most abundant proteins (Fig. 7). Several smaller proteins below 20 kD and larger proteins ranging from 100 to 500 kD in size were also observed. This simple protein profile for the latex of fig tree contrasts with the rubber tree protein profile that contains more than 100 different molecular-sized proteins (Posch et al., 1997).
Figure 7.
Analysis of the proteins in the latex and WRP of fig tree and rubber tree by 12% (a) and 6% (b) to 17% gradient SDS-PAGE. Rubber particle proteins were solubilized by incubating fig tree WRP in a detergent solution containing 0.1% (w/v) Triton X-100 and 1% (w/v) SDS. After electrophoresis, proteins were detected by Coomassie Blue staining. L, Latex; RP-1, first WRP; RP-2, second WRP; H, rubber tree latex; M, marker.
A few proteins are associated with the enzymatically active rubber particles of fig tree (Fig. 7, a and b). Many soluble proteins found in the latex and proteins loosely associated with rubber particles were removed during the extensive centrifugation/washing cycle for preparing WRP. Catalytically active fig tree WRP contained an abundant protein(s) with molecular mass of 25 kD and a weaker band corresponding to 48-kD protein (Fig. 7). The 48-kD protein was hardly visible in the WRP after extensive washing.
Siler and Cornish (1993) reported that a large protein of 376 kD in size was tightly associated with F. elastica rubber particles, and that this protein did not stain with Coomassie Blue, but was easily detected by silver stain. To test whether fig tree rubber particles might contain similar high molecular mass proteins, the WRP proteins were separated on 6% to 17% (w/v) gradient SDS gels. As shown in Figure 7B, several latex proteins with molecular mass above 100 kD were clearly detected with Coomassie Blue. In contrast, fig tree rubber particles did not contain proteins with molecular mass above 100 kD (data not shown).
N-Terminal Amino Acid Sequencing and Immunoinhibition Analyses Reveal Novel Rubber Particle Proteins in Fig Tree
To determine whether the rubber particle proteins in fig tree were similar to other previously characterized rubber transferases, partial N-terminal amino acid sequences of the 25- and 48-kD proteins were determined. The N-terminal amino acid sequence for the 25-kD peptide is DPPAVLDDAGG, and the 48-kD peptide has the apparent N-terminal sequence of LXNPNDLDALVKI. Database searches with these sequences did not identify any other similar protein.
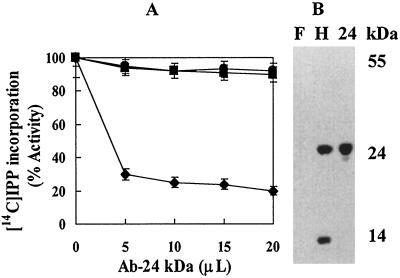
The 25-kD protein is the most abundant protein in the fig tree rubber particle, and is similar in size to the small rubber particle protein (SRPP) in rubber tree that plays a positive role in rubber biosynthesis (Oh et al., 1999). We, therefore, wanted to test whether the fig tree 25-kD protein was related to the SRPP. Western analysis of the rubber particle proteins in fig tree and rubber tree indicated that polyclonal antibody raised against SRPP bound to SRPP in rubber tree. The SRPP antibody also bound to the 14-kD protein that was originally designated as the rubber elongation factor in rubber tree (Dennis and Light, 1989). In contrast, the same antibody did not bind to the 25-kD protein in fig tree (Fig. 8B). This suggested that the most abundant rubber particle protein in fig tree was different from SRPP in rubber tree.
Figure 8.
A, Inhibition of rubber biosynthesis of rubber tree WRP, but not fig tree WRP by an antibody to the SRPP from rubber tree. Reactions were carried out as described in Figure 3 in 50 μL of reaction mixture containing 5 mg WRP and indicated amount of antibody raised against the SRPP from rubber tree. ▪, Fig tree WRP; ●, control serum; ♦, rubber tree WRP. B, Western analysis of rubber particle proteins. F, Fig tree; H, rubber tree; 24, purified SRPP.
To confirm that the 25-kD protein in fig tree was different from the SRPP in rubber tree, immunoinhibition effect of the SRPP-antibody on rubber biosynthesis was investigated. Varying amounts of polyclonal SRPP-antibody were added to the reaction mixtures containing fig tree and rubber tree WRP. Since SRPP-antibody binds to both the SRPP and the 14-kD proteins of rubber tree WRP (Oh et al., 1999), a marked decrease in [14C]IPP incorporation was observed by the addition of SRPP-antibody to the rubber tree WRP (Fig. 8A). In contrast, no decrease in [14C]IPP incorporation was detected by the addition of the same antibody to fig tree WRP. Control rabbit serum also did not inhibit [14C]IPP incorporation.
Natural Rubber Synthesized in Vitro Is Comparable in Size to Endogenous Rubber in Plant
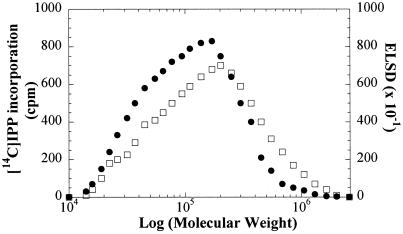
Mr distribution of the in vitro synthesized rubber was determined by reverse-phase thin-layer chromatography (RP-TLC) and gel-permeation chromatography (GPC). Most of the radiolabeled products were retained at the origin, whereas several weak bands migrated with the solvent (data not shown). This suggested that the 14C-polyisoprenes synthesized in vitro were mainly a mixture of long-chain length rubbers. To determine the Mr of the enzymatically synthesized rubber, the rubbers extracted with benzene from reaction mixtures containing WRPs were analyzed by GPC. The Mr distributions of the endogenous rubber traced by an evaporative light scattering detector (ELSD) and of 14C-labeled rubber synthesized in vitro are quite similar (Fig. 9). The peak Mr of the radiolabeled rubber was 180 kD, which is similar in size to that of the natural rubber extracted from the latex of fig tree.
Figure 9.
Mr distribution of endogenous rubber extracted from the latex of fig tree (□) and distribution of 14C-labeled rubber synthesized in vitro (●). Reaction was carried out in 1 mL of reaction mixture containing 200 mg WRP as described in Figure 3. The rubbers synthesized in vitro were extracted with benzene and subjected to a GPC. The eluent was monitored by ELSD and assayed for radioactivity.
DISCUSSION
Although the rubber content in the latex of fig tree is not as high as that in rubber tree and several other plants, its content is comparable to other rubber biosynthesizing temperate plants that accumulate about 1% to 3% rubber (Bowers, 1990). The amounts of latex in plants and rubber content in the latex vary depending on the physiological conditions of the plants. For example, the activity of rubber transferase and the accumulation of rubber were increased in guayule by low temperature (Ji et al., 1993) and water stress (Reddy and Das, 1988). Since we have only analyzed the rubber content of fig tree harvested during the summer, it is not known if this represents the maximum rubber content in the latex of fig tree. Further experiments will be needed to investigate seasonal variation of the amounts of natural rubber in fig tree. The molecular size of rubber is determined by the action of several enzymes and/or factors such as rubber transferase and rubber elongation factor. Since the latex of rubber tree has higher Mr rubber than fig tree, it is possible that the enzymes and/or factors involved in this aspect of rubber biosynthesis in rubber tree are different from those in fig tree.
In addition to these enzymes or factors, a recent paper demonstrated that the rate of rubber initiation and polymerization in vitro, and the final rubber Mr are affected by the concentration of initiator and IPP (Castillon and Cornish, 1999, and refs. therein). Similar to other rubber transferases characterized so far, fig tree rubber transferase has a much higher binding affinity for FPP initiator than for IPP with apparent Kms for FPP and IPP of 2.8 and 228 μm, respectively. These values are comparable with those of rubber tree, guayule, and F. elastica (Cornish and Backhaus, 1990; Cornish, 1993; Cornish and Siler, 1996; Castillon and Cornish, 1999). Therefore, the relationship between the concentration of initiator and IPP and the Mr of rubber in fig tree and other plant species remain critical components to be determined.
All rubber polymerase or rubber transferase (EC 2.5.1.20) have a divalent ion requirement for activity, and the concentration of Mg2+ in the latex serum may serve to affect the activity of rubber transferase. It has been shown that the optimum Mg2+ concentration for [14C]IPP incorporation into rubber in vitro for rubber tree is 2 mm. IPP incorporation decreases at higher magnesium concentrations (Lynen, 1969; Yusof et al., 1998). The present analyses also show that the optimum Mg2+ concentration for [14C]IPP incorporation into rubber for fig tree WRP is 1 mm. This is similar to that required for rubber tree WRP. However, our results clearly indicate that fig tree latex contains seven times more Mg2+ ion than rubber tree latex and this may account for some of the differential responses to EDTA.
Rubber biosynthesis occurs on the surface of rubber particles and the enzymes and/or factors necessary for rubber biosynthesis are tightly associated with the rubber particles in rubber tree (Cornish, 1993), guayule (Benedict et al., 1990; Cornish and Backhaus, 1990), and F. elastica (Cornish and Siler, 1996). However, despite continued efforts to isolate rubber transferase, the structure and the nature of this enzyme have not yet been identified. Based on a series of cross-specific immunoinhibition analyses, a 375-kD protein was suggested to be a rubber transferase in F. elastica (Siler and Cornish, 1993, 1994; Cornish et al., 1994). In guayule a 48.5-kD protein was the most abundant rubber particle protein and was regarded as a rubber transferase candidate (Backhaus et al., 1991). However, this protein was subsequently shown to be a P450 allen oxide synthase (Pan et al., 1995). A 14-kD protein tightly associated with large rubber particles in rubber tree was suggested as a rubber elongation factor (Dennis and Light, 1989), and its gene was subsequently cloned (Attanyaka et al., 1991; Goyvaerts et al., 1991). However, the direct role of these proteins in rubber biosynthesis have not yet been proven. More recently, we have isolated in rubber tree a cDNA encoding a major rubber particle protein tightly associated with small rubber particle, and demonstrated that SRPP plays a positive role in rubber biosynthesis (Oh et al., 1999). Although a direct involvement of these proteins in rubber chain elongation has not been demonstrated, they are associated with biosynthetically active rubber particles. Considering that different plant species produce different polymer sizes of rubber, it is possible that different complements of associated proteins determine ultimate polymer size.
In conclusion, fig tree contains natural rubber comparable to many other rubber biosynthesizing temperate plants. The rubber particle proteins of fig tree are distinct from that of rubber tree, in that the major proteins tightly associated with the particles are unique and not related to any other rubber transferase. The present results also show that different physiological condition including divalent metal ions in the latex serum can be an important factor in determining different rubber biosynthetic activities in fig tree and rubber tree.
MATERIALS AND METHODS
Extraction of the Natural Rubber
The latex of fig tree (Ficus carica) was collected directly from the fig trees grown either in the field or in a greenhouse throughout the year, and the latex of Ficus elastica was collected only from a tree grown in a greenhouse. The latex of rubber tree (Heavea brasiliensis) was obtained from the Rubber Research Institute of Malaysia (Sungei Buloh). A branch of fig tree was fractionated into barks, leaves, and fruits, and cut into small segments. The samples were dried completely in a forced-air oven at 70°C for 1 to 2 d. The rubber was extracted from fig tree, F. elastica, and rubber tree by acetone-benzene extraction method as described elsewhere (Stipanovic et al., 1980, 1982; Ji et al., 1993). The dried samples were homogenized in acetone in a homogenizer. The homogenized sample was centrifuged at 6,000 rpm for 10 min and the acetone supernatant was decanted. The pellet was extracted once more with acetone. Rubber was then extracted from the pellet by homogenizing it with benzene in a homogenizer followed by centrifugation. This procedure was repeated twice. After centrifugation at 7,000 rpm for 10 min, the benzene fraction was subjected to rotary vacuum evaporation to remove the solvent. For the analysis of rubber in the latex, acetone was added directly to the latex sample, and the rubber was extracted using the procedure described above. The percentage of rubber in the plants was determined by weighing the residues from the benzene extraction.
FTIR and 13C NMR Spectroscopy
The pressed discs for IR analysis were prepared by mixing the dried rubber extract with powdered potassium bromide. The mixture was ground thoroughly in a smooth agate mortar and pressed under a pressure of 10,000 to 15,000 psi into a transparent disc. IR spectra were obtained on a FTIR spectrometer (Perkin-Elmer, Norwalk, CT) purged with nitrogen. Four scans were summed in the spectral range of 4,000 to 370 cm−1 at a resolution of 2 cm−1. For each measurement, a blank spectrum was obtained to compensate for moisture in the sample chamber. For NMR measurements, the dried rubber extracts were dissolved in 0.4 mL of 99.5% C6D6 (Sigma, St. Louis). NMR spectra were obtained on a spectrometer (AMX-300 MHz, Bruker, Billerica, MA) at 24°C. About 5,000 scans were accumulated with a spectral width of 20,000 Hz. The spectra were referenced relative to C6D6 (128.5 ppm).
GPC
The rubber samples in tetrahydrofaron were filtered through a membrane of 0.4-μm porosity before GPC analysis. GPC (HPLC, Waters, Milford, MA) was carried out using three columns in series, a mixed-bed polydivinylbenzene column with molecular mass cutoff from 100 to 10,000 kD (Jordi), and two polystyrene-divinyl-benzene copolymer gels having an exclusion limit of 4 × 107 and 6 × 10 4 (Supelco, Bellefonte, PA). Measurements were made at column temperature of 35°C using tetrahydrofaron as eluent at a flow rate of 1 mL min−1, and the rubber was monitored by an ELSD (Alltech, Deerfield, IL). For the analysis of in vitro synthesized rubber, the eluent monitored by an ELSD was collected at 0.5-min intervals and assayed for radioactivity. The weight average Mr of the rubber was estimated by comparing the elution profile of the sample to the Mr distribution profile of standard polystyrene and polyisoprene.
Preparation of WRPs
The latex from fig tree was collected directly into ice-cold buffer containing 100 mm Tris [Tris(hydroxymethyl)-aminomethane]-HCl, pH 7.5, 5 mm MgSO4, 10 mm dithiothreitol (DTT), and 0.1 mm phenylmethylsulfonyl fluoride. Rubber tree latex was obtained from the Rubber Research Institute of Malaysia. The latex was centrifuged at 44,000g for 1 h at 4°C. The top creamy fraction of rubber particle was collected, resuspended in the same buffer containing 12% (w/v) glycerol, and recentrifuged. The supernatant serum fraction and the sedimented heavy particles were discarded. This washing procedure was repeated three times as described (Cornish and Backhaus, 1990; Siler and Cornish, 1993). Fig tree rubber particles sedimented during the washing cycle in the buffer without glycerol, similar to F. elastica rubber particles (Siler and Cornish, 1993; Cornish and Siler, 1996), but different from rubber tree and guayule rubber particles that remained afloat (Siler and Cornish, 1993). The buoyant rubber particles recovered after washing in wash buffer containing 12% (w/v) glycerol were stored at −20°C until use or were suspended in sample buffer containing 100 mm Tris-HCl, pH 7.5, 2 mm MgSO4, and 5 mm DTT.
Isolation and Gel Electrophoresis of the Proteins
The whole latex containing soluble- and rubber particle-associated proteins were analyzed by SDS-PAGE. Rubber particle proteins were solubilized by incubating rubber particles in a detergent solution containing 0.1% (w/v) Triton X-100 and 1% (w/v) SDS. The detergent-treated suspension was centrifuged at 13,000 rpm for 10 min and the supernatant fraction was analyzed by SDS-12% (w/v) PAGE. In other experiments, proteins were precipitated with cold acetone at 4°C for 24 h, and the precipitated proteins were washed with cold acetone and hexane to remove any lipid material and polyisoprene. The pellet was suspended in a gel-loading buffer, heated at 95°C for 5 min, and the denatured proteins were separated on a SDS-12% (w/v) PAGE. After electrophoresis, proteins were detected by visualization with either a Coomassie Blue or a silver staining using Silver Stain Kit (Bio-Rad Laboratories, Hercules, CA).
In Vitro Rubber Biosynthesis Assay
Rubber biosynthetic activity in vitro was determined by the methods previously described (Cornish and Backhaus, 1990; Siler and Cornish, 1993). Latex or WRP of fig tree and rubber tree were incubated in 50 μL of reaction mixture containing 100 mm Tris-HCl, pH 7.5, 0.1 to 1 mm [14C]IPP (55 mCi mmol−1, Amersham, Buckinghamshire, UK) depending on the experiment, 20 μm FPP, 1 mm MgSO4, and 1 mm DTT for 5 h at 25°C. For control experiments, 25 mm of EDTA was added to the reaction mixture to chelate Mg2+ necessary for rubber transferase activity. The reaction was stopped by adding 25 mm of EDTA. The resulting [14C]IPP-incorporated rubber was quantified by using either a filtration- or a benzene-extraction method as described (Cornish and Backhaus, 1990; Siler and Cornish, 1993; Oh et al., 1999). For the filtration method, the reaction mixture was filtered through either a 0.02- or 0.1-μm anodisc membrane (Whatman, Clifton, NJ). The filter was subjected to repeated washing with 1 m HCl and 95% (w/v) ethanol, and the remaining radioactivity on the washed filters was determined by a liquid scintillation counter (Beckman, Fullerton, CA). For the benzene-extraction method, the reaction mixture was extracted three times with two volumes of benzene, the benzene extract was mixed with a Ready Solv HP scintillation cocktail (Beckman), and the radioactivity was determined by a liquid scintillation counter (Oh et al., 1999). The radioactivity determined in the presence of 25 mm EDTA was used to correct for the [14C]IPP non-specifically trapped by WRP.
Western and Immunoinhibition Analyses of Rubber Biosynthesis
The rabbit polyclonal antibody and the mouse monoclonal antibody prepared against the 24-kD SRPP from rubber tree were kindly provided by Dr. H.Y. Yeang (Rubber Research Institute of Malaysia). Rubber particle proteins separated by SDS-12% (w/v) PAGE were transferred to a nitrocellulose membrane and were detected by SRPP-polyclonal antibody as described (Oh et al., 1999). The effect of SRPP-antibody on rubber biosynthesis was analyzed by incubating various amount of the antibody with WRPs as described (Oh et al., 1999).
Analysis of in Vitro Synthesized Rubber
The 14C-labeled rubber in the reaction mixture was extracted with benzene and was concentrated to a small volume. The rubber was subjected to TLC on a precoated silica-60 RP-TLC (Merck, Rahway, NJ) developed with a solvent system of acetone:water (19:1, v/v). The TLC plate was exposed to the image plate and the position of the rubber was monitored by a phosphor imager (Fuji, Tokyo; Tangpakdee et al., 1997). For the analysis of shorter chain-length rubber, polyprenyl pyrophosphates were hydrolyzed to the corresponding alcohols by adding potato acid phosphatase (20 units) in 50 mm acetate buffer, pH 4.7, 60% (w/v) methanol, and 0.1% (w/v) Triton X-100 at 37°C for 24 h (Fujii et al., 1982). The resulting alcohols were analyzed by a RP-TLC developed in acetone:water (19:1, v/v; Tangpakdee et al., 1997). Molecular-size analysis of the in vitro synthesized rubber was achieved by GPC as described above.
ACKNOWLEDGMENTS
We thank Hoong Yeet Yeang at the Rubber Research Institute of Malaysia for providing us with the antibodies. We acknowledge help from Kangmoo Hur at Kwangju Institute of Science and Technology (KJIST, Kwangju, Korea) for assistance with IR spectroscopy, and Jinhee Lee at KJIST with NMR spectroscopy. We thank Gap Chae Chung for helping with the atomic emission spectrometer.
Footnotes
This work was supported in part by Agricultural R&D Promotion Center (grant no. 297066–5) from the Korean Ministry of Agriculture. This is Kumho Life and Environmental Science Laboratory Publication number 35.
LITERATURE CITED
- Ahmed W, Khan AQ, Malik A. Two triterpenes from the leaves of Ficus carica. Planta Med. 1988;54:481. doi: 10.1055/s-2006-962522. [DOI] [PubMed] [Google Scholar]
- Attanyaka DPSTG, Kekwick RGO, Franklin FCH. Molecular cloning and nucleotide sequencing of the rubber elongation factor gene from Hevea brasiliensis. Plant Mol Biol. 1991;16:1079–1081. doi: 10.1007/BF00016080. [DOI] [PubMed] [Google Scholar]
- Backhaus RA. Rubber formation in plants: a mini review. Isr J Bot. 1985;34:283–293. [Google Scholar]
- Backhaus RA, Cornish K, Chen SF, Huang DS, Bess VH. Purification and characterization of an abundant rubber particle protein from guayule. Phytochemistry. 1991;30:2493–2497. [Google Scholar]
- Benedict CR, Madhavan S, Greenblatt GA, Venkatachalam KV, Foster MA. The enzymatic synthesis of rubber polymer in Parthenium argentatum Gray. Plant Physiol. 1990;92:816–821. doi: 10.1104/pp.92.3.816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowers JE. Natural rubber producing plants for the United States. Beltsville, MD: National Agricultural Library; 1990. [Google Scholar]
- Castillon J, Cornish K. Regulation of initiation and polymer molecular weight of cis-1,4-polyisoprene synthesized in vitro by particles isolated from Parthenium argentatum (Gray) Phytochemistry. 1999;51:43–51. [Google Scholar]
- Cornish K. The separate roles of plant cis and trans prenyl transferase in cis-1,4-polyisoprene biosynthesis. Eur J Biochem. 1993;218:267–271. doi: 10.1111/j.1432-1033.1993.tb18374.x. [DOI] [PubMed] [Google Scholar]
- Cornish K, inventor. December 3, 1996. Hypoallergenic natural rubber products from Parthenium argentatum (Gray) and other non-hevea brasiliensis species. Patent no. 5580942
- Cornish K, inventor. February 10, 1998. Hypoallergenic natural rubber products from Parthenium argentatum (Gray) and other non-hevea brasiliensis species. Patent no. 5717050
- Cornish K, Backhaus RA. Rubber transferase activity in rubber particles of guayule. Phytochemistry. 1990;29:3809–3813. [Google Scholar]
- Cornish K, Siler DJ. Characterization of cis-prenyl transferase activity localized in a buoyant fraction of rubber particles from Ficus elastica latex. Plant Physiol Biochem. 1996;34:377–384. [Google Scholar]
- Cornish K, Siler DJ, Grosjean OKK. Immunoinhibition of rubber particle-bound cis-prenyltransferases in Ficus elastica and Parthenium argentatum. Phytochemistry. 1994;35:1425–1428. [Google Scholar]
- Dennis MS, Light DR. Rubber elongation factor from Hevea brasiliensis: identification, characterization, and role in rubber biosynthesis. J Biol Chem. 1989;264:18608–18617. [PubMed] [Google Scholar]
- Duch MW, Grant DM. Carbon-13 chemical shift studies of the 1,4-polybutadiens and the 1,4-polyisoprenes. Macromolecules. 1970;3:165–174. [Google Scholar]
- Fujii H, Koyama T, Ogura K. Efficient enzymatic hydrolysis of polyprenyl pyrophosphates. Biochim Biophys Acta. 1982;712:716–718. [PubMed] [Google Scholar]
- Goyvaerts E, Dennis M, Light D, Chua NH. Cloning and sequencing of the cDNA encoding the rubber elongation factor of Hevea brasiliensis. Plant Physiol. 1991;97:317–321. doi: 10.1104/pp.97.1.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji W, Benedict CR, Foster MA. Seasonal variations in rubber biosynthesis, 3-hydroxy-3-methylglutaryl-coenzyme A reductase, and rubber transferase activities in Parthenium argentatum in the Chihuahuan desert. Plant Physiol. 1993;103:535–542. doi: 10.1104/pp.103.2.535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SS, Lee CH, Oh SL, Chung DH. Chemical components in the two cultivars of Korean figs Ficus carica L. J Korean Agric Chem Soc. 1992;35:51–54. [Google Scholar]
- Light DR, Dennis MS. Purification of a prenyltransferase that elongates cis-polyisoprene rubber from the latex of Hevea brasiliensis. J Biol Chem. 1989;264:18589–18597. [PubMed] [Google Scholar]
- Lynen F. Biochemical problems of rubber synthesis. J Rubb Res Inst Malaya. 1969;21:389–406. [Google Scholar]
- Madhavan S, Benedict CR. Isopentenyl pyrophosphate cis-1,4-polyisoprenyl transferase from Guayule (Parthenium argentatum Gray) Plant Physiol. 1984;75:908–913. doi: 10.1104/pp.75.4.908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh SK, Kang H, Shin DH, Yang J, Chow KS, Yeang HY, Wagner B, Breiteneder H, Han KH. Isolation, characterization, and functional analysis of a novel cDNA clone encoding a small rubber particle protein from Hevea brasiliensis. J Biol Chem. 1999;274:17132–17138. doi: 10.1074/jbc.274.24.17132. [DOI] [PubMed] [Google Scholar]
- Oner MD, Akar B. Separation of the proteolytic enzymes from fig tree latex and its utilization in gaziantep cheese production. Lebensm-Wiss Technol. 1993;26:318–321. [Google Scholar]
- Pan Z, Durst F, Werck-Reinchhart D, Gardner HW, Camara B, Cornish K, Backhaus RA. The major protein of guayule rubber particle is a cytochrome P450: characterization based on cDNA cloning and spectroscopic analysis of the solubilized enzyme and its reaction products. J Biol Chem. 1995;270:8487–8494. doi: 10.1074/jbc.270.15.8487. [DOI] [PubMed] [Google Scholar]
- Posch A, Chen Z, Wheeler C, Dunn MJ, Raulf-Heimsoth M, Baur X. Characterization and identification of latex allergens by two-dimensional electrophoresis and protein microsequencing. J Allergy Clin Immunol. 1997;99:385–395. doi: 10.1016/s0091-6749(97)70057-5. [DOI] [PubMed] [Google Scholar]
- Reddy AR, Das VSR. Enhanced rubber accumulation and rubber transferase activity in guayule under water stress. J Plant Physiol. 1988;133:152–155. [Google Scholar]
- Shiraishi SC, Kawakami K, Widodo SE, Shiraishi M, Kitazaki M. Organic acid profiles in the juice of fig fruits. J Fac Agric Kyushu Univ. 1996;41:29–33. [Google Scholar]
- Siler DJ, Cornish K. A protein from Ficus elastica rubber particles is related to proteins from Hevea brasiliensis and Parthenium argentatum. Phytochemistry. 1993;32:1097–1102. [Google Scholar]
- Siler DJ, Cornish K. Identification of Parthenium argentatum rubber particle proteins immunoprecipitated by an antibody that specifically inhibits rubber transferase activity. Phytochemistry. 1994;36:623–627. [Google Scholar]
- Stipanovic RD, O'Brien DH, Rogers CE, Hanlon KD. Natural rubber from sunflower. J Agric Food Chem. 1980;28:1322–1323. [Google Scholar]
- Stipanovic RD, Seiler GJ, Rogers CE. Natural rubber from sunflower: 2. J Agric Food Chem. 1982;30:611–613. [Google Scholar]
- Tangpakdee J, Tanaka Y, Ogura K, Koyama T, Wititsuwannakul R, Wititsuwannakul D, Asawatreratanakul K. Isopentenyl diphosphate isomerase and prenyl transferase activities in bottom fraction and C-serum from Hevea latex. Phytochemistry. 1997;45:261–267. [Google Scholar]
- Yusof F, Audley BG, Ismail F, Walker JM. A rapid assay for the incorporation of isopentenyl diphosphate in rubber biosynthesis. J Rubb Res. 1998;1:48–66. [Google Scholar]