Abstract
Introduction: Esophageal cancer is one of the most common malignant tumors in the world. Eukaryotic translation initiation factors 3e (eIF3e) makes a notable difference in the initiation of protein synthesis and tumor progression. However, the role of eIF3e in ESCC has not been revealed yet. This study aims to investigate the bio-functional and prognostic role of eIF3e in human ESCC tissues and cells.
Methods: Immunohistochemical staining and Western blot were performed to detect the eIF3e expression in ESCC patients' tissues. The Kaplan-Meier product limit method and Cox regression were conducted to analyze the association between eIF3e expression, together with other related clinical/pathological features, and patients' prognosis. In the analysis of bio-functional role of eIF3e, CCK-8 and Transwell assay were performed to compare the proliferative and migrative ability after knockdown of eIF3e.
Results: Up-regulation of eIF3e were demonstrated in ESCC tissues compared with the corresponding para-cancerous tissues. Overexpression of eIF3e was associated with deep tumor depth, lymph nodes metastasis, and advanced TNM stage. Importantly, the patients with high eIF3e expression suffered shorter overall and disease-free survival. Lymph node metastasis and histological grade served as independent prognostic predictors in patients' prognosis. Knockdown of eIF3e could inhibit cell proliferation and migration, in vitro.
Conclusions: The eIF3e expression, or combined with other members of eIF3 complex, might predict poor prognosis of ESCC and serve as a potential breakthrough in the multimodality therapy of ESCC.
Keywords: Esophageal squamous cell carcinoma, Eukaryotic translation initiation factors 3e, prognosis
Introduction
Esophageal cancer is one of the most common malignant tumors with high morbidity and mortality all over the world. China, Eastern Asia, where esophageal squamous cell carcinoma (ESCC) are markedly more than esophageal adenocarcinoma, suffer from the highest rates of incidence 1. For lack of effective targeted therapy for ESCC, despite surgery, radiotherapy and chemotherapy, the prognosis of patients remains poor. Therefore, it is a meaningful trend to investigate the underlying molecular and cellular processes of carcinogenesis, which might offer novel therapeutic targets for treatments in the future 1, 2.
Protein synthesis is a regular procedure to maintain body metabolism and function. Also, during the course of carcinogenesis and progression, protein synthesis plays an important role 3. In the initiation phase of protein synthesis, Eukaryotic translation initiation factors (eIFs) are the vital components for the formation of an initiation complex. The eIF3 is an 800-kDa molecular, which is the largest factor in the eIF family, made up of 13 subunits that are named from eIF3a to eIF3m 4. Through binding Met-tRNAi to the 40S ribosome and facilitating ribosome recruitment to mRNA, eIF3 makes a notable difference in the initiation of protein synthesis. Among the 13 subunits of eIF3, eIF3e is the p48 subunit and contributes to the stability of the whole eIF3 complex 5. However, the exact role of eIF3e in malignant tumor is still controversial in different types of tumors. In breast and lung cancer, eIF3e expression was down-regulated and served as a tumor suppressor6. On the contrary, in glioblastoma and colon cancer, eIF3e was demonstrated to overexpress and facilitate the tumor progression 7, 8. To date, the role of eIF3e in ESCC has not been revealed yet.
This study aimed to investigate the bio-functional and prognostic role of eIF3e in human ESCC tissues and cells. We demonstrated that eIF3e was up-regulated in ESCC tissues and high level eIF3e negatively correlated with the survival of patients. We also revealed that knockdown of eIF3e expression with small interfering RNA (siRNA) could significantly inhibit the proliferation and migration of ESCC cells, which indicated the tumor-promoting role of eIF3e in ESCC.
Materials and Methods
Tissue samples and follow-up
Up to 241 patients who underwent radical esophagectomy in the Department of Thoracic Surgery, Zhongshan Hospital, Fudan University, China, from 2008 to 2009, were enrolled in the study. All of the primary ESCC and adjacent tissues were collected after surgery and embedded with paraffin. For the follow-up, each patient was required to take chest CT, abdominal ultrasonography and serum tumor markers every three month during the first year after surgery. Then, the period of the follow-up was extended to twice a year. Patients' clinical/pathological data were collected in details, including sex, age, tumor pathology, tumor size, tumor depth, lymph node metastasis, tumor-node-metastasis (TNM) stage (according to American Joint Committee on Cancer, eighth edition) 9, histological grade, and overall survival (OS, defined as the interval from surgery to death) and disease free survival (DFS, defined as the time elapsed from the date of surgery to the discovery of local relapse or distant metastasis). In the Statistical analysis of DFS, the exact span of 22 paitents' DFS could not be figured out during the follow-up. Therefore, the rest 219 patients were collected for the analysis of DFS. The whole follow-up was finished at July 1th, 2017. The study has got the approval of the ethics committee on human research of Zhongshan Hospital, Fudan University.
Immunohistochemical staining (IHC) and evaluation
Immunohistochemical (IHC) staining was performed as previously described 10. The eIF3e antibody (Abcam 36766, Cambridge, UK) was used to detect the expression of the related protein. Two experienced pathologists, who had no idea of the patients' information and the purpose of the research, were invited to read the biopsies and scored the immunoreaction of the eIF3e expression. Each section was semi-quantitatively scored for the percentage of immunoreactions as follows: 0 (0% immunoreactive cells), 1 (1-10 % immunoreactive cells), 2 (11-50% immunoreactive cells) or 3 (>50% immunoreactive cells). In addition, we also semi-quantitatively scored the staining intensity of each section as follows: 0 (negative), 1 (weak, +), 2 (intermediate, ++), or 3 (strong, +++). The total immunoreaction score was calculated as the sum of both two factors. All the samples were classified into negative (0), weak (1-2), moderate (3-4) and strong (5-6) staining groups, respectively. For statistical purposes, the total immunoreaction scores of moderate and strong groups were classified as eIF3ehigh group, while the negative and weak groups were classified as eIF3elow group.
Cell lines culture, transfection and Western blot assay
The human ESCC cell line EC109 and KYSE510 were selected for research and cultured in DMEM, supplemented with 10% fetal bovine serum and 100 IU/ml penicillin/streptomycin in a humidified incubator, under 95% air and 5% CO2 at 37℃.
Lipofectamine 2000 (Invitrogen, Carlsbad, USA) was applied for transient transfection. Predesigned siRNA duplexes were purchased from Sangon Company. The sequences of siRNA-eIF3e-1 are 5'-GGAAUUAUUACAAGGUAAA-3' (F) and 5'- UUUACCUUGUAAUAAUUCC-3' (R). The sequences of siRNA-eIF3e-2 are 5'-GGAAGACCUUACACGGUUA-3' (F) and 5'- UAACCGUGUAAGGUCUUCC-3' (R). The sequences of siRNA-eIF3e-3 are 5'-CAAACAAGGAUGUUCGAAA-3' (F) and 5'- UUUCGAACAUCCUUGUUUG-3' (R). The knockdown group was cells transfected with siRNA-eIF3e sequence. The normal control group was cells transfected with a non-targeted scramble sequence. The transfection and Western blot assay were performed as previously described 11. The eIF3e antibody (Abcam 36766, Cambridge, UK) was used to detect the expression of the related protein.
RNA extraction and qRT-PCR analysis
The methods of total RNA extraction and qRT-PCR analysis were consistent to the previous research 11. The primers of eIF3e are 5'-ACAAGGCAAATGCAGTCAACCAGG-3' (F) and 5'-TGCTGCTCCTGAGTAATTCCCACA-3' (R). The primers of β-actin are 5'-TGACGTGGACATCCGCAAAG-3' (F) and 5'-CTGGAAGGTGGACAGCGAGG-3' (R).
CCK-8 assay and Transwell assay
CCK-8 assay was performed as previously described 12. After transfection, the OD values were detected at 24, 48, 72, 96 hours thereafter, respectively. Transwell assay was also performed as previously described 12. Each experiment was repeated 3 times.
Statistical analysis
Statistical analysis was performed with the SPSS 17.0. The correlations between the expression of eIF3e and clinical/pathological characteristics were analyzed by the Chi-square test. The DFS and OS curves were drawn with the Kaplan-Meier product limit method and analyzed with the log-rank test. The correlations between risk factors (related clinical/pathological characteristics) and DFS/OS were conducted with Cox proportional-hazards regression. The risk factors (covariates) included tumor size, tumor depth, lymph node metastasis, histological grade, TNM stage and eIF3e expression. The way of “mean ± standard deviation” was applied to note the data gathered in experiments. The comparison of individual variables were performed with Student's t-test, χ2 test, Fisher's exact test or Spearman coefficients to analyze the difference between each subgroup. Two-tailed tests at p<0.05 were considered statistically significant.
Results
High eIF3e expression negatively correlated with the ESCC patients' prognosis
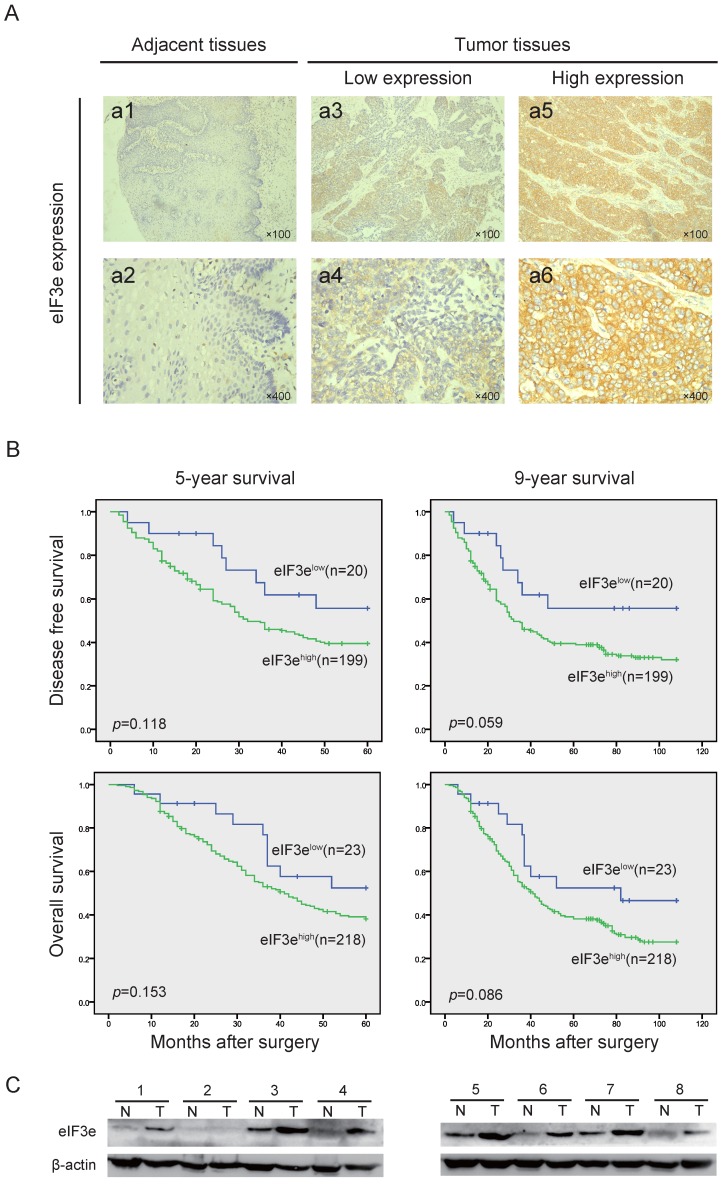
In the 241 ESCC tissues, we applied IHC to detect the eIF3e expression and found that cytoplasm was the main site for eIF3e staining (Fig. 1A). According to the scoring criteria described in the methods and materials, 23 patients were incorporated into low expression group (10%), while 218 patients were incorporated into high expression group (90%). Then, Chi-square test was applied to analyze the associations between eIF3e expression and clinical/pathologic features. As shown in Table 1, high eIF3e expression was correlated with deeper tumor depth (p=0.032), lymph nodes metastasis (p=0.023) and more advanced TNM stage (p=0.002), positively. In addition, with the progression of histological differentiation level, the percentage of high eIF3e expression, though not reaching the criterion of statistical significance, was gradually up-regulated (Table 1). However, no significant difference was found in the correlation between eIF3e and other factors, such as sex, age, tumor size, etc.
Fig 1.
The eIF3e expression is overexpressed in ESCC and negatively associated with patents' prognosis. (A), IHC analysis of eIF3e expression in ESCC tissues from the enrolled patients. The typical images of eIF3e expression level in adjacent tissues and tumor tissues were exhibited (original magnification × 100 for a1, 3, 5 and original magnification × 400 for a2, 4, 6). (B), 5-year and 9-year DFS and OS curves of enrolled ESCC patients, obtained by Kaplan-Meier product limit method, were compared according to eIF3e expression level. (C), Western blot assay was performed to detect the eIF3e expression in ESCC tumors and corresponding para-cancerous tissues. β-actin was used as an internal reference.
Table 1.
Analysis of the associations between eIF3e expression and clinical/pathologic features
| Clinicopathologic features | eIF3e expression | P value | ||
|---|---|---|---|---|
| Low | High | |||
| Total | 241 | 23(10) | 218(90) | |
| Age | ||||
| ≤60years | 128 | 13(10) | 115(90) | 0.730 |
| >60years | 118 | 10(9) | 103(91) | |
| Sex | ||||
| Male | 199 | 16(8) | 180(92) | 0.215 |
| Female | 47 | 7(16) | 38(84) | |
| Tumor size (cm) | ||||
| ≤3 | 148 | 13(9) | 132(91) | 0.707 |
| >3 | 98 | 10(10) | 86(90) | |
| Tumor depth | ||||
| T1 | 27 | 6(22) | 21(78) | 0.032 |
| T2 | 51 | 6(12) | 44(88) | |
| T3+4a | 166 | 11(7) | 153(93) | |
| Lymph node metastasis | ||||
| 0 | 150 | 19(13) | 127(87) | 0.023 |
| ≥1 | 53 | 4(4) | 91(96) | |
| TNM stage | ||||
| ⅠA+ⅠB | 26 | 7(27) | 19(73) | 0.002 |
| ⅡA+ⅡB | 136 | 13(10) | 119(90) | |
| ⅢA+ⅢB+ⅢC | 84 | 3(4) | 80(97) | |
| Histological grade | ||||
| Ⅰ | 5 | 1(20) | 4(80) | 0.620 |
| Ⅱ | 130 | 13(10) | 113(90) | |
| Ⅲ | 65 | 9(8) | 101(92) | |
a Bold values indicate statistical significance (p < 0.05).
Abbreviation: ESCC: Esophageal squamous cell carcinoma; eIF3e: Eukaryotic translation initiation factors 3e.
To explore the correlation between eIF3e expression and patients' prognosis, we employed Kaplan-Meier product limit method to draw the 5-year and 9-year DFS and OS curves. At the terminal of follow-up, 160 patients died of recurrence or metastasis, and 41 patients were lost to follow-up. The 5-year DFS and OS were 39.5% and 40.5%, respectively. The 9-year DFS and OS were 29.3% and 34.2%, respectively. As shown in Table 2 and 3, univariate analysis revealed that older age, larger tumor size, deeper tumor depth, lymph node metastasis, advanced TNM stage and higher histological grade were significantly correlated with poorer 5-year and 9-year DFS and OS (p<0.05).
Table 2.
Univariate analysis of ESCC patients' DFS
| Variables | 5-year DFS | 9-year DFS | ||||
|---|---|---|---|---|---|---|
| Events/patients (%) | Median DFS (mo) | P value | Events/patients (%) | Median DFS (mo) | P value | |
| Age | ||||||
| ≤60 years | 59/116(51) | 48 | 0.013 | 67/116(58) | 48 | 0.031 |
| >60 years | 66/103(64) | 28 | 69/103(67) | 28 | ||
| Sex | ||||||
| Male | 104/178(58) | 35 | 0.356 | 113/178(63) | 35 | 0.361 |
| Female | 21/41(51) | 43 | 23/41(56) | 43 | ||
| Tumor size | ||||||
| <3cm | 69/133(52) | 45 | 0.024 | 76/133(57) | 45 | 0.021 |
| ≥3cm | 56/86(65) | 29 | 60/86(70) | 29 | ||
| Tumor depth | ||||||
| T1 | 8/23(35) | NR | 0.015 | 8/23(35) | NR | 0.007 |
| T2 | 25/48(52) | 48 | 29/48(60) | 48 | ||
| T3+T4 | 92/148(62) | 29 | 99/148(67) | 29 | ||
| Lymph node metastasis | ||||||
| No | 55/131(42) | NR | 0.000 | 64/131(49) | 88 | 0.000 |
| Yes | 70/88(79) | 18 | 72/88(82) | 18 | ||
| TMN stage | ||||||
| ⅠA+ⅠB | 6/22(17) | NR | 0.000 | 7/22(32) | NR | 0.000 |
| ⅡA+ⅡB | 58/121(48) | NR | 66/121(54) | 61 | ||
| ⅢA+ⅢB+ⅢC | 61/76(80) | 18 | 63/76(83) | 18 | ||
| Histological grade | ||||||
| Ⅰ | 0/4(0) | NR | 0.000 | 0/4(0) | NR | 0.000 |
| Ⅱ | 56/115(49) | 49 | 64/115(56) | 49 | ||
| Ⅲ | 69/100(69) | 24 | 72/100(72) | 24 | ||
| eIF3e expression | ||||||
| Low | 8/20(40) | NR | 0.118 | 8/20(40) | NR | 0.059 |
| High | 117/199(59) | 32 | 128/199(64) | 32 | ||
a Bold values indicate statistical significance (p < 0.05).
Abbreviation: ESCC: Esophageal squamous cell carcinoma; DFS: disease free survival; eIF3e: Eukaryotic translation initiation factors 3e. NR, not reached.
Table 3.
Univariate analysis of ESCC patients' OS
| Variables | 5-year OS | 9-year OS | ||||
|---|---|---|---|---|---|---|
| Events/patients (%) |
Median DFS (mo) | P value | Events/patients (%) |
Median DFS (mo) | P value | |
| Age | ||||||
| ≤60 years | 70/128(55) | 49 | 0.035 | 80/128(56) | 49 | 0.027 |
| >60 years | 72/113(64) | 35 | 81/113(72) | 35 | ||
| Sex | ||||||
| Male | 118/196(60) | 41 | 0.551 | 134/196(68) | 41 | 0.524 |
| Female | 24/45(53) | 45 | 27/45(60) | 45 | ||
| Tumor size | ||||||
| <3cm | 79/145(54) | 48 | 0.031 | 91/145(63) | 48 | 0.026 |
| ≥3cm | 63/96(66) | 36 | 70/96(73) | 36 | ||
| Tumor depth | ||||||
| T1 | 9/27(33) | NR | 0.009 | 12/27(44) | NR | 0.009 |
| T2 | 29/50(58) | 50 | 34/50(68) | 50 | ||
| T3+T4 | 104/164(62) | 36 | 115/164(70) | 36 | ||
| Lymph node metastasis | ||||||
| No | 63/146(43) | NR | 0.000 | 79/146(54) | 78 | 0.000 |
| Yes | 79/95(83) | 25 | 82/95(86) | 25 | ||
| TMN stage | ||||||
| ⅠA+ⅠB | 7/26(27) | NR | 0.000 | 10/26(38) | NR | 0.000 |
| ⅡA+ⅡB | 66/132(50) | 53 | 79/132(60) | 53 | ||
| ⅢA+ⅢB+ⅢC | 69/83(83) | 25 | 72/83(87) | 25 | ||
| Histological grade | ||||||
| Ⅰ | 1/5(20) | NR | 0.000 | 1/5(20) | NR | 0.000 |
| Ⅱ | 64/126(51) | 53 | 77/126(61) | 53 | ||
| Ⅲ | 77/110(70) | 31 | 83/110(75) | 21 | ||
| eIF3e expression | ||||||
| Low | 10/23(43) | NR | 0.153 | 11/23(48) | 82 | 0.086 |
| High | 132/218(61) | 41 | 150/218(69) | 41 | ||
a Bold values indicate statistical significance (p < 0.05).
Abbreviation: ESCC: Esophageal squamous cell carcinoma; OS: overall survival; eIF3e: Eukaryotic translation initiation factors 3e; NR: not reached.
Importantly, though not reaching statistical significant difference, the patients in eIF3ehigh group suffered lower survival and shorter median survival than the patients in eIF3elow group (5-year DFS: 39.5% vs. 55.7%, median 5-year DFS: 32 months vs. not reached. 9-year DFS: 32.0% vs. 55.7%, median 9-year DFS: 32 months vs. not reached. 5-year OS: 37.0% vs. 52.4%, median 5-year OS: 41 months vs. not reached. 9-year OS: 27.5% vs. 46.6%, median 9-year OS: 41 months vs. 82 months.) (Fig. 1B). In multivariate analysis, Cox proportional-hazards regression results revealed that “lymph node metastasis” and “histological grade” (p<0.05) served as independent prognostic predictors in patients' prognosis (Table 4 and 5).
Table 4.
Multivariate analysis of ESCC patients' DFS
| Variables | 5-year DFS | 9-year DFS | |||||||
|---|---|---|---|---|---|---|---|---|---|
| HR | 95%CI | P value | HR | 95%CI | P value | ||||
| Tumor size | 0.066 | 0.060 | |||||||
| <3cm | Reference | Reference | |||||||
| ≥3cm | 0.712 | 0.496 | 1.023 | 0.066 | 0.716 | 0.505 | 1.014 | 0.060 | |
| Tumor depth | 0.456 | 0.505 | |||||||
| T1 | Reference | Reference | |||||||
| T2 | 0.772 | 0.273 | 2.180 | 0.625 | 0.616 | 0.217 | 1.750 | 0.364 | |
| T3+4 | 0.721 | 0.430 | 1.208 | 0.214 | 0.797 | 0.494 | 1.286 | 0.352 | |
| Lymph node metastasis | 0.007 | 0.016 | |||||||
| 0 | Reference | Reference | |||||||
| ≥1 | 0.354 | 0.165 | 0.757 | 0.007 | 0.407 | 0.196 | 0.843 | 0.016 | |
| TNM stage | 0.997 | 0.937 | |||||||
| ⅠA+ⅠB | Reference | Reference | |||||||
| ⅡA+ⅡB | 0.942 | 0.204 | 4.341 | 0.939 | 0.967 | 0.232 | 4.035 | 0.963 | |
| ⅢA+ⅢB+ⅢC | 0.986 | 0.454 | 2.144 | 0.972 | 0.890 | 0.420 | 1.886 | 0.762 | |
| Histological grade | 0.017 | 0.035 | |||||||
| Ⅰ | Reference | Reference | |||||||
| Ⅱ | 0.000 | 0.000 | 2.76×10201 | 0.961 | 0.000 | 0.000 | 3.07×10180 | 0.956 | |
| Ⅲ | 0.588 | 0.413 | 0.408 | 0.004 | 0.632 | 0.447 | 0.895 | 0.010 | |
| eIF3e expression | 0.704 | 0.456 | |||||||
| Low | Reference | Reference | |||||||
| High | 0.866 | 0.412 | 1.819 | 0.704 | 0.755 | 0.361 | 1.581 | 0.456 | |
a Bold values indicate statistical significance (p < 0.05).
Abbreviation: ESCC: Esophageal squamous cell carcinoma; DFS: disease free survival; HR: hazard ratio; CI: confidence interval; eIF3e: Eukaryotic translation initiation factors 3e.
Table 5.
Multivariate analysis of ESCC patients' OS
| Variables | 5-year OS | 9-year OS | |||||||
|---|---|---|---|---|---|---|---|---|---|
| HR | 95%CI | P value | HR | 95%CI | P value | ||||
| Tumor size | 0.054 | 0.035 | |||||||
| <3cm | Reference | Reference | |||||||
| ≥3cm | 0.718 | 0.512 | 1.006 | 0.054 | 0.709 | 0.516 | 0.976 | 0.035 | |
| Tumor depth | 0.721 | 0.882 | |||||||
| T1 | Reference | Reference | |||||||
| T2 | 0.754 | 0.275 | 2.072 | 0.584 | 0.829 | 0.329 | 2.090 | 0.691 | |
| T3+4 | 0.840 | 0.521 | 1.354 | 0.475 | 0.918 | 0.594 | 1.419 | 0.701 | |
| Lymph node metastasis | 0.003 | 0.016 | |||||||
| 0 | Reference | Reference | |||||||
| ≥1 | 0.336 | 0.164 | 0.691 | 0.003 | 0.431 | 0.217 | 0.855 | 0.016 | |
| TNM stage | 0.824 | 0.847 | |||||||
| ⅠA+ⅠB | Reference | Reference | |||||||
| ⅡA+ⅡB | 0.760 | 0.172 | 3.354 | 0.717 | 0.676 | 0.178 | 2.564 | 0.565 | |
| ⅢA+ⅢB+ⅢC | 1.061 | 0.507 | 2.218 | 0.876 | 0.885 | 0.436 | 1.794 | 0.734 | |
| Histological grade | 0.023 | 0.029 | |||||||
| Ⅰ | Reference | Reference | |||||||
| Ⅱ | 0.395 | 0.050 | 3.137 | 0.380 | 0.227 | 0.036 | 2.157 | 0.220 | |
| Ⅲ | 0.628 | 0.447 | 0.884 | 0.008 | 0.669 | 0.486 | 0.920 | 0.013 | |
| eIF3e expression | 0.766 | 0.550 | |||||||
| Low | Reference | Reference | |||||||
| High | 0.904 | 0.465 | 1.758 | 0.766 | 0.824 | 0.437 | 1.554 | 0.550 | |
a Bold values indicate statistical significance (p < 0.05).
Abbreviation: ESCC: Esophageal squamous cell carcinoma; OS: overall survival; HR: hazard ratio; CI: confidence interval; eIF3e: Eukaryotic translation initiation factors 3e.
eIF3e is up-regulated in ESCC tissues
We picked up 8 pairs of fresh primary ESCC tumors and the corresponding para-cancerous tissues to detect the eIF3e expression with Western blot. As shown in Fig. 1C, eIF3e expression in ESCC tissues was significantly higher than that in para-cancerous ones in 7 pairs of tissues. The other 1 pair of tissues showed no eIF3e expression both in tumor and para-cancerous tissues. Therefore, we could arrive at the conclusion that eIF3e expression was up-regulated in ESCC tissues.
eIF3e promotes the cell proliferation and migration of ESCC
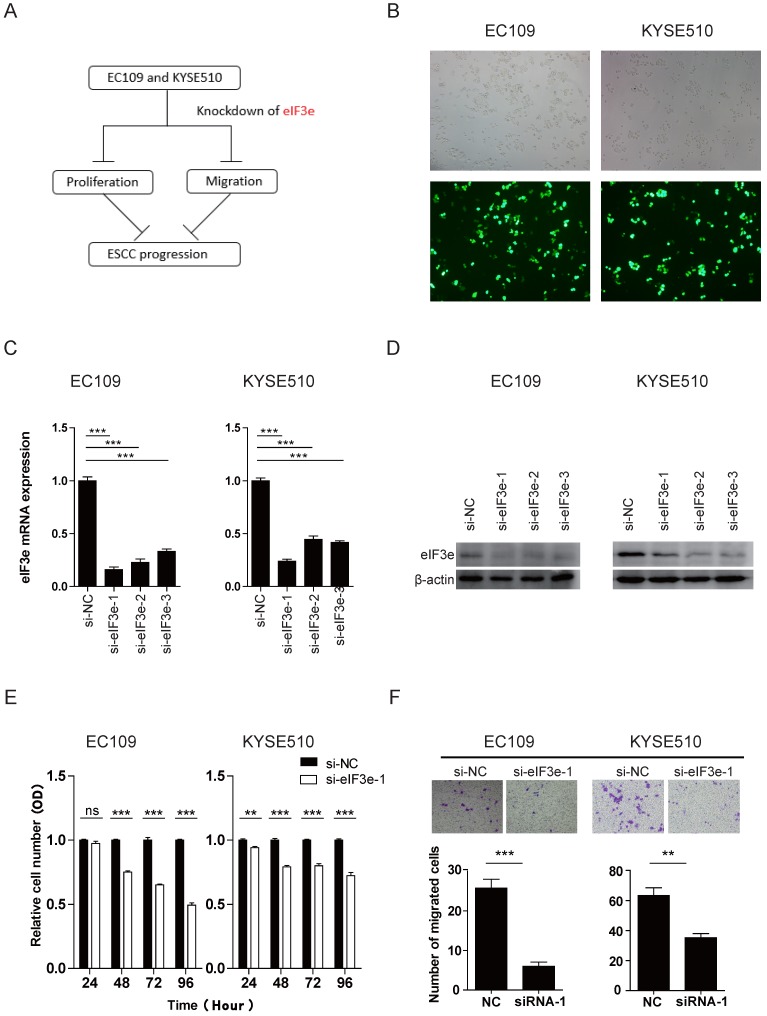
To explore the way of eIF3e to influence the bio-function of ESCC, we designed bio-functional assay (Fig. 2A). We constructed 3 fragments of siRNA to knock down the eIF3e expression in EC109 and KYSE510 cell lines. The efficiency of the transfection was high in both two cell lines, according to the Cy3-modified expression under light microscope and fluorescence microscope (Fig. 2B). The effect of the knockdown was validated through qRT-PCR and Western blot. As shown in the Fig. 2C and 2D, all the 3 fragments of siRNA showed great effect of knockdown in both two cell lines and we selected eIF3e-siRNA-1 and eIF3e-siRNA-2 for further study.
Fig 2.
eIF3e promotes the proliferative and migrative ability of ESCC. (A), the graph summarized the design of bio-functional assay. (B), the effeciency of the knockdown was validated through the Cy3-modified expression under light microscope and fluorescence microscope. (C and D), the effect of the knockdown was validated through qRT-PCR and Western blot analyses. β-actin was used as an internal reference. (E), the proliferative ability was assessed with CCK-8 assay at 24, 48, 72, and 96 hours after transfection. (F), the migrative ability was assessed with Transwell assay. (ns: no significance, * p <0.05, **p <0.01, ***p < 0.001).
CCK-8 assay was performed to study whether eIF3e made a difference in tumor proliferation. As shown in Fig. 2E and S1A, in both two cell lines, the proliferative level of knock-down groups were significantly lower than that of normal control (NC) groups after 24 hours after transfection (p < 0.05), which indicated that eIF3e promoted the proliferation of ESCC (Fig. 2E and S1B). To investigate whether eIF3e also influence the migration ability of ESCC cells, we performed Transwell assay. The results in Fig.2F showed that the numbers of migration cells in knock-down group were significantly less than those in NC group (p < 0.05), which indicated that eIF3e promoted the migration of ESCC.
To explore the potential mechanism behind these bio-functional change, we applied Western blot to detect some potential related signaling pathway, including AKT, epithelial-mesenchymal transition (EMT), p53. The data showed that after knockdown of eIF3e, no significant change appeared in these pathway related markers level (Fig S2A). In the future, we will continue exploring the deeper mechanism behind the biological phenomena of eIF3e promoting the proliferation and migration of ESCC.
Discussion
The function of eIF3 complex on tumor was investigated widely, among which eIF3e subunit was an important factor that made a difference in the initiation and progression of cancers 13. It has been reported that eIF3e binds directly to eIF4G, through interaction, to play an important role in recruiting the ribosome to the mRNA 14. In this study, we detected eIF3e expression in ESCC tissues and proved that eIF3e expression was up-regulated in cancer tissues compared with para-cancerous tissues. Through survival analysis, we demonstrated that high eIF3e expression correlated with poor prognosis. To explore the bio-function of eIF3e, we knocked down eIF3e expression and found that eIF3e promoted the cell proliferation and migration ability of ESCC, which might act together to promote the progression of ESCC and lead to poor prognosis. These data was the first step in our journey to explore the carcinogenesis role of eIF3 complex and the hidden mechanisms.
The earliest reports of eIF3e expression in tumors were in breast and non-small cell lung carcinoma 6, 15. Both two studies demonstrated that eIF3e expression was down-regulated and might serve as a tumor suppressor. Later, Desnoyers and colleagues further proved that decreased eIF3e expression in lung and breast epithelial cells leads to epithelial-to-mesenchymal transition (EMT) through activation of the TGF-β signaling pathway 16. However, in the study of glioblastoma, Sesen and colleagues demonstrated that eIF3e expression was elevated and inhibition of eIF3e expression led into decreased proliferation by the ways of cell cycle arrest and increased apoptosis 7. Similarly, in colon cancer, high expression of eIF3e was detected and contributed to tumor progression and predict poor prognosis 8. Therefore, the conflicts in the altered expression level of eIF3e in different tumors arouse our interests. We tried to detect the biological role of eIF3e in ESCC, in which no related study had been involved. Here, our data provided evidence that eIF3e over-expressed in ESCC tissues, accelerated tumor proliferation and migration ability, and correlated with poor prognosis. The results were consistent with the findings in glioblastoma and colon cancer, but contrary to the phenomena in breast and lung cancer. The altered expressional level between different cancers might be caused by the different pathologic patterns, which influenced the biological characteristics of each cancer.
The potential mechanisms of eIF3e in tumorigenesis were still under exploration and far from understanding. Grzmil and colleagues created an in vivo model of eIF3e function in zebrafish and demonstrated that eIF3e could modulate MEK-ERK signaling pathway, of which disorder would lead into abnormal proliferation of cells and tumorigenesis 17. Chen and colleagues found that eIF3e could down-regulated hypoxia inducible factor 2 alpha by hypoxia and pVHL independent regulation 18. In breast cancer, Morris and colleagues demonstrated eIF3e served as a regulator of DNA double-strand breaks through RNF8-dependent ubiquitylation pathway 19. These results unveiled the hidden mechanism of eIF3e and facilitates further study.
In the survival analysis, the patients in eIF3ehigh suffered lower survival and shorter median survival than the patients in eIF3elow group (5-year DFS: 39.5% vs. 55.7%, median 5-year DFS: 32 months vs. not reached. 9-year DFS: 32.0% vs. 55.7%, median 9-year DFS: 32 months vs. not reached. 5-year OS: 37.0% vs. 52.4%, median 5-year OS: 41 months vs. not reached. 9-year OS: 27.5% vs. 46.6%, median 9-year OS: 41 months vs. 82 months.) (Fig. 1B). Though the difference did not reach the criterion of statistical significance, the tendency of the difference could be discovered. For the reason that eIF3e was one of the 13 subunits of eIF3 complex, which should perform biological characteristics as an entirety, the bio-function was weakened when we detected the role of eIF3e alone 20. In our previous study, we demonstrated that eIF3b showed significant correlation with poor prognosis 2. In addition, considerable studies concerns the biological role of each subunits of eIF3 complex. Similar individual up-regulation of the subunits in specific cancers were reported for the 3a, 3b, 3c, 3d, 3h, 3i and 3m subunits 2, 21-26, while down-regulation is seen for the 3f subunit, which were also correlated with prognosis, significantly or insignificantly 27. The limitations of the current studies lied in the detection of an eIF3 subunit alone without assessing the status of other eIF3 subunits or other translational components 3. Further study would enroll much more patients and other subunits of eIF3 complex into the study, which could contribute to more comprehensive understanding of the prognostic role of eIF3e in ESCC.
To conclude, we have demonstrated that eIF3e was up-regulated in ESCC tissues, and high levels of eIF3e plays an important role in ESCC cells proliferation and migration. Up-regulation of eIF3e was correlated with deep tumor depth, lymph nodes metastasis, and advanced TNM stage, which predicted poor prognosis of ESCC. Also, eIF3e might serve as a potential breakthrough in the multimodality therapy of ESCC and offers a valuable guide for further investigation. In future, we will continue investigating the carcinogenesis mechanism of eIF3e, together with other subunits of eIF3 complex, to explore novel therapy for ESCC.
Supplementary Material
Supplementary figures.
Acknowledgments
This work was supported by the National Natural Science Foundation (81302099, 81372313 and 81401876), the Doctoral Fund for Young scholar of Ministry of Education of China (20110071120065), the Doctoral Fund for new teacher, Personnel Training Plan of Zhongshan Hospital, Fudan University and Outstanding study plan of Fudan University.
Abbreviations
- ESCC
Esophageal squamous cell carcinoma
- DFS
disease free survival
- OS
overall survival
- HR
hazard ratio
- CI
confidence interval
- eIF3e
Eukaryotic translation initiation factors 3e
- NR
not reached
- TMN
tumor-node-metastasis.
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA: a cancer journal for clinicians. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Xu F, Xu CZ, Gu J, Liu X, Liu R, Huang E. et al. Eukaryotic translation initiation factor 3B accelerates the progression of esophageal squamous cell carcinoma by activating beta-catenin signaling pathway. Oncotarget. 2016;7:43401–11. doi: 10.18632/oncotarget.9726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hershey JW. The role of eIF3 and its individual subunits in cancer. Biochimica et biophysica acta. 2015;1849:792–800. doi: 10.1016/j.bbagrm.2014.10.005. [DOI] [PubMed] [Google Scholar]
- 4.Hinnebusch AG. eIF3: a versatile scaffold for translation initiation complexes. Trends in biochemical sciences. 2006;31:553–62. doi: 10.1016/j.tibs.2006.08.005. [DOI] [PubMed] [Google Scholar]
- 5.Chiluiza D, Bargo S, Callahan R, Rhoads RE. Expression of truncated eukaryotic initiation factor 3e (eIF3e) resulting from integration of mouse mammary tumor virus (MMTV) causes a shift from cap-dependent to cap-independent translation. The Journal of biological chemistry. 2011;286:31288–96. doi: 10.1074/jbc.M111.267294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marchetti A, Buttitta F, Pellegrini S, Bertacca G, Callahan R. Reduced expression of INT-6/eIF3-p48 in human tumors. International journal of oncology. 2001;18:175–9. doi: 10.3892/ijo.18.1.175. [DOI] [PubMed] [Google Scholar]
- 7.Sesen J, Cammas A, Scotland SJ, Elefterion B, Lemarie A, Millevoi S. et al. Int6/eIF3e is essential for proliferation and survival of human glioblastoma cells. International journal of molecular sciences. 2014;15:2172–90. doi: 10.3390/ijms15022172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li Z, Lin S, Jiang T, Wang J, Lu H, Tang H. et al. Overexpression of eIF3e is correlated with colon tumor development and poor prognosis. International journal of clinical and experimental pathology. 2014;7:6462–74. [PMC free article] [PubMed] [Google Scholar]
- 9.Rice TW, Ishwaran H, Ferguson MK, Blackstone EH, Goldstraw P. Cancer of the Esophagus and Esophagogastric Junction: An Eighth Edition Staging Primer. Journal of thoracic oncology: official publication of the International Association for the Study of Lung Cancer. 2017;12:36–42. doi: 10.1016/j.jtho.2016.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lu CL, Guo J, Gu J, Ge D, Hou YY, Lin ZW. et al. CXCR4 heterogeneous expression in esophageal squamous cell cancer and stronger metastatic potential with CXCR4-positive cancer cells. Diseases of the esophagus: official journal of the International Society for Diseases of the Esophagus / ISDE. 2014;27:294–302. doi: 10.1111/dote.12100. [DOI] [PubMed] [Google Scholar]
- 11.Gu J, Ding JY, Lu CL, Lin ZW, Chu YW, Zhao GY. et al. Overexpression of CD88 predicts poor prognosis in non-small-cell lung cancer. Lung cancer. 2013;81:259–65. doi: 10.1016/j.lungcan.2013.04.020. [DOI] [PubMed] [Google Scholar]
- 12.Zhao GY, Lin ZW, Lu CL, Gu J, Yuan YF, Xu FK. et al. USP7 overexpression predicts a poor prognosis in lung squamous cell carcinoma and large cell carcinoma. Tumour biology: the journal of the International Society for Oncodevelopmental Biology and Medicine. 2015;36:1721–9. doi: 10.1007/s13277-014-2773-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen L, Uchida K, Endler A, Shibasaki F. Mammalian tumor suppressor Int6 specifically targets hypoxia inducible factor 2 alpha for degradation by hypoxia- and pVHL-independent regulation. The Journal of biological chemistry. 2007;282:12707–16. doi: 10.1074/jbc.M700423200. [DOI] [PubMed] [Google Scholar]
- 14.LeFebvre AK, Korneeva NL, Trutschl M, Cvek U, Duzan RD, Bradley CA. et al. Translation initiation factor eIF4G-1 binds to eIF3 through the eIF3e subunit. The Journal of biological chemistry. 2006;281:22917–32. doi: 10.1074/jbc.M605418200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Buttitta F, Martella C, Barassi F, Felicioni L, Salvatore S, Rosini S. et al. Int6 expression can predict survival in early-stage non-small cell lung cancer patients. Clinical cancer research: an official journal of the American Association for Cancer Research. 2005;11:3198–204. doi: 10.1158/1078-0432.CCR-04-2308. [DOI] [PubMed] [Google Scholar]
- 16.Desnoyers G, Frost LD, Courteau L, Wall ML, Lewis SM. Decreased eIF3e Expression Can Mediate Epithelial-to-Mesenchymal Transition through Activation of the TGFbeta Signaling Pathway. Molecular cancer research: MCR. 2015;13:1421–30. doi: 10.1158/1541-7786.MCR-14-0645. [DOI] [PubMed] [Google Scholar]
- 17.Grzmil M, Whiting D, Maule J, Anastasaki C, Amatruda JF, Kelsh RN. et al. The INT6 cancer gene and MEK signaling pathways converge during zebrafish development. PloS one. 2007;2:e959. doi: 10.1371/journal.pone.0000959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen L, Endler A, Uchida K, Horiguchi S, Morizane Y, Iijima O. et al. Int6/eIF3e silencing promotes functional blood vessel outgrowth and enhances wound healing by upregulating hypoxia-induced factor 2alpha expression. Circulation. 2010;122:910–9. doi: 10.1161/CIRCULATIONAHA.109.931931. [DOI] [PubMed] [Google Scholar]
- 19.Morris C, Tomimatsu N, Burma S, Jalinot P. INT6/EIF3E Controls the RNF8-Dependent Ubiquitylation Pathway and Facilitates DNA Double-Strand Break Repair in Human Cells. Cancer research. 2016;76:6054–65. doi: 10.1158/0008-5472.CAN-16-0723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.des Georges A, Dhote V, Kuhn L, Hellen CU, Pestova TV, Frank J. et al. Structure of mammalian eIF3 in the context of the 43S preinitiation complex. Nature. 2015;525:491–5. doi: 10.1038/nature14891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dong Z, Zhang JT. EIF3 p170, a mediator of mimosine effect on protein synthesis and cell cycle progression. Molecular biology of the cell. 2003;14:3942–51. doi: 10.1091/mbc.E02-12-0784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li T, Li S, Chen D, Chen B, Yu T, Zhao F. et al. Transcriptomic analyses of RNA-binding proteins reveal eIF3c promotes cell proliferation in hepatocellular carcinoma. Cancer science. 2017;108:877–85. doi: 10.1111/cas.13209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang F, Xiang S, Cao Y, Li M, Ma Q, Liang H. et al. EIF3D promotes gallbladder cancer development by stabilizing GRK2 kinase and activating PI3K-AKT signaling pathway. Cell death & disease. 2017;8:e2868. doi: 10.1038/cddis.2017.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cappuzzo F, Varella-Garcia M, Rossi E, Gajapathy S, Valente M, Drabkin H. et al. MYC and EIF3H Coamplification significantly improve response and survival of non-small cell lung cancer patients (NSCLC) treated with gefitinib. Journal of thoracic oncology: official publication of the International Association for the Study of Lung Cancer. 2009;4:472–8. doi: 10.1097/JTO.0b013e31819a5767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Qi J, Dong Z, Liu J, Zhang JT. EIF3i promotes colon oncogenesis by regulating COX-2 protein synthesis and beta-catenin activation. Oncogene. 2014;33:4156–63. doi: 10.1038/onc.2013.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goh SH, Hong SH, Hong SH, Lee BC, Ju MH, Jeong JS. et al. eIF3m expression influences the regulation of tumorigenesis-related genes in human colon cancer. Oncogene. 2011;30:398–409. doi: 10.1038/onc.2010.422. [DOI] [PubMed] [Google Scholar]
- 27.Doldan A, Chandramouli A, Shanas R, Bhattacharyya A, Cunningham JT, Nelson MA. et al. Loss of the eukaryotic initiation factor 3f in pancreatic cancer. Molecular carcinogenesis. 2008;47:235–44. doi: 10.1002/mc.20379. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figures.