Abstract
Homeostatic signaling systems ensure stable, yet flexible neural activity and animal behavior1–4. Defining the underlying molecular mechanisms of neuronal homeostatic signaling will be essential in order to establish clear connections to the causes and progression of neurological disease. Presynaptic homeostatic plasticity (PHP) is a conserved form of neuronal homeostatic signaling, observed in organisms ranging from Drosophila to human1,5. Here, we demonstrate that Semaphorin2b (Sema2b) is target-derived signal that acts upon presynaptic PlexinB (PlexB) receptors to mediate the retrograde, homeostatic control of presynaptic neurotransmitter release at the Drosophila neuromuscular junction. Sema2b-PlexB signaling regulates the expression of PHP via the cytoplasmic protein Mical and the oxoreductase-dependent control of presynaptic actin6,7. During neural development, Semaphorin-Plexin signaling instructs axon guidance and neuronal morphogenesis8–10. Yet, Semaphorins and Plexins are also expressed in the adult brain11–16. Here we demonstrate that Semaphorin-Plexin signaling controls presynaptic neurotransmitter release. We propose that Sema2b-PlexB signaling is an essential platform for the stabilization of synaptic transmission throughout life.
Semaphorins are a large family of secreted or membrane-associated signaling proteins and Plexins serve as signal transducing Semaphorin receptors8–10. Sema-Plexin signaling was initially described as mediating growth cone collapse8,9. But, Sema-Plexin signaling is far more diverse10,11. Importantly, Semaphorins and Plexins continue to be expressed in the mature brain where their function remains largely unknown12–16. Semaphorins, have been documented as synaptic signaling proteins, but this activity has been limited to the control of neuroanatomical synapse formation and elimination15–17. Here, we demonstrate that Sema-Plexin signaling achieves a retrograde, trans-synaptic control of presynaptic neurotransmitter release and homeostatic plasticity.
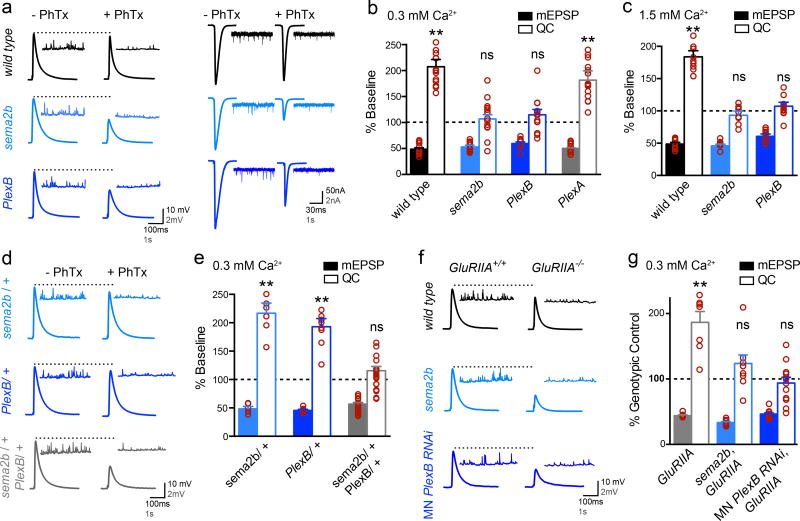
We use a well-documented assay to induce PHP, applying a sub-blocking concentration of the glutamate receptor antagonist philanthotoxin-433 (PhTx; 15µM) to significantly decrease average miniature excitatory postsynaptic potential (mEPSP; 0.3 mM [Ca2+]e) or miniature excitatory postsynaptic current (mEPSC; 1.5mM [Ca2+]e) amplitudes. This postsynaptic perturbation induces a significant increase in presynaptic neurotransmitter release (quantal content; QC) that offsets the postsynaptic perturbation and restores normal muscle excitation (Fig. 1a–c: raw values and sample sizes for all normalized data are presented in Supp Table 1). This is diagnostic of PHP1. When we repeat this assay in a null mutation of either the sema2b gene (sema2bC4) or the PlexB gene (PlexBKG00878), PHP is blocked (Fig. 1a–c; see also Extended Data Fig. 1 for further description of the gene mutations used in this study). Consistent with this being a loss-of-function phenotype, heterozygous mutations (either sema2b/+ or PlexB/+) express normal PHP (Fig. 1d, e). Remarkably, a double heterozygous mutant combination of sema2b/+ and PlexB/+ blocks PHP, consistent with both genes acting in concert to drive the expression of PHP (Fig. 1 d, e).
Figure 1. Sema2b and PlexinB are necessary for presynaptic homeostatic plasticity.
a) Representative traces (left at 0.3mM [Ca2+]e and right at 1.5mM [Ca2+]e). b) Average mEPSP and quantal content (QC) expressed as a percent change in the presence of PhTx compared to each genotype in the absence of PhTx. c) Data as in (b) at 1.5mM [Ca2+]e. d) Representative traces as in (a). e) Average data as in (b). f) Representative traces as in (a) for indicated genotypes alone (GluRIIA+/+) or as double mutants with GluRIIA (GluRIIA−/−). g) Average data for each genotype compared to the genotypic control in the absence of the GluRIIA mutation. ** p<0.01 student’s t-test (2-tail), pairwise comparison to genotypic control. Data are mean ± standard error. Individual data points are overlaid.
We then explored the long-term maintenance of PHP and the involvement of other semaphorin or Plexin gene family members. Deletion of a non-essential glutamate receptor subunit (GluRIIA) induces a long-lasting form of PHP1. We find that long-term PHP is blocked in a sema2b, GluRIIA double mutant as well as in GluRIIA animals expressing transgenic RNAi to knockdown PlexB selectively in motoneurons (Fig. 1f, g; see also below). Next, we tested mutations in all of the remaining semaphorin and Plexin genes encoded in the Drosophila genome (Extended Data Fig. 1b). The sema2b and PlexB mutants are the only ones that disrupt PHP.
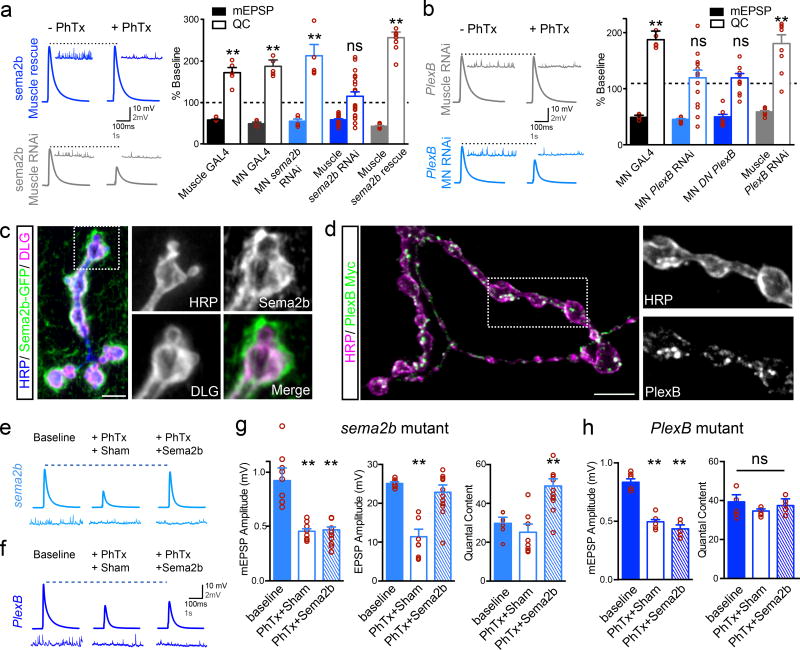
We, then, performed tissue-specific RNAi and transgenic rescue experiments. Expression of UAS-Sema2b-RNAi in motoneurons (OK371-Gal4) had no effect on PHP, whereas expression in muscle (BG57-Gal4) blocked PHP (Fig. 2a). In addition, expression of UAS-sema2b in muscle rescues PHP in the sema2b mutant background (Fig. 2a; Supp Table 1 for additional controls). Consistent with these data, we find that sema2b is expressed in muscle and Sema2b protein, expressed under endogenous promoter sequences, concentrates at postsynaptic membranes (Extended Data Fig. 2). Next, we show that motoneuron-specific expression of UAS-PlexB-RNAi blocks PHP, whereas muscle-specific expression does not (Fig 2b). We also show that motoneuron-specific expression of a previously characterized UAS-PlexBDN dominant-negative transgene, lacking the intracellular signaling domain, blocks PHP (Fig 2b). RNAseq analysis of purified MNs demonstrates PlexB expression in MNs (data not shown). Finally, motoneuron-specific expression of a PlexB-myc transgene shows that PlexB traffics to the presynaptic nerve terminal (Fig 2d). Taken together, our data argue that Sema2b is a ligand originating in muscle that acts via presynaptic PlexB to drive expression of PHP.
Figure 2. Sema2b and PlexinB function as a retrograde trans-synaptic signal.
a) Representative traces (0.3 mM [Ca2+]e). Quantification of mEPSP and QC as in Fig.1b. ‘Muscle Gal4’ is BG57-Gal4/+. ‘MN-Gal4’ is OK371-Gal4/+. ‘MN sema2b RNAi’ is Ok371-Gal4/+; UAS-sema2b-RNAi/+. ‘Muscle sema2b RNAi’ is BG57-Gal4/+, UAS-sema2b-RNAi/+. ‘Muscle sema2b rescue’ is sema2b; BG57-Gal4/ UAS-sema2b. b) Traces, data graphed as in (a). ‘MN PlexB RNAi’ is Ok371-Gal4/+; UAS-PlexB-RNAi/+. ‘MN DN PlexB’ is Ok371-Gal4/+ expressing UAS-PlexB-DN. ‘Muscle PlexB RNAi’ is BG57-Gal4/+, UAS-PlexB-RNAi/+. c) Muscle-specific expression (BG57-Gal4/+) of a UAS-Sema2b-CD8-GFP. Anti-HRP (blue) labels neuronal membrane. Anti-Dlg (pink) labels postsynaptic membranes. d) UAS-PlexB-GFP (green) expressed in motoneurons (OK371-Gal4/+). e) Traces for sema2b mutant in the absence (baseline) and presence of PhTx. ‘Sham’ is incubation in saline lacking Sema2b. The “+Sema2b” indicates addition of Sema2b (100nM). f) Traces as in (e) for PlexB mutant. g) Quantification for conditions in (e). h) Quantification of data in (f). Significance ** = p<0.01; Student’s t-test (2-tail), pairwise to genotypic control (a and b), pairwise comparison to wild type (g and h). Data are mean ± standard error; data points are overlaid.
If Sema2b is a retrograde signal that acts upon the presynaptic PlexB receptor, then it should be possible to reconstitute this retrograde signaling by acute application of Sema2b protein. Purified Sema2b protein was acutely applied to the sema2b mutant NMJ following PhTx treatment to induce PHP. We find that Sema2b protein (~100nM) completely restores PHP in the sema2b mutant, but fails to restore PHP in a PlexB mutant (Fig. 2e–h) (see Extended Data Fig. 3 for additional controls). In addition, application of Sema2b is sufficient to potentiate baseline release, and this effect is also dependent upon PlexB (Extended Data Fig 3). Finally, a membrane tethered UAS-sema2b transgene, expressed in muscle, fails to rescue PHP (Extended Data Fig. 4), even though it concentrates to the postsynaptic membranes (Fig. 2c). Together, these data argue that Sema2b is a secreted postsynaptic ligand that acts upon presynaptic PlexB to enable the expression of PHP. We acknowledge the possibility that PlexB could require a presynaptic co-receptor of, as yet, unknown identity.
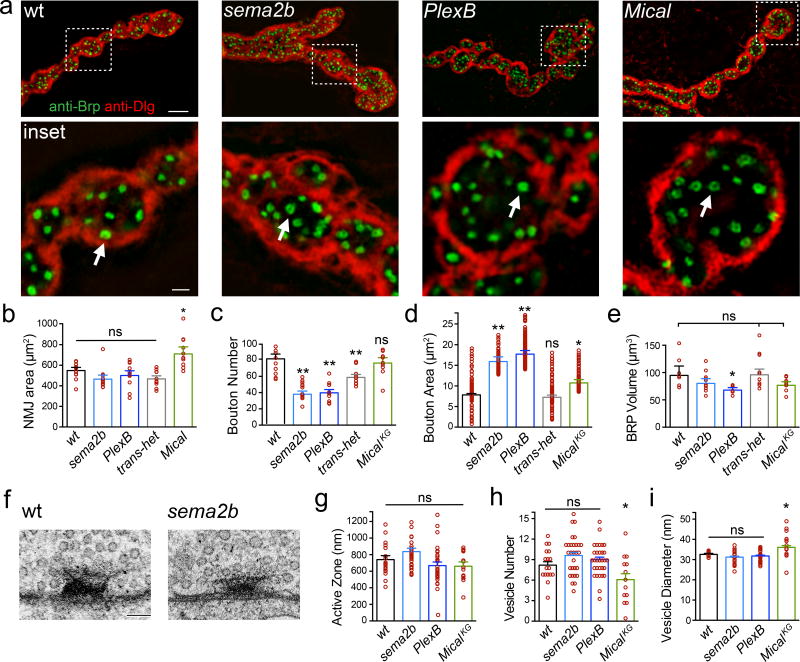
Given that acute application of Sema2b rescues PHP in the sema2b mutant, the failure of PHP in sema2b animals cannot be a secondary consequence of altered NMJ development. None-the-less, Sema2b-Plexin signaling is required for normal NMJ growth. Axon-targeting errors are rare at muscles 6/7, analyzed at the third instar larval stage (Extended Data Table 1). We then demonstrate that sema2b and PlexB NMJ are composed of fewer, larger synaptic boutons (Fig. 3a–d) with no change in total NMJ area (Fig. 3b). The abundance of the active-zone associated protein Bruchpilot (Brp) is unaltered in the sema2b mutant and the sema2b/+;;PlexB/+ double heterozygous animals (Fig. 3e, ‘transhet’), both of which block PHP (Fig. 1a, d, e). There is a significant decrease in total Brp staining in PlexB, an effect of unknown consequence (Fig. 3e; see also below). Qualitatively, the ring-like organization of Brp staining was similar across all genotypes, indicative of normal active zone organization (Fig. 3a inset, arrows). Finally, there is no consistent difference in synapse ultrastructure across genotypes (Fig. 3f–i). Thus, the Sema2b-PlexinB-dependent control of bouton size may be a separable function of Sema2B-PlexB signaling, analogous to anatomical regulation by semaphorins in mammalian systems10–12,15,18.
Figure 3. Altered NMJ growth with normal active zone number and integrity.
a) Structured illumination microscopy (SIM) images of NMJ. Inset: single confocal sections, arrows indicate Brp rings. Scale 5µm. Inset scale 0.5µm. b–e) Quantification of morphology; n=12 except MicalKG n=10. f) Representative active zones. Scale = 70nm. g–i) Quantification of ultrastructure. * = p<0.05, ** = p<0.01 Student’s t-test (2-tail), pairwise comparison to wild type. 2 animals per genotype; wt n=16 active zones (AZ), sema2b n=29 AZ, PlexB n=30 AZ and Mical n=13 AZ. Data are mean ± standard error; data points are overlaid.
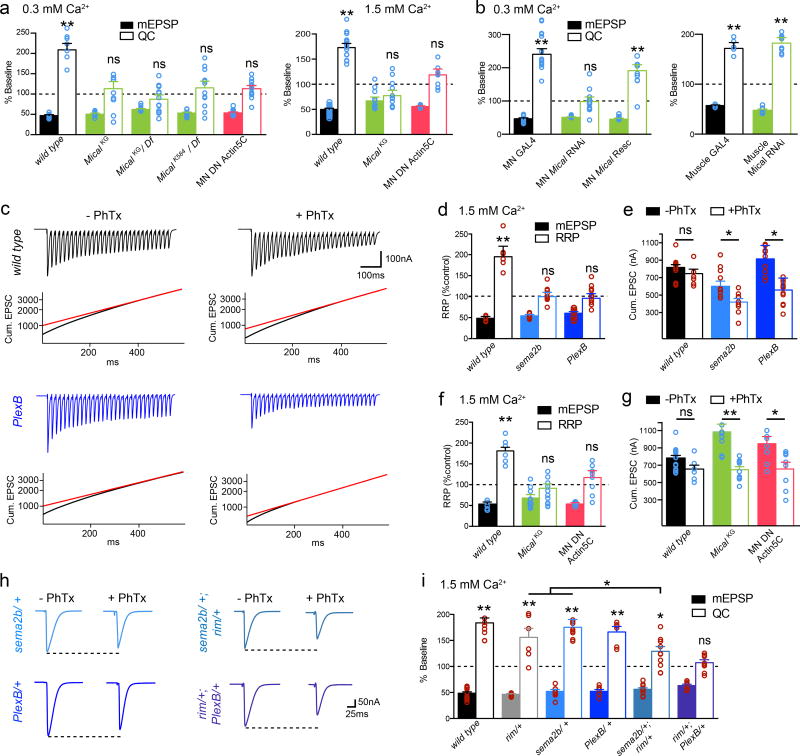
PHP is achieved by potentiation of the readily releasable pool (RRP) of synaptic vesicles1 (see methods). Application of PhTx induces a doubling of the apparent RRP in wild type, an effect that is disrupted in both sema2b and PlexB mutants (Fig. 4). Failure to potentiate the RRP is also revealed as a failure to maintain the cumulative EPSC amplitude following PhTx application (Fig. 4c–e). We then demonstrate a strong genetic interaction with a mutation in the presynaptic scaffolding gene rab3 interacting molecule (rim), a presynaptic homeostatic plasticity gene1. Heterozygous mutations in either rim, or sema2b or PlexB have no effect on PHP (Fig. 4h–i). However, double heterozygous combinations of rim/+ with either sema2b/+ or PlexB/+ strongly impaired the expression of PHP (sema2b/+, rim/+) or abolished PHP (rim/+;; PlexB/+) (Fig. 4h–i). These data do not, however, reflect direct signaling between PlexB and RIM (see also Extended Data Fig. 3e).
Figure 4. Sema2b, PlexinB and Mical control the homeostatic potentiation of the RRP.
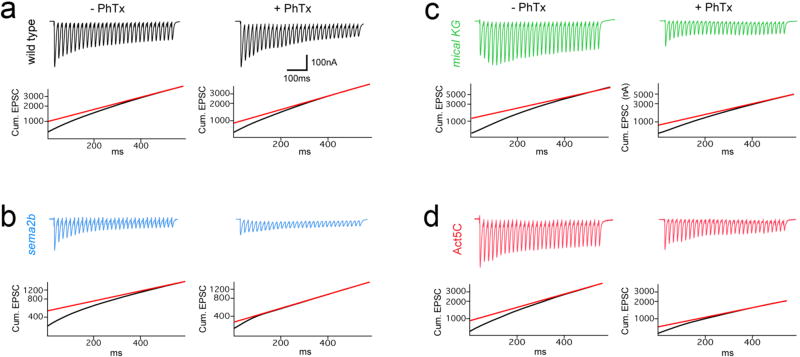
a) The mical mutants (green) at 0.3 (left) and 1.5 (right) [Ca2+]e and Mical-resistant UAS-Act5C transgene (red) block PHP. b) Expression of UAS-mical in motoneurons (MN Mical Rescue) rescues PHP. mical RNAi from MNs blocks PHP. mical RNAi in muscle is without effect (right). c) Traces from wild type (black) and PlexB (blue) (1.5 mM [Ca2+]e) +/− PhTx (stimulation frequency 60Hz, 30 stimuli). Cumulative EPSC and back extrapolation from steady state (red line) is shown below each trace. d) Quantification of the % change in RRP (open) and mEPSP (filled) in the presence of PhTx compared to baseline for each genotype in the absence of PhTx. Sample size: wt (−PhTx)=14, (+PhTx)=9; sema2b (−PhTx)=9, (+PhTx)=9; PlexB (−PhTx)=10, (−PhTx)=11. e) Quantification of cumulative EPSC for recordings in (d). Primary data (red circles). f) Quantification as in (e) for wild type (-PhTx)=6, (+PhTx)=6, the mical mutant (−PhTx)=12 (+PhTx)=11 and in animals overexpressing dominant negative Act5C (−PhTx)=8 (+PhTx)=8. g) Quantification of cumulative EPSC. h) Representative traces. i) Genetic interactions of sema2b/+ or PlexB/+ with the rim/+. Significance * = p<0.05; ** = p<0.01. Data are mean ± standard error; data points are overlaid.
To define how PlexB could modulate the RRP, we tested known downstream signaling elements. We discovered that mical is necessary for PHP (Fig. 4). In Drosophila a single mical gene encodes a highly conserved, multi-domain cytoplasmic protein that mediates actin depolymerization, achieved through redox modification of a specific methionine residue (Met44) in actin7,19. Notably, prior genetic evidence has placed Mical downstream of both PlexA and PlexB signaling during axon guidance20.
An analysis of multiple mical mutations as well as transgenic rescue animals demonstrates that mical is necessary presynaptically for PHP (Fig. 4a,b,f,g). Mical protein is present presynaptically (Extended Data Fig. 5) and presynaptic expression of a Mical-resistant UAS-Actin5C transgene7, which interferes with Mical-mediated actin depolymerization, blocks PHP (Fig. 4a, b). This transgenic protein also concentrates within presynaptic boutons (Extended Data Fig 5). Additional experiments reveal that the homeostatic expansion of the RRP is blocked in mical and when Mical-resistant UAS-Act5 is expressed presynaptically (Fig. 4f,g; Extended Data Fig. 6). We find strong genetic interactions between mical and both the PlexB and rim mutants (Extended Data Fig. 4b–d). Finally, anatomical experiments demonstrate that active zones are normal in the mical mutant, including both light and electron microscopy (Fig. 3). We propose that Mical connects PlexB to the control of the RRP via regulation of presynaptic actin.
For half a century, evidence has underscored the importance of target-derived, retrograde signaling that controls presynaptic neurotransmitter release16. Gene discovery, based on forward genetics, indicates that PHP is controlled by the coordinated action of at least three parallel signaling systems (see schematic and discussion, Extended Data Fig. 7). If our data regarding Sema2b, PlexinB and Mical can be generalized, then Sema-Plexin signaling could represent a platform for retrograde, trans-synaptic, homeostatic control of presynaptic release, thereby stabilizing synaptic transmission and information transfer throughout the nervous systems of organisms ranging from Drosophila to human.
METHODS
Fly Stocks and Genetics
In all experiments, the w1118 strain of Drosophila melanogaster was used as the wild type control. Male and female animals were used. Animals were maintained at 22°C. When performing rescue, RNAi, or over expression experiments with the Gal4/UAS expression system, progeny were raised at 25°C. The following Drosophila stocks were used: sema-2bC4 (21), UAS-sema2b RNAi (Bloomington stock 28932), UAS-sema-2b-TM-GFP (21), UAS-sema2b flies were a gift from Dr. Alex Kolodkin (Johns Hopkins University), P{GawB}Sema-2bNP0592 (Kyoto stock 112237), Sema2bT:Ivir\HA1 (Bloomington stock 65752), P{SUPor-P}PlexBKG00878 (22), UAS-MYC-PlexBEcTM (21), UAS-MYC-PlexB (22), PlexAEY16548 (Bloomington stock 23097), UAS-PlexB-RNAi (18), PlexADf(4)C3 (Bloomington stock 7083), GluRIIA (23), OK371-Gal4 (24), rim103 (25), micalK584 (6), Df(3R)swp2mical mical deficiency (19), micalKG06518 (6), UAS-mical-RNAi (Bloomington stock 31148), UAS-MicalmCherry (26), Mical-GFP Protein Trap (Bloomington stock number 60203), UAS-Act5CGFP M44L (7).
Electrophysiology
Recordings were made from muscle 6 in abdominal segments 2 and 3 of male and female third-instar larvae in current clamp (0.3mM [Ca2+]e) or voltage clamp (1.5mM [Ca2+]e) as indicated, without randomization25,27,28. Haemolymph-like (HL3) saline was used (70 mM NaCl, 5 mM KCl, 10 mM MgCl2, 10 mM NaHCO3, 115 mM sucrose, 4.2 mM trehalose, 5 mM HEPES). Quantal content (QC) was calculated by dividing the average EPSP amplitude by the average mEPSP amplitude for each muscle recording (EPSP/ mEPSP) and averages were made across muscles for a given genotype. For acute pharmacological induction of PHP, larvae were incubated in philanthotoxin-433 (PhTx; 10–20 µM; Sigma-Aldrich) for 10 min according to previously published methods25,27. Two-electrode voltage-clamp (TEVC) recordings were done as previously described in 1.5mM [Ca2+]e HL3 saline25. EPSC analyses were conducted using custom-written routines for Igor Pro 5.0 (Wavemetrics) available as published29, and mEPSPs were analyzed using Mini Analysis 6.0.0.7 (Synaptosoft). Recordings were excluded if the RMP was more depolarized than −60mV. Each experiment was repeated from at least two independent crosses. Key experiments demonstrating a block of synaptic homeostasis were performed blind to genotype by an independent investigator. All controls and experimental genotypes were independently replicated for each experiment in each figure. Experimental sample sizes equal to or greater than 7 were considered sufficiently powered to detect a block in homeostatic plasticity, an effect of size of ~80–120% compared to controls.
Anatomical analyses
Third-instar larval preparations (muscles 6/7) were filleted and fixed in 4% paraformaldehyde, washed, and incubated overnight at 4°C with primary antibodies. Secondary antibodies were applied at room temperature for 2 hr. The following primary antibodies were used: anti-Myc (9E10 Santa Cruz), anti-GFP 3E6 (1:500; mouse; Life Technologies), anti-NC82 (1:100; mouse; Developmental Studies Hybridoma Bank), anti-HA antibody (1:1000; Rabbit; cell signaling technology) and anti-DLG (1:10,000; rabbit). Alexa-conjugated secondary (488, 555) antibodies and Cy5-conjugated goat ant-HRP were used at 1:500 (Life Technologies; Molecular Probes). Larval preparations were mounted in Vectashield (Vector) and imaged with an Axiovert 200 (Zeiss) inverted microscope, a 100X Plan Apochromat objective (1.4 NA) and a cooled charge-coupled device camera (Coolsnap HQ, Roper). Slidebook 5.0 Intelligent Imaging Innovations (3I) software was used to capture, process and analyze images. Structured illumination microscopy imaging was performed using the N-SIM Nikon system, consisting of a Nikon Ti-E Microscope equipped with a Apo TIRF 100×/1.49 Oil objective and an Andor DU897 Camera.
Sema2b ligand generation and application
We utilized Drosophila S2 expression system to expressed Sema2b AP ligand21 for the bath application of Sema2b ligand to the Drosophila NMJ for in vivo electrophysiological recordings. Drosophila S2 cells (obtained from the Vale laboratory, UCSF with additional source information at http://flybase.org/reports/FBtc9000006.html). Cells are mycoplasma negative, tested by MycoAlert (2017). We co-transfected UAS-sema2B-AP and actin-Gal4 plasmids in S2 Cells using effectene (QIAGEN) and incubate the cells at 27° C for four days in serum free Schneider’s media. We then collected the media and diluted the Sema2b-AP ligand in HL3 (0.3 mM Ca2+) to 100nM concentration. To record from the NMJ in the presence of bath applied Sema2b AP ligand, we prepared the larval fillet as described above and incubated the prep in Sema2b-AP HL3 for 10 minutes. To assay Sema2b ligand rescue, we bath applied HL3 (0.3 mM Ca2+) containing both PhTx (15µM) and Sema2b ligand (100nM) to the preparation for a 10 minute incubation. Next, we removed the PhTx/ Sema2b ligand HL3 saline and replaced it with HL3 containing only 100nM Sema2b ligand to record in the presence of Sema2b.
Transmission Electron Microscopy
Transmission electron microscopy for all third instar larvae were prepared and imaged according to methods previously published30.
Data Availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
Extended Data
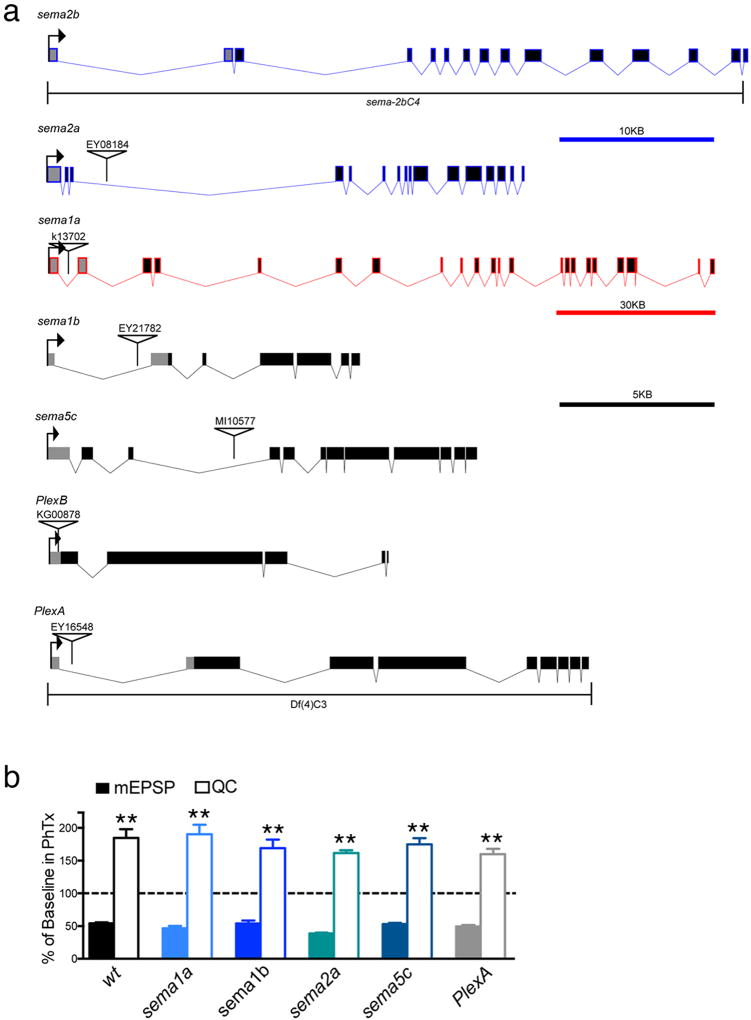
Extended Data Figure 1. Mutations in additional sema and Plexin gene family members do not alter the rapid induction of PHP.
a) Gene diagrams indicating the mutations used in this study. Colors match scale bars. b) Quantification of the percent change in mEPSP (solid bars) and quantal content (open bars) in the presence of PhTx for the following genotypes: wild type (w1118), sema-1ak13702, sema-1bEY21782, sema-2aEY08184, sema-5cMI10577, and PlexAEY1654. Each was either previously described as, or predicted to be, a strong LOF mutant: sema-1ak13702 (17), sema-1bEY21782 (31), sema-2aEY08184 (32), sema-5cMI10577 (33) and PlexAEY1654 (31). See supplemental references. All recordings made at 0.3 mM Ca2+.
Extended Data Figure 2. Evidence for sema2b gene expression in larval muscle.
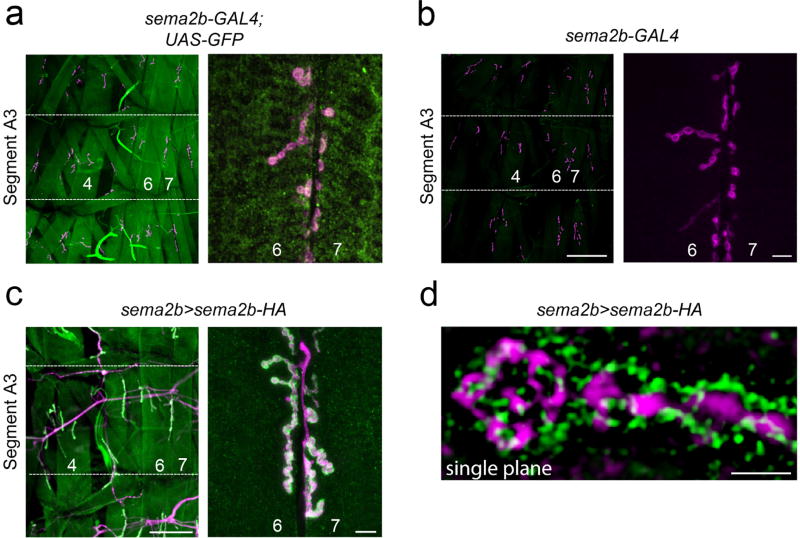
a) At left, a sema2b promoter-Gal4 fusion driving UAS-cd8-GFP at low magnification to reveal multiple NMJ in the peripheral musculature, all expressing GFP. Muscles 4, 6 and 7 are labeled. Segmental boundaries are indicated by the horizontal lines and the middle segment is indicated as abdominal segment 3 (A3). At right, a higher magnification image taken at muscle 6/7, the muscles at which all recordings are made in this study, revealing expression of sema2b promoter-Gal4. The NMJ is labeled with anti-Dlg (pink). Muscle identity is indicated. b) Images taken at an identical exposure to those in (a) revealing no background GFP immunofluorescence in the absence of the UAS-cd8-GFP reporter as a control. Scale bar left: 200µm, right: 10µm. c) Data displayed as in (a). Expression of sema2b-HA is controlled by endogenous promoter sequences. Scale bar left: 200µm, right: 10µm. d) Structured illumination, super-resolution microscopy (SIM) was used to image Sema2b-HA expressed as in (c). A single optical section (single plane) is shown revealing close proximity between Sema2b (green; anti-HA) and the presynaptic membrane (purple) labeled by anti-HRP. Scale bar 2µm.
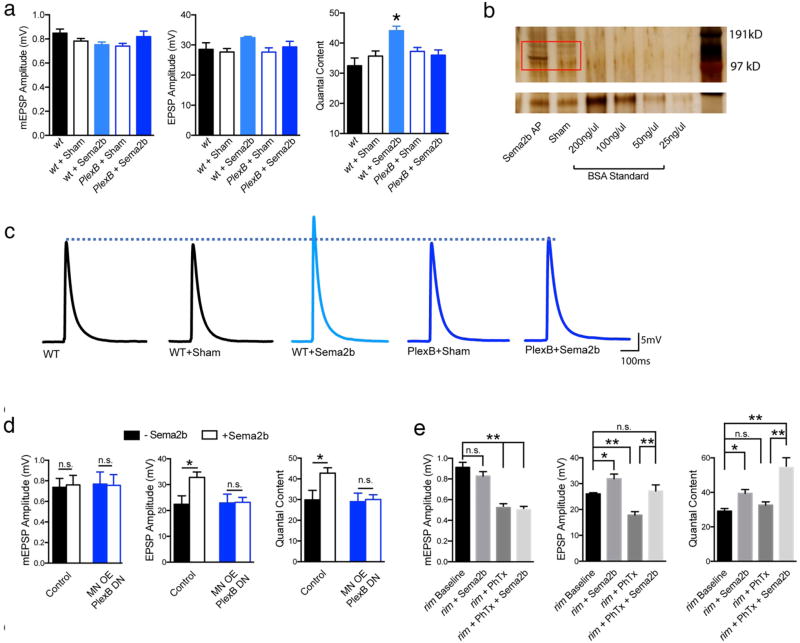
Extended Data Figure 3. Effects of exogenous application of Sema2b protein on baseline transmission.
a) Raw data and analysis of additional control genotypes for the electrophysiological analysis of the effects of application of exogenous Sema2b protein (0.3 mM [Ca2+]e). b) A silver stained protein gel of supernatant collected from S2 cells transfected with both Actin-Gal4 and UAS:Sema2b-AP, or the Actin-Gal4 plasmid alone (sham). The red box highlights Sema2b-AP ligand present at the proper size when both plasmids were transfected together, but absent when Actin-Gal4 plasmid is transfected alone (Sham lane). Bottom, BSA standards. c) Representative traces (0.3mM [Ca2+]e). d) Raw data (0.2 mM [Ca2+]e) for indicated genotypes in the absence (filled bars) and following application of exogenous Sema2b protein (open bars; ~100nM protein). Application of Sema2b protein causes a 40% increase in quantal content in controls (n=6) and this effect is blocked in animals that over-express a PlexB dominant negative transgene in motor neurons (OE MN PlexB DN) (control sample size is n=6, and OE MN PlexB DN sample size is n=14). e) Effects of applying Sema2b protein to the rim103 mutant (compare rim Baseline to rim + Sema2). Experiments performed in 0.4 mM [Ca2+]e to achieve comparable levels of absolute baseline vesicle release to experiments shown in (a). Sema2b protein has no effect on mEPSP amplitudes, but potentiates both the average EPSP and quantal content in rim103, demonstrating a significant (p<0.05) potentiation of release. Sema2b protein rescues the block of PHP observed in the rim103 null mutant. Application of PhTx reduces mEPSP amplitudes in rim103 (p<0.01) and there no significant increase in quantal content (n.s.) resulting in average EPSP amplitudes being smaller than baseline. When Sema 2b is co-applied with PhTx (rim + PhTx + Sema2b), the homeostatic potentiation of quantal content is significantly potentiated (p<0.01) consistent with a rescue of PHP. (rim103 baseline n=6; rim103 + Sema2b, n=13; rim103 + PhTx, n=7; rim103 + PhTx + Sema2b n=10).
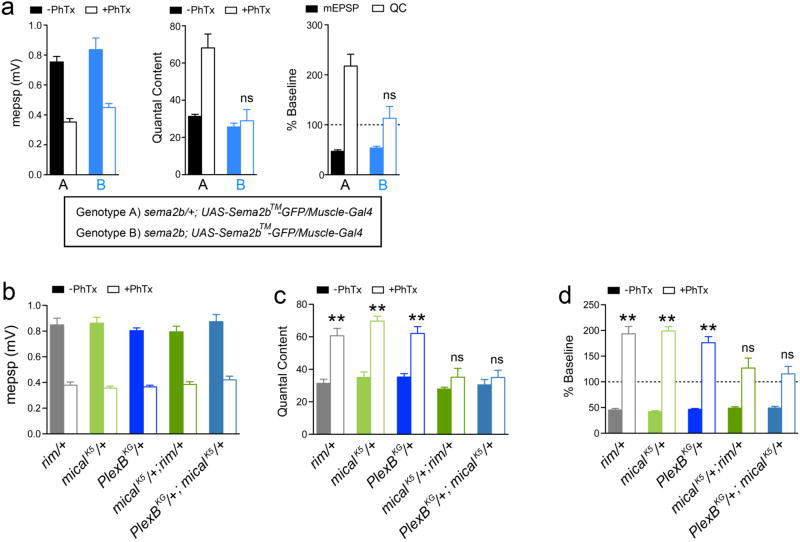
Extended Data Figure 4. Genetic interactions.
a) Averaged mEPSP and quantal content in the absence and presence of PhTx for the indicated genotypes. Both genotypes (A and B) expressed the membrane tethered UAS-sema2b-GFP (UAS-sema2b™-GFP) in muscle (BG57-GAL4). Expression of membrane tethered UAS-Sema2b-GFP has no deleterious effects on neurotransmission or the expression of PHP in control animals harboring a heterozygous mutation in the sema2b gene (sema2b/+) (n=8 minus PhTx and n=8 with PhTx; genotype A). Muscle expression of membrane tethered UAS-sema2b-GFP in the sema2b homozygous mutant background failed to rescue PHP (n=8, n=9; genotype B). This is in contrast to the observation that expression of wild type UAS-sema2b in muscle fully restores PHP in the sema2b mutant background (see Fig. 1 in main text). We conclude that a membrane tethered Sema2b protein is unable to signal to the presynaptic terminal without being secreted from the postsynaptic membranes. These data are consistent with Sema2b being a secreted ligand, originating in muscle, for the induction and expression of PHP. b–d) Averaged mEPSP and quantal content in the absence and presence of PhTx for the indicated genotypes. Heterozygous mutations in rim/+ (n=8, n=8; minus and plus PhTx respectively), PlexB/+ (n=9, n=9) and micalK584/+ (n=8, n=8; note micalK584 shortened to micalK5 in figure) express normal PHP following PhTx-dependent inhibition of mEPSP amplitudes. Double heterozygous combinations of rim/+ with micalK584 /+ (n=9, n=11) or PlexB/+ with micalK584 /+ (n=8, n=13) results in a complete block of PHP. These genetic interactions indicate that mical, rim, and PlexB all participate in a common process that is directly required for PHP.
Extended Data Figure 5. Synaptic localization of Mical and Act5C.
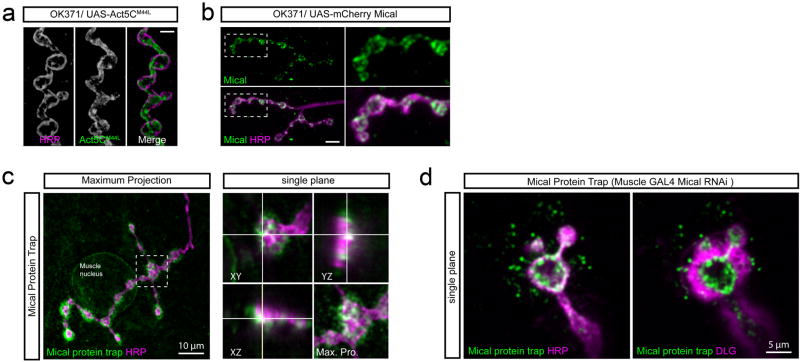
a) Transgenic expression of UAS-Act5CM44L (middle; green in merge at right) localizes throughout the presynaptic terminal marked with anti-HRP (left and magenta). Scale bar 5 um. b) The transgenic expression of UAS-mical-mCherry (green), used to rescue PHP in the mical mutant (see main text), localizes to the presynaptic boutons of motor neurons labeled with anti-HRP (magenta). Scale bar 10 um. c) Image of a Mical-Protein trap (see methods) reveals endogenous localization of Mical protein in postsynaptic muscle and enrichment at the NMJ labeled by anti-HRP (magenta). Projections in the z-plane (y-z or x-z planes) indicate the presence of Mical protein within the presynaptic bouton of the protein trap. d) To selectively image presynaptic Mical-GFP in the protein trap background, UAS-mical-RNAi was selectively expressed in muscle (BG57-GAL4) in the protein trap-background, greatly reducing muscle Mical-GFP and revealing strong presynaptic Mical-GFP originating from the protein trap at the endogenous gene locus. The NMJ is defined by anti-HRP (magenta, left panel) and by anti-Dlg (magenta, right panel).
Extended Data Figure 6. Representative data showing that homeostatic expansion of the RRP fails in sema2b, mical and following expression of mutant UAS-Act5C.
Representative traces and graphs indicating cumulative EPSC and back extrapolation from steady state (red line) for the indicated genotypes. a) Data are shown for wild type. b) Data are shown for sema2b. c) Data are shown for micalKG. d) Data are shown for animals expressing UAS-DN-Act5C. We note that sema2b mutants have a baseline synaptic transmission defect, with a smaller baseline initial EPSC and correspondingly smaller RRP compared to both wild type and PlexB mutants (Fig. 4, main text). Since there is no change in the number of active zone-associated vesicles in sema2b mutants (Fig. 3, main text), this defect must reflect a change in the allocation of vesicles to the RRP at baseline that parallels the failure to homeostatically potentiate the RRP during PHP. However, since PlexB mutants also block PHP without a change in baseline RRP, it seems unlikely that there is a causal link between reduced baseline RRP and failed PHP in the sema2b mutant.
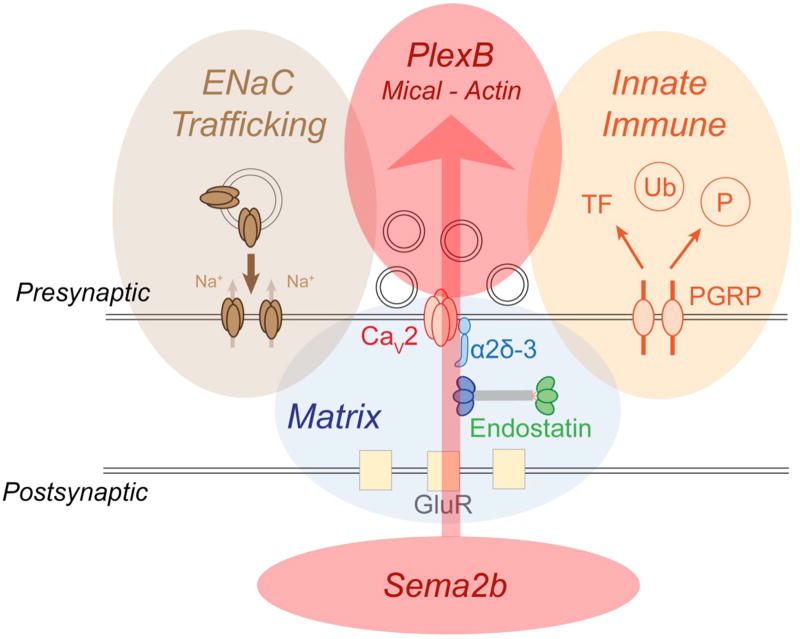
Extended Data Figure 7. A schematic of retrograde, trans-synaptic Sema2b-PlexB signaling.
Signaling is schematized in the context of other mechanisms that have been recently demonstrated to be necessary for presynaptic homeostatic plasticity. Sema2b-PlexB signaling (red shading) is a coherent trans-synaptic, retrograde signaling system that is conveyed, via Mical, to modify presynaptic actin and potentiate the readily releasable synaptic vesicle pool. Other genes have been shown to be necessary for PHP, but none can be connected into a coherent, trans-synaptic signaling cascade. In brown, presynaptic Deg/ENaC channels are inserted into the presynaptic plasma membrane, causing sodium leak and potentiation of presynaptic calcium influx through presynaptic calcium channels (CaV2.1)35. In blue, two components residing in the synaptic extracellular matrix have been implicated in PHP. The α2δ3 auxiliary subunit of the presynaptic calcium channel is necessary for PHP36. The matrix-derived signaling protein Endostatin, a cleavage product of the collagen homologue Multiplexin, is also necessary for PHP37. In orange shading, the innate immune receptor, peptidoglycan recognition protein (PGRP), is essential for PHP30. Signaling downstream of PGRP is hypothesized to reach the neuronal nucleus and could, thereby, mediate the long-term maintenance of PHP. A major task for the future will be to define how these diverse signaling mechanisms participate in a coordinated response that rapidly, accurately and persistently regulates presynaptic neurotransmitter release following disruption of postsynaptic glutamate receptor function.
Extended Data Table 1.
Axon guidance defects for type 1s and type 1b MNs.
| Genotype | 1s Guidance defects | 1b Guidance defects | n |
|---|---|---|---|
| Sema2b4C | 2 | 1 | 20 |
| Sema2b4C/ + | 1 | 0 | 20 |
| PlexB | 0 | 0 | 20 |
| PlexB/ + | 0 | 0 | 20 |
Note: The small number of guidance defects at third instar NMJ may represent the action of compensatory or overlapping mechanisms that promote correct target selection.
Supplementary Material
Acknowledgments
The authors thank Shan Meltzer for consultation throughout and members of the Davis laboratory for comments on an earlier version of this manuscript. Supported by NIH Grant number R01NS39313 and R35NS097212 to GWD.
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Authorship Contributions: B.O.O conducted all experiments and analyses inclusive of genetics, electrophysiology, light microscopy and wrote the text. R.D.F performed electron microscopy. G.W.D helped to analyze electron micrographs and wrote the text.
The authors declare no competing financial interests.
Literature Cited
- 1.Davis GW. Homeostatic signaling and the stabilization of neural function. Neuron. 2013;80:718–28. doi: 10.1016/j.neuron.2013.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Marder E. Variability, compensation, and modulation in neurons and circuits. Proc. Natl. Acad. Sci. U. S. A. 2011;108:15542–8. doi: 10.1073/pnas.1010674108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Turrigiano G. Too many cooks? Intrinsic and synaptic homeostatic mechanisms in cortical circuit refinement. Annu. Rev. Neurosci. 2011;34:89–103. doi: 10.1146/annurev-neuro-060909-153238. [DOI] [PubMed] [Google Scholar]
- 4.Ramocki MB, Zoghbi HY. Failure of neuronal homeostasis results in common neuropsychiatric phenotypes. Nature. 2008;455:912–918. doi: 10.1038/nature07457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cull-Candy S, Miledi R, Uchitel O. On the release of transmitter at normal, myasthenia gravis and myasthenic syndrome affected human end-plates. J. Physiol. 1980:621–638. doi: 10.1113/jphysiol.1980.sp013145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hung RJ, et al. Mical links semaphorins to F-actin disassembly. Nature. 2010;463:823–827. doi: 10.1038/nature08724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hung R-J, Pak CW, Terman JR. Direct Redox Regulation of F-actin Assembly and Disassembly by Mical. Science. 2011;334:1710–1713. doi: 10.1126/science.1211956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kolodkin AL, et al. Fasciclin IV: Sequence, expression, and function during growth cone guidance in the grasshopper embryo. Neuron. 1992;9:831–845. doi: 10.1016/0896-6273(92)90237-8. [DOI] [PubMed] [Google Scholar]
- 9.Luo Y, Raible D, Raper JA. Collapsin: A protein in brain that induces the collapse and paralysis of neuronal growth cones. Cell. 1993;75:217–227. doi: 10.1016/0092-8674(93)80064-l. [DOI] [PubMed] [Google Scholar]
- 10.Koropouli E, Kolodkin AL. Semaphorins and the dynamic regulation of synapse assembly, refinement, and function. Curr. Opin. Neurobiol. 2014;27:1–7. doi: 10.1016/j.conb.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tran TS, et al. Secreted semaphorins control spine distribution and morphogenesis in the postnatal CNS. Nature. 2009;462:1065–9. doi: 10.1038/nature08628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.De Wit J, Verhaagen J. Role of semaphorins in the adult nervous system. Prog. Neurobiol. 2003;71:249–267. doi: 10.1016/j.pneurobio.2003.06.001. [DOI] [PubMed] [Google Scholar]
- 13.Sahay A. Secreted Semaphorins Modulate Synaptic Transmission in the Adult Hippocampus. J. Neurosci. 2005;25:3613–3620. doi: 10.1523/JNEUROSCI.5255-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bouzioukh F, et al. Semaphorin3A regulates synaptic function of differentiated hippocampal neurons. Eur. J. Neurosci. 2006;23:2247–2254. doi: 10.1111/j.1460-9568.2006.04783.x. [DOI] [PubMed] [Google Scholar]
- 15.Uesaka N, et al. Retrograde semaphorin signaling regulates synapse elimination in the developing mouse brain. Science. 2014;344:1020–3. doi: 10.1126/science.1252514. [DOI] [PubMed] [Google Scholar]
- 16.Carrillo RA, Olsen DP, Yoon KS, Keshishian H. Presynaptic activity and CaMKII modulate retrograde semaphorin signaling and synaptic refinement. Neuron. 2010;68:32–44. doi: 10.1016/j.neuron.2010.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Syed DS, Gowda SBM, Reddy OV, Reichert H, VijayRaghavan K. Glial and neuronal Semaphorin signaling instruct the development of a functional myotopic map for Drosophila walking. Elife. 2016;5:1–18. doi: 10.7554/eLife.11572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meltzer S, et al. Epidermis-Derived Semaphorin Promotes Dendrite Self-Avoidance by Regulating Dendrite-Substrate Adhesion in Drosophila Sensory Neurons. Neuron. 2016;89:741–755. doi: 10.1016/j.neuron.2016.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Terman JR, Mao T, Pasterkamp RJ, Yu H, Kolodkin AL. MICALs, a Family of Conserved Flavoprotein Oxidoreductases, Function in Plexin-Mediated Axonal Repulsion. Cell. 2002;109:887–900. doi: 10.1016/s0092-8674(02)00794-8. [DOI] [PubMed] [Google Scholar]
- 20.Ayoob JC, Terman JR, Kolodkin AL. Drosophila Plexin B is a Sema-2a receptor required for axon guidance. Development. 2006;133:2125–35. doi: 10.1242/dev.02380. [DOI] [PubMed] [Google Scholar]
- 21.Wu Z, Sweeney L, Ayoob J, Chak K. A Combinatorial Semaphorin Code Instructs the Initial Steps of Sensory Circuit Assembly in the Drosophila CNS. Neuron. 2011;70:281–298. doi: 10.1016/j.neuron.2011.02.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ayoob JC, Terman JR, Kolodkin AL. Drosophila Plexin B is a Sema-2a receptor required for axon guidance. Development. 2006;133:2125–2135. doi: 10.1242/dev.02380. [DOI] [PubMed] [Google Scholar]
- 23.Petersen Sa, Fetter RD, Noordermeer JN, Goodman CS, DiAntonio A. Genetic analysis of glutamate receptors in drosophila reveals a retrograde signal regulating presynaptic transmitter release. Neuron. 1997;19:1237–1248. doi: 10.1016/s0896-6273(00)80415-8. [DOI] [PubMed] [Google Scholar]
- 24.Mahr A, Aberle H. The expression pattern of the Drosophila vesicular glutamate transporter: A marker protein for motoneurons and glutamatergic centers in the brain. Gene Expr. Patterns. 2006;6:299–309. doi: 10.1016/j.modgep.2005.07.006. [DOI] [PubMed] [Google Scholar]
- 25.Muller M, Liu KSY, Sigrist SJ, Davis GW. RIM Controls Homeostatic Plasticity through Modulation of the Readily-Releasable Vesicle Pool. J. Neurosci. 2012;32:16574–16585. doi: 10.1523/JNEUROSCI.0981-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hung R-J, Spaeth CS, Yesilyurt HG, Terman JR. SelR/MsrB Reverses Mical-mediated Oxidation of Actin to Regulate F-actin Dynamics. Nat. Cell Biol. 2013;15:1445–1454. doi: 10.1038/ncb2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Frank CA, Kennedy MJ, Goold CP, Marek KW, Davis GW. Mechanisms Underlying the Rapid Induction and Sustained Expression of Synaptic Homeostasis. Neuron. 2006;52:663–677. doi: 10.1016/j.neuron.2006.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Frank CA, Pielage J, Davis GW. A Presynaptic Homeostatic Signaling System Composed of the Eph Receptor, Ephexin, Cdc42, and CaV2.1 Calcium Channels. Neuron. 2009;61:556–569. doi: 10.1016/j.neuron.2008.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gaviño Ma, Ford KJ, Archila S, Davis GW. Homeostatic synaptic depression is achieved through a regulated decrease in presynaptic calcium channel abundance. Elife. 2015;4:1–19. doi: 10.7554/eLife.05473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Harris N, et al. The Innate Immune Receptor PGRP-LC Controls Presynaptic Homeostatic Plasticity. Neuron. 2015;88:1157–1164. doi: 10.1016/j.neuron.2015.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bellen HJ, et al. The BDGP Gene Disruption Project: Single Transposon Insertions Associated With 40 % of Drosophila Genes. Genetics. 2004;781167:761–781. doi: 10.1534/genetics.104.026427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Franciscovich AL, Mortimer ADV, Freeman AA, Gu J, Sanyal S. Overexpression Screen in Drosophila Identifies Neuronal Roles of GSK-3 b / shaggy as a Regulator of AP-1-Dependent Developmental Plasticity. Genetics. 2008;180:2057–2071. doi: 10.1534/genetics.107.085555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Venken KJT, et al. MiMIC: a highly versatile transposon insertion resource for engineering Drosophila melanogaster genes. Nat. Methods. 2011;8:737–743. doi: 10.1038/nmeth.1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yu H, Araj HH, Ralls SA, Kolodkin AL. The Transmembrane Semaphorin Sema I Is Required in Drosophila for Embryonic Motor and CNS Axon Guidance. Neuron. 1998;20:207–220. doi: 10.1016/s0896-6273(00)80450-x. [DOI] [PubMed] [Google Scholar]
- 35.Younger M, Müller M, Tong A, Pym E, Davis G. A presynaptic ENaC channel drives homeostatic plasticity. Neuron. 2013;79:1183–1196. doi: 10.1016/j.neuron.2013.06.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang T, ting, et al. α2δ-3 Is Required for Rapid Transsynaptic Homeostatic Signaling. Cell Rep. 2016:2875–2888. doi: 10.1016/j.celrep.2016.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang T, Hauswirth AG, Tong A, Dickman DK, Davis GW. Endostatin Is a Trans-Synaptic Signal for Homeostatic Synaptic Plasticity. Neuron. 2015;83:616–629. doi: 10.1016/j.neuron.2014.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.