Abstract
It is evident that p53 activity is critical for tumour prevention and stress response through its transcriptional activation of genes affecting cellular senescence, apoptosis, cellular metabolism, and DNA repair. The regulation of p53 is highly complex, and MDM2 and MDMX are thought to be critical for deciding the fate of p53, both through inhibitory binding and post-translational modification. Many mouse models have been generated to study the regulation of p53 in vivo, and they have altered our interpretations of how p53 is regulated by MDM2 and MDMX. Although MDM2 is absolutely required for p53 regulation, certain functions are dispensable under unstressed conditions, including the ability of MDM2 to degrade p53. MDMX, on the other hand, may only be required in select situations, like embryogenesis. These models have also clarified how cellular stress signals modify the p53-inhibiting activities of MDM2 and MDMX in vivo. It is clear that more work will need to be performed to further understand the contexts for each of these signals and the requirements of various MDM2 and MDMX functions. Here, we will discuss what we have learned from mouse modelling of MDM2 and MDMX and underscore the ways in which these models could inform future therapies.
Keywords: p53, MDM2, MDMX, E3 ubiquitin ligase, cancer
Introduction
The role of p53 as a tumour-suppressing transcription factor is abundantly clear, and it is well known that p53 is frequently mutated or inactivated in various cancers (Muller and Vousden, 2013). It is also apparent that p53 regulation is highly complex, but two proteins are critically important for proper control of p53: MDM2 and MDMX (also known as MDM4) (Wade et al., 2010). p53 transcription and translation are thought to occur ubiquitously, while MDM2 and MDMX cooperate to control both the post-translational stability and activity of p53 (Hu et al., 2007; Wade et al., 2010). MDM2 is also a transcriptional target of p53 (Barak et al., 1993), which contributes to a feedback loop of regulation.
MDM2, but not MDMX, harbours E3 ubiquitin ligase activity towards p53 (Haupt et al., 1997; Honda et al., 1997; Kubbutat et al., 1997; Jackson and Berberich, 2000), and both proteins can directly bind to the p53 transactivation domain and inhibit transcription (Chen et al., 1993; Shvarts et al., 1996). MDM2 and MDMX interact to form a heterodimer (Tanimura et al., 1999), which is thought to promote more efficient p53 inhibition. Although these activities have been clearly demonstrated in vitro, the relative importance of MDM2−p53 and/or MDMX−p53 binding, MDM2–MDMX heterodimer formation, or MDM2 E3 ligase activity towards in vivo p53 activity has been incompletely understood. For instance, it was previously thought that MDM2 E3 ligase activity was essential for basal p53 regulation, but evidence from mouse models suggests that MDM2 E3 ligase activity is dispensable under normal conditions (Tollini et al., 2014).
The mechanisms of p53 regulation are still being elucidated. Studies in mouse models have both confirmed existing hypotheses and often challenged widely held beliefs about how MDM2 and MDMX function together to regulate p53. This review will address how in vitro and in vivo evidences have conflicted. We will first discuss what MDM2 and MDMX knockout mouse models have told us about how p53 is differentially regulated during embryogenesis and adulthood. Then, we will explore how knockin mouse models have clarified the mechanistic cooperation of MDM2 and MDMX and the upstream signals that regulate their inhibition of p53. Last, we will comment on how mouse models could inform the discovery of novel drug targets or treatment strategies to fight cancer.
Temporal and tissue-specific roles for MDM2 and MDMX: lessons from knockout mice
Mdm2 knockout mice
In the following section, we will review work from whole body MDM2 and MDMX knockout studies. For a more comprehensive discussion of tissue-specific deletion studies, please refer to an accompanying review by Guillermina Lozano and her colleagues (Moyer et al., 2017) in this special issue.
Early in vitro work demonstrated that MDM2 could bind to p53 and mask p53 transactivation activity (Chen et al., 1993; Oliner et al., 1993). However, the degree of MDM2 importance to p53 regulation was not fully appreciated until the creation of Mdm2 deletion alleles in the mouse (Montes de Oca Luna et al., 1995; Jones et al., 1995). Interestingly, mice deficient for p53 are viable, but tend to develop tumours (typically lymphomas) and die by 6 months of age (Donehower et al., 1992). Surprisingly, mice deficient for MDM2 die between embryonic days 4.5–6.5, with pronounced levels of apoptosis. This embryonic lethality caused by loss of MDM2 is rescued by concomitant loss of p53, suggesting that the primary function of MDM2 during embryogenesis is to inhibit undue p53 activation or accumulation. These studies also established that MDM2 and p53 are expressed ubiquitously during embryonic development.
It is also apparent that although MDM2 expression is found throughout the embryo and required during embryogenesis in the presence of p53, MDM2-mediated p53 regulation remains essential in the adult mouse as a whole. The p53-dependent embryonic lethality caused by MDM2 deficiency renders the study of MDM2 in p53 regulation difficult in vivo. To address this, Christophorou et al. (2005) developed a mouse model expressing the hormone-binding domain of a modified oestrogen receptor placed at the 3′ end of the p53 coding sequence, therefore generating a switchable chimeric p53 protein (p53ER hereafter) able to be rendered inactive or active by withdrawal or addition of tamoxifen or 4-hydroxytamoxifen, respectively. The p53ER protein behaves like a null allele in the absence of tamoxifen, which allows for the generation of MDM2-deficient mice and the study of MDM2-dependent p53 regulation in the adult mouse. Ringshausen et al. (2006) crossed p53ER/− mice with Mdm2+/− mice to generate Mdm2−/−;p53ER/− mice. Then, they injected tamoxifen into these mice, rendering p53ER able to be active. Strikingly, all Mdm2−/−;p53ER/− mice died within 5−6 days after a single tamoxifen injection, presenting severe anaemia and bone marrow ablation, suggesting that p53 regulation is most critical in radio-sensitive tissues. Several proliferative tissues were also severely atrophied, including small intestine and colon tissue. On the other hand, classically radio-insensitive tissues such as the heart and kidney appeared normal following tamoxifen treatment. However, in all tissues analysed, p53 was more transcriptionally active, though not to a level necessarily causing extensive cell death, suggesting that the loss of MDM2 allows for spontaneous p53 activation throughout the body (Ringshausen et al., 2006). Interestingly, only Mdm2−/−;p53ER/− mice, but not Mdm2−/−;p53ER/ER mice, were recovered from these crosses, which suggests that the p53ER protein may have ‘leaky’ activity.
In a similar study, Zhang et al. (2014a) used a conditional Mdm2 deletion allele (Mdm2FM) (Grier et al., 2002) coupled with a whole body, tamoxifen-inducible, Cre-mediated recombination allele (CreER) to study the effects of whole body Mdm2 loss at various stages of aging, since p53 activity has been shown to decline with age (Feng et al., 2007). Similar to Mdm2−/−;p53ER/− mice, 2 to 4-month-old Mdm2FM/−;CreER mice experience p53-mediated morbidity within a few days after tamoxifen injection. Mdm2FM/−;CreER mice also display extensive levels of apoptosis and atrophy in the kidney and liver, radio-insensitive tissues, in addition to extensive damage to radio-sensitive tissues. Loss of Mdm2 results in p53 stabilization and activation in most organs, including the brain, spleen, kidney, liver, and heart. Interestingly, MDM2 is still required for viability in aged mice, but p53 activation and stabilization is less severe in radio-insensitive tissues.
Several other studies (Table 1) have generated tissue-specific deletions of Mdm2 using conditional Mdm2 deletion alleles combined with tissue-specific Cre expression, including expression in differentiated intestinal smooth muscle cells, erythroid, and cardiac tissue (Boesten et al., 2006; Grier et al., 2006; Xiong et al., 2006; Maetens et al., 2007). Others have coupled whole body Mdm2 deletions with tissue-specific reintroduction of p53 (Francoz et al., 2006). Most tissues in which Mdm2 has been deleted, especially those that are highly proliferative, exhibit substantially increased levels of apoptosis, advocating that MDM2-mediated p53 regulation is critical in nearly all tissues in the mouse.
Table 1.
Reduction of Mdm2 or MdmX expression in mice in various tissues and stages.
| Tissue | MDM model | p53 alleles | Cre transgene | Phenotypes and p53 responses | References |
|---|---|---|---|---|---|
| Whole body | Mdm2puro | Wild type | N/A | Decreased body weight, haematopoietic defects, increased apoptosis, increased p53 activity | Mendrysa et al. (2003) |
| MdmXΔEx6 (truncation) | Wild type | N/A | Embryonic lethality, increased p53 activity on p53ΔP/ΔP background | Bardot et al. (2015) | |
| Central nervous system | Mdm2−/− | p53LSL/− | Nestin-Cre | Embryonic lethality, increased p53 protein levels and activity, increased apoptosis | Francoz et al. (2006) |
| MdmX−/− | p53LSL/− | Nestin-Cre | Microcephaly, growth retardation, increased p53 activity and cell cycle arrest | ||
| Mdm2FM/FM | Wild type | Nestin-Cre | Neonatal lethality, hydrancephaly, increased p53 protein levels and activity, aberrant apoptosis and proliferation | Grier et al. (2002), Xiong et al. (2006) | |
| MdmXFX/FX | Wild type | Nestin-Cre | Neonatal lethality, porencephaly, increased p53 activity, aberrant apoptosis and proliferation | ||
| Mdm2−/− | p53ER/− | N/A | No discernable phenotypes | Ringshausen et al. (2006) | |
| MdmX−/− | p53ER/− | N/A | Increased p53 activity, increased apoptosis in subventricular zones | Garcia et al. (2011) | |
| Bone | Mdm2F11-12 | Wild type | Col3.6-Cre | E19.5 lethality, skeletal defects, elevated p53 activity but not protein levels, reduced proliferation | Lengner et al. (2006) |
| Intestine | Mdm2FM/FM | Wild type | Villin-Cre | Normal lifespan, intestinal abnormalities with eventual recovery, increased p53 activity and protein levels | Valentin-Vega et al. (2008) |
| MdmXFX/FX | Wild type | Villin-Cre | No major defects, increased p53-dependent apoptosis and activity in proliferating cells | Valentin-Vega et al. (2009) | |
| Mdm2−/− | p53ER/− | N/A | Atrophy of villi and crypts, increased apoptosis | Ringshausen et al. (2006) | |
| MdmX−/− | p53ER/− | N/A | Increased apoptosis | Garcia et al. (2011) | |
| Mdm2FM/− | Wild type | CAG-Cre (Tamoxifen) | Atrophy in villi and increased apoptosis in crypts of 2−4 months old mice, no phenotypes in 16−18 months old mice | ||
| Heart | Mdm2FM/− | Wild type | αMyhc-Cre | E13.5 lethality, severe defects, increased p53 protein and apoptosis | Grier et al. (2006) |
| MdmXFX/− | Wild type | αMyhc-Cre | Normal, with some premature death at 12 months of age | ||
| Mdm2FM/− | Wild type | CAG-Cre (Tamoxifen) | Tissue fibrosis, increased p53 activity and protein levels | ||
| Mdm2−/− | p53ER/− | N/A | No discernable phenotypes | Ringshausen et al. (2006) | |
| MdmX−/− | p53ER/− | N/A | No discernable phenotypes | Garcia et al. (2011) | |
| Endothelium | Mdm2FM/FM | Wild type | Tie2-Cre | Embryonic lethality, severe vascular defects, increased p53 activity | Zhang et al. (2012) |
| Skin | Mdm2F11-12 | Wild type | K5-Cre | Progressive hair loss and decreased skin elasticity, increased p53 protein levels and activity, increased senescence | Gannon et al. (2011) |
| Smooth muscle | Mdm2FM/FM | Wild type | Sm22-CreERT2 | Death within 12 days after tamoxifen injection, increased p53 protein levels and activity, increased apoptosis | Boesten et al. (2006) |
| MdmXFX/FX | Wild type | Sm22-CreERT2 | No discernable phenotypes | ||
| Red blood cells | Mdm2FM/FM | Wild type | EpoR-GFP-Cre | E13 lethality, defects in erythropoiesis, increased p53 activity | Maetens et al. (2007) |
| MdmXFX/FX | Wild type | EpoR-GFP-Cre | Death between E12.5 and 21 days after birth, anaemia, increased p53 activity | ||
| Lens epithelial cells | Mdm2FM/FM | Wild type | Le-Cre | Defects in lens development, normal birth ratios but hyperglycaemia and neonatal lethality (1 week) present, increased p53 levels and apoptosis, decreased cell proliferation | Zhang et al. (2014b) |
| MdmXFX/FX | Wild type | Le-Cre | Eyeless, normal birth ratios and survival into adulthood, increased p53 levels and apoptosis, decreased cell proliferation |
In contrast to Mdm2 deletion or reduction, transgenic Mdm2 overexpression in the mouse supports increased tumour development, presumably because of increased p53 inhibition (Jones et al., 1998). This, in combination with Mdm2 deletion studies, strengthens the importance of MDM2 to proper p53 regulation at all stages of development.
MdmX knockout mice
Similar to MDM2, loss of MDMX in the mouse has also proven to be embryonic lethal, with concomitant p53 deletion rescuing the lethality (Parant et al., 2001), suggesting that MDM2 and MDMX play non-redundant roles in the inhibition of p53 activation or stabilization. Interestingly, overexpression of an MDM2 transgene (Mdm2Tg/+) can rescue MDMX deletion (Steinman et al., 2005), hinting that MDM2 is perhaps capable of restraining undue p53 activity in vivo but its efficiency is compromised by MDMX loss. From these studies, it is possible to speculate that MDMX serves to either directly enhance MDM2 inhibitory functions or enhance its stability.
It also appears that MDMX is less important to p53 regulation in the adult mouse than MDM2. Garcia et al. (2011) combined MdmX+/− mice with the p53ER model to generate MdmX−/−;p53ER/− mice and tested whether, like MDM2, MDMX is critical to p53 suppression in the adult mouse. Surprisingly, MdmX−/−;p53ER/− mice injected with tamoxifen daily live an average of 29 days. Spontaneous p53 activity was also observed in select tissues. Six hours after tamoxifen injection and in the absence of MDMX, the mRNA expression of p53 cell cycle target gene cdkn1a (p21) was significantly increased in almost all tissues examined. However, the mRNA expression of puma, a p53 apoptotic target, was only significantly increased in radio-sensitive tissues. The expression of cdkn1a following p53ER restoration correlated with decreased proliferation in tissues, while the expression of puma correlated with increased apoptosis. In contrast to Mdm2−/−;p53ER/− mice, which die 5–6 days after a single tamoxifen injection (Ringshausen et al., 2006), MdmX−/−;p53ER/− mice are remarkably tolerant to temporary p53ER restoration. After daily injections of tamoxifen for 1 week, MdmX−/−;p53ER/− mice displayed significant loss of cell proliferation in the spleen, bone marrow, and thymus tissue, but following withdrawal of tamoxifen, the mice were able to recover without long-term adverse consequences. Tissue-specific p53 restoration studies in MdmX−/− mice and tissue-specific deletions of MdmX have further indicated that the necessity of MDMX in p53 regulation is context dependent; conversely, many conditional deletion studies support the idea that MDM2 is critical in the suppression of basal p53 in almost all situations.
Consistent with this idea, several groups have suggested that MDMX serves to enhance MDM2-mediated p53 degradation (Badciong and Haas, 2002; Gu et al., 2002; Linke et al., 2008). The relatively better survival of MdmX−/−;p53ER/− mice compared to Mdm2−/−;p53ER/− mice in the presence of transient p53ER restoration suggests that MDM2 is at least capable of restraining p53 on its own for short periods of time, but it is conceivable that efficient MDM2-mediated p53 inhibition or degradation is required for long-term viability. Indeed, the levels of p53 are increased in MdmX−/−;p53ER/− mouse embryonic fibroblasts (MEFs) compared to MEFs containing MDMX (Garcia et al., 2011), supporting the idea that MDMX plays some role in regulating p53 stability in vivo. It is possible that in MdmX−/−;p53ER/− mice, p53ER could continue to accumulate. Theoretically, stably elevated p53 levels could eventually mandate MDMX enhancement of MDM2-mediated p53 inhibition, indicated by the eventual lethality of continuous tamoxifen injection in MdmX−/−;p53ER/− mice.
It appears that splice variations of MDMX may differentially play a role in its regulation of p53. Recently, Bardot et al. (2015) modelled a conserved splice variant of MDMX, generating mice with an allele of MdmX that obligatorily skips exon 6 (MdmXΔE6), preventing the expression of full-length MDMX and increasing the mRNA expression of a short allele of MdmX (MdmX-S). High expression of the MdmX-S splice variant is correlated with poor survival in several cancers (Bartel et al., 2005; Prodosmo et al., 2008; Lenos et al., 2012), and overexpression-based studies have suggested that MDMX-S may be a more potent p53 inhibitor than MDMX (Rallapalli et al., 1999, 2003). Although MdmX-S mRNA expression was vastly increased in MdmX+/ΔE6 mice, MDMX-S protein levels were low, suggesting that it might be quickly degraded by the proteasome. It also appears that in vivo MDMX-S is much less efficient than MDMX at controlling p53 activity. Bardot et al. (2015) propose that the upregulation of MdmX-S that is observed in cancers could instead serve to prevent the expression of full-length MDMX, and tumours containing overexpression of MdmX-S would likely correlate with mutated p53.
Overall, MDM2 and MDMX deletion models have suggested the following notions about MDM2- and MDMX-mediated p53 regulation: (i) MDM2 is the master regulator of p53 and is necessary to prevent p53-dependent cell death at all stages following embryonic day 5; (ii) MDMX may serve to enhance MDM2-mediated p53 inhibition and/or degradation in a developmental and tissue-specific manner.
Mechanisms of MDM2- and MDMX-mediated p53 regulation: lessons from knockin mice
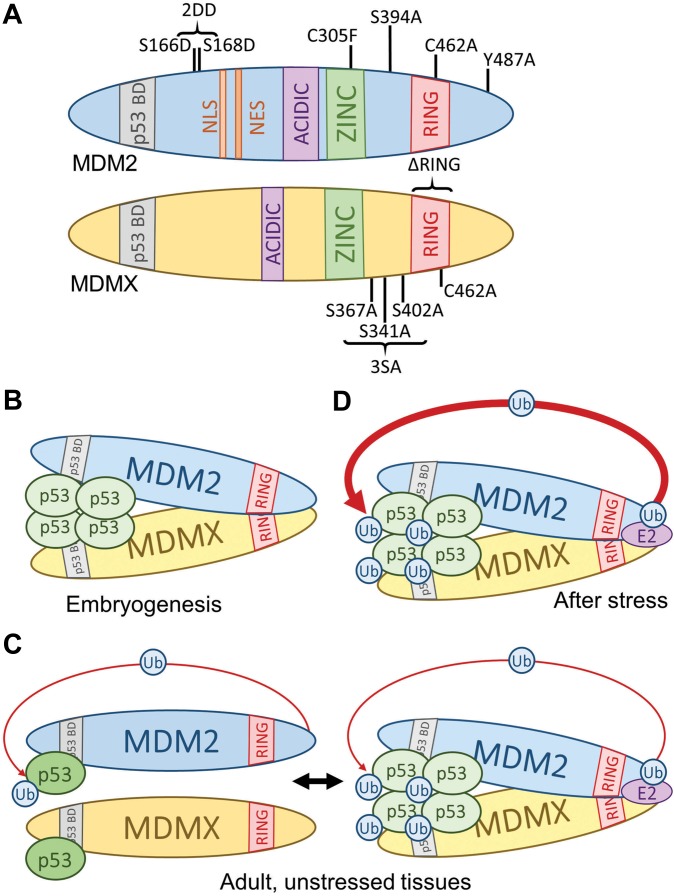
Previous in vitro studies suggested that the primary mechanism of MDM2- and MDMX-dependent p53 inhibition was mediated through direct MDM2 and MDMX binding to the p53 transactivation domain, causing disruption of p53 activity. These studies also revealed that MDM2 could act as an E3 ubiquitin ligase towards p53, causing its degradation by the proteasome (Haupt et al., 1997; Honda et al., 1997). Shortly after this discovery, it was observed that MDM2 harboured autoinhibitory ubiquitination activity, causing its destabilization in the presence of DNA damage (Honda and Yasuda, 2000; Stommel and Wahl, 2004) and allowing for further p53 stabilization. MDM2 and MDMX were also found to be homologous, sharing highly similar p53-binding domains and RING domains (Shvarts et al., 1996), but unlike MDM2, MDMX does not harbour E3 ubiquitin ligase activity (Jackson and Berberich, 2000). Some in vitro studies have suggested that through their respective RING domains (Tanimura et al., 1999), MDMX serves to facilitate MDM2-mediated p53 ubiquitination (Linares et al., 2003). This facilitation could occur indirectly, meaning that MDMX could redirect presumable MDM2 autoinhibitory ubiquitination unto itself, or it could occur directly, meaning that MDMX could directly enhance the transfer of ubiquitin to p53. Several mouse models (Figure 1A and Table 2) have helped to clarify the mechanisms of MDM2- and MDMX-mediated p53 regulation.
Figure 1.
p53 regulation requirements are context dependent. (A) Schematic of MDM2 and MDMX protein modifications that have been generated by knockin mouse models. (B) During embryogenesis, both MDM2 and MDMX are required for proper control of p53 activity. The formation of an MDM2–MDMX heterodimer is also required to restrain p53, but MDM2 E3 ligase activity is dispensable at this time. (C) In unstressed adult tissues, the necessity of MDMX or MDM2–MDMX heterodimer formation for proper p53 control is tissue-dependent. MDM2-mediated p53 ubiquitination may still occur in these tissues, which may require MDMX. (D) After stress, such as DNA damage, MDM2 E3 ligase activity is required to return p53 protein to basal levels and control p53 activity. This may or may not require MDM2–MDMX heterodimer formation. p53 BD, p53-binding domain; NLS, nuclear localization signal; NES, nuclear export signal; ACIDIC, acidic domain; ZINC, zinc finger domain; RING, RING finger domain; Ub, ubiquitin; E2, E2 ubiquitin-conjugating enzyme.
Table 2.
Mdm2 and MdmX knockin mice.
| MDM model | Modification | Phenotypes and p53 responses | References |
|---|---|---|---|
| Mdm2C462A | Disrupts RING domain and MDMX interaction | Embryonic lethal, increased p53 stability and activity | Itahana et al. (2007) |
| Mdm2S394A | Disrupts ataxia-telangiectasia mutated (ATM) phosphorylation | Radioresistant, accelerated spontaneous and MYC-induced tumour formation, resistance to radiation-induced lymphoma | Gannon et al. (2012), Carr et al. (2016) |
| Mdm2C305F | Disrupts ribosomal protein (RP) interaction | Decreased p53 stabilization and activity following ribosomal stress, increased MYC-induced tumours, increased adenomatous polyposis coli (APC) loss-induced colon tumours | Macias et al. (2010), Meng et al. (2015), Liu et al. (2016) |
| Mdm2Y487A | Disrupts E3 ligase function | Increased p53 stability, increased p53 activity after irradiation, increased radiosensitivity | Tollini et al. (2014) |
| Mdm2SNP309G | Increases Mdm2 expression | Increased spontaneous tumourigenesis, reduced p53 levels | Post et al. (2010) |
| Mdm2P2 | Disrupts p53-mediated Mdm2 transcription | Prolonged p53 activity after DNA damage, no apparent change in p53 stability, increased radiosensitivity | Pant and Lozano (2014) |
| Mdm22DD | Mimics constitutive protein kinase B (AKT) phosphorylation in mammary tissue | Accelerated ERBB2-induced tumours, decreased p53 expression | Cheng et al. (2010) |
| MdmXΔRING | Removes RING domain functions | Embryonic lethal, increased p53 activity | Pant et al. (2011) |
| MdmXC462A | Disrupts RING domain and MDM2 binding | Embryonic lethal, increased p53 activity and protein levels | Huang et al. (2011) |
| MdmX3SA | Disrupts AKT, ATM, and Chk2 phosphorylation | Radioresistant, accelerated MYC-induced tumour formation, decreased p53 protein levels and activity | Wang et al. (2009) |
MDM2/MDMX−p53 binding and MDM heterodimer formation
To directly test whether or not MDM2/MDMX−p53 binding alone could restrain p53 activity in vivo, Itahana et al. (2007) created mice carrying a mutation in the MDM2 RING domain (MDM2C462A), thus disrupting MDM2 E3 ligase activity and MDMX binding. Homozygous MDM2C462A mutation results in p53-dependent embryonic lethality before embryonic day 7.5, suggesting that MDM2/MDMX−p53 interaction alone is not sufficient to permit embryonic development. Unpublished observations in our laboratory also suggest that MDM2−p53 or MDMX−p53 interaction may not be sufficient for p53 suppression in the adult mouse. In our hands, Mdm2C462A/C462A;p53ER/− mice die within 4–6 days of tamoxifen injection, which is similar to results obtained from Mdm2−/−;p53ER/− mice, suggesting that MDM2–MDMX heterodimer formation and/or MDM2 E3 ligase activity, rather than MDM−p53 transactivation domain binding, may be the primary mechanisms for MDM-mediated p53 suppression in vivo.
Studies in MdmX knockin mice also appear to corroborate that MDM2/MDMX−p53 binding is insufficient for p53 inhibition, particularly during embryogenesis. Pant et al. (2011) generated an allele carrying an in-frame deletion of the MDMX RING domain (MdmXΔRING). At the same time, Huang et al. (2011) generated an allele carrying a point mutation in the MDMX RING domain (MdmXC462A). Both of these alleles disrupt MDMX–MDM2 interaction without altering MDM2. However, mice homozygous for either MdmXΔRING or MdmXC462A exhibit p53-dependent embryonic lethality. In the presence of MDMXΔRING, MDM2 E3 ligase activity appears to remain intact in MEFs (Pant et al., 2011), suggesting that MDM2-mediated ubiquitination of p53 and MDM2–p53 or MDMX–p53 binding in the absence of heterodimer formation is not sufficient to permit embryonic development. Although these two mouse models both disrupt MDMX–MDM2 binding and present p53-dependent homozygous embryonic lethality, there are several observations in apparent contradiction. First, when combined with the p53neo allele, which expresses ~15% of wild type p53 levels, MEFs containing MDMXΔRING appear to display greater p53 activity with no difference in p53 stabilization compared to MEFs containing wild type MDMX, suggesting that MDMX does not necessarily contribute to MDM2-mediated p53 degradation. On the other hand, MdmXC462A/C462A embryos present both increased p53 abundance and activity. These observations suggest that the MDM2–MDMX interaction is required for efficient p53 inhibition, but may or may not be required for p53 degradation during embryogenesis.
Complicating things further, Pant et al. (2011) also generated a Cre-inducible MDMX RING deletion allele (MDMXflxRING) and crossed these mice with mice containing a tamoxifen-dependent Cre allele (CreER). When adult MdmXflxRING/ΔRING;CreER mice were injected with tamoxifen to generate recombined MdmXΔRING/ΔRING mice, the mice appeared healthy. Most p53 target genes, with the exception of p21, showed little change in expression, suggesting that MDM2–MDMX heterodimer formation is dispensable for the regulation of p53 activity in adult mice. Whether p53 stability is affected by this loss has not been determined and remains an interesting question.
Then, what is the contribution of MDMX to MDM2-mediated p53 regulation in vivo? It is clear that the necessity of MDMX is context specific. During embryogenesis, it appears that MDM2–MDMX heterodimer formation is critical for p53 suppression, but the mechanism of this inhibition is incompletely understood. Several possibilities exist, including that the MDM2–MDMX heterodimer facilitates more efficient MDM2/MDMX−p53 binding and transcriptional inhibition than either protein alone. In support of this idea, Pant et al. (2011) observed somewhat decreased binding of MDMXΔRING to p53R172H, which harbours a missense mutation rendering it transcriptionally inactive (Lang et al., 2004). We have also noticed that MDM2/MDMX−p53 binding is impaired in Mdm2−/−;p53ER/− and MdmX−/−;p53ER/− MEFs, respectively, compared to p53ER/− MEFs (our unpublished data). On the other hand, it appears that MDM2–MDMX binding is dispensable to p53 inhibition in the adult mouse (Pant et al., 2011).
MDM2 E3 ligase activity
The recently developed MDM2Y487A mouse model (Tollini et al., 2014) has provided insight into both basal and stress-dependent p53 regulation by MDM2 and MDMX. As an extension of the MDM2C462A model, in which both MDM2 E3 ligase activity and MDM2–MDMX interaction are disrupted, the MDM2Y487A mutation disrupts MDM2 E3 ubiquitin ligase activity while maintaining MDM2–MDMX interaction. Surprisingly, unlike MDM2C462A, MDMXΔRING, or MDMXC462A mice, MDM2Y487A mice survive into adulthood, with little phenotypic difference from wild type mice under normal, unstressed conditions. This clearly indicates that MDM2 E3 ligase activity is not essential for regulating p53 during embryonic development. No degradation of p53 is observed in MEFs, and although MDMX levels are also increased, p53 activity is greater than in wild type. This perhaps suggests that either MDM2–p53 or MDMX–p53 binding is not sufficient for complete p53 activity suppression, or that without E3 ligase-mediated degradation by MDM2, increased levels of p53 are also spontaneously more active.
Although Mdm2Y487A/Y487A mice appear normal under unstressed conditions, these mice are highly sensitive to even sub-lethal doses of ionizing radiation (IR), dying within ~20 days after exposure due to p53-dependent haematopoietic failure, indicating that the MDM2 E3 ligase activity is necessary for p53 degradation and suppression during DNA damaging conditions.
MDM2 has been shown to inhibit p53 acetylation by p300 (Kobet et al., 2000; Ito et al., 2001; Jin et al., 2002). Tollini et al. (2014) also compared the total and acetylated p53 levels of Mdm2C462A/C462A;p53ER/− and Mdm2Y487A/Y487A;p53ER/− MEFs and found that although total p53 levels were equivalent between the two, p53 acetylation levels were much greater in Mdm2C462A/C462A;p53ER/− MEFs. In addition, p53–p300 binding was increased in the absence of the MDM2–MDMX heterodimer, possibly indicating that the MDM2–MDMX heterodimer is more efficient than MDM2 alone in inhibiting p53 acetylation by p300, suggesting another mechanism through which the heterodimer could inhibit p53 activity in vivo.
MDM2–MDMX heterodimerization appears to be particularly important for suppressing chronic, basal levels of p53 activation, such as what might occur during embryogenesis, while MDM2 E3 ligase activity is dispensable under these conditions. However, under stressed conditions where p53 is acutely activated, such as DNA damaging conditions, the MDM2–MDMX heterodimer appears to be insufficient for restraining p53 in the adult mouse. These conditions appear to require the further degradation of p53, mandating use of MDM2 E3 ligase activity.
p53–Mdm2 feedback
In addition to regulating p53 stability and activity, Mdm2 is a p53 target gene (Barak et al., 1993). This feedback loop of regulation is thought to be important for returning p53 to basal levels and activity following a p53-activating insult. To directly address the importance of the p53–MDM2 feedback loop to p53 regulation in vivo, Pant and Lozano (2014) generated the Mdm2P2 allele, in which point mutations were introduced to two p53-binding sites within the Mdm2 promoter region. p53 stabilization in response to several stresses occurred in a similar manner to wild type Mdm2 mice, but p53 activity persisted longer in Mdm2P2 mice and MEFs, suggesting that basal levels of MDM2 are sufficient for p53 regulation in unstressed cells, but the p53–MDM2 feedback loop is required for restraining stress-induced p53 in vivo. In addition, the heterozygous deletion of MdmX appeared to enhance p53 stability in Mdm2P2/P2;MdmX+/− MEFs, suggesting that MDMX may enhance the degradation of stress-induced p53.
Disrupting upstream p53 signalling through MDM2 and MDMX mutation
DNA damage
Knockin mouse models have allowed us to appreciate the complex interactions of MDM2 and MDMX in p53 regulation, but they have also been used by several groups to determine the contributions of various upstream signals to p53 activation (Table 2). Activation of p53 requires transient inhibition of the activities of MDM2 and/or MDMX, which is thought to be mediated through upstream signalling factors. For instance, in vitro studies have shown that in response to DNA damage, ATM phosphorylates MDM2, inhibiting MDM2 E3 ligase activity and RING domain-dependent oligomer formation (Cheng et al., 2009, 2011). To test the importance of MDM2 phosphorylation at serine 394 (serine 395 in human), Gannon et al. (2012) generated the MDM2S394A mouse, replacing serine 394 with an alanine and disrupting MDM2 phosphorylation in vivo. Basal p53 levels and activity were unchanged in these mice. In response to lethal doses of IR, MDM2S394A mice experience reduced p53 stabilization and activation, translating to increased survival compared to wild type mice, indicating that MDM2 serine 394 phosphorylation is an important event preceding the propagation of p53 stabilization and activation following IR-mediated DNA damage. Conversely, Gannon et al. (2012) also generated mice containing a substitution of serine 394 with a phosphomimetic aspartate residue (MDM2S394D). Basal p53 levels and activity were unchanged compared to wild type mice, indicating that phosphorylation at serine 394 is not sufficient for p53 stabilization. Following IR, however, p53 stabilization and activation was greater and persisted longer in MDM2S394D mice, suggesting perhaps that the serine 394 phosphorylation mark is responsible for maintaining activation of p53 or is typically removed shortly after p53 activation.
MDMX is also phosphorylated following DNA damage, and these phosphorylation events are thought to be important for MDMX degradation following IR treatment (see next section). ATM phosphorylates MDMX serine 403 (402 in mouse) (Pereg et al., 2005), while Chk2 can phosphorylate serine 342 and serine 367 (341 and 367 in mouse) (Chen et al., 2005; Okamoto et al., 2005; LeBron et al., 2006; Pereg et al., 2006). To study the importance of MDMX phosphorylation to p53 activation following DNA damage, Wang et al. (2009) generated MDMX3SA mice, in which serine 341, serine 367, and serine 402 of MDMX are replaced with alanine residues. Upon loss of MDMX phosphorylation capability, MDMX3SA appears to be stabilized at basal levels. Following IR treatment, MDMX3SA remains stable compared to MDMX, and p53 protein levels and transcriptional activity appear to be lower in MDMX3SA MEFs and thymuses. In addition, MDMX3SA mice are resistant to lethal IR treatment and sensitive to MYC-induced lymphomagenesis. These results are in congruence with the reduced basal and DNA damage-induced p53 activity observed in MDMX3SA mice, suggesting that MDMX phosphorylation and subsequent degradation is important for proper p53 activation. These results also suggest that basal MDMX phosphorylation could be required for basal levels of p53 activity.
Oncogene activation
It is known that p53 responds to a variety of stresses in order to perform various pro-survival or pro-apoptotic functions, but the upstream signals of p53 activation are still being elucidated. MDM2S394A mice are somewhat susceptible to tumour formation, indicating that ATM-mediated MDM2 phosphorylation is likely important for allowing proper p53 activation in response to endogenous, cancer-causing DNA damage events. Carr et al. (2016) analysed this propensity for tumourigenesis in mice harbouring the MDM2S394A mutation by performing a regimen of repeated low doses of IR and by crossing Mdm2S394A mice with Eµ-Myc mice. MDM2S394A mice are resistant to IR-induced lymphomagenesis yet are highly susceptible to c-MYC-induced tumourigenesis, suggesting that the role of MDM2 phosphorylation in p53 regulation and tumour prevention is highly context- and stress type-specific.
Although oncogenes can invoke p53 stabilization by inducing DNA damage, it has been recently appreciated that the accelerated growth of cancer cells can also invoke p53 stabilization. For instance, accelerated cell growth mandates the increased production of ribosomes. The c-MYC oncogene is a master regulator of ribosomal biogenesis and directly upregulates the transcription of many RPs (Van Riggelen et al., 2010). Several RPs have been found in vitro to bind to the central zinc finger domain of MDM2 and prevent p53 inhibition (Zhang et al., 2003; Dai and Lu, 2004; Dai et al., 2004; Chen et al., 2007; Zhang and Lu, 2009). In order to directly test the role of RP–MDM2 interaction towards p53 activation in vivo, Macias et al. (2010) generated the MDM2C305F mouse. The MDM2C305F mutation resides in the region of RPL11 and RPL5 binding and thus prevents their interaction with MDM2. Mice with the MDM2C305F mutation display normal responses to DNA damage, but are highly susceptible to c-MYC overexpression- and adenomatous polyposis coli loss-induced lymphomagenesis and colorectal tumourigenesis, respectively (Macias et al., 2010; Meng et al., 2015; Liu et al., 2016). On the other hand, MDM2C305F mice are surprisingly resistant to HRASG12V-mediated melanomagenesis (Meng et al., 2016), possibly due to increased MDM2–RPL23 interaction mediated by the MDM2C305F mutation.
Nutrient availability
It has become increasingly apparent that MDM2 and MDMX also serve to regulate p53 in response to nutrient availability. This is not surprising, as it is clear that p53 itself regulates cellular energy homoeostasis (Vousden and Ryan, 2009; Zhang et al., 2010). It has also been reported that changes in nutrient abundance can drastically alter ribosomal biogenesis (Boulon et al., 2010). Altered ribosomal biogenesis induces the binding of RPs to MDM2 (Zhang and Lu, 2009), which inhibits MDM2-mediated p53 degradation, activating p53-induced (or dependent) metabolic alterations. Consistently, the MDM2C305F mouse, with its impaired RP–MDM2 binding, is deficient in p53-mediated fatty acid oxidation in response to fasting (Liu et al., 2014) and p53-mediated fat storage in response to sustained high-fat diet feeding (unpublished data). Although fasting or high-fat diet treatments appear to activate p53 signalling through the RP–MDM2 interaction, p53 activation in response to glucose deprivation appears to be MDMX-dependent (He et al., 2014). He et al. (2014) showed that glucose deprivation enhances 5′ adenosine monophosphate-activated protein kinase (AMPK) phosphorylation of MDMX at serine 342 (serine 341 in mouse). They suggest that MDMX S342 phosphorylation reduces its activity against p53 by enhancing MDMX interaction with 14-3-3, which allows p53 to become stabilized. Using the MDMX3SA mouse, this study suggested that loss of MDMX phosphorylation is correlated with reduced p53 stability and activity in response to AMPK induction.
MDM2 and MDMX stability regulation in vivo
Previous hypotheses proposed that MDM2 E3 ligase activity was important not only for p53 regulation, but also for stability of MDM2 and MDMX. In fact, in vitro mutations in the MDM2 RING domain result in increased stability of overexpressed MDM2 protein (Honda and Yasuda, 2000). However, mouse models have opposed these observations. The MDM2C462A and MDM2Y487A mouse models have specifically challenged the notion that MDM2 autoubiquitination occurs in vivo.
The MDM2C462A and MDM2Y487A mutations disrupt MDM2 E3 ligase activity in vivo, yet the half-life and ubiquitination levels of MDM2 do not change in these mice (Itahana et al., 2007; Tollini et al., 2014), suggesting that in the live mouse, MDM2 stability is mediated by the activity of other E3 ligases. However, some discrepancies exist between these models and other knockin mice. For example, the MDM2C462A mutation disrupts MDM2–MDMX interaction but does not affect MDM2 degradation (Itahana et al., 2007). Yet, the MDMXC462A mutation also disrupts MDM2–MDMX interaction without directly altering MDM2, and in the absence of MDM2–MDMX binding, Huang et al. (2011) observed that MDM2 ubiquitination was disrupted. However, since the MDM2C462A mutation and the MDMXC462A mutation reside in the RING domains of MDM2 or MDMX, which is thought to be important for the structure of these proteins (Poyurovsky et al., 2007), it is possible that these mutations alter the functions of the proteins beyond simple loss of RING domain function. In addition, mice containing MDMXΔRING, which also does not interact with MDM2, display no difference in MDM2 half-life compared to mice containing wild type MDMX (Pant et al., 2011). Resolving these conflicting observations is important to understand MDM2 stability in vivo. The MDM2Y487A mouse has provided some clarification of this problem, because the MDM2Y487A mutation does not occur in the RING domain of MDM2, and would not likely alter MDM2 structure greatly. Lacking MDM2 E3 ligase activity and maintaining MDM2–MDMX binding, MDM2Y487A is degraded equally quickly compared to MDM2 (Tollini et al., 2014). Two independent E3 ligase activity-disrupting mutations of MDM2 have shown that MDM2 E3 ligase activity is not required for basal MDM2 degradation in vivo (Itahana et al., 2007; Tollini et al., 2014), although whether or not MDM2–MDMX interaction is required for MDM2 ubiquitination is still unclear in the present mouse models. In vitro MDMX overexpression has been shown to stabilize MDM2, and this stabilization is dependent on the RING domain of each protein (Tanimura et al., 1999; Linares et al., 2003). Conversely, knockdown of MDMX has resulted in reduced MDM2 expression (Gu et al., 2002). Because of this, it was previously proposed that MDMX could redirect MDM2 E3 ligase activity from MDM2 unto itself and stabilize MDM2, but if MDM2 autoubiquitination does not truly occur in vivo, this may not be the case.
In vivo models have advocated that MDM2 does in fact control MDMX stability. For example, MDM2Y487A mice lacking MDM2 E3 ligase activity have increased protein levels of MDMX (Tollini et al., 2014), which is in line with in vitro studies suggesting that MDM2 E3 ligase activity acts to ubiquitinate MDMX (Kawai et al., 2003; Pan and Chen, 2003). In addition, MDMXΔRING, which does not have the ability to interact with MDM2, is not degraded compared to MDMX in untreated or IR-treated MEFs (Pant et al., 2011).
MDM2 and MDMX degradation following IR has been observed by many groups in cell culture (Kawai et al., 2003; Stommel and Wahl, 2004). This regulation has been recapitulated in several mouse models. For example, MDM2S394A, which cannot be phosphorylated by ATM, appears to be resistant to IR-induced degradation (Carr et al., 2016). MDMX3SA is also more stable than MDMX in multiple tissues and is resistant to DNA damage-induced degradation (Wang et al., 2009). This indicates both that MDMX may have some level of constitutive phosphorylation that is important for its normal degradation in vivo and that DNA damage-induced phosphorylation is necessary for proper regulation of MDMX stability. Following IR treatment, MDMX3SA also interacts with MDM2 similarly to MDMX, which suggests that DNA damage-induced phosphorylation does not hinder MDM2–MDMX interaction.
Using mouse models to inform future therapies
p53 mutation occurs in ~50% of human cancers, and p53 is often functionally inactivated in tumours harbouring wild type p53, due to aberrantly expressed MDM2 or MDMX (Tovar et al., 2006). A cancer-associated human single nucleotide polymorphism (SNP) (Bond et al., 2004) in the second promoter of Mdm2 contributing to increased Mdm2 transcription was recently modelled in the mouse (Post et al., 2010). Mice containing a T to G human SNP (SNP 309) were susceptible to decreased p53 function and increased tumourigenesis. This model suggests that even naturally occurring MDM2 ‘overexpression’ (as opposed to transgenic overexpression) does in fact contribute to p53 functional inactivation.
A growing number of studies have suggested targeting mutant p53 or restoring wild type p53 as cancer treatment strategy (Burgess et al., 2016; Soragni et al., 2016). Many drugs specifically targeting MDM2–p53 interaction, MDMX–p53 interaction, or MDM2-mediated ubiquitination of p53 have been developed (Vassilev et al., 2004; Wade et al., 2013; Burgess et al., 2016). For example, MDMX loss in c-MYC-driven tumours extends survival after p53ER restoration (Garcia et al., 2011). In addition, CreER-mediated p53neo restoration in transplanted MDM2-overexpressing tumours also appears to extend survival in mice (Li et al., 2014). However, so far these treatment strategies have enjoyed limited efficacy in the clinic.
In vivo studies have also suggested that other approaches could be taken to restore p53 function in human cancers harbouring wild type p53, such as inhibiting MDM2–MDMX binding or inhibiting MDM2 E3 ligase activity. The inhibition of MDM2 E3 ligase activity may be especially attractive as a treatment strategy, because the MDM2Y487A mouse model shows that genetic ablation of MDM2 E3 ligase activity is tolerated by the adult mouse as well as the developing embryo (Tollini et al., 2014), which suggests that this strategy could avoid toxicity issues. In response to p53-activating stimuli, cells containing MDM2Y487A demonstrate increased p53 stability and activity. Observations from our laboratory support this strategy in principle, as homozygous MDM2Y487A mutation appears to allow for prolonged survival in response to c-MYC-induced tumourigenesis (our unpublished data). Although several inhibitors of MDM2 E3 ligase activity have been identified and shown to stabilize p53 (Yang et al., 2005; Herman et al., 2011; Roxburgh et al., 2012), their activity and specificity may not yet be sufficient for human use. To our knowledge, MDM2 E3 ligase inhibitors have not been tested in humans, but several other small-molecule MDM2 antagonists are currently in Phase I trials (Burgess et al., 2016).
Mouse models have also suggested that complete restoration of p53 function in the presence of radiation should be used with caution, as abundant p53 activity is especially toxic to radio-sensitive tissues (Ringshausen et al., 2006; Tollini et al., 2014; Zhang et al., 2014a). It is possible that tissue-targeted therapies will need to be used in combination with any p53-reactivating therapies to avoid this problem.
Concluding remarks
Mouse models altering MDM2 and MDMX have given us a clearer understanding of the in vivo roles of MDM2 and MDMX in p53 regulation (Figure 1) and established that MDM2 and MDMX proteins are master p53 regulators. However, several questions remain. Although several in vitro studies suggest that MDMX may facilitate MDM2-mediated p53 degradation, we still do not have a clear understanding of whether this occurs in vivo. We still do not completely understand how MDM2 is degraded. In addition, MDM2 and MDMX appear to have differing activities in p53 transcriptional inhibition, but we do not understand how or why this may occur. Although many questions remain, the tools presented in this review are indicative of the importance of in vivo modelling and point to a bright future of continued research in the MDM2/MDMX–p53 field.
Acknowledgements
The authors would like to thank Hui Tian, Jing Yang, and Derek Franklin (Department of Radiation Oncology, University of North Carolina at Chapel Hill) for their helpful discussions of this manuscript. The authors apologize if they failed to cite any relevant articles.
Funding
This review was supported by grants from the National Institutes of Health (CA127770, CA 100302, and CA167637), the Natural Science Foundation of China (NSFC) to Y.Z., and the National Institute of General Medical Sciences (5T32 GM007092) to N.R.T.
Conflict of interest: none declared.
References
- Badciong J.C., and Haas A.L. (2002). MdmX is a RING finger ubiquitin ligase capable of synergistically enhancing Mdm2 ubiquitination. J. Biol. Chem. 277, 49668–49675. [DOI] [PubMed] [Google Scholar]
- Barak Y., Juven T., Haffner R., et al. (1993). mdm2 expression is induced by wild type p53 activity. EMBO J. 12, 461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardot B., Bouarich-Bourimi R., Leemput J., et al. (2015). Mice engineered for an obligatory Mdm4 exon skipping express higher levels of the Mdm4-S isoform but exhibit increased p53 activity. Oncogene 34, 2943–2948. [DOI] [PubMed] [Google Scholar]
- Bartel F., Schulz J., Böhnke A., et al. (2005). Significance of HDMX-S (or MDM4) mRNA splice variant overexpression and HDMX gene amplification on primary soft tissue sarcoma prognosis. Int. J. Cancer 117, 469–475. [DOI] [PubMed] [Google Scholar]
- Boesten L., Zadelaar S., De Clercq S., et al. (2006). Mdm2, but not Mdm4, protects terminally differentiated smooth muscle cells from p53-mediated caspase-3-independent cell death. Cell Death Differ. 13, 2089–2098. [DOI] [PubMed] [Google Scholar]
- Bond G.L., Hu W., Bond E.E., et al. (2004). A single nucleotide polymorphism in the MDM2 promoter attenuates the p53 tumor suppressor pathway and accelerates tumor formation in humans. Cell 119, 591–602. [DOI] [PubMed] [Google Scholar]
- Boulon S., Westman B.J., Hutten S., et al. (2010). The nucleolus under stress. Mol. Cell 40, 216–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess A., Chia K.M., Haupt S., et al. (2016). Clinical overview of MDM2/X-targeted therapies. Front. Oncol. 6, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carr M.I., Roderick J.E., Gannon H.S., et al. (2016). Mdm2 phosphorylation regulates its stability and has contrasting effects on oncogene and radiation-induced tumorigenesis. Cell Rep. 16, 2618–2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D., Zhang Z., Li M., et al. (2007). Ribosomal protein S7 as a novel modulator of p53–MDM2 interaction: binding to MDM2, stabilization of p53 protein, and activation of p53 function. Oncogene 26, 5029–5037. [DOI] [PubMed] [Google Scholar]
- Chen J., Marechal V., and Levine A.J. (1993). Mapping of the p53 and mdm-2 interaction domains. Mol. Cell. Biol. 13, 4107–4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Gilkes D.M., Pan Y., et al. (2005). ATM and Chk2-dependent phosphorylation of MDMX contribute to p53 activation after DNA damage. EMBO J. 24, 3411–3422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Q., Chen L., Li Z., et al. (2009). ATM activates p53 by regulating MDM2 oligomerization and E3 processivity. EMBO J. 28, 3857–3867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Q., Cross B., Li B., et al. (2011). Regulation of MDM2 E3 ligase activity by phosphorylation after DNA damage. Mol. Cell. Biol. 31, 4951–4963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng X., Xia W., Yang J.-Y., et al. (2010). Activation of murine double minute 2 by Akt in mammary epithelium delays mammary involution and accelerates mammary tumorigenesis. Cancer Res. 70, 7684–7689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christophorou M.A., Martin-Zanca D., Soucek L., et al. (2005). Temporal dissection of p53 function in vitro and in vivo. Nat. Genet. 37, 718–726. [DOI] [PubMed] [Google Scholar]
- Dai M.-S., and Lu H. (2004). Inhibition of MDM2-mediated p53 ubiquitination and degradation by ribosomal protein L5. J. Biol. Chem. 279, 44475–44482. [DOI] [PubMed] [Google Scholar]
- Dai M.-S., Zeng S.X., Jin Y., et al. (2004). Ribosomal protein L23 activates p53 by inhibiting MDM2 function in response to ribosomal perturbation but not to translation inhibition. Mol. Cell. Biol. 24, 7654–7668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donehower L.A., Harvey M., Slagle B.L., et al. (1992). Mice deficient for p53 are developmentally normal but susceptible to spontaneous. Nature 356, 19. [DOI] [PubMed] [Google Scholar]
- Feng Z., Hu W., Teresky A.K., et al. (2007). Declining p53 function in the aging process: a possible mechanism for the increased tumor incidence in older populations. Proc. Natl Acad. Sci. USA 104, 16633–16638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francoz S., Froment P., Bogaerts S., et al. (2006). Mdm4 and Mdm2 cooperate to inhibit p53 activity in proliferating and quiescent cells in vivo. Proc. Natl Acad. Sci. USA 103, 3232–3237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gannon H.S., Donehower L.A., Lyle S., et al. (2011). Mdm2–p53 signaling regulates epidermal stem cell senescence and premature aging phenotypes in mouse skin. Dev. Biol. 353, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gannon H.S., Woda B.A., and Jones S.N. (2012). ATM phosphorylation of Mdm2 Ser394 regulates the amplitude and duration of the DNA damage response in mice. Cancer Cell 21, 668–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia D., Warr M.R., Martins C.P., et al. (2011). Validation of MdmX as a therapeutic target for reactivating p53 in tumors. Genes Dev. 25, 1746–1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grier J.D., Xiong S., Elizondo-Fraire A.C., et al. (2006). Tissue-specific differences of p53 inhibition by Mdm2 and Mdm4. Mol. Cell. Biol. 26, 192–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grier J.D., Yan W., and Lozano G. (2002). Conditional allele of mdm2 which encodes a p53 inhibitor. Genesis 32, 145–147. [DOI] [PubMed] [Google Scholar]
- Gu J., Kawai H., Nie L., et al. (2002). Mutual dependence of MDM2 and MDMX in their functional inactivation of p53. J. Biol. Chem. 277, 19251–19254. [DOI] [PubMed] [Google Scholar]
- Haupt Y., Maya R., Kazaz A., et al. (1997). Mdm2 promotes therapid degradation of p53. Nature 387, 296–299. [DOI] [PubMed] [Google Scholar]
- He G., Zhang Y.W., Lee J.H., et al. (2014). AMP-activated protein kinase induces p53 by phosphorylating MDMX and inhibiting its activity. Mol. Cell. Biol. 34, 148–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman A.G., Hayano M., Poyurovsky M.V., et al. (2011). Discovery of Mdm2-MdmX E3 ligase inhibitors using a cell-based ubiquitination assay. Cancer Discov. 1, 312–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda R., Tanaka H., and Yasuda H. (1997). Oncoprotein MDM2 is a ubiquitin ligase E3 for tumor suppressor p53. FEBS Lett. 420, 25–27. [DOI] [PubMed] [Google Scholar]
- Honda R., and Yasuda H. (2000). Activity of MDM2, a ubiquitin ligase, toward p53 or itself is dependent on the RING finger domain of the ligase. Oncogene 19, 1473–1476. [DOI] [PubMed] [Google Scholar]
- Hu B., Gilkes D.M., and Chen J. (2007). Efficient p53 activation and apoptosis by simultaneous disruption of binding to MDM2 and MDMX. Cancer Res. 67, 8810–8817. [DOI] [PubMed] [Google Scholar]
- Huang L., Yan Z., Liao X., et al. (2011). The p53 inhibitors MDM2/MDMX complex is required for control of p53 activity in vivo. Proc. Natl Acad. Sci. USA 108, 12001–12006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itahana K., Mao H., Jin A., et al. (2007). Targeted inactivation of Mdm2 RING finger E3 ubiquitin ligase activity in the mouse reveals mechanistic insights into p53 regulation. Cancer Cell 12, 355–366. [DOI] [PubMed] [Google Scholar]
- Ito A., Lai C.H., Zhao X., et al. (2001). p300/CBP-mediated p53 acetylation is commonly induced by p53-activating agents and inhibited by MDM2. EMBO J. 20, 1331–1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson M.W., and Berberich S.J. (2000). MdmX protects p53 from Mdm2-mediated degradation. Mol. Cell. Biol. 20, 1001–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y., Zeng S.X., Dai M.-S., et al. (2002). MDM2 inhibits PCAF (p300/CREB-binding protein-associated factor)-mediated p53 acetylation. J. Biol. Chem. 277, 30838–30843. [DOI] [PubMed] [Google Scholar]
- Jones S.N., Hancock A.R., Vogel H., et al. (1998). Overexpression of Mdm2 in mice reveals a p53-independent role for Mdm2 in tumorigenesis. Proc. Natl Acad. Sci. USA 95, 15608–15612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones S.N., Roe A.E., Donehower L.A., et al. (1995). Rescue of embryonic lethality in Mdm2-deficient mice by absence of p53. Nature 378, 206–208. [DOI] [PubMed] [Google Scholar]
- Kawai H., Wiederschain D., Kitao H., et al. (2003). DNA damage-induced MDMX degradation is mediated by MDM2. J. Biol. Chem. 278, 45946–45953. [DOI] [PubMed] [Google Scholar]
- Kobet E., Zeng X., Zhu Y., et al. (2000). MDM2 inhibits p300-mediated p53 acetylation and activation by forming a ternary complex with the two proteins. Proc. Natl Acad. Sci. USA 97, 12547–12552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubbutat M.H.G., Jones S.N., and Vousden K.H. (1997). Regulation of p53 stability by Mdm2. Nature 387, 299–303. [DOI] [PubMed] [Google Scholar]
- Lang G.A., Iwakuma T., Suh Y.-A., et al. (2004). Gain of function of a p53 hot spot mutation in a mouse model of Li-Fraumeni syndrome. Cell 119, 861–872. [DOI] [PubMed] [Google Scholar]
- LeBron C., Chen L., Gilkes D.M., et al. (2006). Regulation of MDMX nuclear import and degradation by Chk2 and 14-3-3. EMBO J. 25, 1196–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengner C.J., Steinman H.A., Gagnon J., et al. (2006). Osteoblast differentiation and skeletal development are regulated by Mdm2–p53 signaling. J. Cell Biol. 172, 909–921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenos K., Grawenda A.M., Lodder K., et al. (2012). Alternate splicing of the p53 inhibitor HDMX offers a superior prognostic biomarker than p53 mutation in human cancer. Cancer Res. 72, 4074–4084. [DOI] [PubMed] [Google Scholar]
- Li Q., Zhang Y., El-Naggar A.K., et al. (2014). Therapeutic efficacy of p53 restoration in Mdm2-overexpressing tumors. Mol. Cancer Res. 12, 901–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linares L.K., Hengstermann A., Ciechanover A., et al. (2003). HdmX stimulates Hdm2-mediated ubiquitination and degradation of p53. Proc. Natl Acad. Sci. USA 100, 12009–12014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linke K., Mace P., Smith C., et al. (2008). Structure of the MDM2/MDMX RING domain heterodimer reveals dimerization is required for their ubiquitylation in trans. Cell Death Differ. 15, 841–848. [DOI] [PubMed] [Google Scholar]
- Liu S., Tackmann N., Yang J., et al. (2016). Disruption of the RP-MDM2-p53 pathway accelerates APC loss-induced colorectal tumorigenesis. Oncogene, 10.1038/onc.2016.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., He Y., Jin A., et al. (2014). Ribosomal protein–Mdm2–p53 pathway coordinates nutrient stress with lipid metabolism by regulating MCD and promoting fatty acid oxidation. Proc. Natl Acad. Sci. USA 111, E2414–E2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macias E., Jin A., Deisenroth C., et al. (2010). An ARF-independent c-MYC-activated tumor suppression pathway mediated by ribosomal protein-Mdm2 Interaction. Cancer Cell 18, 231–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maetens M., Doumont G., De Clercq S., et al. (2007). Distinct roles of Mdm2 and Mdm4 in red cell production. Blood 109, 2630–2633. [DOI] [PubMed] [Google Scholar]
- Mendrysa S.M., McElwee M.K., Michalowski J., et al. (2003). mdm2 is critical for inhibition of p53 during lymphopoiesis and the response to ionizing irradiation. Mol. Cell. Biol. 23, 462–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X., Carlson N., Dong J., et al. (2015). Oncogenic c-Myc-induced lymphomagenesis is inhibited non-redundantly by the p19Arf–Mdm2–p53 and RP–Mdm2–p53 pathways. Oncogene 34, 5709–5717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X., Tackmann N.R., Liu S., et al. (2016). RPL23 links oncogenic RAS signaling to p53-mediated tumor suppression. Cancer Res. 76, 5030–5039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montes de Oca Luna R., Wagner D.S., and Lozano G. (1995). Rescue of early embryonic lethality in mdm2-deficient mice by deletion of p53. Nature 378, 203–206. [DOI] [PubMed] [Google Scholar]
- Moyer S.M., Larsson C.A., and Lozano G. (2017). Mdm proteins: critical regulators of embryogenesis and homoeostasis. J. Mol. Cell Biol. 10.1093/jmcb/mjx004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller P.A., and Vousden K.H. (2013). p53 mutations in cancer. Nat. Cell Biol. 15, 2–8. [DOI] [PubMed] [Google Scholar]
- Okamoto K., Kashima K., Pereg Y., et al. (2005). DNA damage-induced phosphorylation of MdmX at serine 367 activates p53 by targeting MdmX for Mdm2-dependent degradation. Mol. Cell. Biol. 25, 9608–9620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliner J.D., Pietenpol J.A., Thiagalingam S., et al. (1993). Oncoprotein MDM2 conceals the activation domain of tumour suppressor p53. Nature 362, 857–860. [DOI] [PubMed] [Google Scholar]
- Pan Y., and Chen J. (2003). MDM2 promotes ubiquitination and degradation of MDMX. Mol. Cell. Biol. 23, 5113–5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pant V., and Lozano G. (2014). Dissecting the p53-Mdm2 feedback loop in vivo: uncoupling the role in p53 stability and activity. Oncotarget 5, 1149–1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pant V., Xiong S., Iwakuma T., et al. (2011). Heterodimerization of Mdm2 and Mdm4 is critical for regulating p53 activity during embryogenesis but dispensable for p53 and Mdm2 stability. Proc. Natl Acad. Sci. USA 108, 11995–12000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parant J., Chavez-Reyes A., Little N.A., et al. (2001). Rescue of embryonic lethality in Mdm4-null mice by loss of Trp53 suggests a nonoverlapping pathway with MDM2 to regulate p53. Nat. Genet. 29, 92–95. [DOI] [PubMed] [Google Scholar]
- Pereg Y., Lam S., Teunisse A., et al. (2006). Differential roles of ATM-and Chk2-mediated phosphorylations of Hdmx in response to DNA damage. Mol. Cell. Biol. 26, 6819–6831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereg Y., Shkedy D., de Graaf P., et al. (2005). Phosphorylation of Hdmx mediates its Hdm2-and ATM-dependent degradation in response to DNA damage. Proc. Natl Acad. Sci. USA 102, 5056–5061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Post S.M., Quintás-Cardama A., Pant V., et al. (2010). A high-frequency regulatory polymorphism in the p53 pathway accelerates tumor development. Cancer Cell 18, 220–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poyurovsky M.V., Priest C., Kentsis A., et al. (2007). The Mdm2 RING domain C-terminus is required for supramolecular assembly and ubiquitin ligase activity. EMBO J. 26, 90–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prodosmo A., Giglio S., Moretti S., et al. (2008). Analysis of human MDM4 variants in papillary thyroid carcinomas reveals new potential markers of cancer properties. J. Mol. Med. 86, 585–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rallapalli R., Strachan G., Cho B., et al. (1999). A novel MDMX transcript expressed in a variety of transformed cell lines encodes a truncated protein with potent p53 repressive activity. J. Biol. Chem. 274, 8299–8308. [DOI] [PubMed] [Google Scholar]
- Rallapalli R., Strachan G., Tuan R.S., et al. (2003). Identification of a domain within MDMX-S that is responsible for its high affinity interaction with p53 and high-level expression in mammalian cells. J. Cell. Biochem. 89, 563–575. [DOI] [PubMed] [Google Scholar]
- Ringshausen I., O'Shea C.C., Finch A.J., et al. (2006). Mdm2 is critically and continuously required to suppress lethal p53 activity in vivo. Cancer Cell 10, 501–514. [DOI] [PubMed] [Google Scholar]
- Roxburgh P., Hock A.K., Dickens M.P., et al. (2012). Small molecules that bind the Mdm2 RING stabilize and activate p53. Carcinogenesis 33, 791–798. [DOI] [PubMed] [Google Scholar]
- Shvarts A., Steegenga W., Riteco N., et al. (1996). MDMX: a novel p53-binding protein with some functional properties of MDM2. EMBO J. 15, 5349. [PMC free article] [PubMed] [Google Scholar]
- Soragni A., Janzen D.M., Johnson L.M., et al. (2016). A designed inhibitor of p53 aggregation rescues p53 tumor suppression in ovarian carcinomas. Cancer Cell 29, 90–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinman H.A., Hoover K.M., Keeler M.L., et al. (2005). Rescue of Mdm4-deficient mice by Mdm2 reveals functional overlap of Mdm2 and Mdm4 in development. Oncogene 24, 7935–7940. [DOI] [PubMed] [Google Scholar]
- Stommel J.M., and Wahl G.M. (2004). Accelerated MDM2 auto-degradation induced by DNA-damage kinases is required for p53 activation. EMBO J. 23, 1547–1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanimura S., Ohtsuka S., Mitsui K., et al. (1999). MDM2 interacts with MDMX through their RING finger domains. FEBS Lett. 447, 5–9. [DOI] [PubMed] [Google Scholar]
- Tollini L.A., Jin A., Park J., et al. (2014). Regulation of p53 by Mdm2 E3 ligase function is dispensable in embryogenesis and development, but essential in response to DNA damage. Cancer Cell 26, 235–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tovar C., Rosinski J., Filipovic Z., et al. (2006). Small-molecule MDM2 antagonists reveal aberrant p53 signaling in cancer: implications for therapy. Proc. Natl Acad. Sci. USA 103, 1888–1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentin-Vega Y., Okano H., and Lozano G. (2008). The intestinal epithelium compensates for p53-mediated cell death and guarantees organismal survival. Cell Death Differ. 15, 1772–1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentin-Vega Y.A., Box N., Terzian T., et al. (2009). Mdm4 loss in the intestinal epithelium leads to compartmentalized cell death but no tissue abnormalities. Differentiation 77, 442–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Riggelen J., Yetil A., and Felsher D.W. (2010). MYC as a regulator of ribosome biogenesis and protein synthesis. Nat. Rev. Cancer 10, 301–309. [DOI] [PubMed] [Google Scholar]
- Vassilev L.T., Vu B.T., Graves B., et al. (2004). In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science 303, 844–848. [DOI] [PubMed] [Google Scholar]
- Vousden K.H., and Ryan K.M. (2009). p53 and metabolism. Nat. Rev. Cancer 9, 691–700. [DOI] [PubMed] [Google Scholar]
- Wade M., Li Y.-C., and Wahl G.M. (2013). MDM2, MDMX and p53 in oncogenesis and cancer therapy. Nat. Rev. Cancer 13, 83–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade M., Wang Y.V., and Wahl G.M. (2010). The p53 orchestra: Mdm2 and Mdmx set the tone. Trends Cell Biol. 20, 299–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y.V., Leblanc M., Wade M., et al. (2009). Increased radioresistance and accelerated B cell lymphomas in mice with Mdmx mutations that prevent modifications by DNA-damage-activated kinases. Cancer Cell 16, 33–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong S., Van Pelt C.S., Elizondo-Fraire A.C., et al. (2006). Synergistic roles of Mdm2 and Mdm4 for p53 inhibition in central nervous system development. Proc. Natl Acad. Sci. USA 103, 3226–3231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Ludwig R.L., Jensen J.P., et al. (2005). Small molecule inhibitors of HDM2 ubiquitin ligase activity stabilize and activate p53 in cells. Cancer Cell 7, 547–559. [DOI] [PubMed] [Google Scholar]
- Zhang Q., He X., Chen L., et al. (2012). Synergistic regulation of p53 by Mdm2 and Mdm4 is critical in cardiac endocardial cushion morphogenesis during heart development. J. Pathol. 228, 416–428. [DOI] [PubMed] [Google Scholar]
- Zhang X.-D., Qin Z.-H., and Wang J. (2010). The role of p53 in cell metabolism. Acta Pharmacol. Sin. 31, 1208–1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., and Lu H. (2009). Signaling to p53: ribosomal proteins find their way. Cancer Cell 16, 369–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Wolf G.W., Bhat K., et al. (2003). Ribosomal protein L11 negatively regulates oncoprotein MDM2 and mediates a p53-dependent ribosomal-stress checkpoint pathway. Mol. Cell. Biol. 23, 8902–8912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Xiong S., Li Q., et al. (2014. a). Tissue-specific and age-dependent effects of global Mdm2 loss. J. Pathol. 233, 380–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Zhang X., and Lu H. (2014. b). Aberrant activation of p53 due to loss of MDM2 or MDMX causes early lens dysmorphogenesis. Dev. Biol. 396, 19–30. [DOI] [PMC free article] [PubMed] [Google Scholar]