Abstract
The co-translational targeting or insertion of secretory and membrane proteins into the endoplasmic reticulum (ER) is a key biological process mediated by the signal recognition particle (SRP). In eukaryotes, the SRP68–SRP72 (SRP68/72) heterodimer plays an essential role in protein translocation. However, structural information on the two largest SRP proteins, SRP68 and SRP72, is limited, especially regarding their interaction. Herein, we report the first crystal structures of human apo-SRP72 and the SRP68/72 complex at 2.91Å and 1.7Å resolution, respectively. The SRP68-binding domain of SRP72 contains four atypical tetratricopeptide repeats (TPR) and a flexible C-terminal cap. Apo-SRP72 exists mainly as dimers in solution. To bind to SRP68, the SRP72 homodimer disassociates, and the indispensable C-terminal cap undergoes a pronounced conformational change to assist formation of the SRP68/72 heterodimer. A 23-residue polypeptide of SRP68 is sufficient for tight binding to SRP72 through its unusually hydrophobic and extended surface. Structural, biophysical, and mutagenesis analyses revealed that cancer-associated mutations disrupt the SRP68–SRP72 interaction and their co-localization with ER in mammalian cells. The results highlight the essential role of the SRP68–SRP72 interaction in SRP-mediated protein translocation and provide a structural basis for disease diagnosis, pathophysiology, and drug design.
Keywords: SRP72, SRP68, protein translocation, crystal structures, cancer, protein–protein interaction, signal recognition particle
Introduction
Co-translational targeting of secretory and membrane proteins to the endoplasmic reticulum (ER) is a functionally conserved and key biological process mediated by signal recognition particle (SRP). SRP is a ribonucleoprotein complex that recognizes the signal sequence of the nascent polypeptide emerging from the ribosomal exit tunnel and directs the ribosome nascent chain (RNC) complex to the translocation channel via GTP-dependent interaction with SRP receptors (SR; SRαβ in eukaryotes, FtsY in bacteria) (Egea et al., 2005; Akopian et al., 2013).
Mammalian SRP is composed of six protein subunits (SRP9, SRP14, SRP19, SRP54, SRP68, and SRP72, named based on molecular weight) assembled hierarchically on a 7S RNA molecule of ~300 nt (Doudna and Batey, 2004). It can be divided into an Alu domain comprising the terminal RNA regions and SRP9/SRP14 protein subunits, and an S domain comprising the central RNA region and the other four protein subunits. The Alu domain binds to the RNC complex and arrests ribosomal elongation, whereas the S domain recognizes the signal peptide and targets the complex to translocon (Weichenrieder et al., 2000; Halic et al., 2004). SRP RNA is synthesized in the nucleus and requires the binding of SRP68 and SRP72 for nuclear export (van Nues et al., 2008). Recruitment of the universally conserved SRP54 in the cytosol is the final step of the assembly of the SRP complex. Unlike mammalian SRP, bacterial SRP only contains a 4.5S or 6S RNA and Ffh, a homolog of the SRP54. Consistent with the role in nuclear export of eukaryotic SRP, SRP68 and SRP72 are detected in cytosol as well as the nucleolus (Politz et al., 2000) and are highly conserved in organisms ranging from yeast to human; however, no homologs have been found in bacterial or archaeal genomes and they are thus considered specific to eukaryotic SRPs (Zwieb and Eichler, 2002; Buskiewicz et al., 2004; Andersen et al., 2006). The two proteins form a stable heterodimer that mainly binds to the 3-way junction of helices 5–8 of the 7S RNA (Halic et al., 2004, 2006a). The SRP68/72 subunits add another layer of regulation of the mammalian SRP, compared with the simpler bacterial SRP, and are essential for the proper functioning of mammalian SRP. Reconstituted SRP lacking the SRP68/72 heterodimer failed to recognize signal peptide (Grosshans et al., 2001), to direct translocation, or to arrest elongation (Siegel and Walter, 1985).
Many cryo-electron microscopy studies at low resolution have been reported for the eukaryotic SRP-RNC targeting complex, which helped to illuminate its biological function. The structures of all subunits except SRP68/72 have been rigorously studied (Halic et al., 2006b; Voorhees and Hegde, 2015; Jomaa et al., 2016). Recently, the crystal structure of a 28 kDa RNA-binding domain (RBD) of SRP68 in complex with SRP RNA and SRP19 revealed how SRP68 binding remodels the RNA in the S domain and affects the recruitment of SRP54 and RNC (Grotwinkel et al., 2014). However, structural details of the domains of SRP68 and SRP72, the two largest SRP proteins, remain elusive, except for the SRP68 RBD. And how the two proteins form a heterodimer is also unknown. Furthermore, mutations in SRP68 and SRP72 have been observed in many cancers (Supplementary Table S1). It remains unclear how these mutations correlate with tumorigenesis.
In the present study, we report the first crystal structures of the human SRP72 N-terminal tetratricopeptide repeat (TPR) domain in apo form and in complex with SRP68 at 2.91Å and 1.7Å resolution, respectively. The SRP68-binding domain of SRP72 contains four atypical TPR repeats (TPR1–4) and an intrinsically flexible C-terminal ‘cap’ essential for tight target protein binding. Apo-SRP72 exists mainly as dimers in solution. Upon binding to SRP68, the C-terminus of the SRP72 dimer disassociates and undergoes a pronounced conformational change. SRP68 binds tightly to the hydrophobic concave surface of the SRP72 TPR domain. Furthermore, by combining structural, biophysical, and mutagenesis analyses and in vivo localization assays, we show that a minimum fragment of SRP68 is necessary and sufficient to bind to SRP72. We also reveal the key residues involved in the interaction and the rationale of cancer-associated mutations. Our results provide the first structural insight into the SRP68–SRP72 interaction and a basis for understanding the pathology of diseases related to SRP-mediated protein translocation.
Results
Structure determination of apo-SRP72 and the SRP68/72 complex
Previous studies indicated that the C-terminal region of SRP68 binds to the N-terminal domain of SRP72 (Iakhiaeva et al., 2006). Various constructs of the C-terminal region of SRP68 were expressed in E. coli but found to form inclusion bodies, which prevented us from obtaining significant amounts of soluble protein. Therefore, we co-expressed a set of SRP72 and SRP68 fragments in E. coli and successfully purified the complex using immobilized metal affinity chromatography. We also generated more than a dozen SRP68–SRP72 mutants based on surface entropy reduction mutagenesis (Derewenda and Vekilov, 2006). After extensive crystallization trials, one mutant of the complex (SRP721–163–SRP68509–614 E608A Q609A K610A) successfully yielded crystals suitable for diffraction experiments. Because the sequence homology between SRP72 and any homologous protein with a known three-dimensional structure was <10%, we determined the structure using the single-wavelength anomalous diffraction (SAD) method using Hg-derived complex crystals. The structure was refined to a resolution of 1.7Å in the space group P6322, with an Rwork of 16.3% and an Rfree of 18.9% (Table 1). To investigate the conformational changes of SRP72 upon binding to SRP68, we also obtained the structure of SRP72 in the apo form. Crystals of apo-SRP72 were easily grown in a reservoir solution containing ammonium sulfate as the precipitant, but the diffraction quality of these crystals was poor, even after extensive optimization of the crystallization conditions and various post-crystallization treatments such as annealing and dehydration. Fortunately, high-quality diffracting crystals for apo-SRP72 were eventually obtained using dimethylethylammonium propane sulfonate as a detergent. The apo-SRP721–163 structure was solved at 2.91 Å in the space group P212121 by molecular replacement using the above determined SRP68/72 complex structure as a template (Table 1).
Table 1.
Data collection and refinement statistics of apo-SRP72 and the SRP68/72 complex.a
| Hg2+-SRP68/72 | SRP68/72 complex (PDB: 5WRV) | Apo-SRP72 (PDB: 5WRW) | |
|---|---|---|---|
| Data collection | |||
| Wavelength | 1.0085 | 0.9792 | 0.9792 |
| Space group | P6322 | P6322 | P212121 |
| Cell dimensions | |||
| a, b, c (Å) | 122.22, 122.22, 78.78 | 120.60, 120.60, 80.00 | 83.44,123.72, 150.72 |
| α, β, γ (˚) | 90, 90, 120 | 90, 90, 120 | 90, 90, 90 |
| Resolution (Å) | 50.00–2.45 (2.49–2.45) | 50.00–1.70 (1.73–1.70) | 50.00–2.91 (2.96–2.91) |
| Rsym (%) | 10.0 (67.9) | 5.9 (61.2) | 9.8 (63.1) |
| I/σ | 46.7 (2.6) | 62.2 (2.07) | 26.2 (2.3) |
| Completeness (%) | 100 (100) | 99.5 (99.9) | 99.9 (100) |
| Total no. of reflections | 794160 | 1454874 | 534438 |
| Unique reflections | 13423 | 38126 | 35059 |
| Redundancy | 29.7 (18.7) | 13.2 (12.0) | 7.8 (7.8) |
| Refinement | |||
| Resolution (Å) | 50.00–1.70 (1.75–1.70) | 50.00–2.91 (2.99–2.91) | |
| No. of reflections | 36033 (2407) | 33221 (2418) | |
| Rwork/Rfree (%) | 16.3/18.9 | 23.1/26.6 | |
| No. of atoms | |||
| Protein | 1434 | 6149 | |
| Ligand/ions | 49 | 10 | |
| Water | 226 | 89 | |
| B-factors (Å2) | |||
| Protein | 34.8 | 97.3 | |
| Ligand/ion | 49.2 | 90.3 | |
| Water | 52.7 | 70.4 | |
| rms deviations | |||
| Bond lengths (Å) | 0.007 | 0.009 | |
| Bond angles (°) | 1.007 | 1.200 | |
| Ramachandran plot (%)b | 89.8/10.2/0/0 | 88.6/11.1/0.3/0 | |
aThree crystal experiments for each structure.
bResidues in most favored, additional allowed, generously allowed, and disallowed regions of the Ramachandran plot.
Rsym= ΣhΣi|Ih,i−Ih|/ΣhΣlIh,i, where Ih is the mean intensity of the i observations of symmetry related reflections of h.
Overall structures of apo-SRP72 and the SRP68/72 complex reveal an atypical TPR fold
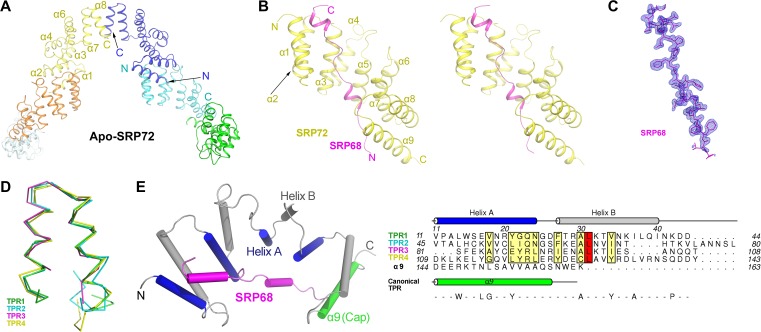
The final structural model of apo-SRP72 contains six protein molecules in the asymmetric unit (Figure 1A and Table 1). Each apo-SRP72 protomer is folded into a unique hook-like shape that is nearly identical with an overall root-mean-square deviation (RMSD) of 1.0 Å for all Cα atoms. Residues 1–8 and 139–163 of each apo-SRP72 protomer could not be modeled, because the electron density was disordered (Supplementary Figure S1A), indicating conformational flexibility. Each protomer had eight long antiparallel α-helices (α1−α8) and a short inserted α′-helix between α4 and α5.
Figure 1.
Crystal structures of human apo-SRP72 and the SRP68/72 complex. (A) Overall structure of apo-SRP72. Six protomers are shown in different colors. (B) Stereoview of the SRP68/72 complex. SRP72 and SRP68 are colored yellow and magenta, respectively. (C) The composite simulated-annealing Fo-Fc ‘omit’ electron density map of SRP68 in the complex, contoured at 3.0 σ. (D) Structural overlay of the four TPRs in the SRP68/72 complex shows an atypical TPR and a same packing angle of the two helices. (E) Structure-based sequence alignment of all four TPRs in human SRP72 reveals inconsistence with canonical TPR motif. The canonical TPR sequence is shown at the bottom, and the conserved residues of SRP72 are shown in boxes.
In the structure of the complex, the final refined model contains one SRP72 molecule (residues 9–162), one SRP68 molecule (residues 588–610), two sulfate ions, two acetate ions, five glycerol molecules, and 226 water molecules in the asymmetric unit (Figure 1B and Table 1). Surprisingly, a smaller than expected fragment of SRP68 was visible in the complex structure (Figure 1B, C and Supplementary Movie S1), presumably resulting from degradation during crystallization (Supplementary Figure S2B). SRP68 adopts an extended conformation containing two short helices. Intriguingly, SRP72 is composed of nine long antiparallel α-helices in the complex. The first eight of these helices adopt four unusually continuous TPR folds, namely TPR1 (α1 and α2), TPR2 (α3 and α4), TPR3 (α5 and α6), and TPR4 (α7 and α8), despite their deviation from the canonical TPR consensus sequence (Figure 1D and E). TPR superhelix usually consists of two layers of A and B α-helices (Figure 1E), giving rise to a structure with concave and convex surfaces. The SRP68 peptide binds tightly to the concave surface (buried surface area = 1165 Å2) of the SRP72 structure in a head-to-tail configuration (Figure 1B). Sequence alignment of SRP72 from different species reveals that residues in the concave surface are highly conserved, as residues of SRP68 that are involved in the interaction with this surface (Supplementary Figure S1). This suggests that the interaction between SRP68 and SRP72 is evolutionarily conserved.
A DALI search (Holm and Rosenström, 2010) of the SRP72 structure identified the TPR-containing protein magnetosome protein MamA (PDB ID 3vty, RMSD = 3.1 Å, Z = 16.4) (Zeytuni and Zarivach, 2012) as the closest structural homolog (Supplementary Figure S3). Surprisingly, further inspection of the SRP72 TPRs reveals that their sequence length varies from 28 to 36 residues (Figure 1D and E), whereas canonical TPR motifs consist of 34 residues (D'Andrea and Regan, 2003). Moreover, very low sequence conservation, if any, is observed between the SRP72 TPRs and the canonical TPR consensus sequence (D'Andrea and Regan, 2003) (Figure 1E). The identification of each TPR motif does not conform to the previous studies (Iakhiaeva et al., 2006, 2009), neither were predicted using the profile-based algorithm TPRpred (Karpenahalli et al., 2007). The four TPR motifs in SRP72 were assigned entirely based on structure similarity and denoted as atypical.
Apo-SRP72 exists mainly as a dimer in solution
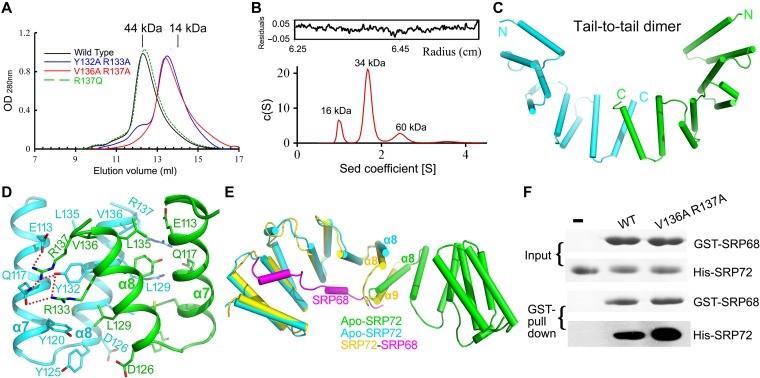
TPR domains mediate protein−protein interactions, molecular recognition, and the assembly of multiprotein complexes, including self-assembly (D'Andrea and Regan, 2003). To investigate the oligomeric state of SRP72, we first analyzed the apo-SRP72 structure. In the asymmetric unit, six apo-SRP72 molecules can be grouped into three identical dimers. To determine whether a dimer is the major species in solution, we performed size exclusion chromatography (SEC) and analytical ultracentrifugation (AUC) experiments. SEC analysis suggested that apo-SRP72 did exist mainly as a dimer in solution (Figure 2A, black). However, a broad peak in the SEC profile suggested that several oligomeric states might exist in solution. Indeed, sedimentation velocity (SV) AUC analysis revealed that while apo-SRP72 exists mainly as a dimer in solution, significant populations of monomer and tetramer are also present in solution (Figure 2B).
Figure 2.
Apo-SRP72 mainly exists as a dimer in solution. (A) SEC of SRP72 on a Superdex 75 10/300 GL column. The SEC profiles of the wild-type, Y132A/R133A, V136A/R137A double-mutants, and R137Q single mutant are colored black, blue, red, and green, respectively. Peak positions for two standard proteins are indicated as black lines on the top. (B) Analytical ultracentrifugation of the wild-type apo-SRP72. The c (S) distribution from SV analysis is shown. (C) Schematic representation of the apo-SRP72 as a tail-to-tail homodimer. The two molecules are colored cyan and green. The N- and C-termini of each monomer are labeled. (D) The interaction details of the tail-to-tail homodimer. The dimerization interface has two symmetric interaction networks, formed between two TPR4. Note that only one interaction interface is shown. (E) Structural overlay of SRP72 in the SRP68/72 complex (yellow) and in the tail-to-tail homodimer of apo-SRP72 (cyan and green). (F) The SRP72 monomeric mutant greatly increases the binding of GST-tagged SRP68 to SRP72.
Interestingly, structural analysis indicates two possible dimer configurations, a tail-to-tail dimer with an interface area of 585 Å2 (Figure 2C and Supplementary Figure S4A) or a head-to-head dimer (Supplementary Figure S4B) with an interface area of 427 Å2. In a tail-to-tail dimer (Figure 2D), two symmetric interaction networks are present in the interface formed by helices α7 and α8 of TPR4 of each monomer, and the two monomers are tied together by several hydrogen bonds, cation-π, electrostatic, and hydrophobic interactions. The interacting residues include Glu113, Gln117, Tyr120, Leu129, Tyr132, Arg133, Val136, and Arg137. In a head-to-head dimer (Supplementary Figure S4B), most interactions occur between helices α1 and α2 of each monomer. The number of interactions is less than that in a tail-to-tail dimer, and the strength is likely to be weaker based on the distance between interacting atoms. This suggests that the tail-to-tail configuration is more probable.
To further confirm the oligomeric state of apo-SRP72 in solution, we performed small-angle X-ray scattering analysis (SAXS), which is a powerful tool for structure validation and quantitative analysis of flexible systems and is highly complementary to high-resolution methods such as X-ray crystallography and NMR. We carried out SAXS with apo-SRP721–163 at various concentrations (Supplementary Table S2). At 2 mg/ml, the tail-to-tail dimer fitted best to the SAXS profile, followed by the head-to-head dimer, while a tetramer or monomer fitted less well (Supplementary Figure S4). Moreover, a minimal ensemble search (MES) (Krissinel and Henrick, 2007) was performed, since this can be very useful for analyzing mixtures in solution. A subset of conformation ensembles containing both monomer and dimer was selected to fit the experimental data. An ensemble containing both types of dimer fitted the data significantly better than a single type of dimer (Supplementary Figure S4F). In the ensemble mixture, the tail-to-tail dimer was the major component. The decisive evidence came from the mutagenesis analysis. Two double mutants in helix α8, Y132A/R133A and V136A/R137A, both eluted as monomers in SEC experiments (Figure 2A), confirming that the dimerization of apo-SRP72 observed in the crystals also occurs in solution. The results of SEC, AUC, SAXS, and mutagenesis analyses collectively suggest that apo-SRP72 exists predominantly as a tail-to-tail dimer in solution.
A minimum fragment of SRP68 is enough for binding to SRP72
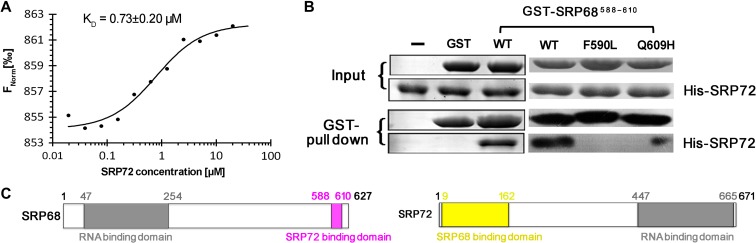
Due to the facile degradation and instability of SRP68 fragments, determination of the minimum fragment of SRP68 required for binding would benefit functional studies and the design of peptidomimetic inhibitors. Previous studies narrowed down the peptide region at the C-terminus of SRP68 that is involved in binding with SRP72 to residues 530–620 or 570–605 (Iakhiaeva et al., 2006, 2009). However, our preliminary experiments indicated that these fragments were either prone to degradation or not soluble, and thus were not suitable for structural studies. We opted for a co-expression strategy to obtain pure, stable SRP72–SRP68 heterodimers.
SEC and corresponding SDS-PAGE analysis indicated that several protein complexes were purified to a high level and stable in vitro. The construct that yielded the complex structure was SRP721–163–SRP68509–614 E608A Q609A K610A. Intriguingly, only residues SRP68588–610 were visible in the electron density of the complex structure (Figure 1B, C and Supplementary Movie S1). SDS-PAGE analysis of crystals of the complex revealed that SRP68 was degraded during crystallization by a trace amount of an unknown E. coli protease that survived purification (Supplementary Figure S2B). In this minimal 23-residue fragment of SRP68, three prolines were observed clearly in the electron density (Figure 1C and Supplementary Figure S1B). This finding is consistent with the observation that proline-rich motifs are often involved in protein−protein interactions (Zarrinpar et al., 2003).
To validate the binding of the 23-residue peptide of SRP68 with SRP72, we also performed microscale thermophoresis (MST) and GST pull-down experiments (Figure 3A and B). Both experiments showed that these two fragments can interact with each other effectively, and MST revealed that SRP68 binds to SRP72 tightly with a KD of 0.73 ± 0.20 μM. Moreover, GST pull-down showed that mutations close to both termini of the SRP68 peptide (F590L or Q609H) disrupted or compromised the interaction, which also confirmed that this 23-residue peptide is necessary for binding (Figure 3B, right panel). Detailed interaction analysis also confirmed that almost all residues in this peptide are involved in its interaction with SRP72 (Supplementary Figure S5). Therefore, we successfully narrowed down the binding regions of the SRP68–SRP72 interaction (Figure 3C) and redefined the minimum fragment of the SRP68-derived peptide using a structural approach.
Figure 3.
The minimum fragment of SRP68 required to bind to SRP72. (A) MST measurements of the binding affinity of SRP68 for SRP72. The resulting binding curve from plotting the FNorm (‰) versus concentration was fit using a hyperbolic function to yield a KD of 0.73 ± 0.20 μM. (B) Residues 588−610 of SRP68 is enough for SRP72 binding. (C) Localization of the interaction regions in SRP68 and SRP72. A schematic diagram shows that both SRP68 and SRP72 consist of a RBD and a SRP72- or SRP68-binding domain.
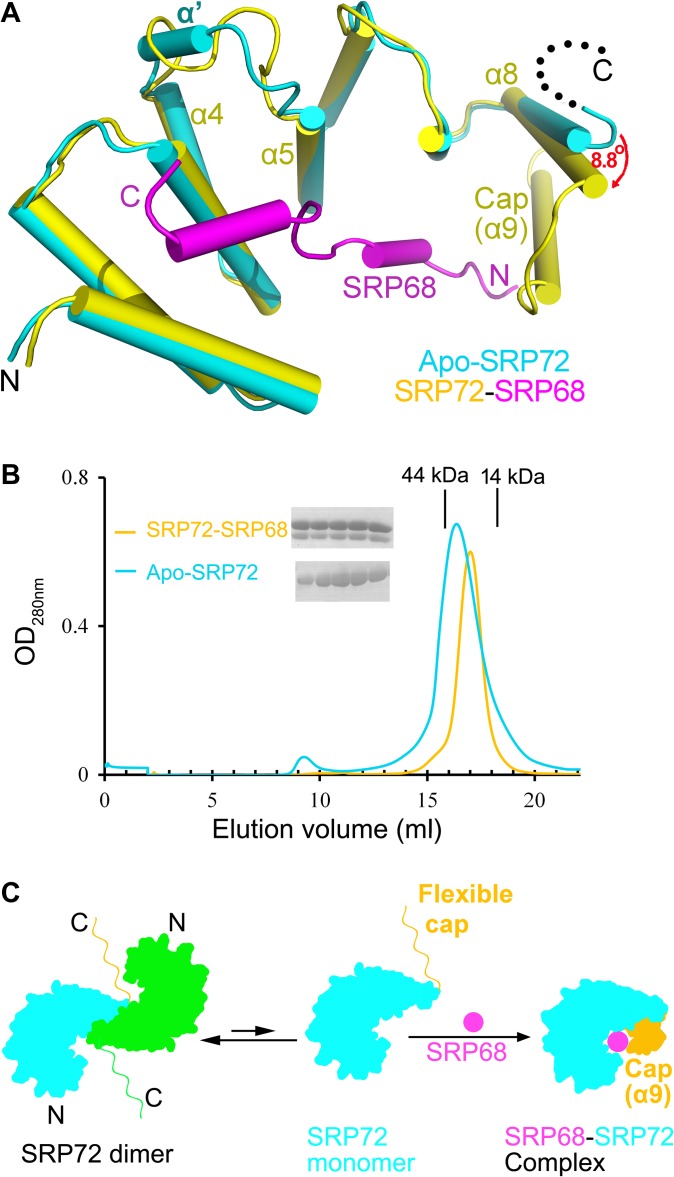
SRP72 undergoes pronounced conformational changes upon binding to SRP68
To uncover the conformational changes undergone by SRP72 upon binding to SRP68, we compared the structures of SRP72 in the apo- and SRP68-bound forms. Structural superposition (Figure 4A) showed that the overall conformation of the four TPRs was similar, with an RMSD of 1.2 Å for all Cα atoms. It was noticeable that residues 72–76 (TKVLA) between α4 and α5 that form a small α′-helix in the apo form adopt a loop conformation in the complex. The most substantial differences occurred at the C-terminus (Figure 4A).
Figure 4.
SRP72 undergoes conformational changes upon binding to SRP68. (A) Structural comparison of apo-SRP72 (cyan) and the SRP72 (yellow)-SRP68 (magenta) complex. The α8 of the TPR4 rotated 8.8° towards (red arrow) to move closer to SRP68 when apo-SRP72 and SRP68-bound SRP72 were superimposed. (B) SEC profile of the SRP72 in the absence (cyan) or presence (yellow) of SRP68 (Superdex 200 10/300 GL column). Elution volumes of the molecular mass standards are marked at the top of the panel. The factions of the peaks were detected. The peak widths are significantly reduced, and the peaks become sharper after SRP68 binding. (C) SRP68–SRP72 binding model. A characteristic change occurred after SRP68 is bound to the concave surface of SRP72. The two molecules of apo-SRP72 are colored cyan and green. SRP68 and the cap are shown in magenta and orange, respectively.
Upon SRP68 binding, helix α8 of TPR4 rotates 8.8° and moves ~2.8 Å closer to SRP68. Moreover, in the absence of SRP68, residues 139–163 (cap) of apo-SRP72 were not traceable in the electron density map, owing to high flexibility rather than degradation, as confirmed by the SDS-PAGE of apo-SRP72 crystals (Supplementary Figure S2A). The conformational flexibility of the C-terminus of apo-SRP72 is consistent with the fact that it was very difficult to obtain high-resolution crystals of apo-SRP72. Intriguingly, these missing residues formed an additional α-helix (α9) upon binding to SRP68. The complex structure clearly reveals that α9 of SRP72 plays a critical role as a ‘cap’ in binding to SRP68 via hydrophobic and hydrophilic interactions. This flexibility reflects the capability of the cap to adopt a variety of conformations, of which we have now seen two extremes in the compact and the fully extended forms. The motion of the cap is functionally significant for target recognition by SRP72. The importance of the cap was confirmed by a mutagenesis experiments showing that its deletion led to a significant loss in binding affinity between SRP72 and SRP68 (Supplementary Figure S6). This is consistent with a previous report (Iakhiaeva et al., 2009). Thus, the cap is essential for SRP68 binding.
Furthermore, a detailed structural comparison of the SRP68/72 complex and apo-SRP72 revealed that the α8 helix from the second molecule in the apo-SRP72 homodimer would sterically clash with the α9 helix and hinder the binding of SRP68 (Figure 2E). SEC results (Figure 4B) showed that the SRP68/72 complex forms a heterodimer (~30 kDa, expected molecular weight = 30.3 kDa), and apo-SRP72 exists mainly as a homodimer (~40 kDa, expected monomer molecular weight = 18.5 kDa) in solution. Therefore, the apo-SRP72 homodimer must disassociate before binding to SRP68. Furthermore, the peak width on the SEC profile was significantly reduced and became sharper following SRP68 binding, suggesting the conformation of the complex was more homogeneous than the apo form. To further explore the correlation between apo-SRP72 disassociation and SRP68 binding, we measured the binding of a monomeric double mutant of SRP72 (V136A/R137A) to SRP68 using GST-pull down assays. Unsurprisingly, the energy barrier to dimerization was broken in the double mutant, which bound to SRP68 much stronger than did the wild-type protein (Figure 2F). Taken together, the results suggest that the fragment comprising residues 9–162 is the minimal SRP72 construct that is necessary and sufficient for tight binding to SRP68, and the conformational changes in the C-terminus of SRP72 are critical (Supplementary Movie S1).
SRP68 binds to the unusually hydrophobic concave surface of SRP72
The determined complex structure revealed that SRP68 binds to the concave TPR surface of SRP72 (Figure 1E). Furthermore, multiple sequence alignment of SRP72 from different species (Supplementary Figure S1) revealed that many residues located in the binding interfaces are conserved. Most of the highly conserved residues of SRP72, mainly distributed in TPR3 and TPR4, were restricted to the inner helix A, which forms the concave surface of the TPR superhelix.
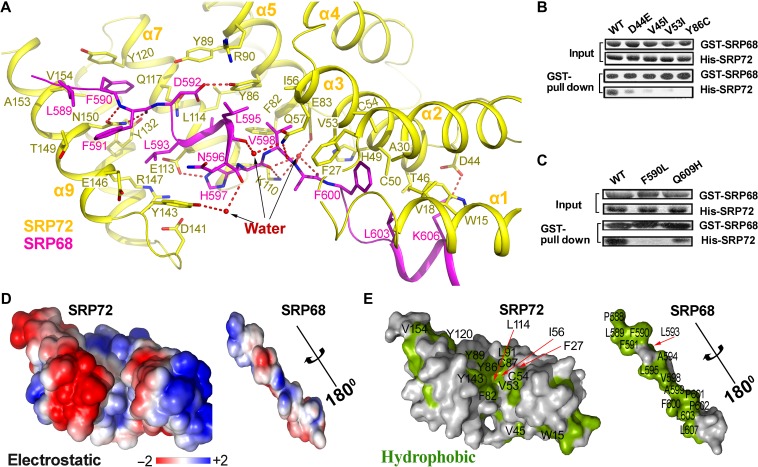
Most TPR proteins studied to date have a moderately positively or highly negatively charged concave surface that is used for binding to protein partners (D'Andrea and Regan, 2003; Zeytuni and Zarivach, 2012). However, for SRP72, surface analyses revealed a large hydrophobic zone lining the groove (Figure 5 and Supplementary Figure S5), which differs significantly from other TPR proteins such as MamA and Hsp70/Hsp90 (Scheufler et al., 2000; Zeytuni et al., 2011). The hydrophobic groove of SRP72 includes Trp15, Val18, Cys50, Val53, Cys54, Ile56, Phe82, Tyr86, Tyr89, Tyr132, and V154 (Figure 5E, left panel). These hydrophobic residues are highly conserved across different species and are identical in many species (Supplementary Figure S1). SRP68 contains complementary aromatic and aliphatic residues on its surface that bind to SRP72, including Leu589, Phe590, Phe591, Leu593, Ala594, Leu595, Val598, Phe600, and Leu603 (Figure 5E, right panel). These residues are highly conserved across different species and cover most of the surface. In addition to interactions involving these hydrophobic residues, several hydrophilic interactions exist, including direct or water-mediated hydrogen bonds and electrostatic interactions involving Asp44, Gln57, Tyr86, Arg90, Lys110, Glu113, Asn150 in SRP72 and Phe591, Asp592, His597, Val598, Lys606 in SRP68 (Figure 5 and Supplementary Figure S5). Overall, the interactions between SRP68 and SRP72 are mainly more hydrophobic, rather than hydrophilic. This is consistent with the observation that the complex remains stable in the presence of 1 M NaCl (Iakhiaeva et al., 2009).
Figure 5.
The extensive interactions between SRP72 and SRP68. (A) Detailed representation of interactions between SRP72 (yellow) and SRP68 (magenta). Interacting residues are shown sticks. Water molecules involved in binding are represented as red spheres. (B and C) As shown by GST pull-down assays, cancer-associated mutations in SRP72 and SRP68 impair the interaction between SRP68 and SRP72. (D) The electrostatic potential (±2kBT) of the binding interface. The surface potential is displayed as a color gradient ranging from red (negative) to blue (positive). Note that SRP68 is rotated 180° around the axis to show the interface. (E) A surface representation of the binding interface, with hydrophobic residues in green. Residues contributing to the hydrophobic groove are labeled.
Cancer-associated mutations disrupt the interaction between SRP72 and SRP68 in vitro
Heterodimerisation of SRP68 and SRP72 plays an essential role in the SRP pathway, and many mutations have been linked to human diseases. For example, mutation of SRP72 causes familial aplasia and myelodysplasia (Kirwan et al., 2012). We searched in the COSMIC cancer database and summarized cancer-associated mutations that occur within the minimal SRP72 and SRP68 constructs in Supplementary Table S1. SRP72 mutations (D44E, V45I, V53I, and Y86C) and SRP68 mutations (F590L and Q609H) have been observed in human cancers, underscoring the importance of these residues in tumorigenesis. Interestingly, in SRP68/72 complex structure, all these residues are located at the SRP68–SRP72 interface (Figure 5 and Supplementary Figure S5). Remarkably, these residues are highly conserved in most species, particularly Tyr86 in SRP72 and Phe590 and Asp592 in SRP68 (Supplementary Figure S1). In the D44E, V45I, V53I (SRP72), and Q609H (SRP68) substitutions, smaller residues were replaced by larger residues. Based on the structure, these larger side chains would be predicted to induce strong steric hindrance and thus disrupt the adjacent hydrophobic interactions and the electrostatic interactions between Asp44 (SRP72) and Lys606 (SRP68). Therefore, these mutations could greatly reduce the binding of SRP68 to SRP72. To test this hypothesis, D44E, V45I (SRP72), and Q609H (SRP68) mutations were generated. As expected, the interactions between mutant forms of SRP68 or SRP72 with their wild-type counterparts were significantly reduced (Figure 5B, C and Supplementary Figure S7). However, for the Y86C (SRP72) and F590L (SRP68) mutations, the scenario was different. Tyr86 of SRP72 lies in the center of the hydrophobic groove mediating the interaction (Figure 5E), and is also involved in an electrostatic interaction with Arg90 in SRP72 and a hydrogen bond with Asp592 in SRP68 (Figure 5A). The Y86C substitution in SRP72 likely abolishes all these interactions. Phe590 in SRP68 forms a π–π interaction with Tyr120 in SRP72 (Figure 5A). Similarly, changing Phe590 in SRP68 to Leu would likely disrupt the interaction. Furthermore, in vitro GST pull-down and MST assays showed that both Y86C and F590L mutations resulted in a complete loss of binding (Figure 5B, C and Supplementary Figure S7). To ascertain whether the mutations disrupt the structure of the mutants, circular dichroism (CD) spectroscopy was used to probe the secondary structure of the mutants. All the mutants had secondary structure nearly identical with that of the wild-type protein (Supplementary Figure S8).
Cancer-associated mutations disrupt the localization of SRP72 and SRP68 in vivo
To explore the effects of the above disease-associated mutations in vivo, we performed cellular co-localization assays in HeLa cells following a published protocol (Kirwan et al., 2012). Both wild-type SRP72 and SRP68 showed uniform coincidence with an ER marker (Figure 6A). However, the SRP72 V53I and Y86C mutants, as well as the SRP68 F590L mutant, showed diminished co-localization with ER and presented a more diffuse distribution pattern in HeLa cells (Figure 6A). This altered cellular distribution of the mutants might be due to the disruption of the SRP72−SRP68 interaction. Consistent with this observation, in HCT116 cells, these mutants also severely compromised or abolished the binding ability compared with wild-type proteins (Figure 6B). Moreover, the results of the in vivo experiments corroborate those of the in vitro biochemical assays (Figure 5B and C). In summary, the mislocalisation of SRP68 and SRP72 mutants might be due to the loss of the SRP68–SRP72 interaction, resulting in the failure to bind to the other SRPs.
Figure 6.
Mutations of key residues in SRP72–SRP68 interface impair the binding and localization. (A) Cellular co-localization between the ER and wild-type (WT) or mutants. Fixed cell images were obtained by using HeLa cells. SRP68 F590L mutation and SRP72 V53I, Y86C mutations showed diminished co-localization with ER compared to wild-type SRP68 and SRP72, respectively. Experiments were performed three times independently with similar results. Scale bar, 10 μm. (B) Binding abilities of the SRP72 and SRP68 mutants. Flag-tagged WT SRP68 or its mutants were from HCT116 cell lysate.
Discussion
SRP68 and SRP72, the two largest subunits of the eukaryotic SRP ribonucleoprotein complex, form a stable heterodimer in solution (Scoulica et al., 1987; Politz et al., 2000). Many studies have highlighted the essential role of SRP68/72 heterodimerisation in SRP assembly and transport (Siegel and Walter, 1985; Grosshans et al., 2001). However, structural details and the molecular mechanism of their interaction remain poorly understood. Herein, we solved the first SRP68/72 complex structure at atomic resolution (1.7 Å). In the final phase of manuscript submission, a similar structure of a SRP68/72 complex was reported (Becker et al., 2017). In our study, we identified the minimal domains of both SRP68 and SRP72 that are necessary and sufficient for their interaction, and found a C-terminal capping helix in SRP72 that is required for tight binding. The key residues involved in binding were confirmed using both in vitro and in vivo experiments. Our results provide structural insight into the regulation of protein translocation by the SRP68/72 complex.
Peptide-based drugs ranging in size from 5 to 50 amino acids are gaining increasing attention from the pharmaceutical industry in recent years due to their exceptional specificity in engaging targets, compared with small-molecule compounds, and their good bioavailability (Craik et al., 2013; Fosgerau and Hoffmann, 2015). Due to insolubility and proneness to degradation, previously reported SRP68 fragments (Iakhiaeva et al., 2006, 2009) are very difficult for use in vitro. Based on our complex structure, we defined a minimum 23-residue peptide of SRP68 that binds to SRP72 tightly, which was confirmed by MST and GST pull-down experiments. This short peptide fragment may be used as a tool to disrupt the binding of SRP68 and SRP72 in vivo and to study the function of the SRP68/72 complex.
Based on structural homology, we found that the N-terminal domain of SRP72 contains four TPR-like motifs. However, each motif has slight variations in sequence length and does not conform to the canonical TPR consensus sequence. Only a Leu residue is completely conserved in helix B of all four TPR motifs, which form the convex surface of the TPR superhelix (Figure 1E). The SRP68 peptide binds tightly to this concave surface of SRP72 (Figure 1B). The concave surface of TPR proteins is usually charged, but that of SRP72 is hydrophobic and therefore atypical. Thus, our findings also reveal the versatility of this basic protein fold.
Most TPR proteins contain an extra helix that is essential for the solubility or stability of these isolated domains (D'Andrea and Regan, 2003). The extra helix of SRP72, however, is not critical for the solubility or stability of the TPR domain, since deletion of the cap does not destabilize the protein. A previous mutagenesis study revealed the importance of the sequence following the four TPRs of SRP72 for target protein recognition and tight binding to SRP68 (Iakhiaeva et al., 2009). Our structural study revealed that this part of the SRP72 structure is disordered and does not form a helix until it binds to SRP68. This region effectively ‘caps’ the SRP68 peptide to enhance the binding affinity. The flexibility and conformational changes of the C-terminal cap of SRP72 might be unique among all TPR structures solved to date. While in other proteins the superhelix undergoes conformational changes upon binding to different protein partners, as observed in the case of the helical HEAT-repeat protein importin β (Cingolani et al., 1999; D'Andrea and Regan, 2003; Fukuhara et al., 2004), in SRP72, it is the C-terminal cap, but not the four TPR motifs, that undergoes a major conformational change upon binding to the partner SRP68. These conformational changes of the cap region are accompanied by the dissociation of the apo-SRP72 homodimer.
The TPR domain is a known protein−protein interaction module. Mutations in TPR-containing proteins may disrupt target protein recognition and have been associated with a variety of human diseases, such as Leber's congenital amaurosis (Sohocki et al., 2000) and chronic granulomatous disease (Grizot et al., 2001). Unsurprisingly, cancer-associated mutations in the SRP68–SRP72 interface disrupted the interaction between SRP72 and SRP68 both in vitro and in vivo (Figures 5B, C and 6). These results highlighted the importance of the residues involved in the interaction between SRP72 and SRP68.
Many studies have highlighted the importance of the interaction between SRP68 and SRP72 in SRP-mediated assembly and transport (Siegel and Walter, 1985; Grosshans et al., 2001). Our structure reveals a total buried surface area between SRP68 and SRP72 of 1165 Å2. The large size of this interface area and the extensive interactions involved (Figure 5) are consistent with the results of a previous study that suggested a strong interaction between SRP68 and SRP72, based on stability in 2 M urea (Iakhiaeva et al., 2009).
Both SRP68 and SRP72 play a key role in SRP-mediated protein targeting. It has been reported previously that SRP72 is involved in SRP receptor binding and bearing of the signal sequence in newly translated proteins into the lumen of the ER (Siegel and Walter, 1988). When SRP72 fails to bind to SRP68, it can result in improper translocation of proteins destined for the cell membrane or extracellular space. All cancer-associated mutants tested in the present study showed diminished co-localization with ER in mammalian cells (Figure 6A). Thus, cancer-associated mutations disrupt the formation of the SRP68/72 heterodimer and affect their localization in vivo. The results underscore the importance of the SRP68–SRP72 interaction in protein transport and processing. Our study suggests that the inability of SRP72 to bind to SRP68 might be functionally important in SRP72- and SRP68-associated human cancers.
Together, our results provide the first structural insight into SRP68/72 heterodimerisation and the pathophysiology of protein translocation-related diseases involving mutations in SRP68/72. Most importantly, identification of the predominantly hydrophobic concave SRP68 binding surface in SRP72 offers an opportunity to develop molecules for target validation in cancer therapeutics. The design and development of chemical compounds or peptides that mimic the 23-residue minimal SRP68 fragment could yield promising drug candidates.
Materials and methods
Cloning, protein expression, and purification
Various fragments of DNA coding for wild-type human SRP72 or SRP68 (a gift of Prof. Jiemin Wong) were PCR-amplified and subcloned into pET28a (for N-terminal His6-tag) or pGEX-4T-2 (for N-terminal GST-tag) vectors via the BamHI/XhoI restriction sites with a tobacco etch virus (TEV) protease cleavage site as previous publication (Deng et al., 2015; Gai et al., 2016; Ni et al., 2016) for recombinant protein expressions. For fluorescence imaging and cell culture experiments, the DNA fragments encoding SRP72 (residues 1−671) and SRP68 (residues 1−627) were cloned into pEGFP-C1 (kindly provided by Prof. Inderjeet Dokal) and pCMV-flag vectors, respectively. All the point mutations of SRP68 and SRP72 used in this study were created using the standard PCR-based mutagenesis method and were verified by DNA sequencing.
For the expression of the SRP68/72 complex, pET28-SRP721–163 and pGEX-SRP68509–614 plasmids were co-transfected into BL21 (DE3) cells (Merck). Cells were grown at 37°C until OD600 reached 0.8−1.0 and induced at 18°C by the addition of 0.5 mM isopropyl β-D-thiogalactoside for another 12 h. Cells were harvested by centrifugation at 4000× g for 10 min, resuspended in buffer A (20 mM Tris pH 8.0, 500 mM NaCl) supplemented with 0.1% Triton X-100 and 1 mM PMSF, and disrupted by sonication. The cells were clarified at 18300× g for 30 min, and the supernatant was filtered with a 0.4 μm filter membrane before being loaded onto a His affinity column (GE Healthcare). The column was washed with three column volumes of buffer A supplemented with 20 mM imidazole and eluted in buffer A supplemented with 300 mM imidazole. TEV protease was added to the eluate at a 1:10 (w/w, protease/protein) ratio for 6 h at 4°C to remove His tag and GST tag. The SRP68/72 elution was further purified by ion exchange (Q sepharose, GE Healthcare) and SEC in buffer A. Peak fractions were collected and analyzed by SDS-PAGE, the target proteins were pooled and concentrated to 10−20 mg/ml for crystallization with buffer A supplemented with 5 mM tris (2-carboxyethyl) phosphine before setting up crystallization. All the protein purification procedures were performed at 4°C.
Crystallization and data collection
Crystallization of apo-SRP72 and the SRP68/72 complex were performed using the hanging drop vapor diffusion method and various commercially crystallization kits in 24-well plates. We generated more than a dozen different surface entropy reduction mutants of the SRP68/72 complex. Only the complex containing the E608A/Q609A/K610A mutation in SRP68, which was predicted in a solvent exposed loop, yielded better crystals. SRP68/72 complex crystals were observed in the well solution containing 1.26 M (NH4)2SO4, 0.1 M Tris-HCl pH 8.0, and 0.2 M Li2SO4 at room temperature. For crystals of apo-SRP72, the diffraction was poor. The diffraction qualities were not improved after extensive optimization by varying concentrations and species of precipitants, buffer, salt, additives, and detergents. After the longstanding trials, final crystals were grew at 4°C with solution containing 0.1 M Tris-HCl pH 8.5, 1.8 M (NH4)2SO4, 0.1 M NaAc, and 0.05 M dimethylethylammonium propane sulfonate. All of the crystals were cryoprotected in mother liquor containing 25% (v/v) glycerol and flash-frozen in liquid nitrogen. To solve the structure, the SRP68/72 complex crystal was soaked in the crystallization well solution with 2 mM Hg(Ac)2 for 30 min. The dataset for the Hg(Ac)2 derivative was collected at 100 K on beamline BL1A at the Photon Factory (KEK) with λ = 1.0085 Å. The data of apo-SRP72 and the SRP68/72 complex were collected on beamlines BL17U1 and BL19U1 at the Shanghai Synchrotron Radiation Facility or 3W1A beamline at the Beijing synchrotron radiation facility (BSRF). All data were processed using HKL2000 (Winn et al., 2011). Data collection and processing statistics are summarized in Table 1.
Cell culture with transfection and fluorescence imaging
HeLa cells were cultured in Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 10% fetal bovine serum (Invitrogen). Transfections of EGFP-SRP72, EGFP-SRP68, or related mutant plasmids were performed with Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions and the previously published method (Kirwan et al., 2012; Zhang et al., 2014). After 24 h, cells were treated (15 min) with 1 μM ER-Tracker Red (Beyotime Institute of Biotechnology, Haimen, China) and fixed with 4% paraformaldehyde for 2 min. The images were acquired with a fluorescence microscope.
Supplementary material
Supplementary material is available at Journal of Molecular Cell Biology online.
Supplementary Material
Acknowledgements
We thank Prof. Inderjeet Dokal (University of London, UK) and Prof. Jiemin Wong (East China Normal University, China) for kindly providing us cDNA, Prof. Jiafu Long (Nankai University, China) for help with analytical ultracentrifugation experiments, and Prof. Hening Lin (Cornell University, USA) for critical reading. We would like to thank the staff at the beamlines BL19U1, BL19U2, and BL17U at Shanghai Synchrotron Radiation Facility, beamline BL1A of Photon Factory at Tsukuba, and beamlines 3W1A and 4B8 at the Beijing Synchrotron Radiation Facility for the excellent technical assistance during data collection.
Funding
This work was supported financially by the National Natural Science Foundation of China (31370720, 31570725, and 91519332), the National Key Research and Development Program of China (2016YFC1200400), and the Extramural Scientists of State Key Laboratory of Agrobiotechnology Grant 2017SKLAB6-3.
Conflict of interest: none declared.
References
- Akopian D., Shen K., Zhang X., et al. (2013). Signal recognition particle: an essential protein-targeting machine. Annu. Rev. Biochem. 82, 693–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen E.S., Rosenblad M.A., Larsen N., et al. (2006). The tmRDB and SRPDB resources. Nucleic Acids Res. 34, D163–D168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker M.M., Lapouge K., Segnitz B., et al. (2017). Structures of human SRP72 complexes provide insights into SRP RNA remodeling and ribosome interaction. Nucleic Acids Res. 45, 470–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buskiewicz I., Deuerling E., Gu S.Q., et al. (2004). Trigger factor binds to ribosome-signal-recognition particle (SRP) complexes and is excluded by binding of the SRP receptor. Proc. Natl Acad. Sci. USA 101, 7902–7906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cingolani G., Petosa C., Weis K., et al. (1999). Structure of importin-β bound to the IBB domain of importin-α. Nature 399, 221–229. [DOI] [PubMed] [Google Scholar]
- Craik D.J., Fairlie D.P., Liras S., et al. (2013). The future of peptide-based drugs. Chem. Biol. Drug Des. 81, 136–147. [DOI] [PubMed] [Google Scholar]
- D'Andrea L.D., and Regan L. (2003). TPR proteins: the versatile helix. Trends Biochem. Sci. 28, 655–662. [DOI] [PubMed] [Google Scholar]
- Deng Z., Wang Q., Liu Z., et al. (2015). Mechanistic insights into metal ion activation and operator recognition by the ferric uptake regulator. Nat. Commun. 6, 7642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derewenda Z.S., and Vekilov P.G. (2006). Entropy and surface engineering in protein crystallization. Acta Crystallogr. D Biol. Crystallogr. 62, 116–124. [DOI] [PubMed] [Google Scholar]
- Doudna J.A., and Batey R.T. (2004). Structural insights into the signal recognition particle. Annu. Rev. Biochem. 73, 539–557. [DOI] [PubMed] [Google Scholar]
- Egea P.F., Stroud R.M., and Walter P. (2005). Targeting proteins to membranes: structure of the signal recognition particle. Curr. Opin. Struct. Biol. 15, 213–220. [DOI] [PubMed] [Google Scholar]
- Fosgerau K., and Hoffmann T. (2015). Peptide therapeutics: current status and future directions. Drug Discov. Today 20, 122–128. [DOI] [PubMed] [Google Scholar]
- Fukuhara N., Fernandez E., Ebert J., et al. (2004). Conformational variability of nucleo-cytoplasmic transport factors. J. Biol. Chem. 279, 2176–21681. [DOI] [PubMed] [Google Scholar]
- Gai Z., Chu W., Deng W., et al. (2016). Structure of the TBC1D7-TSC1 complex reveals that TBC1D7 stabilizes dimerization of the TSC1 C-terminal coiled coil region. J. Mol. Cell Biol. 8, 411–425. [DOI] [PubMed] [Google Scholar]
- Grizot S., Fieschi F., Dagher M.C., et al. (2001). The active N-terminal region of p67phox. Structure at 1.8 A resolution and biochemical characterizations of the A128V mutant implicated in chronic granulomatous disease. J. Biol. Chem. 276, 21627–21631. [DOI] [PubMed] [Google Scholar]
- Grosshans H., Deinert K., Hurt E., et al. (2001). Biogenesis of the signal recognition particle (SRP) involves import of SRP proteins into the nucleolus, assembly with the SRP-RNA, and Xpo1p-mediated export. J. Cell Biol. 153, 745–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grotwinkel J.T., Wild K., Segnitz B., et al. (2014). SRP RNA remodeling by SRP68 explains its role in protein translocation. Science 344, 101–104. [DOI] [PubMed] [Google Scholar]
- Halic M., Becker T., Pool M.R., et al. (2004). Structure of the signal recognition particle interacting with the elongation-arrested ribosome. Nature 427, 808–814. [DOI] [PubMed] [Google Scholar]
- Halic M., Blau M., Becker T., et al. (2006. a). Following the signal sequence from ribosomal tunnel exit to signal recognition particle. Nature 444, 507–511. [DOI] [PubMed] [Google Scholar]
- Halic M., Gartmann M., Schlenker O., et al. (2006. b). Signal recognition particle receptor exposes the ribosomal translocon binding site. Science 312, 745–747. [DOI] [PubMed] [Google Scholar]
- Holm L., and Rosenström P. (2010). Dali server: conservation mapping in 3D. Nucleic Acids Res. 38, W545–W549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iakhiaeva E., Bhuiyan S.H., Yin J., et al. (2006). Protein SRP68 of human signal recognition particle: identification of the RNA and SRP72 binding domains. Protein Sci. 15, 1290–1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iakhiaeva E., Hinck C.S., Hinck A.P., et al. (2009). Characterization of the SRP68/72 interface of human signal recognition particle by systematic site-directed mutagenesis. Protein Sci. 18, 2183–2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jomaa A., Boehringer D., Leibundgut M., et al. (2016). Structures of the E. coli translating ribosome with SRP and its receptor and with the translocon. Nat. Commun. 7, 10471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpenahalli M.R., Lupas A.N., and Söding J. (2007). TPRpred: a tool for prediction of TPR-, PPR- and SEL1-like repeats from protein sequences. BMC Bioinformatics 8, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirwan M., Walne A.J., Plagnol V., et al. (2012). Exome sequencing identifies autosomal-dominant SRP72 mutations associated with familial aplasia and myelodysplasia. Am. J. Hum. Genet. 90, 888–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krissinel E., and Henrick K. (2007). Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797. [DOI] [PubMed] [Google Scholar]
- Ni X., Ru H., Ma F., et al. (2016). New insights into the structural basis of DNA recognition by HINa and HINb domains of IFI16. J. Mol. Cell Biol. 8, 51–61. [DOI] [PubMed] [Google Scholar]
- Politz J.C., Yarovoi S., Kilroy S.M., et al. (2000). Signal recognition particle components in the nucleolus. Proc. Natl Acad. Sci. USA 97, 55–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheufler C., Brinker A., Bourenkov G., et al. (2000). Structure of TPR domain–peptide complexes: critical elements in the assembly of the Hsp70–Hsp90 multichaperone machine. Cell 101, 199–210. [DOI] [PubMed] [Google Scholar]
- Scoulica E., Krause E., Meese K., et al. (1987). Disassembly and domain structure of the proteins in the proteins in the signal-recognition particle. Eur. J. Biochem. 163, 519–528. [DOI] [PubMed] [Google Scholar]
- Siegel V., and Walter P. (1985). Elongation arrest is not a prerequisite for secretory protein translocation across the microsomal membrane. J. Cell Biol. 100, 1913–1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel V., and Walter P. (1988). Each of the activities of signal recognition particle (SRP) is contained within a distinct domain: analysis of biochemical mutants of SRP. Cell 52, 39–49. [DOI] [PubMed] [Google Scholar]
- Sohocki M.M., Bowne S.J., Sullivan L.S., et al. (2000). Mutations in a new photoreceptor-pineal gene on 17p cause Leber congenital amaurosis. Nat. Genet. 24, 79–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Nues R.W., Leung E., McDonald J.C., et al. (2008). Roles for Srp72p in assembly, nuclear export and function of the signal recognition particle. RNA Biol. 5, 73–83. [DOI] [PubMed] [Google Scholar]
- Voorhees R.M., and Hegde R.S. (2015). Structures of the scanning and engaged states of the mammalian SRP-ribosome complex. Elife 4, e07975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weichenrieder O., Wild K., Strub K., et al. (2000). Structure and assembly of the Alu domain of the mammalian signal recognition particle. Nature 408, 167–173. [DOI] [PubMed] [Google Scholar]
- Winn M.D., Ballard C.C., Cowtan K.D., et al. (2011). Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67, 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarrinpar A., Bhattacharyya R.P., and Lim W.A. (2003). The structure and function of proline recognition domains. Sci. STKE 2003, RE8. [DOI] [PubMed] [Google Scholar]
- Zeytuni N., Ozyamak E., Ben-Harush K., et al. (2011). Self-recognition mechanism of MamA, a magnetosome-associated TPR-containing protein, promotes complex assembly. Proc. Natl Acad. Sci. USA 108, E480–E487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeytuni N., and Zarivach R. (2012). Structural and functional discussion of the tetra-trico-peptide repeat, a protein interaction module. Structure 20, 397–405. [DOI] [PubMed] [Google Scholar]
- Zhang H., Chang Y.C., Brennan M.L., et al. (2014). The structure of Rap1 in complex with RIAM reveals specificity determinants and recruitment mechanism. J. Mol. Cell Biol. 6, 128–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwieb C., and Eichler J. (2002). Getting on target: the archaeal signal recognition particle. Archaea 1, 27–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.