Abstract
Most E26 transformation-specific (ETS) transcription factors are involved in the pathogenesis and progression of cancer. This is in part due to the roles of ETS transcription factors in basic biological processes such as growth, proliferation, and differentiation, and also because of their regulatory functions that have physiological relevance in tumorigenesis, immunity, and basal cellular homoeostasis. A member of the E74-like factor (ELF) subfamily of the ETS transcription factor family—myeloid elf-1-like factor (MEF), designated as ELF4—has been shown to be critically involved in immune response and signalling, osteogenesis, adipogenesis, cancer, and stem cell quiescence. ELF4 carries out these functions as a transcriptional activator or through interactions with its partner proteins. Mutations in ELF4 cause aberrant interactions and induce downstream processes that may lead to diseased cells. Knowing how ELF4 impinges on certain cellular processes and how it is regulated in the cells can lead to a better understanding of the physiological and pathological consequences of modulated ELF4 activity.
Keywords: myeloid elf-1-like factor (MEF), ELF4, ETS transcription factors, immune regulation, cancer, transcriptional regulation
Introduction
The E26 transformation-specific (ETS) transcription factors are recognized as having an 85-amino acid DNA-binding domain—the ETS domain—that binds to the DNA sequence 5′-GGA(A/T)-3′, and are evolutionarily conserved throughout the metazoa (Sharrocks, 2001). The ETS family has 28 members found in human and 27 members in mice, with human ETV7/TEL2 having no mouse orthologue (Foos and Hauser, 2004; Hollenhorst et al., 2011). The ETS factors are grouped into 12 subfamilies according to the degree of similarity in their structures. Some subfamilies have only one member such as GABPA, SPDEF, and ETV2, while most have 2−3 members within the subfamily (Gutierrez-Hartmann et al., 2007; Hollenhorst et al., 2011). The ETS factors are mostly involved in crucial biological processes such as development, differentiation, angiogenesis, growth, proliferation, senescence, and cell death (Oikawa and Yamada, 2003). Unsurprisingly, many ETS transcription factors are implicated in the emergence and progression of cancer (Seth and Watson, 2005; Kar and Gutierrez-Hartmann, 2013). Aside from their roles in malignancy, the ETS factors are also involved in cellular homoeostasis and immune regulation (Gallant and Gilkeson, 2006; Garrett-Sinha, 2013; Liu et al., 2016).
The E74-like factor (ELF) subfamily members can regulate immune responses and the development of immune-related cells (Yamada et al., 2010; Choi et al., 2011). The members of this subgroup are ELF1, ELF2/NERF, and ELF4/MEF. ELF3 and ELF5 do not belong to the ELF subfamily, but rather belong to the epithelium-specific ETS (ESE) subfamily along with ESE-3/EHF, and are also known as ESE-1 and ESE-2, respectively. The ESE factors contain a pointed domain (Feldman et al., 2003; Oliver et al., 2012), which is not possessed by members of the ELF subfamily. ELF1, the founding member of the ELF family, is suggested to have important regulatory roles in erythroid maturation and the development of natural killer (NK), NK-T, and T cells (Wurster et al., 1994; Calero-Nieto et al., 2010; Choi et al., 2011). ELF2/NERF (Oettgen et al., 1996) has been implicated in the processes of invasion and metastasis, as well as in blood vessel development (Gaspar et al., 2002; Zhang et al., 2007).
General features of ELF4
Twenty years ago, a new member of the ETS family of transcription factors was discovered. The gene was cloned from a human megakaryocytic cell line. It has some structural similarity with the human ELF-1 gene, and was named ‘myeloid elf-1-like factor’ (MEF) (Miyazaki et al., 1996). There is 94% homology in the ETS region between MEF and ELF-1, making MEF a member of the ELF subfamily of ETS factors, and has been designated as ‘ELF4’. Despite the high homology of the DNA-binding domain between ELF-1 and ELF-4, they do not transactivate the same genes (Miyazaki et al., 1996), and in fact, they have an overall similarity of only 46% (Aryee et al., 1998). ELF4, which is located in chromosome Xq26, was previously reported to be highly expressed in peripheral blood and present in all major tissues except the brain (Miyazaki et al., 1996; Aryee et al., 1998). Recent and more advanced methods of analyses have refined our knowledge on the gene expression pattern of ELF4. The extremely useful information presented in http://biogps.org/gene/2000 confirmed that human ELF4 has a high expression in whole blood. It is also detected in the brain tissues, albeit at comparatively low level. Moreover, human ELF4 is highly expressed in a variety of hematopoietic cells, such as NK cells, myeloid cells, T cells, and monocytes, and is moderately expressed in lung, placenta, thymus, and bone marrow. Mouse Elf4 gene is highly expressed in mast cells, follicular B cells, lymph nodes, bone marrow, and pancreas (http://biogps.org/gene/2000). The high expression level of ELF4 in cells of hematopoietic lineage suggests a role of ELF4 in immunity and in blood cell disorders.
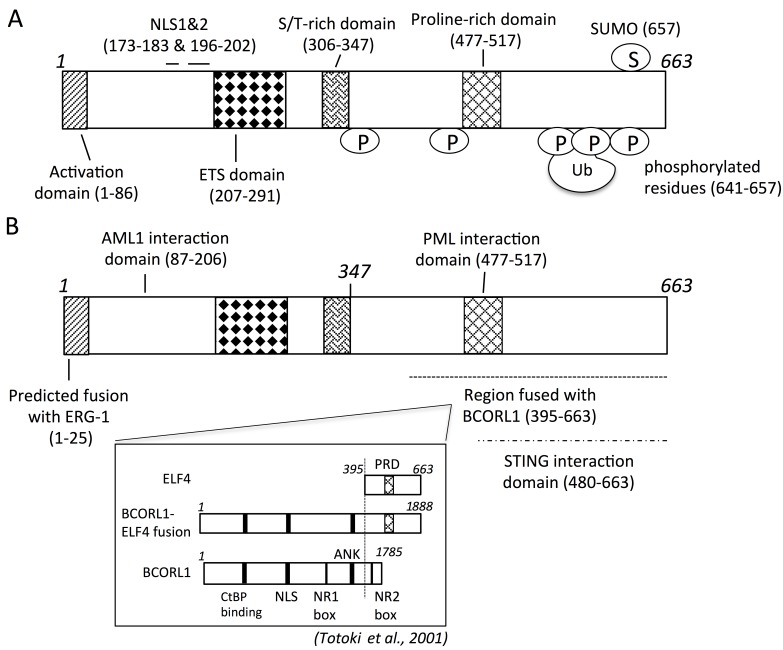
The ELF4 gene encodes a 663-amino acid protein that is composed of an activation domain at the N-terminal region, an ETS domain that binds to DNA consensus sequence, a serine/threonine-rich region, and a proline-rich region (Miyazaki et al., 1996; Suico et al., 2002) (Figure 1A). It contains two nuclear localization signals at 173−183 and 196−202 amino acids, is mostly nuclear localizing, and can undergo ubiquitination and degradation in the nucleus (Suico et al., 2002, 2014; You et al., 2013). ELF4 is phosphorylated at the C-terminal region, mostly at the serine and threonine residues from 641−657 amino acids (Miyazaki et al., 2001; Liu et al., 2006) (Figure 1A). ELF4 can interact with various proteins at either the N-terminal or C-terminal region (Figure 1B) as discussed below. Elf4 knockout (Elf4–/–) mice have no neonatal defects and develop normally, indicating that ELF4 does not play an essential role in embryonic development and that other ETS members may be able to compensate for its absence. However, Elf4–/– mice have deficiencies in NK, NK-T, and T cell functions and impaired expansion of central and effector memory T cells to recall responses (Lacorazza et al., 2002; Mamonkin et al., 2014). This review aims to take stock of what is known about ELF4, especially in terms of how it is regulated and what biological processes and factors it regulates. The genes that are regulated by ELF4 are listed in Table 1, and the transcriptional and post-translational regulations of ELF4 are listed in Table 2. Despite the many facets of ELF4 being uncovered over the years, much more remain unknown, and it is hoped that the past lessons will provide a sound basis for future discoveries.
Figure 1.
Functional, interacting, and modified residues of ELF4 protein. (A) ELF4 has 663 amino acid residues. Numbers are amino acid residues. P, phosphorylation; S, SUMOylation; Ub, ubiquitination; NLS, nuclear localization signal. (B) ELF4 interacts or is fused with various proteins at the indicated amino acid residues. AML1, PML, and STING proteins interact with ELF4. Fusions with ERG-1 and BCORL1 result from chromosomal translocation and inversion, respectively. Inset, fused regions of BCORL1 and ELF4. The C-terminal region of ELF4 (395−663 amino acids) is fused with most of BCORL1 (1−1618 amino acids). PRD, proline-rich domain; ANK, tandem ankryin repeats; NR boxes, nuclear receptor recruitment motifs. The indicated domains are not drawn to scale.
Table 1.
Genes regulated by ELF4.
| Genes | Functions | Effect of ELF4 | References |
|---|---|---|---|
| GM-CSF | Growth factor, immunity | Activation | Miyazaki et al. (1996) |
| IL-3 | Growth factor, immunity | Activation | Miyazaki et al. (1996) |
| LYZ | Antibacterial, immunity | Activation | Kai et al. (1999) |
| DEFB4A | Antimicrobial, immunity | Activation | Lu et al. (2004) |
| IL-8 | Chemokine, immunity | Activation, repression | Seki et al. (2002), Hedvat et al. (2004) |
| PRF1 | Cytolytic, immunity | Activation | Lacorazza et al. (2002) |
| IFNB1 | Cytokine, immunity | Activation | You et al. (2013) |
| IFNA | Cytokine, immunity | Activation | You et al. (2013) |
| Klf2 | Transcription factor, development | Activation | Yamada et al. (2009) |
| Klf4 | Transcription factor, differentiation | Activation | Yamada et al. (2009) |
| Msx2 | Inhibitor of osteoblast differentiation | Activation | Kim et al. (2007) |
| Mab21L1 | Transcriptional repressor | Activation | Kim et al. (2007), Kim et al. (2011) |
| Dlx5 | Positive mediator of Bmp signalling | Repression | Kim et al. (2007) |
| PPAR-γ | Activator of adipocyte differentiation | Activation | Baek et al. (2012), Baek and Baek (2013) |
| MMP-9 | Degrades extracellular matrix | Repression | Seki et al. (2002) |
| MDM2 | Ubiquitin E3 ligase | Activation | Sashida et al. (2009) |
| Sox2 | Transcription factor, stem cell pluripotency | Activation | Bazzoli et al. (2012) |
| DLX3 | Homeobox transcription factor, inhibitor | Activation | Ando et al. (2016) |
| HRK | Pro-apoptotic | Activation | Ando et al. (2016) |
Table 2.
The regulations of ELF4.
| Regulation | Regulatory factors | Effect | References |
|---|---|---|---|
| Transcription | SP1 | Activation | Koga et al. (2005) |
| Transcription | E2F1 | Activation | Taura et al. (2011) |
| Transcription | HIF-1α | Activation | Suico et al. (2016) |
| Transcription/enhancer | PU.1, FLI-1, ERG | Activation | Smith et al. (2012) |
| Transcription | GFI1B | Repression | Smith et al. (2012) |
| Phosphorylation | CYCLIN A/CDK2 | Restriction of ELF4 activity to G1 phase | Miyazaki et al. (2001) |
| Phosphorylation | ATM kinase | Protein degradation | Sashida et al. (2011) |
| Phosphorylation | TBK1 | Activation | You et al. (2013) |
| Phosphorylation | MKK4/JNK1 | Activation | Kim et al. (2011) |
| Ubiquitination | SCF/SKP2 | Protein degradation | Liu et al. (2006) |
| Ubiquitination | MDM2 | Protein degradation | Suico et al. (2014) |
| SUMOylation | SUMO-1/-2 | Repression | Suico et al. (2006) |
| Interactions/fusions | Partners | ||
| Protein interaction | PML | Activation | Suico et al. (2004a, b) |
| Protein interaction | STING | Activation | You et al. (2013) |
| Protein interaction | RUNX2 | Repression, activation | Kim et al. (2007), Lee et al. (2010) |
| Protein interaction | AML1B | Activation | Mao et al. (1999) |
| Protein interaction | AML1/ETO fusion protein | Repression | Mao et al. (1999) |
| Protein interaction | Wild-type and mutant nucleophosmin | Repression and activation, respectively | Ando et al. (2013) |
| Translocation/fusion | ERG | Not determined | Moore et al. (2006) |
| Inversion/fusion | BCORL1 | Repression | Totoki et al. (2011) |
ELF4 in immunity
One of the first elucidated roles of ELF4 was the activation of the immune-related molecules interleukin (IL)-3 and granulocyte macrophage-colony stimulating factor (GM-CSF) in myeloid and T cell lines (Miyazaki et al., 1996). At the time that Miyazaki et al. (1996) reported the cloning of ELF4, our laboratory was also attempting to clone this gene (unpublished data). We found that in epithelial cells, ELF4 strongly activates the lysozyme gene, an antimicrobial protein that is important in host defence at mucosal surfaces and in leukocytes. Among the ETS factors tested such as ELF-1, ESE-1, ETS-1, ETS-2, and PEA-3, only ELF4 was able to upregulate the lysozyme promoter activity (Kai et al., 1999). Another antimicrobial molecule that is regulated by ELF4 is the human beta defensin 2 (HβD2). ELF4 binds to the promoter of HβD2 to activate the gene in human lung adenocarcinoma cell line A549 (Lu et al., 2004). In hematopoietic cells, ELF4, but not other ETS factors, can activate IL-8, a CXC chemokine expressed in macrophage, epithelial, and endothelial cells (Hedvat et al., 2004). IL-8 is an important mediator of immune response by recruiting leukocytes to sites of infection.
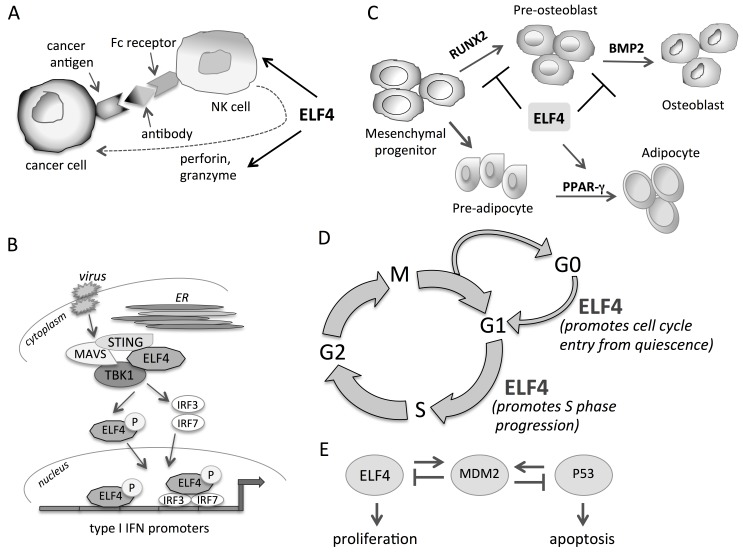
A study on Elf4–/– mice primarily revealed that ELF4 is essential in the development and function of NK and NK-T cells (Lacorazza et al., 2002). Elf4–/– NK cells are incapable of lysing tumour cell targets due to a defective perforin gene expression and minimal secretion of interferon (IFN)-γ. ELF4 binds to the perforin promoter and activates basal perforin transcription (Lacorazza et al., 2002) (Figure 2A). A highly significant study done by Fikrig's group demonstrated that Elf4–/– mice were more susceptible to West Nile virus infection, subsequently revealing a critical function of ELF4 in the induction of type I IFNs Ifn-α and Ifn-β (You et al., 2013). The C-terminal region of ELF4 (480−663 amino acids; Figure 1B) directly interacts with stimulator of interferon genes (STING), an adaptor protein in the type I IFN signalling pathway, and is recruited to the STING−MAVS−TBK1 complex (Figure 2B). This interaction activates ELF4 by phosphorylation at serine 331 residue, induces the nuclear translocation of ELF4, and results in the binding of ELF4 to IFN promoters. ELF4 stimulates IFN promoters in cooperation with the interferon regulatory factors IRF-3 and IRF-7 (Figure 2B). In addition, ELF4 is activated by virus infection (You et al., 2013). This work established ELF4 as a critical factor in type I IFN responses (Szabo and Rajnavolgyi, 2014).
Figure 2.
The roles of ELF4. (A) ELF4 is necessary for the development and function of NK cells. (B) During viral infection, ELF4 interacts with the immune signalling adaptor STING and is recruited to the STING−MAVS−TBK1 complex. TBK1 phosphorylates ELF4, resulting in the translocation of ELF4 to the nucleus where it binds to and activates type I IFN promoters cooperatively with IRF-3 and IRF-7. (C) ELF4 inhibits RUNX2-mediated and BMP-2 signalling-dependent differentiation of osteoblasts. ELF4 enhances PPAR-γ-induced differentiation of adipocytes. (D) ELF4 drives hematopoietic stem cells (HSCs) from quiescent state (G0) to G1 phase and promotes the cell cycle progression from G1 to S phase. (E) ELF4 and P53 transcriptionally activate MDM2, which is the E3 ligase of both ELF4 and P53.
In another study, Lacorazza's group clearly revealed the modulatory role of ELF4 in the proliferation of naïve CD8+ T cells and the homing of T cells by activating the zinc-finger transcription factors Krüppel-like factor (Klf)-4 and Klf2. Elf4–/– mice displayed higher steady-state proliferation rate of naïve CD8+ T cells, deregulated T cell population expansion after immunization, and redistribution of cells to non-lymphoid tissues, suggesting the importance of ELF4 in controlling the homoeostasis, activation, and homing of T cells (Yamada et al., 2009). Consistent with these deficiencies in T cell function in Elf4–/– mice, lower number of effector memory CD8+ T cells was produced in the absence of Elf4 during Listeria monocytogenes infection, indicating the importance of Elf4 in the expansion of both central and effector memory CD8+ T cells (Mamonkin et al., 2014). Importantly, Lee et al. (2014) showed that the absence of ELF4 resulted in more severe disease phenotypes of experimental autoimmune encephalomyelitis in mice. Elf4–/– mice had higher level of IL-17 due to enhanced Th17 differentiation of CD4+ T cells. This suggested that ELF4 is necessary in restricting the Th17 response to infection by limiting CD4+ T cell differentiation to Th17 lineage. Collectively, these studies demonstrated the importance of ELF4 in the regulation of adaptive and innate immunity.
The clinical significance of ELF4 in terms of its role in immunity is highlighted by the reports of Nelson's group showing that mutation in ELF4 may have caused the X-linked hypogammaglobulinemia with growth hormone deficiency (XLH-GHD), a rare immunodeficiency disorder (Stewart et al., 2005). Patients affected with this disease carry a mutation of a base change at 1487 from T to C, resulting in a change at codon 369 from serine to proline (Stewart et al., 2008). Although serine 369 had not been shown as a critical phosphorylation site in other studies (Miyazaki et al., 2001; Liu et al., 2006), the authors hypothesized that a mutation in this site will interfere with ELF4 interaction with other proteins or its ability to bind to DNA, or the mutation may disrupt the proper folding of ELF4 protein (Stewart et al., 2008). XLH-GHD patients present varied symptoms such as chronic infection, tissue inflammation, and arthritis, and share many other clinical features with the more common X-linked agammaglobulinemia (XLA), which is caused by mutations in the Bruton's tyrosine kinase (BTK) gene (Lopez-Herrera et al., 2014). It is interesting to note that a recent report showed that the severity of disease in patients with common variable immunodeficiency (CVID), including hypogammaglobulinemia, inversely correlated with the circulating NK cell count. The lower the number of circulating NK cells, the more severe the disease phenotypes (Ebbo et al., 2016). It is tempting to speculate that in these patients, the ELF4 gene and/or protein is dysfunctional, leading to reduced NK cell function and the consequent immune defects. Further investigations on ELF4, especially the effects of ELF4 mutation on its function in immune response, and additional clinical data linking ELF4 mutation with human diseases will establish ELF4 as an important player in human immune defence.
ELF4 in development and differentiation
The members of the ETS family of transcription factors are well known for their functions in lineage development and cellular differentiation (Maroulakou and Bowe, 2000; Sato, 2001; Meadows et al., 2011), and ELF4 is no exception. As mentioned above, ELF4 is necessary for the development of NK and NK-T cells, and has a growing significance in hematopoiesis (Lacorazza and Nimer, 2003). ELF4 has also been reported to exert a suppressive effect on osteogenic differentiation (Kim et al., 2007). In vitro studies revealed that ELF4 expression is highest in the early stage of differentiation of MC3T3-E1 cells (Kim et al., 2007), a murine osteogenic cell line capable of differentiating into osteoblasts. Elf4 mRNA expression is downregulated by bone morphogenic protein-2 (BMP-2), and ELF4 in turn antagonizes the BMP-2 signalling. The ability of ELF4 to suppress bone formation is via interaction with RUNX2 protein (Kim et al., 2007), a molecule downstream of BMP-2 and critical regulator of osteoblast differentiation (Ryoo et al., 2006). Interaction of ELF4 with RUNX2 prevents RUNX2 from binding to osteoblast-specific element OSE2 present in osteocalcin gene. In addition, ELF4 inhibits the transcription of Dlx5, an effector of BMP-2 for bone formation, and induces the transcription of Msx2, a negative regulator of osteoblast differentiation (Kim et al., 2007). The in vitro data were verified by in vivo studies of transgenic mice overexpressing Elf4 specifically in osteoblasts under the control of the Col1α1 promoter (Col1α1-Elf4 Tg mice). These mice displayed osteopenia in vertebrae, more porous cortical bone, and fewer trabeculae in long bones due to the suppression of osteoblast maturation and increase of osteoclast differentiation induced by ELF4 (Seul et al., 2011). Collectively, these findings demonstrated that ELF4 inhibits bone formation (Figure 2C). Interestingly, the Col1α1-Elf4 Tg mice have high bone marrow adiposity while displaying low bone mass (Baek et al., 2012). Both osteoblasts and adipocytes are derived from mesenchymal stem cells, and by having a shared progenitor there is a degree of plasticity between these cell lineages (Berendsen and Olsen, 2014). Primary bone marrow cells derived from Col1α1-Elf4 Tg mice have higher potential for adipogenic differentiation compared with that from wild-type mice. The overexpression of Elf4 also enhanced the adipogenic differentiation of MC3T3-E1 cells (Baek et al., 2012). ELF4 binds to and transactivates the peroxisome proliferator-activated receptor γ (PPARγ), a crucial factor that drives adipocyte differentiation. Upregulation of PPARγ activity results in bone loss and an increase in bone marrow adiposity (Zhuang et al., 2016). Therefore, ELF4 induces adipogenic differentiation while dampening bone formation by suppressing the factors involved in osteoblast formation and activating the factors necessary for adipocyte differentiation (Baek and Baek, 2013) (Figure 2C). Despite the clear presentation of the role of ELF4 in osteogenic and adipogenic differentiation and the genes it affects, some questions remain, such as how is ELF4 regulated and how deeply it is involved in the cellular fate decision between adipogenesis and osteogenesis. How is ELF4 activated to generate its downstream effects, and at what point in the cascade downstream of mesenchyme does ELF4 begin to get involved? Importantly, does it participate in the pathogenesis and progression of bone diseases such as osteoporosis? Is it involved in metabolic disease? The answers to these questions could prove to be crucial not only to clarify the depth of the involvement of ELF4 in mesenchymal lineage differentiation, but also to understand its role in health and disease.
ELF4 in cell homoeostasis
It has been documented that ELF4 drives the cell cycle progression from G1 to S phase (Miyazaki et al., 2001) (Figure 2D). This is one of the earliest known functions of ELF4, and is in line with the role of ETS transcription family members in regulating cell cycle and cell proliferation (Feldman et al., 2003; Lunardi et al., 2015). The sequential phosphorylation, ubiquitination, and degradation of ELF4 that restrict the activity of ELF4 in the G1 phase lead to the transition from G1 to S phase and promote cell proliferation (Liu et al., 2006). But while the involvement of ELF4 in G1/S phase progression was clearly elucidated, the underlying mechanism of how ELF4 exerts this effect is less clear. Also lacking is the information of the cell cycle-related genes that are targeted by ELF4 to induce this phase transition. Consistent with being able to regulate the cell cycle progression, ELF4 facilitates the entry of quiescent (G0) HSCs into the cell cycle and induces HSC proliferation (Lacorazza et al., 2006) (Figure 2D). Elf4–/– HSCs have increased population residing in the G0 phase compared with wild-type counterparts. The quiescent state of HSCs is necessary to preserve the genomic integrity of the cells over its lifetime and protect the stem cell pool against stress and injury. Cell quiescence is a reversible, ‘poised’ state of the cells that allows for rapid activation when expansion and differentiation are needed (Cheung and Rando, 2013). ELF4 is one of the factors that promote the shift of HSCs from quiescent to an active, cycling state. Moreover, Lacorazza et al. (2006) showed that Elf4–/– HSCs are resistant to the effects of chemotherapy and radiation, suggesting that ELF4 may also regulate the quiescence and chemoresistance in leukaemia stem cells. ELF4 was also shown to control the quiescence of endothelial cells (Sivina et al., 2011). The ability of ELF4 to modulate HSC quiescence is partly due to its suppressive effect on p53, which maintains HSC quiescence, via the transcriptional activation of the p53 E3 ligase MDM2 (Liu et al., 2009; Sashida et al., 2009). That ELF4 influences the activity of the tumour suppressor p53, thereby affecting cell cycle and cellular state and fate, indicates the involvement of ELF4 in maintaining the homoeostasis of cells (Figure 2E). Its absence could result in the imbalance of cell proliferation and quiescence.
ELF4 is implicated in the cellular DNA damage response to γ-irradiation (Sashida et al., 2011). The loss of ELF4 hastens the repair of damaged DNA by abrogating DNA damage response but consequently, this gives rise to radio-resistant cell phenotype. In Elf4–/– cells, the activity of p53 was dampened in response to γ-irradiation, and less apoptotic cells were detected, indicating that ELF4 is necessary for inducing p53 function under γ-irradiation condition (Sashida et al., 2011). How ELF4 regulates p53 activation during γ-irradiation is less clear, but the dual nature of ELF4 in terms of its relation to p53 (inhibitory and activating) is intriguing and needs further investigation to understand the role of ELF4 in normal and stressed cells.
ELF4 in cancer
Aberrant interaction of ELF4 with the oncofusion protein acute myeloid leukaemia (AML1)/ETO is one of the indications of the involvement of ELF4 in cancer (Mao et al., 1999). ELF4 and AML1 interaction occurs at the N-terminal portion of ELF4 (Figure 1B) and the runt homology domain of AML1 proteins. AML1B interaction with ELF4 enhances the transcriptional activity of ELF4. However, the oncoprotein AML1/ETO−ELF4 interaction abolished the transactivating ability of ELF4. This implies that interference of ELF4 activity and function by this aberrant interaction may partly contribute to the dysregulation of myeloid differentiation and subsequent pathogenesis of t(8;21) AML (Mao et al., 1999). A chromosomal translocation in AML involving ELF4 was characterized in a patient wherein ELF4 was fused with another ETS transcription factor, ERG-1 (Moore et al., 2006). This rare chromosomal translocation t(Xq25−26;21q22) occurs within intron 2 of ELF4 and intron 1 of ERG-1, resulting in the fusion of exon 2 of ELF4 and exon 2 of ERG-1. The N-terminal region of ELF4 (amino acid residues 1−25; Figure 1B) was predicted to fuse with ERG-1. Although the function of this fusion protein has not been characterized, it was noted that because the ELF4 promoter drives the ELF4−ERG fusion, the pattern of expression of this fusion protein would be similar to ELF4. However, the oncofusion is composed predominantly of amino acids from ERG-1 C-terminus, pointing to the possibility that the fusion protein would have the function and activity of ERG including its interactions with other proteins (Moore et al., 2006). How exactly ELF4 interferes with the normal roles of ERG-1 in this setting (and vice versa) is still unclear. Sashida et al. (2010) had emphasized that ELF4 and ERG could act as oncogenes. Indeed, the high level of ELF4 in the FAB subtype of AML correlated with poor prognosis in AML patients (Fukushima et al., 2003). It has also been shown that while the wild-type nucleophosmin (NPM1), a nucleolar phosphoprotein, interacts with ELF4 and inhibits its activity, mutant NPM1 does not interact with ELF4, which may result in increased ELF4 transactivation potential (Ando et al., 2013). Importantly, NPM1 is mutated in 50% of AML patients with normal karyotype (Falini et al., 2005).
The association of ELF4 with cancer has been reported not only in hematopoietic transformation, but also in solid tumours. A fusion transcript of BCORL1−ELF4 was found by parallel sequencing of primary hepatitis C virus (HCV)-positive hepatocellular carcinoma derived from a patient (Totoki et al., 2011). BCORL1 is a transcriptional co-repressor that associates with class II histone deacetylase and CtBP1. Interestingly, BCORL1 is recurrently mutated in AML (Tiacci et al., 2012). The fusion protein BCORL1−ELF4 arose from the combination of exons 1−11 of BCORL1 and exon 8 of ELF4 through intra-chromosomal inversion (Xq25 and 11q12). BCORL1−ELF4 has drastically reduced repression activity compared with wild-type BCORL1 protein (Totoki et al., 2011), while ELF4 C-terminus, which is the ELF4 fragment fused to BCORL1 (Figure 1B, inset), has even lower repressor activity. The transcriptional-activation domain and DNA-binding domain of ELF4 reside in the N-terminus (residues 1−86) and at residues 207−291, respectively (Miyazaki et al., 1996; Suico et al., 2002). The C-terminal region of ELF4 is necessary for its interaction with other proteins (Suico et al., 2004b). This fusion may interfere with the repressor activity of BCORL1. But how significantly it contributes to the HCV-positive hepatic carcinogenesis remains to be determined, because there is a total of 22 validated chromosomal rearrangements in the examined tumour tissue (Totoki et al., 2011), and tumour development is a complex process that involves myriad pathways and numerous proteins.
ELF4 is highly expressed in primary human ovarian tumours and ovarian cell lines, although there was no correlation of ELF4 expression with tumour stage, grade, and patient survival (Yao et al., 2007). The knockdown of ELF4 suppressed the malignant potential of ovarian cancer cells, indicating a role of ELF4 in promoting tumorigenic traits (Yao et al., 2007). ELF4 has also been implicated in promoting the stem-like characteristic in gliomas due to the ability of ELF4 to activate Sox2 (Bazzoli et al., 2012), which is one of the transcription factors necessary to maintain stem cell traits. It was shown that ELF4 expression level is significantly higher in human gliomas compared with non-tumour brain tissue, and that Elf4–/– mice have impaired neurosphere formation and slower growth of gliomas (Bazzoli et al., 2012). One proposed mechanism for the oncogenic effect of ELF4 is its potential to activate MDM2 (Sashida et al., 2009), the E3 ligase of the tumour suppressor p53. ELF4 binds to and activates the MDM2 promoter. Furthermore, ELF4 can inhibit the oncogene-induced activation of p16 (Sashida et al., 2009), a modulator of cell cycle. That ELF4 can drive cell cycle progression may also be a factor in its ability to induce aberrant proliferation under certain cellular conditions.
In contrast to several works describing ELF4 as an oncogenic protein, a number of studies showed that ELF4 could act as tumour suppressor in epithelial tissues. Seki et al. (2002) first determined that ELF4 suppressed the growth of lung adenocarcinoma in nude mice injected with lung adenocarcinoma A549 cells. ELF4 expression is downregulated in a panel of cancer cell lines, and the overexpression of ELF4 in A549 cells inhibited the growth and invasiveness of A549 cells. ELF4 exerted its effect most likely via the inhibition of matrix metalloproteinase (MMP)-9 and IL-8. But how ELF4 suppresses MMP-9 and IL-8 expression was not investigated (Seki et al., 2002). Genomic profiling of prostate tumour initiating cells or ‘prostatospheres’ revealed that ELF4 gene was downregulated in these cells (Duhagon et al., 2010). Interestingly, in a recent study, Asai et al. (2016) was able to generate a murine model of multiple myeloma in Elf4-deficient hypomorphic-Rad50 mutant mice (Elf4–/–Rad50s/s). ELF4 deficiency ameliorated the hematopoietic failure in Rad50s/s mice, but surprisingly, 70% of these mice developed plasma cell neoplasms or multiple myeloma. These mice had abnormal proliferation of plasma cells, increased production of monoclonal sera, progressive anaemia, and osteoporosis (Asai et al., 2016). These phenotypes indicate that ELF4 is critically involved in the proper regulation of not only the myeloid cells but also cells of the lymphoid lineage. In a recent study by Ando et al. (2016), ELF4 was demonstrated to be a tumour suppressor in a variety of tumour cell lines. Forced expression of wild-type ELF4 inhibited the proliferation of cancer cells, but mutant ELF4 (L211M), whose DNA-binding potential (and thus, its transactivation capacity) is lost, had no anti-proliferative effect (Ando et al., 2016). The BCORL1−ELF4 fusion protein, which had significant attenuation in its transactivation capacity compared to wild-type ELF4, was also unable to inhibit proliferation. Induction of wild-type ELF4 expression resulted in more tumour cells residing in the G0−G1 phase and less in the S phase, an observation that is not consistent with previously known function of ELF4 in promoting the cell cycle to the S phase (Liu et al., 2006). By genome-wide screening for binding sites of ELF4, it was revealed that ELF4 targets the genes associated with anti-proliferation, such as DLX3 and HRK (Ando et al., 2016). This study clearly delineated the anti-tumour function of ELF4.
Considering these findings together, the question of how we reconcile the contrasting roles of ELF4 arises. Because Elf4–/– mice have defective NK and NK-T cells that are incapable of lysing tumour targets, it is not surprising that ELF4 loss of function may generate an altered immune surveillance, resulting in a cellular environment that predisposes towards aberrant cell growth. Elf4–/– mice do not spontaneously develop tumours (Lacorazza et al., 2002), so that ELF4 may not be considered a bona fide tumour suppressor. But its dysregulated expression and function, by overexpression, silencing, and/or chromosomal translocation, could compromise the balance between cell proliferation, differentiation, quiescence, and death. More than 40 mutations of ELF4 protein, distributed throughout the protein structure, have been found in human cancers (Ando et al., 2016). How these mutations affect the function of ELF4 in relation to cell fate remains to be elucidated. It could be that some of these mutations confer a pro-tumorigenic effect while some anti-tumorigenic, others may silence ELF4 or over-activate it, and the final effect and contribution of ELF4 towards the cell fate will depend on environmental cues and cellular context. By itself, ELF4 may not be strictly a tumour suppressor or an oncogene, rather it is a molecule that is a critical player in cellular homoeostasis, and a disruption of its functions creates a milieu that makes cells more vulnerable to cellular insults.
Transcriptional regulation of ELF4
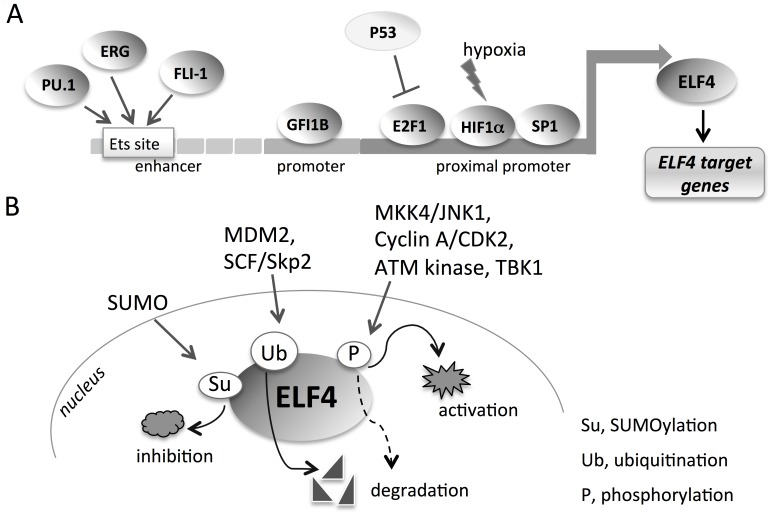
To gain insight into how ELF4 transcription is regulated, we first cloned the 2-kb upstream of the transcription start site. We characterized the ELF4 promoter as being responsive to the transcription factor SP1. SP1 binds to the ELF4 promoter and induces ELF4 activity (Koga et al., 2005). We also found that ELF4 transcription is activated by the transcription factor E2F1 (Taura et al., 2011). The specific binding of E2F1 to ELF4 proximal promoter (~200 bp upstream of start site) was abrogated by the interaction between E2F1 and p53 (Figure 3A). In doxorubicin-induced senescent cells, wherein p53 is activated, p53 directly interacts with E2F1, sequestering E2F1 away from the ELF4 promoter, and preventing E2F1 binding to consensus site. This is one way in which p53 negatively regulates ELF4 (Taura et al., 2011). ELF4 is also transcriptionally upregulated by hypoxia-inducible factor-1α (HIF-1α) (Suico et al., 2016) (Figure 3A). Under hypoxic condition, activated HIF-1α binds to the ELF4 promoter and activates ELF4 transcription, which leads to an increase in ELF4 function (Suico et al., 2016). These investigations of ELF4 transcriptional regulation were carried out mainly in epithelial cells. Whether ELF4 is similarly regulated by these transcription factors in hematopoietic tissues remains to be elucidated.
Figure 3.
The regulations of ELF4. (A) The transcriptional regulation of ELF4. ELF4 is targeted by various transcription factors in its promoter and enhancer regions. (B) The post-translational regulations of ELF4. ELF4 protein is subject to modifications that lead to its activation, dampening of activity (inhibition), or degradation. Phosphorylation by ATM kinase leads to the degradation of ELF4 (broken arrow).
Smith et al. (2012) analysed the transcriptional control of ELF4 in hematopoietic and endothelial cells and identified five distinct regulatory regions, including three promoter regions and two enhancer regions, in the ELF4 locus. These active promoter and enhancer regions, characterized by chromatin modification of trimethylated lysine 4 of histone 3 (H3Me3K4), were mapped to kb −39, −30, and −2 for promoter regions and −16 and −10 for enhancer regions in hematopoietic and endothelial cells (Smith et al., 2012). The enhancer region contains ETS sites wherein the ETS factors PU.1, ERG, and FLI-1 bind and induce ELF4 activity (Figure 3A). Importantly, the study revealed that ELF4 is suppressed by the transcriptional repressor GFI1B, which binds to the ELF4 promoter at −30 kb, and this repression is necessary for the maturation of erythroid cells (Smith et al., 2012). This makes ELF4 a key target of GFI1B, which is an important repressor during erythropoiesis (Garcon et al., 2005), although little is known about other genes that GFIB has to suppress. Interestingly, GFI-1, the close relative of GFI1B, is induced by p53 to promote HSC quiescence, whereas ELF4 can inhibit p53 (Liu et al., 2009). Piecing together these findings generates a network of transcription factors that incorporates ELF4 in the regulation of HSCs. Although ELF4 is important in driving stem cells out of quiescence into expansion, its presence may prevent erythroid differentiation. Thus, tight regulation of ELF4 expression and function through various factors is necessary.
Post-translational regulation of ELF4
ELF4 is a substrate for phosphorylation of the cyclin-dependent kinase (CDK) cyclin A1/Cdk2 and cyclin D/Cdk4 at the serine and threonine residues at 641−657 amino acids. Following this post-translational modification, ELF4 is ubiquitinated by the E3 ubiquitin ligase SCF/SKP2 (Miyazaki et al., 2001; Liu et al., 2006) (Figure 3B). These modifications limit the activity of ELF4 in the G1 phase of cell cycle. Non-phosphorylatable ELF4 mutant cannot be ubiquitinated by SCF/SKP2, underscoring the sequential post-translational modifications as necessary for ELF4 degradation (Liu et al., 2006). In addition to SCF/SKP2, MDM2 is also an E3 ubiquitin ligase that targets ELF4 for degradation regardless of the phosphorylation status of ELF4 (Suico et al., 2014). The overexpression of MDM2 decreased the protein expression level of ELF4, while MDM2 knockdown increased ELF4 expression. Because ELF4 transcriptionally activates MDM2, this provides a critical self-regulatory feedback mechanism of ELF4 regulation (Sashida et al., 2009; Suico et al., 2014) similar to that of p53 and MDM2. The MDM2-induced decrease of ELF4 protein can occur in the absence of p53, indicating a p53-independent mechanism for ELF4 autoregulatory inhibition (Suico et al., 2014) (Figures 2E and 3B). However, the presence of p53 enhanced the suppressive effect of MDM2, suggesting that p53 may also participate in the degradation of ELF4 through its ability to transcriptionally activate MDM2. This triad of p53−MDM2−ELF4 provides an intriguing interplay of reciprocal negative regulation between p53 and ELF4 via MDM2 (Suico et al., 2014) (Figure 2E). In response to DNA damage, ELF4 is phosphorylated by the ATM kinase at T70, S369, and S472 residues, leading to the degradation of ELF4 (Sashida et al., 2011). ELF4 is also phosphorylated by the MKK4/JNK signalling pathway at amino acid residue 641 to enhance the transactivating function of ELF4 on the MAB21L1 promoter in the context of parathyroid hormone-induced bone resorption (Kim et al., 2011).
The conjugation of small ubiquitin-related modifier (SUMO) to ELF4 protein also occurs as another post-translational modification for ELF4 (Figure 3B) (Suico et al., 2006). Specifically, lysine 657 in the C-terminal region of ELF4 is conjugated to SUMO. The consequence of this modification is the suppression of the transactivating function of ELF4 (Suico et al., 2006). Chromatin immunoprecipitation assay revealed that SUMO-conjugated ELF4 had diminished recruitment to the lysozyme promoter, which is one of the ELF4 target genes. In addition to phosphorylation, ubiquitination, and sumoylation, ELF4 may also be acetylated, being lysine-rich with numerous lysine residues that are predicted to be acetylated (unpublished data). The post-translational changes that ELF4 can undergo are important, because these modifications clearly affect the functions and activities of ELF4 (Figure 3B), and thereby affecting the signalling pathways downstream of ELF4. Not only protein modification, but also protein−protein interaction has been reported for ELF4. Promyelocytic leukaemia protein (PML) interacts with ELF4 at the C-terminal region (Figure 1B), and induces the localization of ELF4 to PML nuclear bodies. This leads to the enhancement of ELF4 transcriptional function on the lysozyme and HβD2 promoters (Suico et al., 2004a, b). But whether the ELF4−PML interaction has physiological or pathophysiological relevance is unknown. Further discoveries of ELF4 interaction with other proteins, such as AML1 (Mao et al., 1999) and nucleophosmin (Ando et al., 2013) among others, could provide better understanding of how functions of ELF4 are modulated by proteins that ELF4 comes in contact with.
Concluding remarks
As a transcription factor, ELF4 may be able to activate or repress its target genes depending on certain cellular context and conditions. The majority of the works on ELF4 had demonstrated that ELF4 is a strong transcriptional activator, and only a couple of studies had shown ELF4 as a repressor. While the activating function of ELF4 has been mechanistically explained and proved through deletion studies of its transactivation domain, the mechanism of how ELF4 exerts its repressing capability has not been clearly elucidated. It is possible that ELF4 can recruit certain co-repressors to the promoter region of its target genes. Future studies may clarify this possibility.
As a protein that is part of a broader network, ELF4 acts in conjunction with its partner proteins. An aberration of ELF4 activity due to mutation, cell damaging stimuli, or dysregulated cellular signalling upstream of ELF4 could result in the loss of cellular homoeostasis, which may lead to a diseased cell state. The lessons learned about the roles and regulations of ELF4 may be applied to other ETS transcription factors as well and could open an avenue towards better understanding and strategies of therapies for bone diseases, immunity, metabolism, and cancer.
Funding
This work has been supported by the Grants-in-Aid for Science (Kakenhi #23590082 to M.A.S. and #22390015 to H.K.) provided by the Ministry of Education, Culture, Sports, Science and Technology (MEXT) of Japan.
Conflict of interest: none declared.
References
- Ando K., Tsushima H., Matsuo E., et al. (2013). Mutations in the nucleolar phosphoprotein, nucleophosmin, promote the expression of the oncogenic transcription factor MEF/ELF4 in leukemia cells and potentiates transformation. J. Biol. Chem. 288, 9457–9467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ando M., Kawazu M., Ueno T., et al. (2016). Mutational landscape and antiproliferative functions of ELF transcription factors in human cancer. Cancer Res. 76, 1814–1824. [DOI] [PubMed] [Google Scholar]
- Aryee D.N., Petermann R., Kos K., et al. (1998). Cloning of a novel human ELF-1-related ETS transcription factor, ELFR, its characterization and chromosomal assignment relative to ELF-1. Gene 210, 71–78. [DOI] [PubMed] [Google Scholar]
- Asai T., Hatlen M.A., Lossos C., et al. (2016). Generation of a novel, multi-stage, progressive, and transplantable model of plasma cell neoplasms. Sci. Rep. 6, 22760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek K., and Baek J.H (2013). The transcription factors myeloid elf-1-like factor (MEF) and distal-less homeobox 5 (Dlx5) inversely regulate the differentiation of osteoblasts and adipocytes in bone marrow. Adipocyte 2, 50–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek K., Cho J.Y., Hwang H.R., et al. (2012). Myeloid Elf-1-like factor stimulates adipogenic differentiation through the induction of peroxisome proliferator-activated receptor γ expression in bone marrow. J. Cell. Physiol. 227, 3603–3612. [DOI] [PubMed] [Google Scholar]
- Bazzoli E., Pulvirenti T., Oberstadt M.C., et al. (2012). MEF promotes stemness in the pathogenesis of gliomas. Cell Stem Cell 11, 836–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berendsen A.D., and Olsen B.R (2014). Osteoblast-adipocyte lineage plasticity in tissue development, maintenance and pathology. Cell. Mol. Life Sci. 71, 493–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calero-Nieto F.J., Wood A.D., Wilson N.K., et al. (2010). Transcriptional regulation of Elf-1: locus-wide analysis reveals four distinct promoters, a tissue-specific enhancer, control by PU.1 and the importance of Elf-1 downregulation for erythroid maturation. Nucleic Acids Res. 38, 6363–6374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung T.H., and Rando T.A (2013). Molecular regulation of stem cell quiescence. Nat. Rev. Mol. Cell Biol. 14, 329–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi H.J., Geng Y., Cho H., et al. (2011). Differential requirements for the Ets transcription factor Elf-1 in the development of NKT cells and NK cells. Blood 117, 1880–1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duhagon M.A., Hurt E.M., Sotelo-Silveira J.R., et al. (2010). Genomic profiling of tumor initiating prostatospheres. BMC Genomics 11, 324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebbo M., Gerard L., Carpentier S., et al. (2016). Low circulating natural killer cell counts are associated with severe disease in patients with common variable immunodeficiency. EBioMedicine 6, 222–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falini B., Mecucci C., Tiacci E., et al. (2005). Cytoplasmic nucleophosmin in acute myelogenous leukemia with a normal karyotype. N. Engl. J. Med. 352, 254–266. [DOI] [PubMed] [Google Scholar]
- Feldman R.J., Sementchenko V.I., and Watson D.K (2003). The epithelial-specific Ets factors occupy a unique position in defining epithelial proliferation, differentiation and carcinogenesis. Anticancer Res. 23, 2125–2131. [PubMed] [Google Scholar]
- Foos G., and Hauser C.A (2004). The role of Ets transcription factors in mediating cellular transformation In: Gossen M., Kaufmann J., and Triezenberg S.J. (eds). Transcription Factors. Berlin: Springer, 259–276. [Google Scholar]
- Fukushima T., Miyazaki Y., Tsushima H., et al. (2003). The level of MEF but not ELF-1 correlates with FAB subtype of acute myeloid leukemia and is low in good prognosis cases. Leuk. Res. 27, 387–392. [DOI] [PubMed] [Google Scholar]
- Gallant S., and Gilkeson G (2006). ETS transcription factors and regulation of immunity. Arch. Immunol. Ther. Exp. (Warsz) 54, 149–163. [DOI] [PubMed] [Google Scholar]
- Garcon L., Lacout C., Svinartchouk F., et al. (2005). Gfi-1B plays a critical role in terminal differentiation of normal and transformed erythroid progenitor cells. Blood 105, 1448–1455. [DOI] [PubMed] [Google Scholar]
- Garrett-Sinha L.A. (2013). Review of Ets1 structure, function, and roles in immunity. Cell. Mol. Life Sci. 70, 3375–3390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaspar J., Thai S., Voland C., et al. (2002). Opposing functions of the Ets factors NERF and ELF-1 during chicken blood vessel development. Arterioscler. Thromb. Vasc. Biol. 22, 1106–1112. [DOI] [PubMed] [Google Scholar]
- Gutierrez-Hartmann A., Duval D.L., and Bradford A.P (2007). ETS transcription factors in endocrine systems. Trends Endocrinol. Metab. 18, 150–158. [DOI] [PubMed] [Google Scholar]
- Hedvat C.V., Yao J., Sokolic R.A., et al. (2004). Myeloid ELF1-like factor is a potent activator of interleukin-8 expression in hematopoietic cells. J. Biol. Chem. 279, 6395–6400. [DOI] [PubMed] [Google Scholar]
- Hollenhorst P.C., McIntosh L.P., and Graves B.J. (2011). Genomic and biochemical insights into the specificity of ETS transcription factors. Annu. Rev. Biochem. 80, 437–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kai H., Hisatsune A., Chihara T., et al. (1999). Myeloid ELF-1-like factor up-regulates lysozyme transcription in epithelial cells. J. Biol. Chem. 274, 20098–20102. [DOI] [PubMed] [Google Scholar]
- Kar A., and Gutierrez-Hartmann A (2013). Molecular mechanisms of ETS transcription factor-mediated tumorigenesis. Crit. Rev. Biochem. Mol. Biol. 48, 522–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim B.G., Park Y.J., Libermann T.A., et al. (2011). PTH regulates myleoid ELF-1-like factor (MEF)-induced MAB-21-like-1 (MAB21L1) expression through the JNK1 pathway. J. Cell. Biochem. 112, 2051–2061. [DOI] [PubMed] [Google Scholar]
- Kim Y.J., Kim B.G., Lee S.J., et al. (2007). The suppressive effect of myeloid Elf-1-like factor (MEF) in osteogenic differentiation. J. Cell. Physiol. 211, 253–260. [DOI] [PubMed] [Google Scholar]
- Koga T., Suico M.A., Nakamura H., et al. (2005). Sp1-dependent regulation of Myeloid Elf-1 like factor in human epithelial cells. FEBS Lett. 579, 2811–2816. [DOI] [PubMed] [Google Scholar]
- Lacorazza H.D., Miyazaki Y., Di Cristofano A., et al. (2002). The ETS protein MEF plays a critical role in perforin gene expression and the development of natural killer and NK-T cells. Immunity 17, 437–449. [DOI] [PubMed] [Google Scholar]
- Lacorazza H.D., and Nimer S.D (2003). The emerging role of the myeloid Elf-1 like transcription factor in hematopoiesis. Blood Cells Mol. Dis. 31, 342–350. [DOI] [PubMed] [Google Scholar]
- Lacorazza H.D., Yamada T., Liu Y., et al. (2006). The transcription factor MEF/ELF4 regulates the quiescence of primitive hematopoietic cells. Cancer Cell 9, 175–187. [DOI] [PubMed] [Google Scholar]
- Lee J.M., Libermann T.A., and Cho J.Y (2010). The synergistic regulatory effect of Runx2 and MEF transcription factors on osteoblast differentiation markers. J. Periodontal Implant Sci. 40, 39–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee P.H., Puppi M., Schluns K.S., et al. (2014). The transcription factor E74-like factor 4 suppresses differentiation of proliferating CD4+ T cells to the Th17 lineage. J. Immunol. 192, 178–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M., Gao W., van Velkinburgh J.C., et al. (2016). Role of Ets proteins in development, differentiation, and function of T-cell subsets. Med. Res. Rev. 36, 193–220. [DOI] [PubMed] [Google Scholar]
- Liu Y., Elf S.E., Miyata Y., et al. (2009). p53 regulates hematopoietic stem cell quiescence. Cell Stem Cell 4, 37–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Hedvat C.V., Mao S., et al. (2006). The ETS protein MEF is regulated by phosphorylation-dependent proteolysis via the protein-ubiquitin ligase SCFSkp2. Mol. Cell. Biol. 26, 3114–3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Herrera G., Vargas-Hernandez A., Gonzalez-Serrano M.E., et al. (2014). Bruton's tyrosine kinase--an integral protein of B cell development that also has an essential role in the innate immune system. J. Leukoc. Biol. 95, 243–250. [DOI] [PubMed] [Google Scholar]
- Lu Z., Kim K.A., Suico M.A., et al. (2004). MEF up-regulates human β-defensin 2 expression in epithelial cells. FEBS Lett 561, 117–121. [DOI] [PubMed] [Google Scholar]
- Lunardi A., Varmeh S., Chen M., et al. (2015). Suppression of CHK1 by ETS family members promotes DNA damage response bypass and tumorigenesis. Cancer Discov 5, 550–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mamonkin M., Puppi M., and Lacorazza H.D (2014). Transcription factor ELF4 promotes development and function of memory CD8(+) T cells in Listeria monocytogenes infection. Eur. J. Immunol. 44, 715–727. [DOI] [PubMed] [Google Scholar]
- Mao S., Frank R.C., Zhang J., et al. (1999). Functional and physical interactions between AML1 proteins and an ETS protein, MEF: implications for the pathogenesis of t(8;21)-positive leukemias. Mol. Cell. Biol. 19, 3635–3644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maroulakou I.G., and Bowe D.B. (2000). Expression and function of Ets transcription factors in mammalian development: a regulatory network. Oncogene 19, 6432–6442. [DOI] [PubMed] [Google Scholar]
- Meadows S.M., Myers C.T., and Krieg P.A (2011). Regulation of endothelial cell development by ETS transcription factors. Semin. Cell Dev. Biol. 22, 976–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazaki Y., Boccuni P., Mao S., et al. (2001). Cyclin A-dependent phosphorylation of the ETS-related protein, MEF, restricts its activity to the G1 phase of the cell cycle. J. Biol. Chem. 276, 40528–40536. [DOI] [PubMed] [Google Scholar]
- Miyazaki Y., Sun X., Uchida H., et al. (1996). MEF, a novel transcription factor with an Elf-1 like DNA binding domain but distinct transcriptional activating properties. Oncogene 13, 1721–1729. [PubMed] [Google Scholar]
- Moore S.D., Offor O., Ferry J.A., et al. (2006). ELF4 is fused to ERG in a case of acute myeloid leukemia with a t(X;21)(q25-26;q22). Leuk. Res. 30, 1037–1042. [DOI] [PubMed] [Google Scholar]
- Oettgen P., Akbarali Y., Boltax J., et al. (1996). Characterization of NERF, a novel transcription factor related to the Ets factor ELF-1. Mol. Cell. Biol. 16, 5091–5106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oikawa T., and Yamada T (2003). Molecular biology of the Ets family of transcription factors. Gene 303, 11–34. [DOI] [PubMed] [Google Scholar]
- Oliver J.R., Kushwah R., and Hu J. (2012). Multiple roles of the epithelium-specific ETS transcription factor, ESE-1, in development and disease. Lab. Invest. 92, 320–330. [DOI] [PubMed] [Google Scholar]
- Ryoo H.M., Lee M.H., and Kim Y.J (2006). Critical molecular switches involved in BMP-2-induced osteogenic differentiation of mesenchymal cells. Gene 366, 51–57. [DOI] [PubMed] [Google Scholar]
- Sashida G., Bae N., Di Giandomenico S., et al. (2011). The MEF/Elf4 transcription factor fine tunes the DNA damage response. Cancer Res. 71, 4857–4865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sashida G., Bazzoli E., Menendez S., et al. (2010). The oncogenic role of the ETS transcription factors MEF and ERG. Cell Cycle 9, 3457–3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sashida G., Liu Y., Elf S., et al. (2009). ELF4/MEF activates MDM2 expression and blocks oncogene-induced p16 activation to promote transformation. Mol. Cell. Biol. 29, 3687–3699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato Y. (2001). Role of ETS family transcription factors in vascular development and angiogenesis. Cell Struct. Funct. 26, 19–24. [DOI] [PubMed] [Google Scholar]
- Seki Y., Suico M.A., Uto A., et al. (2002). The ETS transcription factor MEF is a candidate tumor suppressor gene on the X chromosome. Cancer Res. 62, 6579–6586. [PubMed] [Google Scholar]
- Seth A., and Watson D.K. (2005). ETS transcription factors and their emerging roles in human cancer. Eur. J. Cancer 41, 2462–2478. [DOI] [PubMed] [Google Scholar]
- Seul K.J., Cho H.S., Heo S.H., et al. (2011). Osteoblast-specific expression of MEF induces osteopenia through downregulation of osteoblastogenesis and upregulation of osteoclastogenesis. J. Bone Miner. Res. 26, 341–350. [DOI] [PubMed] [Google Scholar]
- Sharrocks A.D. (2001). The ETS-domain transcription factor family. Nat. Rev. Mol. Cell Biol. 2, 827–837. [DOI] [PubMed] [Google Scholar]
- Sivina M., Yamada T., Park C.S., et al. (2011). The transcription factor E74-like factor controls quiescence of endothelial cells and their resistance to myeloablative treatments in bone marrow. Arterioscler. Thromb. Vasc. Biol. 31, 1185–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith A.M., Calero-Nieto F.J., Schutte J., et al. (2012). Integration of Elf-4 into stem/progenitor and erythroid regulatory networks through locus-wide chromatin studies coupled with in vivo functional validation. Mol. Cell. Biol. 32, 763–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart D.M., Tian L., Notarangelo L.D., et al. (2005). Update on X-linked hypogammaglobulinemia with isolated growth hormone deficiency. Curr. Opin. Allergy Clin. Immunol. 5, 510–512. [DOI] [PubMed] [Google Scholar]
- Stewart D.M., Tian L., Notarangelo L.D., et al. (2008). X-linked hypogammaglobulinemia and isolated growth hormone deficiency: an update. Immunol. Res. 40, 262–270. [DOI] [PubMed] [Google Scholar]
- Suico M.A., Fukuda R., Miyakita R., et al. (2014). The transcription factor MEF/Elf4 is dually modulated by p53-MDM2 axis and MEF-MDM2 autoregulatory mechanism. J. Biol. Chem. 289, 26143–26154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suico M.A., Koyanagi T., Ise S., et al. (2002). Functional dissection of the ETS transcription factor MEF. Biochim. Biophys. Acta 1577, 113–120. [DOI] [PubMed] [Google Scholar]
- Suico M.A., Lu Z., Shuto T., et al. (2004. a). The regulation of human β-defensin 2 by the ETS transcription factor MEF (myeloid elf-1-like factor) is enhanced by promyelocytic leukemia protein. J. Pharmacol. Sci. 95, 466–470. [DOI] [PubMed] [Google Scholar]
- Suico M.A., Nakamura H., Lu Z., et al. (2006). SUMO down-regulates the activity of Elf4/myeloid elf-1-like factor. Biochem. Biophys. Res. Commun. 348, 880–888. [DOI] [PubMed] [Google Scholar]
- Suico M.A., Taura M., Kudo E., et al. (2016). The ETS factor myeloid elf-1-like factor (MEF)/Elf4 is transcriptionally and functionally activated by hypoxia. Biol. Pharm. Bull. 39, 641–647. [DOI] [PubMed] [Google Scholar]
- Suico M.A., Yoshida H., Seki Y., et al. (2004. b). Myeloid Elf-1-like factor, an ETS transcription factor, up-regulates lysozyme transcription in epithelial cells through interaction with promyelocytic leukemia protein. J. Biol. Chem. 279, 19091–19098. [DOI] [PubMed] [Google Scholar]
- Szabo A., and Rajnavolgyi E. (2014). Finding a fairy in the forest: ELF4, a novel and critical element of type I interferon responses. Cell. Mol. Immunol. 11, 218–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taura M., Suico M.A., Fukuda R., et al. (2011). MEF/ELF4 transactivation by E2F1 is inhibited by p53. Nucleic Acids Res. 39, 76–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiacci E., Grossmann V., Martelli M.P., et al. (2012). The corepressors BCOR and BCORL1: two novel players in acute myeloid leukemia. Haematologica 97, 3–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Totoki Y., Tatsuno K., Yamamoto S., et al. (2011). High-resolution characterization of a hepatocellular carcinoma genome. Nat. Genet. 43, 464–469. [DOI] [PubMed] [Google Scholar]
- Wurster A.L., Siu G., Leiden J.M., et al. (1994). Elf-1 binds to a critical element in a second CD4 enhancer. Mol. Cell. Biol. 14, 6452–6463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada T., Gierach K., Lee P.H., et al. (2010). Cutting edge: expression of the transcription factor E74-like factor 4 is regulated by the mammalian target of rapamycin pathway in CD8+ T cells. J. Immunol. 185, 3824–3828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada T., Park C.S., Mamonkin M., et al. (2009). Transcription factor ELF4 controls the proliferation and homing of CD8+ T cells via the Kruppel-like factors KLF4 and KLF2. Nat. Immunol. 10, 618–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao J.J., Liu Y., Lacorazza H.D., et al. (2007). Tumor promoting properties of the ETS protein MEF in ovarian cancer. Oncogene 26, 4032–4037. [DOI] [PubMed] [Google Scholar]
- You F., Wang P., Yang L., et al. (2013). ELF4 is critical for induction of type I interferon and the host antiviral response. Nat. Immunol. 14, 1237–1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B., Tomita Y., Qiu Y., et al. (2007). E74-like factor 2 regulates valosin-containing protein expression. Biochem. Biophys. Res. Commun. 356, 536–541. [DOI] [PubMed] [Google Scholar]
- Zhuang H., Zhang X., Zhu C., et al. (2016). Molecular mechanisms of PPAR-γ governing MSC osteogenic and adipogenic differentiation. Curr. Stem Cell Res. Ther. 11, 255–264. [DOI] [PubMed] [Google Scholar]