Abstract
Glioma is a complex disease with limited treatment options. Recent advances have identified isocitrate dehydrogenase (IDH) mutations in up to 80% lower grade gliomas (LGG) and in 76% secondary glioblastomas (GBM). IDH mutations are also seen in 10%–20% of acute myeloid leukemia (AML). In AML, it was determined that mutations of IDH and other genes involving epigenetic regulations are early events, emerging in the pre-leukemic stem cells (pre-LSCs) stage, whereas mutations in genes propagating oncogenic signal are late events in leukemia. IDH mutations are also early events in glioma, occurring before TP53 mutation, 1p/19q deletion, etc. Despite these advances in glioma research, studies into other molecular alterations have lagged considerably. In this study, we analyzed currently available databases. We identified EZH2, KMT2C, and CHD4 as important genes in glioma in addition to the known gene IDH1/2. We also showed that genomic alterations of PIK3CA, CDKN2A, CDK4, FIP1L1, or FUBP1 collaborate with IDH mutations to negatively affect patients’ survival in LGG. In LGG patients with TP53 mutations or IDH1/2 mutations, additional genomic alterations of EZH2, KMC2C, and CHD4 individually or in combination were associated with a markedly decreased disease-free survival than patients without such alterations. Alterations of EZH2, KMT2C, and CHD4 at genetic level or protein level could perturb epigenetic program, leading to malignant transformation in glioma. By reviewing current literature on both AML and glioma and performing bioinformatics analysis on available datasets, we developed a hypothetical model on the tumorigenesis from premalignant stem cells to glioma.
Keywords: glioma, epigenetics, bioinformatics, prognosis, gene mutation
Introduction
Molecular mutations are frequent in both acute myeloid leukemia (AML) and lower grade glioma (LGG). The convenience of separating individual cells of AML combined with marker-based fluorescence-activated cell sorting (FACS) purification of hematopoietic stem cells (HSCs, Lin−CD34+CD38−CD99−TIM3−) and leukemia cells (CD99+TIM3+) from blood samples of individual patients has allowed the establishment of cell clones from single HSCs where tracking of molecular mutations in clonal evolution over time is possible (Corces-Zimmerman et al., 2014). This technological advance has enabled the identification of sequential molecular mutations during clonal evolution and the concept of pre-leukemic stem cell (LSC). It was found that gene mutations occur in a sequential pattern in AML where some mutations occur early while others are late events (Corces-Zimmerman et al., 2014).
Pre-LSCs have the potential to give rise to multiple normal hematopoietic lineages and are yet able to develop into full-blown leukemia (Sato et al., 2016). On the other hand, the concept of LSCs was demonstrated over 20 years ago (Lapidot et al., 1994). LSCs do not have the potential to produce normal hematopoietic lineages, but they develop into leukemia over time (Sato et al., 2016). The transformation from pre-LSC to LSC occurs as oncogenic mutations accumulate in pre-LSCs. Major molecular mutations identified in AML to date include mutations in IDH1, IDH2, ten eleven translocation methylcytosine dioxygenase 2 (TET2), DNA (cytosine-5-)methyltransferase 3 alpha (DNMT3A), structural maintenance of chromosome 1A (SMC1A), Wilms tumor 1 (WT1), additional sex combs like transcriptional regulator 1 (ASXL1), runt-related transcription factor 1 (RUNX1), nucleophosmin 1 (NPM1), FMS-related tyrosine kinase 3 (FLT3), KRAS/NRAS, etc. (Busque et al., 2012; TCGA, 2013a; Zhang et al., 2013; Corces-Zimmerman et al., 2014). Among these, mutations in IDH1, IDH2, WT1, DNMT3A, TET2, and ASXL1 are well characterized as early, pre-leukemic events, whereas mutations in FLT3 and KRAS/NRAS are thought to be late events, occurring in the leukemic stage (Table 1). Mutation in NPM1 is considered to occur more often in the later leukemic stage than in the pre-leukemic stage (Corces-Zimmerman et al., 2014). Mutations in both IDH1/2 and TET2 result in the DNA hypermethylation phenotypes and they are in the same IDH/WT1/TET2 axis in AML pathogenesis, as WT1 is a binding partner of TET2 (Cimmino et al., 2011). Thus, genes mutated in pre-LSCs share common function and perform epigenetic modification by regulating DNA or histone methylations or histone acetylations or the reversal of methylations and acetylations.
Table 1.
Genes with frequent alterations in AML and glioma and their effects on stem cell function.
| Category by functions | Genes involved | Corresponding proteins | Effects on stem cell function | |
|---|---|---|---|---|
| AML | Glioma | |||
| Epigenetic modifiers and stem cell regulators | ASXL1 | Additional sex combs like 1 | HSCs pool maintenance | |
| DNMT3A | DNA methyltransferase 3A | Important in self-renewal | ||
| IDH1/2 | IDH1/2 | Isocitrate dehydrogenase 1 and 2 | Role in differentiation | |
| TET2 | Ten eleven translocation methylCytosine dioxygenase 2 | Role in differentiation | ||
| EZH2 | EZH2 | Enhancer of zeste homolog 2 | Stem cell maintenance | |
| Signal propagators or proliferation activators | FLT3 | FMS-related tyrosine K 3 | Role in stem cell survival | |
| K/N-Ras | Rat Sarcoma Viral Homolog | Role in differentiation, etc. | ||
| c-KIT | KIT proto-oncogene RTK | |||
| CEBPa | CCAAT/EBPα | Maintain quiescent state | ||
| EGFR | EGF receptor | Role in proliferation | ||
| Tumor suppressors | PI3KCA | Phosphatidylinositol 3-k | Role in self-renewal | |
| TP53 | TP53 | Tumor protein p53 | Role in genome stability | |
| WT1 | WT1 | Wilms tumor 1 | Role in cell growth | |
| CDKN2A/B | CDKN2A/B | CDK inhibitor 2A/B | Role in proliferation | |
| PTEN | Phosphatase & tensin homolog | Maintain quiescent state | ||
| Histone chaperones | NPM1 | Nucleophosmin | Role in genome stability | |
CDK, cyclin-dependent kinase; EBP, enhancer-binding protein; EGF, epidermal growth factor.
Wild-type IDH1/2 protein is an enzyme which catalyzes the conversion of isocitrate into alpha-ketoglutarate (α-KG) via decarboxylation in the presence of NADP+. As the result of this reaction, NADPH is produced. Another enzyme named Tet methylcytosine dioxygenase 2 (TET2) is then able to hydroxylate 5-methylcytosine (5 mc), resulting in the production of 5-hydroxymethylcytosine (5 hmc). This process is α-KG dependent, leading to DNA demethylation (Cimmino et al., 2011). Mutant IDH proteins use isocitrate and NADPH to produce R(−)-2-hydroxyglutarate (2-HG) and NADP+. The product 2-HG is a ‘oncometabolite’ (Dang et al., 2009; Xu et al., 2011; Koivunen et al., 2012; Losman et al., 2013; Pusch et al., 2014). The oncometabolite 2-HG can inhibit the active DNA demethylating functions of TET family proteins with the consequences of impaired DNA demethylation (Xu et al., 2011; Liu et al., 2016). It was revealed that there was strong association between IDH mutation and DNA hypermethylation in glioma (Li et al., 2014). IDH mutations occur as a single amino acid missense mutations at R132 of IDH1 or R172 of IDH2. The functional consequences of mutation at R132 of IDH1 are analogous to that of mutation at R172 of IDH2 in glioma, although IDH1 mutation is more common than IDH2 mutation in both AML and glioma (Cohen et al., 2013; Wang et al., 2016). IDH mutation rate is about 80% in LGG and secondary glioblastomas (GBM) and 5%–7% in primary GBM (Balss et al., 2008; Sanson et al., 2009). Genetic analysis of sequential biopsy samples from the same glioma patient indicated that IDH1 mutation was an early event, occurring well ahead of TP53 mutations or 1p/19q deletion (Watanabe et al., 2009). This result is remarkably similar to that of experiments analyzing clonal evolution in the same patient in AML (Corces-Zimmerman et al., 2014). Based on extensive studies in AML and the clonal evolution model where all the defined gene alterations in pre-LSCs change genomic dynamics by epigenetic perturbation, we speculate that gliomas arising from IDH mutations may evolve in a similar fashion.
Another alteration which frequently occurs in pre-LSC is DNMT3A mutation. Mutations in DNMT3A have been found in 22% of all AML (Ley et al., 2010; Yan et al., 2011). Phenotypic changes from DNMT3A mutation include DNA hypomethylation or loss of DNA methylation (Yan et al., 2011; Jeong et al., 2014). The consequences of hypermethylation or hypomethylation in genomic DNA resulted from mutations of IDH1/2, TET2 or DNMT3A are the disturbance of methylation homeostasis in the stem cell compartment, leading to deregulation of stem cell self-renewal and differentiation and resulting in the development of cancer (Gereige & Mikkola., 2009). In addition to DNA methylation, other epigenetic modifications such as histone methylation or acetylation are also important in stem cell function (Zhang and Zhang, 2017). ASXL1 protein is such an epigenetic modifier of gene transcription through histone regulation. Mutation in ASXL1 is also found in the pre-leukemic phase of AML. Normal function of ASXL1 is crucial in maintaining the hematopoietic stem cell pool and loss-of-function mutation in ASXL1 gene impairs hematopoietic stem cell development (Abdel-Wahab et al., 2013).
In contrast to the extensive studies at genetic level in AML, little is known about other genes with alterations in glioma development besides IDH1/2. Since almost all the genomic alterations found in pre-LSC occur in genes coding for factors with a role in epigenetic modulation, we are interested in whether changes of these factors or genes are also present in glioma and whether there are other driver genes whose mutations or alterations have an impact on patients’ survival among the IDH1/2-mutated glioma patients.
Results
Identification of EZH2, KMT2C, and CHD4 as potential epigenetic modulators involved in LGG with an impact on patient survival
In order to investigate whether genes mutated, deleted, or amplified in pre-LSC are also altered in a similar fashion in glioma, we inquired all the genes reported in the literature with a role in AML development and progression plus additional epigenetic modifiers collected from public databases and additional genes with high-mutation rate and high frequency of copy number alterations in glioma. Among the subset of selected epigenetic modifiers, we checked the frequencies of gene mutations and copy number alterations (CNA) of 171 genes whose protein products are considered epigenetic modifiers in all 283 complete LGG tumors in the dataset. Of 171 genes (Supplementary Table S2), 168 are from dbEM (a database of epigenetic modifiers, website: crdd.osdd.net/raghava/dbem/index.php) (Nanda et al., 2016). The other three genes are from previous reports in the literature and include ASH2L, RIOX1, and KMT5A.
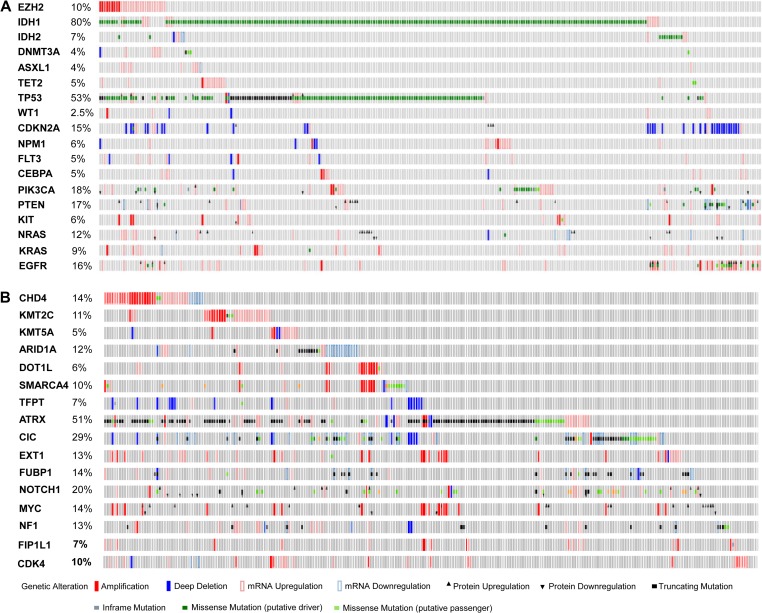
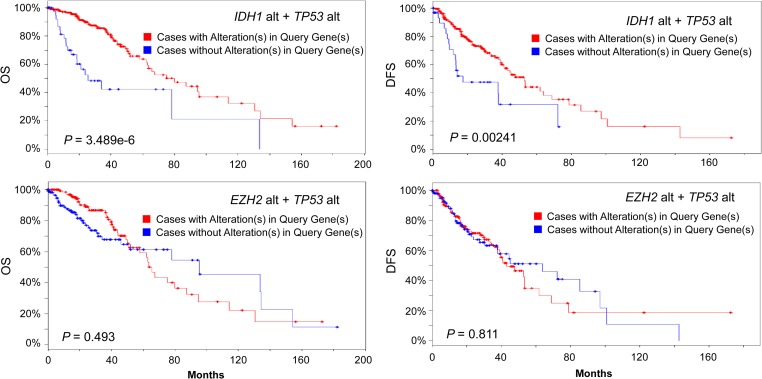
In our analysis of LGG database, following IDH1 (77% mutation rate or 80% alteration rate), TP53 is the second most frequently altered gene with a frequency of 53%, followed by ATRX (51%), CIC (29%), PIK3CA (18%), PTEN (17%), NOTCH1 (17%), EGFR (16%), and CDKN2A (15%), KDM5A (15%), FUBP1 (14%), CHD4 (14%), ARID1A (13%), NF1 (13%), MYC (13%), EXT1 (13%), NRAS (12%), KMT2C (11%), EZH2 (10%), SMARCA4 (10%), HDAC4 (9%), KRAS (9%), TFPT (7%), and DOT1L (6%) (Figure 1A and B). Among these altered genes, nine are epigenetic modifier genes which include ARID1A, CHD4, DOT1L, EZH2, HDAC4, KDM5A, KMT2C, SMARCA4, and TFPT. Frequently mutated genes in pre-LSC of AML such as DNMT3A, TET2, and ASXL1 had a low alteration frequency (<5%) in glioma. After extensive analysis, only three genes CHD4, EZH2, and KMT2C in the epigenetic modifier group, when altered in tumors, showed a significant reduction in disease-free survival in the selected patient group. The other six genes did not have significant effects on either overall survival (OS) or disease-free survival (DFS) in the selected LGG patient group. Presentation of results from the analyses focused on these three genes which included CHD4, EZH2, and KMT2C thereafter. Most of CHD4, EZH2, and KMT2C alterations are gene amplification which is different from many other genes in query such as IDH1/2, DNMT3A, ASXL1, and TET2, where mutations are more frequently seen (Figure 1A and B). Alteration of CHD4, EZH2 and KMT2C gene occurs less frequently in primary GBM (2%, 4%, and 5%, respectively, among 291 all complete GBM tumors; TCGA, 2013b). TP53 mutation (35%) has a lower frequency in GBM while CDKN2A (59%; TCGA, 2013b) has a much higher frequency of alterations (deletions) in GBM (data not shown). We next studied their impact on survival. Consistent with previous reports (Yan et al., 2009), both OS and DFS were significantly better among all 283 complete LGG patients with IDH1/2 mutations alone (data not shown) or IDH1/2 mutations plus TP53 mutations (Figure 2, upper panel). Patients with both EZH2 alterations (gene amplification or mRNA upregulation) and TP53 mutations had no survival advantages compared to unaltered group (P > 0.1) (Figure 2, lower panel). Similarly, patients with both KMT2C alterations and TP53 mutations had no significant difference in survival from unaltered group (Logrank test P-value > 0.1). The same observation was true to patients with both CHD4 alterations and TP53 mutation (data not shown). We then speculated that effects of epigenetic modifier alterations on tumor initiation and patients’ survival could manifest when another important gene was mutated or deleted.
Figure 1.
OncoPrint of genes related to AML and glioma. OncoPrint of 18 genes related to AML and glioma (A) and 16 genes related to glioma (B). Database used: Brain Lower Grade Glioma (TCGA, provisional) 530 samples. All complete tumors (283) were selected for analysis. Alterations include mutations, putative copy number alterations, mRNA expression data, protein expression Z-scores (RPPA).
Figure 2.
OS and DFS in cases with alterations in IDH1 and TP53 (top panel) and in EZH2 and TP53 (lower panel) versus no alteration. Database used: Brain Lower Grade Glioma (TCGA, provisional) 530 samples. Alt, alteration. Blue line: cases without alterations in query genes (n = 45, 35, 125, 112 clockwise). Red line: cases with alterations in query genes (n = 236, 224, 156, 147 clockwise). For EZH2 alt plus TP53 alt, P > 0.05, not significant compared to unaltered group. X-axis: months survival (left panels) or months disease-free (right panels). P-value is from Logrank test.
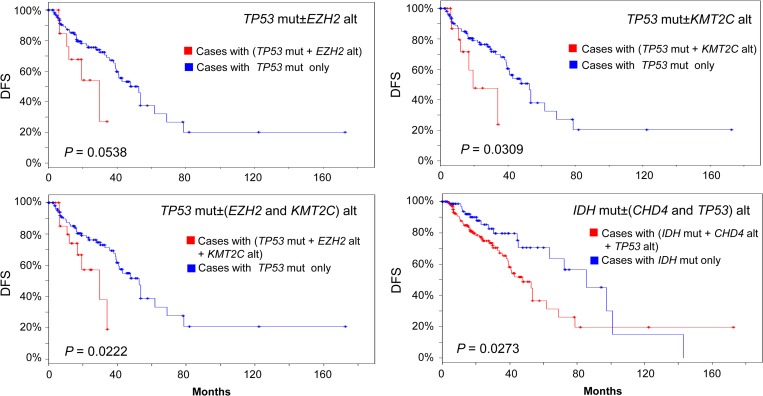
Since over 50% of LGG tumors had TP53 mutations, a subgroup of 146 patients with TP53 mutations were selected from the total of 283 all complete LGG. In these 146 cases, patients with and without alterations of epigenetic modifiers EZH2, KMT2C, or CHD4 were compared for their OS and DFS. We hypothesized that changes in epigenetic modifiers EZH2, KMT2C, and CHD4 act in the premalignant stem cell stage of glioma by epigenetic perturbation to contribute to disease recurrence, and thus decreasing disease-free survival (van Rhenen et al., 2005; Gentles et al., 2010; Wang et al., 2017). We tested this hypothesis by investigating the effect of epigenetic modifier gene alterations in the context of TP53 mutations, an important driver gene for tumor progression. Our results clearly showed that alteration of either EZH2 or KMT2C in the presence of TP53 mutations decreased DFS significantly compared to TP53 mutations only group (Figure 3). The presence of both EZH2 and KMT2C alterations further decreased DFS within the TP53-mutated group. The presence of both IDH mutation and CHD4 alteration in combination with TP53 mutations reduced the DFS within IDH-mutated group compared to IDH1/2 mutation only group (Figure 3), although the presence of CHD4 alteration did not change either OS or DFS among TP53-mutated subgroup (P > 0.5, data not shown). Alteration of EZH2, KMT2C, and CHD4 epigenetic modifiers contributed to disease recurrence, resulting in shortened disease-free survival time without decreasing overall survival significantly. This is by far the strongest evidence implying that epigenetic perturbation by genomic alteration of EZH2, KMT2C, or CHD4 is associated with increased disease recurrence rate. Other genes such as Capicua transcriptional repressor or CIC gene, had 20% mutation rate in LGG (Figure 1B). CIC gene mutations were reported to contribute to oligodendroglioma (Bettegowda et al., 2011). We selected a total of 56 cases with CIC mutations from 283 all complete LGG. Alterations of EZH2 markedly reduced the OS of patients compared to those patients without EZH2 alterations within the selected CIC-mutated group (Logrank test P = 0.00437). In the meantime, alterations of CHD4 significantly decreased the DFS within the same group (Logrank test P = 0.0417). However, accurate interpretation of the results is limited due to a small sample size.
Figure 3.
DFS in the TP53-mutated (n = 146) or IDH-mutated (n = 231) LGG group. Blue line: patients with TP53 mutation (TP53 mut−) or IDH mutation (IDH mut−) only (n = 121, 119, 81, 112 clockwise). Red line: patients with TP53 mut or IDH mut plus other gene alteration (TP53 mut+ or IDH mut+, n = 17, 19, 137, 26 clockwise). Mut, mutated. Alt, alteration. P-value is from Logrank test.
IDH mutations collaborate with PIK3CA, CDKN2A, CDK4, FIP1L1, or FUBP1 gene alterations to negatively affect survival in a subset of LGG patients
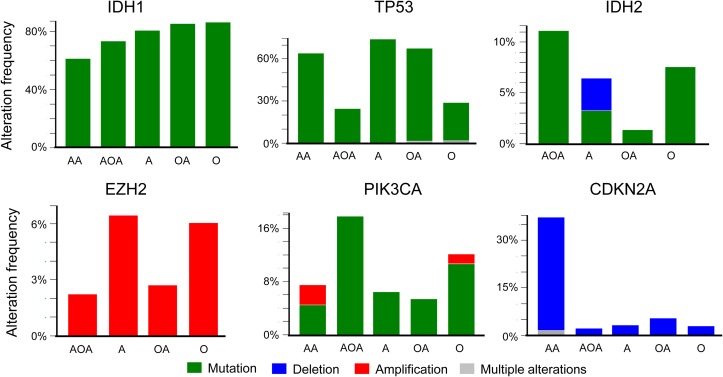
Mutations, deletions, gene amplifications, up- or down-regulations of mRNA and proteins are common events in glioma (Figure 1A and B). Genomic changes in tumor suppressor genes and oncogenes play important roles in tumor initiation, disease progression, and prognosis. IDH1 mutation occurred with a similar high frequency in all LGG subgroups (Figure 4). In contrast, IDH2 mutation occurred more often in anaplastic olioastrocytoma (11%) than in oligodendroglioma (7.4%). Its mutation was present in only 1.2% oligoastrocytoma and 3% astrocytoma. IDH2 gene was deleted in about 3% astrocytoma. No IDH2 mutations were detected in anaplastic astrocytoma (Figure 4). The combined mutation rate of IDH1 and IDH2 is 81% in LGG. Interestingly, mutations of IDH1 and IDH2 are mutually exclusive in LGG, which is consistent with previous report in glioma (Yan et al., 2009) and the findings in AML (Papaemmanuil et al., 2016; Figure 1A), but their mutations share the same consequences that are the generation of ‘oncometabolite’ 2-HG and changed DNA methylation status, as both IDH1 and IDH2 are epigenetic modulators (Dang et al., 2009; Xu et al., 2011; Losman et al., 2013; Feinberg et al., 2016). LGG patients have a better OS and DFS when IDH1/2 mutations are present (Cohen et al., 2013).
Figure 4.
Alterations (mutation in green, deletion in blue, amplification in red, and multi-alterations in gray) of genes in glioma subtypes, from left: anaplastic astrocytoma (AA), anaplastic oligoastrocytoma (AOA), astrocytoma (A), oligoastrocytoma (OA), oligodendroglioma (O). Database used: Brain Lower Grade Glioma (TCGA, provisional) 530 samples.
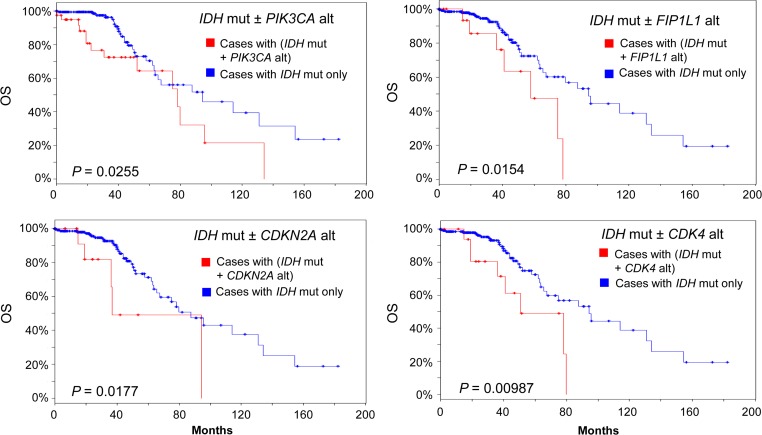
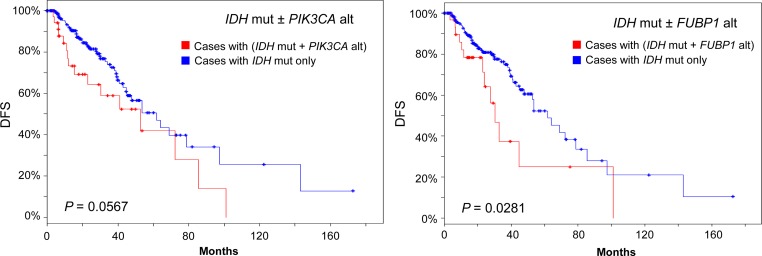
Since the majority of LGG patients have IDH1/2 mutations, we speculated that further stratification of IDH1/2-mutated group may reveal new subsets who may have other driver oncogene mutations or gene amplifications and who may be vulnerable to therapeutic interventions. Thus, we examined the effect of IDH1/2 mutations in combination with other genomic changes on survival in IDH1/2-mutated LGG patient group (231 out of 283 all complete tumors). We tested the effects of 28 other genes whose mutations or copy number alterations (CNA) are relatively common in LGG or GBM or both in the selected 231 patients. These genes include TP53, ATRX, CIC, NOTCH1, PIK3CA, EGFR, FUBP1, NF1, CDKN2A, PTEN, MYC, EXT1, RAD21, PTK2, AGO2, CDK4, PDGFRA, KIT, MLLT3, CHIC2, MDM4, FIP1L1, MDM2, DDIT3, PIK3R1, SPTA1, FLG, and PCLO. We found that genetic alterations of individual gene PIK3CA or CDKN2A, CDK4 or FIP1L1 significantly decreased OS of LGG patients among the selected group (Logrank test P < 0.05; Figure 5). In the meantime, the presence of FUBP1 genetic alterations (mainly mutations for FUBP1 gene) also significantly reduced DFS among the same patient group (Figure 6). Gene alterations of PIK3CA and FIP1L1 had a marginal reduction on DFS (Logrank test P = 0.0567 and P = 0.0723, respectively; Figure 6, data not shown for FIP1L1). The rest of the genes tested did not significantly affect the OS or DFS of the selected patients (data not shown).
Figure 5.
OS within the IDH-mutated LGG group. Sample size n = 231. Blue line: patients with IDH mutation only (IDH mut−, n = 183, 211, 219, 211 clockwise). Red line: patients with IDH mut plus other gene alteration (IDH mut+). Mut, mutated. Alt, alteration. Alterations of PIK3CA (n = 46), FIP1L1 (n = 18), CDKN2A (n = 10), or CDK4 (n = 18) decreased OS in patients with IDH mut. P-value is from Logrank test.
Figure 6.
DFS in the IDH-mutated LGG group. Sample size n = 231. Blue line: patients with IDH mutation only (IDH mut−, n = 177 and 185 from left to right). Red line: patients with IDH mutation plus other gene alteration (IDH mut+). Mut, mutated. Alt, alteration. Alterations of FUBP1 (n = 33) reduced DFS significantly. PIK3CA alt (n = 41) marginally decreased DFS. P-value is from Logrank test.
EZH2 and CHD4 reduce disease-free survival of patients with other collaborating genetic abnormalities in IDH-mutated LGG group
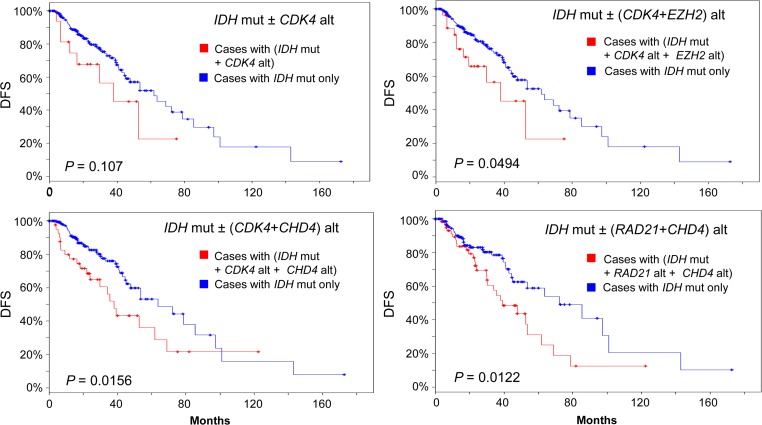
We next examined the effect of altered EZH2 or CHD4 with other collaborating genetic abnormalities on disease-free survival in IDH-mutated LGG patients by analyzing the cases with IDH1/2 mutations (n = 231). Interestingly, alterations of either EZH2 or CHD4 or KMT2C brought down DFS from marginal to a significant level in patients with PIK3CA or FIP1L1 alteration (Logrank test P-value < 0.05). In addition, CDK4 alteration by itself did not change the DFS significantly in IDH-mutated LGG patients compared with those without CDK4 altertion (Figure 7, Logrank test P-value = 0.107), but addition of either EZH2 or CHD4 significantly decreased DFS in CDK4-altered and IDH-mutated patient group, indicating a role of EZH2- or CHD4-mediated epigenetic purterbation in LGG recurrence (Figure 7). Of note, CHD4 or EZH2 alone did not cause significant reduction in DFS without CDK4 alteration in IDH-mutated LGG patients (Logrank test P-value = 0.0859 and 0.0558, respectively). Furthermore, CHD4 alteration collaborated with RAD21 to decrease DFS in IDH-mutated LGG patients compared to those without CHD4 and RAD21 alterations (Figure 7, lower right panel). RAD21 alone was unable to reduce DFS significantly in IDH-mutated LGG patients (Logrank test P-value = 0.203).
Figure 7.
DFS in the IDH-mutated (n = 231) LGG group. Blue line: patients with IDH mutation only (IDH mut−, n = 201, 187, 175, 156 clockwise). Red line: patients with IDH mutation plus alteration of other genes (IDH mut+). CDK4 alt, n = 17; (CDK4+EZH2) alt, n = 31; (CDK4+CHD4) alt, n = 43; (RAD21+CHD4) alt, n = 62. Alt, alteration. Mut, mutated.
Co-occurrence, mutual exclusivity, and their interaction of altered genes in LGG
Next, we performed analyses to explore whether alterations of these epigenetic modifier genes had the tendency to appear concurrently with each other or with other genes in LGG. The analyses covered 18 genes including epigenetic modifiers ARID1A, CHD4, DOT1L, EZH2, HDAC4, KDM5A, KMT2C, SMARCA4, TFPT, and glioma-related genes ATRX, CDKN2A, CIC, EGFR, FUBP1, MYC, NOTCH1, PIK3CA, TP53. Results show that EZH2 and KMT2C have a strong tendency to co-occur in LGG (P < 0.001). EZH2 also tends to co-occur with CDKN2A deletion, TP53 mutation, or EGFR alterations, respectively. KMT2C tends to co-occur significantly with ATRX. CHD4 and KDM5A have a strong tendency to co-occur in LGG (P < 0.001). Alteration of CHD4 also tends to appear with that of TFPT, TP53, ATRX, MYC, respectively (P < 0.05). CHD4 has a significant tendency for mutual exclusivity with EGFR (Supplementary Table S1). In addition, EZH2 amplification tends to occur concurrently with mutated DNMT3A, or ASXL1 (Figure 1A). Besides co-occurrence and mutual exclusivity, alterations of genes could be different across subtypes of LGG. Among 12 genes (IDH1/2, EZH2, TP53, CDKN2A, PIK3CA, KMT2C, CHD4, CDK4, FUBP1, FIP1L1, andCIC) analyzed (Figure 4 and Supplementary Figure S1), no EZH2 mutations were found in LGG of this dataset. However, EZH2 alteration manifested in the form of gene amplification was found in 6.4% astrocytoma and 6% oligodendroglioma patients. Low frequency of EZH2 gene amplification existed in anaplastic oligoastrocytoma (2.2%) and oligoastrocytoma subtypes (2.5%). Interestingly, no EZH2 gene amplification was recorded in anaplastic astrocytoma (Figure 4). TP53 mutations occurred across all subtypes of LGG with some variations in frequency. The majority (99%) of CDKN2A alterations were in the form of deletion. All subtypes of glioma had PIK3CA gene mutations with a varied frequency. Anaplastic astrocytoma and oligodendroglioma also had PIK3CA amplification at 2% or 1%, respectively (Figure 4). KMT2C amplification was seen across five subtypes. Low frequency of KMT2C mutations (1.5%) was also seen in anaplastic astrocytoma, oligoastrocytoma, and oligodendroglioma (Supplementary Figure S1). CHD4 gene amplification was seen across all five subtypes, but was more frequent in anaplastic astrocytoma (10%), astrocytoma (13%), and oligoastrocytoma (9%). A few mutations of CHD4 were seen in anaplastic astrocytoma and oligodendroglioma (Supplementary Figure S1).
CIC was mutated with a high frequency in anaplastic oligoastrocytoma (44%) and oligodendroglioma (38%) subtypes. Low frequency of gene deletion of CIC (<5%) was seen across all subtypes. FUBP1 was also mutated in anaplastic oligoastrocytoma (24%), oligodendroglioma (13%), and oligoastrocytoma (4%). Only 1.6% anaplastic astrocytoma subtype had FUBP1 mutation and no mutation or other alterations of this gene were seen in astrocytoma subtype. Common alteration of FIP1L1 gene was gene amplification seen mainly in two subtypes anaplastic astrocytoma (7.4%) and anaplastic oligoastrocytoma (9%). Low frequency mutation of FIP1L1 was seen in oligodendroglioma (1.4%). No FIP1L1 gene alterations were seen in either astrocytoma or oligoastrocytoma subtypes. CDK4 gene was mostly amplified in anaplastic astrocytoma (12%), with lower gene amplification in oligoastrocytoma (1.2%). No gene amplification or other changes of CDK4 were detected in anaplastic oligoastrocytoma (Supplementary Figure S1). Although the mutation spectrum and frequency differ across subtypes of LGG, the significance of this difference remains a topic for further investigations.
EZH2, KMT2C, CHD4, and TP53 and their networks in LGG
EZH2 is connected to epigenetic regulators such as HDACs, DNMT1 in the EZH2–TP53 network. KMT2C is connected with NCOA2 (nuclear receptor coactivator 2) and others. CHD4 is connected to other epigenetic modifier TAF1A, HDAC, and others such as TATA-box binding protein (TBP), a critical molecule with an important role in gene transcription. Changes in EZH2, KMT2C, or CHD4 genomic status by gene amplification mutation, or expression could have profound effects on its neighbors and connection nodes in the network (Supplementary Figure S2). Changes in the network status could dysregulate gene transcription, leading to oncogenic transformation and activation. These networks show the complexity of changes which may happen to other connection nodes or neighbors when EZH2, KMT2C, CHD4 alterations are combined with TP53 mutation or CDKN2A deletion or others.
Discussion
Gliomas can be categorized into four grades (Grades I–IV) based on World Health Organization (WHO) criteria (Louis et al., 2007). Grade I is generally regarded as benign and curable by surgery. Grades II and III are considered as malignant and invasive. By tradition, Grades II and III are often described as LGG. Grade IV, also called glioblastoma multiforme (GBM), is the most invasive and advanced form. GBM is further divided into primary and secondary GBMs based on clinical presentation at diagnosis. Primary GBMs are often de novo advanced gliomas, whereas secondary GBMs often develop from LGG.
In this study, we showed that LGG patients with somatic IDH1/2 mutations had a worse overall survival when alteration of another gene such as PIK3CA, CDKN2A, CDK4, or FIP1L1 was present. Mutation of PIK3CA, or deletion of CDKN2A, or gene amplification of CDK4 will likely increase the kinase activities of cancer cells, contributing to LGG pathogenesis (Peng et al., 2014; Zhang and Yang, 2014). LGG patients with IDH1/2 mutations and alteration of FUBP1 (mainly mutation for FUBP1) had a significant reduction in disease-free survival as well. Alteration of PIK3CA (mainly mutation) or FIP1L1 (mainly gene amplification) had a marginal reduction in DFS within IDH1/2-mutated patient group. Our findings bring out a very important point. Namely, targeted therapy should be considered in IDH1/2-mutated LGG patients when PIK3CA mutation, CDKN2A deletion, CDK4 gene amplification, FIP1L1 gene amplification, or FUBP1 mutation is detected. For example, PIK3CA inhibitors can be used for patients with PIK3CA mutations in LGG patients who also have somatic IDH1/2 mutations. By the same token, CDK inhibitors can be used for patients with either CDK4 gene amplification or CDKN2A gene deletion. To the clinician’s advantage, CDK4/6 inhibitors palbociclib (PD0332991) and ribociclib (LEE011) have been approved for the treatment of breast cancer (Zhang and Yang, 2014; Xu et al., 2017). Excitingly, PI3K inhibitors Alpelisib (BYL719) and Sonolisib (PX-866) have been developed and are being tested in clinical trials for the treatment of different types of cancers (Pons-Tostivint et al., 2017). Our results indicate that there is an urgent need to test these pathway inhibitors (PI3K inhibitors, CDK4/6 inhibitors, or other inhibitors) in the treatment of LGG when patients are positive for IDH1/2 mutations and have PIK3CA mutation, CDK4 gene amplification, CDKN2A gene deletion, or other gene alterations.
Many studies have been done to link IDH mutations with known oncogenes such as HIF-1α (Zhao et al., 2009). However, subsequent studies demonstrated that IDH mutations may not be sufficient to upregulate HIF 1a (Williams et al., 2011; Koivunen et al., 2012). In our analysis, HIF1A alteration (mainly mRNA upregulation without mutations) was detected in only 4% of cases with IDH1/2 mutations. This upregulation of HIF-1α mRNA did not change either OS or DFS in IDH-mutated patient group (Logrank test P-value = 0.507 for OS and 0.739 for DFS). The drawback from our analysis is that data of HIF1-α protein level in these tumors were not available in the database.
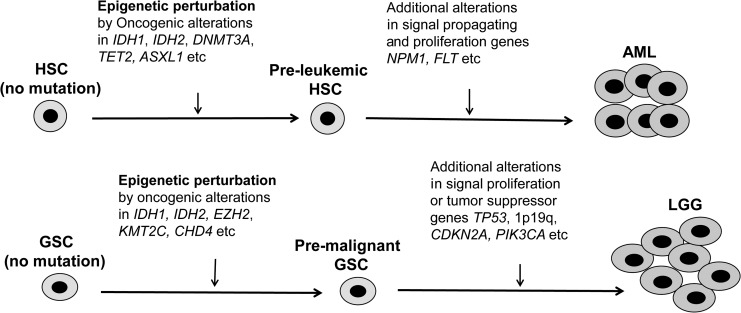
We have also demonstrated that EZH2 is altered in around 10% LGG patients in the form of genomic gene amplification or mRNA upregulation. This alteration frequency is markedly higher than that in GBM (2.2%–4%, data not shown). Alteration of EZH2 can occur concurrently with DNMT3A or ASXL1 mutations, the two mutations frequently seen in pre-LSC. This co-occurrence implies that they have a coordinated role in epigenetic modulation or perturbation and change in glioma stem cell function. We also showed that CHD4 and KMT2C were altered with similar frequency in glioma as EZH2. Similar to DNMT3A and ASXL1 in pre-LSC, alterations of EZH2, KMT2C, and CHD4 are likely present early in pre-malignant glial stem cells (GSC). These pre-malignant GSCs accumulate other genetic mutations such as TP53 mutation, CDKN2A deletion, and mutations in EGFR, PIK3CA, KRAS, or FLT later in clonal evolution. Indeed, alterations of EZH2, CHD4, and KMT2C decreased DFS in either TP53-mutated LGG or IDH1/2-mutated LGG together with other genetic events, strongly implying that they acted in the glioma stem cell stage. Our hypothetical model is illustrated in Figure 8 where similar genetic events starting from stem cells lead to a common disease cancer in different organs. When it happened in HSCs, AML would form. If it occurred in GSCs, then LGG would be the outcome.
Figure 8.
Hypothetical model of malignant transformation of glial stem cells and the development of glioma. HSC, hematopoietic stem cell; AML, acute myeloid leukemia; GSC, glial stem cell; LGG, lower grade glioma.
EZH2 mRNA upregulation was detected in some AML cases, but its gene amplification has never been documented in AML. EZH2 is not a major gene in pre-LSC based on current research in AML. However, both gene amplification and mRNA upregulation of EZH2 were detected in LGG, indicating that this gene likely plays a distinctive role in glioma. The alteration in EZH2 may lead to epigenetic perturbation of the genome in the stem cell compartment and prime pre-malignant GSCs to malignant transformation along the way when additional genetic changes such as TP53 mutation, CDKN2A deletion, or 1p/19q deletion are acquired. Indeed, it was reported that EZH2 is crucial for glioma stem cell maintenance (Suva et al., 2009). Currently, EZH2 as therapeutic target in cancer is under investigation (Kim and Roberts, 2016). Genomic alterations of KMT2C and EZH2 in LGG have a strong tendency to emerge at the same time (Supplementary Table S1). KMT2C (also called MLL3) belongs to the mixed lineage leukemia (MLL) family of histone lysine methyltransferases. The enzyme methylates histone at lysine 4 (Lys-4) position which serves as a tag for epigenetic activation of transcription (Fujimoto et al., 2012). Different somatic mutations of KMT2C may have different or even opposing effects in cancer (Weirich et al., 2015). The biological effects of KMT2C gene amplification in glioma remain an interesting topic to be elucidated.
CHD4 (chromodomain-helicase-DNA-binding protein 4) is part of the histone deacetylase NuRD complex that remodels chromatin through histone deacetylation. CHD4 can recruit DNA methyltransferases to sites of damaged DNA where it methylates DNA de novo. CHD4 plays a role in maintaining DNA hypermethylation-associated transcriptional silencing of tumor suppressor genes (Xia et al., 2017a). CHD4-associated NuRD complex was found to be enriched at nascent DNA in embryonic stem cells, but not in NIH3T3 fibroblasts. Its presence helps maintain protein stability of replication-associated UHRF1 factor (Aranda et al., 2014). CHD4 is important in the maintenance of liver cancer stem cells and is involved in resistance to chemotherapy (Nio et al., 2015). CHD4 was also reported to promote cancer cell proliferation and correlate with a poor prognosis in non-small cell lung cancer patients (Xu et al., 2016). CHD4 interacts with a stem cell transcription factor ZFHX4 to regulate tumor initiation of glioblastoma (Chudnovsky et al., 2014). Somatic CHD4 mutation is not frequent in glioma in our analysis. However, somatic CHD4 mutation is as high as 17% in serous endometrial tumors, implying a possibly critical role in cancer (Gallo et al., 2012). Finally, CHD4 is a coregulator of PAX3–FOXO1 and potential therapeutic target in solid tumor and leukemia as well (Sperlazza et al., 2015; Böhm et al., 2016). RAD21, double-strand-break repair protein rad21 homolog, is a component of the cohesion complex and a target gene of Wnt–β-catenin signaling pathway (Xu et al., 2014). RAD21 is also a critical transcriptional regulator of genes and long interspersed retrotransposons. Elevated gene amplification and expression of RAD21 and its mediation of retrotransposon expression were seen in cancer with unstable genome and dysregulated gene expression (Xu et al., 2014). It is not surprising that epigenetic perturbation by CHD4 and activation of RAD21 can result in reduced DFS in IDH1/2-mutated LGG patients.
Copy number gain or gene amplification of EZH2, KMT2C, and CHD4 correlates with the upregulation of their mRNA expression (Supplementary Figure S3 for EZH2 and KMT2C; Supplementary Figure S4 for CHD4). This is especially apparent for CHD4. Therefore, it is reasonable to speculate that copy number gain or gene amplification of EZH2, KMT2C, or CHD4 will lead to increased expression of their respective mRNA and protein. Increased expression of EZH2, KMT2C, and CHD4 leads to the change of their enzymatic activity, causing epigenetic perturbation and malignant transformation. Accumulation of other genetic changes eventually leads to the formation of glioma. Since alteration of EZH2, KMT2C, or CHD4 epigenetic modifier genes increases recurrence rate in TP53-mutated glioma patients, targeting these epigenetic modifiers could be the treatment of choice in preventing disease recurrence and increasing disease-free survival in glioma.
Though this study did reveal several genes related to the survival time of the LGG patients, it only employed the well-developed online tools for bioinformatics research (Cerami et al., 2012; Gao et al., 2013). In the future, more advanced data mining techniques (Jiang et al., 2011, 2015; Zhang et al., 2014, 2015a, b, 2016; Jiang, 2015; Kim et al., 2015; Melamed et al., 2015; Tanaka and Ogishima, 2015; Wang et al., 2015; Xia et al., 2017b) will be integrated to continue this research.
Materials and methods
We used four datasets which included the following: (i) Brain Lower Grade Glioma (TCGA, provisional) with 530 samples, for which we selected 283 all complete tumors for analysis; (ii) Glioblastoma (TCGA, 2008) with 206 samples, from which only 91 all complete tumors were chosen for analysis from 206 samples; (iii) Glioblastoma (TCGA, 2013b) with 580 samples, from which 291 all complete tumors were selected for analysis out of 580 samples; (iv) AML (TCGA, 2013a) with 200 samples, in which 166 all complete tumors were used for analysis. For these datasets, chosen genomic profiles included mutations, putative copy number alterations, protein expression by reverse phase protein array (RPPA), mRNA expression by RNAseq (V2 RSEM). Genes entered for query are the following: EZH2, CHD4, IDH1, IDH2, DNMT3A, ASXL1, TET2, TP53, WT1, CDKN2A, NPM1, FLT3, CEBPA, PIK3CA, PTEN, KIT, NRAS, KRAS, KMT2C, andEGFR. These chosen genes had frequent alterations either in AML or glioma, or both based on available literature. Occasionally, fewer genes or additional genes were entered in query based on search needs. Data retrieval were done via cBioPortal for Cancer Genomics (website: http://www.cbioportal.org). Data generation was made by using the bioinformatics tool at our disposal (Cerami et al., 2012; Gao et al., 2013).
Supplementary Material
Supplementary material
Supplementary material is available at Journal of Molecular Cell Biology online.
Funding
This work was supported by the Natural Science Foundation of China (61372138), the Chinese Recruitment Program of Global Youth Experts, Chongqing Collaborative Innovation Center for Brain Science, and U.S. National Institutes of Health (U01 CA166886-03) (to L.Z.). Y.L. was supported by the start-up fund from the Vivian L. Smith Department of Neurosurgery, McGovern Medical School, the University of Texas Health Science Center at Houston, and Craig H. Neilsen Foundation (338617). J.Z. was supported by the National Institutes of Health (R01HL093195 and R21CA178513).
Conflict of interest: none declared.
References
- Abdel-Wahab O., Gao J., Adli M., et al. (2013). Deletion of Asx11 results in myelodysplasia and severe developmental defects in vivo. J. Exp. Med. 210, 2641–2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aranda S., Rutishauser D., and Ernfors P. (2014). Identification of a large protein network involved in epigenetic transmission in replicating DNA of embryonic stem cells. Nucleic Acids Res. 42, 6972–6986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balss J., Meyer J., Mueller W., et al. (2008). Analysis of the IDH1 codon 132 mutation in brain tumors. Acta Neuropathol. 116, 597–602. [DOI] [PubMed] [Google Scholar]
- Bettegowda C., Agrawal N., Jiao Y., et al. (2011). Mutations in CIC and FUBP1 contribute to human oligodendroglioma. Science 333, 1453–1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böhm M., Wachtel M., Marques J.G., et al. (2016). Helicase CHD4 is an epigenetic coregulator of PAX3-FOXO1 in alveolar rhabdomyosarcoma. J. Clin. Invest. 126, 4237–4249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busque L., Patel J.P., Figueroa M.E., et al. (2012). Recurrent somatic TET2 mutations in normal elderly individuals with clonal hematopoiesis. Nat. Genet. 44, 1179–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerami E., Gao J., Dogrusoz U., et al. (2012). The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2, 401–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chudnovsky Y., Kim D., Zheng S., et al. (2014). ZFHX4 interacts with the NuRD core member CHD4 and regulates the glioblastoma tumor-initiating cell state. Cell Rep. 6, 313–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimmino L., Abdel-Wahab O., Levine R.L., et al. (2011). TET family proteins and their role in stem cell differentiation and transformation. Cell Stem Cell 9, 193–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen A., Holmen S., and Colman H. (2013). IDH1 and IDH2 mutations in gliomas. Curr. Neurol. Neurosci. Rep. 13, 345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corces-Zimmerman M.R., Hong W., Weissman I.L., et al. (2014). Preleukemic mutations in human acute myeloid leukemia affect epigenetic regulators and persist in remission. Proc. Natl Acad. Sci. USA 111, 2548–2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang L., White D.W., Gross S., et al. (2009). Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature 462, 739–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg A.P., Koldobskiy M.A., and Göndör A. (2016). Epigenetic modulators, modifiers and mediators in cancer aetiology and progression. Nat. Rev. Genet. 17, 284–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimoto A., Totoki Y., Abe T., et al. (2012). Whole-genome sequencing of liver cancers identifies etiological influences on mutation patterns and recurrent mutations in chromatin regulators. Nat. Genet. 44, 760–764. [DOI] [PubMed] [Google Scholar]
- Gallo M.L., O’Hara A.J., Rudd M.L., et al. (2012). Exome sequencing of serous endometrial tumors identifies recurrent somatic mutations in chromatin-remodeling and ubiquitin ligase complex genes. Nat. Genet. 44, 1310–1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J., Aksoy B.A., Dogrusoz U., et al. (2013). Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 6, p11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentles A.J., Plevritis S.K., Majeti R., et al. (2010). Association of a leukemic stem cell gene expression signature with clinical outcomes in acute myeloid leukemia. JAMA 304, 2706–2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gereige L., and Mikkola H.K.A. (2009). DNA methylation is a guardian of stem cell self-renewal and multipotency. Nat. Genet. 41, 1164–1166. [DOI] [PubMed] [Google Scholar]
- Jeong M., Sun D., Luo M., et al. (2014). Large conserved domains of low DNA methylation maintained by Dnmt3a. Nat. Genet. 46, 17–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang R. (2015). Walking on multiple disease-gene networks to prioritize candidate genes. J. Mol. Cell Biol. 7, 214–230. [DOI] [PubMed] [Google Scholar]
- Jiang B., Dai W., Khaliq A., et al. (2015). Novel 3D GPU based numerical parallel diffusion algorithms in cylindrical coordinates for health care simulation. Math. Comput. Simul. 109, 1–19. [Google Scholar]
- Jiang B., Struthers A., Sun Z., et al. (2011). Employing graphics processing unit technology, alternating direction implicit method and domain decomposition to speed up the numerical diffusion solver for the biomedical engineering research. Int. J. Numer. Methods Biomed. Eng. 27, 1829–1849. [Google Scholar]
- Kim T.H., Monsefi N., Song J.H., et al. (2015). Network-based identification of feedback modules that control RhoA activity and cell migration. J. Mol. Cell Biol. 7, 242–252. [DOI] [PubMed] [Google Scholar]
- Kim K.H., and Roberts C.W. (2016). Targeting EZH2 in cancer. Nat. Med. 22, 128–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koivunen P., Lee S., Duncan C.G., et al. (2012). Transformation by the (R)-enantiomer of 2-hydroxyglutarate linked to EGLN activation. Nature 483, 484–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapidot T., Sirard C., Vormoor J., et al. (1994). A cell initiating human acute myeloid leukaemia after transplantation into SCID mice. Nature 367, 645–648. [DOI] [PubMed] [Google Scholar]
- Ley T.J., Ding L., Walter M.J., et al. (2010). DNMT3A mutations in acute myeloid leukemia. N. Engl. J. Med. 363, 2424–2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Chowdhury R., Liu F., et al. (2014). Tumor-suppressive miR-148a is silenced by CpG island hypermethylation in IDH1 mutant gliomas. Clin. Cancer Res. 20, 5808–5822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu A., Hou C., Chen H., et al. (2016). Genetics and epigenetics of glioblastoma: applications and overall incidence of IDH1 mutation. Front. Oncol. 6, 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losman J.A., Looper R.E., Koivunen P., et al. (2013). (R)-2-hydroxyglutarate is sufficient to promote leukemogenesis and its effects are reversible. Science 339, 1621–1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louis D.N., Ohgaki H., Wiesler O.D., et al. (2007). WHO Classification of Tumours of the Central Nervous System (4th edn). Lyon, France: IARC Press. [Google Scholar]
- Melamed R.D., Wang J., Iavarone A., et al. (2015). An information theoretic method to identify combinations of genomic alterations that promote glioblastoma. J. Mol. Cell Biol. 7, 203–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanda J.S., Kumar R., and Raghava G.P. (2016). dbEM: A database of epigenetic modifiers curated from cancerous and normal genomes. Sci. Rep. 6, 19340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nio K., Yamashita T., Okada H., et al. (2015). Defeating EpCAM+ liver cancer stem cells by targeting chromatin remodeling enzyme CHD4 in human hepatocellular carcinoma. J. Hepatol. 63, 1164–1172. [DOI] [PubMed] [Google Scholar]
- Papaemmanuil E., Gerstung M., Bullinger L., et al. (2016). Genomic classification and prognosis in acute myeloid leukemia. N. Engl. J. Med. 374, 2209–2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng H., Peng T., Wen J., et al. (2014). Characterization of p38 MAPK isoforms for drug resistance study using systems biology. Bioinformatics 30, 1899–1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pons-Tostivint E., Thibault B., and Guillermet-Guibert J. (2017). Targeting PI3K signaling in combination cancer therapy. Trends Cancer 3, 454–469. [DOI] [PubMed] [Google Scholar]
- Pusch S., Schweizer L., Beck A.C., et al. (2014). D-2-hydroxyglutarate producing neo-enzymatic activity inversely correlates with frequency of the type of isocitrate dehydrogenase 1 mutations found in glioma. Acta Neuropathol. Commun. 2, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanson M., Marie Y., Paris S., et al. (2009). Isocitrate dehydrogenase 1 codon 132 mutation is an important prognostic biomarker in gliomas. J. Clin. Oncol. 27, 4150–4154. [DOI] [PubMed] [Google Scholar]
- Sato H., Wheat J.C., Steidl U., et al. (2016). DNMT3A and TET2 in the pre-leukemic phase of hematopoietic disorders. Front. Oncol. 6, 187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperlazza J., Rahmani M., Beckta J., et al. (2015). Depletion of the chromatin remodeler CHD4 sensitizes AML blasts to genotoxic agents and reduces tumor formation. Blood 126, 1462–1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suva M., Riggi N., Janiszewska M., et al. (2009). EZH2 is essential for glioblastoma cancer stem cell maintenance. Cancer Res. 69, 9211–9218. [DOI] [PubMed] [Google Scholar]
- Tanaka H., and Ogishima S. (2015). Network biology approach to epithelial-mesenchymal transition in cancer metastasis: three stage theory. J. Mol. Cell Biol. 7, 253–266. [DOI] [PubMed] [Google Scholar]
- TCGA, Cancer Genome Atlas Research Network. (2008). Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 455, 1061–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TCGA, Cancer Genome Atlas Research Network. (2013. a). Genomic and epigenomic landscape of adult de novo acute myeloid leukemia. N. Engl. J. Med. 368, 2059–2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TCGA, Cancer Genome Atlas Research Network. (2013. b). The somatic genomic landscape of glioblastoma. Cell 156, 462–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Rhenen A., Feller N., Kelder A., et al. (2005). High stem cell frequency in acute myeloid leukemia at diagnosis predicts high minimal residual disease and poor survival. Clin. Cancer Res. 11, 6520–6527. [DOI] [PubMed] [Google Scholar]
- Wang Y., Cheng H., Pan Z., et al. (2015). Reconfiguring phosphorylation signaling by genetic polymorphisms affects cancer susceptibility. J. Mol. Cell Biol. 7, 187–202. [DOI] [PubMed] [Google Scholar]
- Wang H.Y., Tang K., Liang T.Y., et al. (2016). The comparison of clinical and biological characteristics between IDH1 and IDH2 mutations in gliomas. J. Exp. Clin. Cancer Res. 35, 86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M., Yang C., Zhang L., et al. (2017). Molecular mutations and their co-occurrences in cytogenetically normal acute myeloid leukemia. Stem Cells Int. 2017, 6962379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T., Nobusawa S., Kleihues P., et al. (2009). IDH1 mutations are early events in the development of astrocytomas and oligodendrogliomas. Am. J. Pathol. 174, 1149–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weirich S., Kudithipudi S., Kycia I., et al. (2015). Somatic cancer mutations in the MLL3-SET domain alter the catalytic properties of the enzyme. Clin. Epigenetics 7, 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams S.C., Karajannis M.A., Chiriboga L., et al. (2011). R132H-mutation of isocitrate dehydrogenase-1 is not sufficient for HIF-1α upregulation in adult glioma. Acta Neuropathol. 121, 279–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia L., Huang W., Bellani M., et al. (2017. a). CHD4 has oncogenic functions in initiating and maintaining epigenetic suppression of multiple tumor suppressor genes. Cancer Cell 31, 653–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia Y., Yang C., Hu N., et al. (2017. b). Exploring the key genes and signaling transduction pathways related to the survival time of glioblastoma multiforme patients by a novel survival analysis model. BMC Genomics 18, 950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu N., Liu F., Zhou J., et al. (2016). CHD4 is associated with poor prognosis of non-small cell lung cancer patients through promoting tumor cell proliferation. Eur. Respir. J. 48, PA2862. [Google Scholar]
- Xu H., Yan Y., Deb S., et al. (2014). Cohesin Rad21 mediates loss of heterozygosity and is upregulated via Wnt promoting transcriptional dysregulation in gastrointestinal tumors. Cell Rep. 9, 1781–1797. [DOI] [PubMed] [Google Scholar]
- Xu W., Yang H., Liu Y., et al. (2011). Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of a-ketoglutarate-dependent dioxygenases. Cancer Cell 19, 17–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H., Yu S., Liu Q., et al. (2017). Recent advances of highly selective CDK4/6 inhibitors in breast cancer. J. Hematol. Oncol. 10, 97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan H., Parsons D.W., Jin G., et al. (2009). IDH1 and IDH2 mutations in gliomas. N. Engl. J. Med. 360, 765–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan X.J., Xu J., Gu Z.H., et al. (2011). Exome sequencing identifies somatic mutations of DNA methyltransferase gene DNMT3A in acute monocytic leukemia. Nat. Genet. 43, 309–315. [DOI] [PubMed] [Google Scholar]
- Zhang H., Alberich-Jorda M., Amabile G., et al. (2013). Sox4 is a key oncogenic target in C/EBPa mutant acute myeloid leukemia. Cancer Cell 24, 575–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C., Liu C., Cao S., et al. (2015. a). Elucidation of drivers of high-level production of lactates throughout a cancer development. J. Mol. Cell Biol. 7, 267–279. [DOI] [PubMed] [Google Scholar]
- Zhang L., Qiao M., Gao H., et al. (2016). Investigation of mechanism of bone regeneration in a porous biodegradable calcium phosphate (CaP) scaffold by combination of multi-scale agent based model and experimental optimization/validation. Nanoscale 8, 14877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Xue Y., Jiang B., et al. (2014). Multiscale agent-based modelling of ovarian cancer progression under the stimulation of the STAT3 pathway. Int. J. Data Min. Bioinform. 9, 235–253. [DOI] [PubMed] [Google Scholar]
- Zhang L., and Yang C. (2014). Promise of cyclin-dependent kinases 4/6 as therapeutic targets in breast cancer. J. Carcinog. Mutagen. 5, 191. [Google Scholar]
- Zhang W., Zeng T., Liu X., et al. (2015. b). Diagnosing phenotypes of single-sample individuals by edge biomarkers. J. Mol. Cell Biol. 7, 231–241. [DOI] [PubMed] [Google Scholar]
- Zhang L., and Zhang S. (2017). Using game theory to investigate the epigenetic control mechanisms of embryo development: Comment on: ‘Epigenetic game theory: How to compute the epigenetic control of maternal-to-zygotic transition’ by Qian Wang et al. Phys. Life Rev. 20, 140–142. [DOI] [PubMed] [Google Scholar]
- Zhao S., Lin Y., Xu W., et al. (2009). Glioma-derived mutations in IDH1 dominantly inhibit IDH1 catalytic activity and induce HIF-1α. Science 324, 261–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.