Key Points
In the era of widespread rotavirus vaccine use, toxigenic Clostridium difficile, diarrheagenic Escherichia coli, and viruses (particularly norovirus) are commonly detected among children with infectious gastroenteritis in the United States by using a multipathogen molecular panel.
Keywords: acute gastroenteritis, children, FilmArray, gastrointestinal illness
Abstract
Background
Diarrheal diseases are a major cause of ambulatory care visits and hospitalizations among children. Because of overlapping signs and symptoms and expensive and inefficient testing methods, the etiology of pediatric diarrhea is rarely established.
Methods
We identified children <18 years of age who were evaluated for diarrhea at Primary Children’s Hospital in Salt Lake City, Utah, between October 2010 and September 2012. Stool specimens submitted for testing were evaluated by using the FilmArray gastrointestinal diagnostic system, which is a rapid multiplex polymerase chain reaction platform that can simultaneously detect 23 bacterial, viral, and protozoal agents.
Results
A pathogen was detected in 561 (52%) of 1089 diarrheal episodes. The most commonly detected pathogens included toxigenic Clostridium difficile (16%), diarrheagenic Escherichia coli (15%), norovirus GI/GII (11%), and adenovirus F 40/41 (7%). Shiga toxin-producing E coli were detected in 43 (4%) specimens. Multiple pathogens were identified in 160 (15%) specimens. Viral pathogens (norovirus, adenovirus, rotavirus, and sapovirus) were more common among children <5 years old than among those 5 to 17 years old (38% vs 16%, respectively; P < .001). Bacterial pathogens were identified most commonly in children 2 to 4 years of age. Children with 1 or more underlying chronic medical conditions were less likely to have a pathogen identified than those without a chronic medical condition (45% vs 60%, respectively; P < .01). Viral pathogens were detected more commonly in the winter, whereas bacterial pathogens were detected more commonly in the summer.
Conclusions
Toxigenic C difficile, diarrheagenic E coli, and norovirus were the leading organisms detected among these children with diarrhea. Viral pathogens are identified frequently among young children with acute gastroenteritis.
BACKGROUND
Infectious diarrhea is an important public health concern, both in resource-poor settings and in industrialized countries [1]. The World Health Organization’s Global Burden of Disease study found that diarrheal diseases were the seventh leading cause of death worldwide in 2010 [2]. According to recent estimates from the US Centers for Disease Control and Prevention, approximately 179 million diarrheal illnesses occur each year in the United States, with more than 473 000 hospitalizations [3]. The impact is greatest among young children. Despite significant reductions after the introduction of the rotavirus vaccine, there are more than 105 000 hospitalizations for gastroenteritis annually among children <5 years of age [4–6].
There have been few comprehensive studies of the etiology of gastroenteritis in the United States [7–10]. Diagnostic tests are either not widely available in clinical laboratories or have limited sensitivities for many of the diarrheal pathogens that infect children [8, 11–14]. With the recent advent of molecular diagnostic tests that detect multiple diarrheal pathogens, it is now possible to characterize more thoroughly the clinical epidemiology of infectious diarrhea among hospitalized children [15]. In this study, we sought to describe the clinical epidemiology of pediatric infectious diarrhea over a 2-year period using the FilmArray gastrointestinal diagnostic system (FilmArray GI panel), a rapid multiplex polymerase chain reaction (PCR)–based testing platform that simultaneously detects 23 different viral, bacterial, and protozoal targets.
METHODS
Human Subjects Protection
This prospective observational study was approved by the University of Utah and Intermountain Healthcare institutional review boards (approval number 45464).
Setting and Study Population
We included all children <18 years of age who had submitted a stool sample for any diagnostic testing at Primary Children’s Hospital. A manual chart review was performed to confirm that the children included in this study had symptoms of acute gastrointestinal illness (eg, fever, vomiting, abdominal pain, and diarrhea). Primary Children’s Hospital is a 297-bed children’s hospital that provides primary care for children in the Salt Lake City, Utah, region and tertiary care for children from Utah and 5 surrounding states. Stool specimens that conformed to the shape of the cup were submitted, and standard laboratory testing was performed as requested by the treating physician. Residual samples were stored at –80°C until they were tested with the FilmArray GI panel. We collected specimens between October 2010 and September 2012.
Multiple specimens were often submitted for testing over several days, which we grouped into episodes. Diarrheal episodes were determined after review by 3 of the study investigators (C. S., E. K. K., and A. T. P.). In brief, diarrheal episodes included those for which care was sought during the week before a stool specimen was obtained and the 2 weeks after collection of a stool specimen. All healthcare encounters were then reviewed manually to determine whether they were related to ongoing diarrheal illness. Diarrheal episodes were classified as outpatient, emergency, or inpatient on the basis of the highest level of care sought during the diarrheal illness episode. Thus, if a patient was hospitalized at any point during the course of his or her diarrheal episode, the episode was classified as an inpatient diarrheal episode. Only the first available stool sample submitted for diagnostic testing within each episode was tested using the FilmArray GI panel.
Chronic medical conditions were classified by using a method described by Feudtner et al [16] on the basis of International Classification of Diseases, Ninth Revision, Clinical Modification discharge diagnosis codes.
FilmArray GI Panel Pathogen Detection
The FilmArray is a rapid multiplex PCR platform with which nucleic acid extraction, nested PCR, and amplicon melt-curve analysis are performed in a closed system in 1 hour [17, 18]. The FilmArray GI pouch (investigational-use-only [IUO] version) used in this study simultaneously detects 23 diarrheagenic bacterial, viral, and protozoal agents by using pathogen-specific virulence genes or species-specific regions in housekeeping genes. The FilmArray GI panel was approved recently by the US Food and Drug Administration (FDA) for in vitro diagnostic use for 22 pathogens [4]. The FDA-cleared and IUO versions are identical except for the inclusion of Aeromonas spp. in the IUO version. Toxigenic Clostridium difficile detection was based on identification of the genes that encode an enterotoxin (tcdA) and a cytotoxin (tcdB). The pathotypes of diarrheagenic Escherichia coli were identified using pathotype-specific genetic markers: Shigatoxigenic E coli (STEC) by the detection of Shiga toxin 1 and/or 2 genes (stx1 or stx2) in association with the intimin gene (eae); enteropathogenic E coli (EPEC) by detection of the intimin gene (eae) in the absence of stx1 and/or stx2; enterotoxigenic E coli (ETEC) by genes encoding heat-labile (lt) or heat-stable (st) enterotoxins; enteroinvasive E coli (EIEC) and Shigella by the invasion plasmid antigen H gene (ipah) (EIEC and Shigella cannot be distinguished); and enteroaggregative E coli (EAEC) by pAA virulence plasmid-carried genes encoding the AAF biogenesis transcription regulator (aggR) or outer membrane protein (aatA). The FilmArray GI panel also identifies Salmonella spp., Aeromonas spp., Cryptosporidium spp., and pathogenic species of Vibrio (V cholerae, V parahaemolyticus, V vulnificus, V mimicus, V alginolyticus, and V fluvialis) and Campylobacter (C jejuni, C coli, and C upsaliensis). The viral pathogens detected include adenovirus F 40/41, astrovirus, norovirus GI/GII, rotavirus A, and sapovirus. Parasites detected include Cryptosporidium spp., Cyclospora cayetanensis, Entamoeba histolytica, and Giardia lamblia. Blinded FilmArray testing was performed in batches, and the results were not returned to the treating physician.
Statistical Analysis
Categorical variables were compared with Yates’ corrected χ2 test or Fisher’s exact test, as appropriate. Continuous variables were compared by using the nonparametric Wilcoxon–Mann–Whitney U test. All statistical tests were 2-sided with significance defined at a P value of <.05. Statistical analyses were performed by using Stata 14.1 (Stata Corp, College Station, Texas) and R 3.2.2 (R Foundation for Statistical Computing, Vienna, Austria).
RESULTS
Demographic Characteristics
Between October 2010 and September 2012, stool samples were collected during 1089 diarrheal illness episodes at Primary Children’s Hospital. Diarrheal episodes were classified as inpatient (58%), outpatient (26%), or emergency (16%) (Table 1; Supplemental Table 1). The median age of the study population was 5 years (interquartile range, 2–11 years), and 47% were <5 years of age. More than half (57%) of the children had an underlying chronic medical condition.
Table 1.
Demographic and Clinical Characteristics of 1089 Children With Infectious Diarrhea Between 2010 and 2012
| Characteristic | n (%) |
|---|---|
| Age group | |
| <1 y | 114 (10.5) |
| 1–4 y | 397 (36.5) |
| 5–10 y | 295 (27.1) |
| 11–17 y | 283 (26.0) |
| Sex | |
| Male | 600 (55.1) |
| Female | 489 (44.9) |
| Race/ethnicity | |
| White | 845 (77.6) |
| Hispanic | 90 (8.3) |
| Black | 32 (2.9) |
| Other | 122 (11.2) |
| Chronic medical conditiona | |
| Any chronic medical condition | 626 (57.5) |
| Cardiovascular | 258 (23.7) |
| Malignancy | 240 (22.0) |
| Gastrointestinal | 192 (17.6) |
| Respiratory (including asthma) | 103 (9.5) |
| Clinical setting | |
| Inpatient | 630 (57.9) |
| Emergency department | 174 (16.0) |
| Outpatient | 285 (26.2) |
aChronic medical conditions include chronic infectious diseases, malignancies and neoplasms, endocrine and metabolic conditions, hematologic conditions and immunodeficiencies, and cardiovascular, respiratory, gastrointestinal, genitourinary, congenital, and perinatal conditions, as defined by Feudtner et al [16].
Pathogen Identification
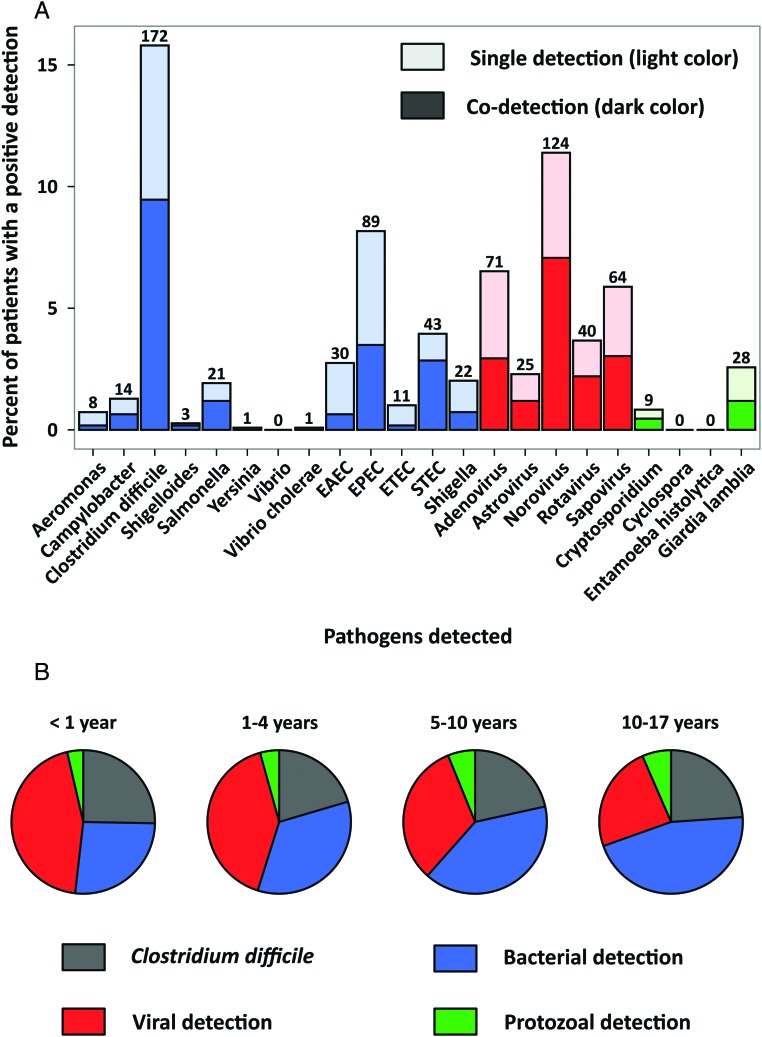
We included 1089 diarrheal episodes among 779 children. A diarrheal pathogen was detected in 561 (52%) of 1089 episodes. A total of 752 pathogens were identified; codetection of 2 or more diarrheal pathogens occurred in 15% of the samples. A bacterial pathogen was detected in 290 (27%) samples. The most common bacterial pathogens were toxigenic C difficile (16%), EPEC (8%), STEC (4%), and ETEC (3%) (Figure 1). Viral pathogens were detected in 26% of the samples. Norovirus GI/GII was the most common virus and was identified in 11% of the specimens. Other viruses were also frequently detected: adenovirus F 40/41 (7%), sapovirus (6%), rotavirus A (4%), and astrovirus (2%). Protozoal pathogens were detected in 4% of the children with diarrheal episodes.
Figure 1.
Detection of 23 infectious diarrhea pathogens among children in Utah, 2010-2012. (A) Numbers and percentages of children with specific diarrheal pathogens detected in all age groups studied. There were 1089 children with a unique diarrheal episode, 561 (52%) of whom had at least 1 pathogen detected. Because the patients could have more than 1 pathogen detected, there were a total of 776 detections. Lighter and darker colors in the histogram indicate single and copathogen detections, respectively. (B) Proportions of detections (single and codetection) according to age group. Abbreviations: EAEC, enteroaggregative E coli; EPEC, enteropathogenic E coli; ETEC, enterotoxigenic E coli; STEC, shigatoxigenic E coli.
The distribution of diarrheal pathogens varied according to age (Table 2). Children <5 years of age were more likely than older children to have 2 or more diarrheal pathogens detected (21% vs 9%, respectively; P < .01). Toxigenic C difficile was detected in 21% of 114 infants <1 year old with a diarrheal episode and in 19% of children 2 to 4 years old as compared with 12% of the older children (P < .01). A second pathogen was identified in 15 (60%) episodes among children <1 year old in whom toxigenic C difficile was detected and in 32 (42%) of 77 episodes among children 2 to 4 years old compared with 6 (19%) of 31 episodes among children 11 to 17 years old with toxigenic C difficile (P < .01 and P = .18, respectively). Bacteria other than toxigenic C difficile were detected more commonly in episodes among children >2 years of age than among the younger children (14% vs 8%, respectively; P = .03). Overall, a virus was detected in 194 (38%) of 511 episodes among children <5 years of age compared with 92 (16%) of 578 episodes among children 5 to 17 years of age (P < .001). Each of the gastrointestinal viruses was significantly more common among diarrheal episodes in children <5 years old than among episodes in children 5 to 17 years old (P < .01 for all). Rotavirus was detected in 8% of episodes among children <1 year old (the target group for live vaccine), 5% of episodes among children 2 to 4 years of age, and 3% of episodes among children 5 to 10 years of age.
Table 2.
Infectious Diarrheal Pathogens Detected From Pediatric Stool Specimens Using the FilmArray Gastrointestinal Panela
| Pathogen | All Ages (n [%]) (n = 1089) | According to Age Group (n [%]) | |||
|---|---|---|---|---|---|
| <1 y (n = 114) | 2–4 y (n = 397) | 5–10 y (n = 295) | 11–17 y (n = 283) | ||
| Bacterial pathogens | 290 (26.6) | 25 (21.9) | 127 (32.0)b | 78 (26.4) | 60 (21.2) |
| Aeromonas | 8 (0.7) | 1 (0.9) | 3 (0.8) | 0 (0) | 4 (1.4) |
| Campylobacter | 14 (1.3) | 0 (0) | 7 (1.8) | 4 (1.4) | 3 (1.1) |
| C difficile | 148 (13.6) | 24 (21.1) | 77 (19.4) | 40 (13.6) | 31 (11.0)b |
| Plesiomonas shigelloides | 3 (0.3) | 1 (0.9) | 1 (0.3) | 0 (0) | 1 (0.4) |
| Salmonella | 21 (1.9) | 1 (0.9) | 4 (1.0) | 5 (1.7) | 11 (3.9) |
| Y enterocolitica | 1 (0.1) | 0 (0) | 0 (0) | 0 (0) | 1 (0.4) |
| Vibrio | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| V cholerae | 1 (0.1) | 0 (0) | 0 (0) | 0 (0) | 1 (0.4) |
| Shigella/EIEC | 22 (2.0) | 0 (0) | 13 (3.3) | 9 (3.1) | 0 (0) |
| Diarrheagenic E coli | 144 (13.2) | 9 (7.9) | 65 (16.4)b | 44 (14.9)b | 26 (9.2) |
| EAEC | 30 (2.8) | 1 (0.9) | 14 (3.5) | 10 (3.4) | 5 (1.8) |
| EPEC | 89 (8.2) | 8 (7.0) | 42 (10.6) | 27 (9.2) | 12 (4.2) |
| ETEC | 11 (1.0) | 0 (0) | 4 (1.0) | 3 (1.0) | 4 (1.4) |
| All STEC | 43 (3.9) | 1 (0.9) | 19 (4.8) | 15 (5.1) | 8 (2.8) |
| E coli O157 | 21 (1.9) | 0 (0) | 9 (2.3) | 7 (2.4) | 5 (1.8) |
| Non-O157 | 22 (2.0) | 1 (0.9) | 10 (2.5) | 8 (2.7) | 3 (1.1) |
| Viral pathogens | 286 (26.3) | 42 (36.8) | 152 (38.3) | 61 (20.7)b | 31 (11.0)b |
| Adenovirus F 40/41 | 71 (6.5) | 11 (9.6) | 37 (9.3) | 14 (4.7) | 9 (3.2)b |
| Astrovirus | 25 (2.3) | 4 (3.5) | 14 (3.5) | 6 (2.0) | 1 (0.4)b |
| Norovirus GI/GII | 124 (11.4) | 16 (14.0) | 70 (17.6) | 23 (7.8) | 15 (5.3)b |
| Rotavirus A | 40 (3.7) | 9 (7.9) | 19 (4.8) | 9 (3.1)b | 3 (1.1)b |
| Sapovirus | 64 (5.9) | 8 (7.0) | 37 (9.3) | 14 (4.7) | 5 (1.8)b |
| Protozoal pathogens | 36 (3.3) | 3 (2.6) | 14 (3.5) | 11 (3.7) | 8 (2.8) |
| Cryptosporidium | 9 (0.8) | 0 (0) | 3 (0.8) | 3 (1.0) | 3 (1.1) |
| C cayetanensis | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| E histolytica | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| G lamblia | 28 (2.6) | 3 (2.6) | 12 (3.0) | 8 (2.7) | 5 (1.8) |
| Codetection | 160 (14.7) | 23 (20.2) | 85 (21.4) | 36 (12.2)b | 16 (5.7)b |
| Negative | 537 (49.3) | 66 (57.9) | 139 (35.0)b | 150 (50.8) | 182 (64.3) |
Abbreviations: EAEC, enteroaggregative E coli; EIEC, enteroinvasive E coli; EPEC, enteropathogenic E coli; ETEC, enterotoxigenic E coli; STEC, shigatoxigenic E coli.
aIn each cell are the number of positive specimens (percentage for diarrheal pathogen). Because multiple pathogens were detected in some children, numbers add up to >100%.
bStatistically significant difference in the frequency of detection relative to diarrheal episodes among children <1 year of age (P < .05).
A diarrheal pathogen was identified more often among diarrheal episodes in children who presented to the emergency department (68%) than among those in an outpatient (48%) or inpatient (47%) setting (P < .01) (Table 3). Viral pathogens were identified more commonly among those seen in the emergency department than in those seen in other settings (45% vs 23%, respectively; P < .001). Norovirus GI/GII was the most commonly detected pathogen among emergency department episodes (21% vs 10%; P < .001). Among diarrheal episodes in children ≥1 year of age, toxigenic C difficile was detected among 8% of emergency department episodes compared with 16% of outpatient episodes and 15% of inpatient episodes (P = .01 and .02, respectively). It was notable that norovirus GI/GII was the second most commonly detected pathogen (9%) among inpatient episodes after toxigenic C difficile (14%). Protozoa were rarely detected in inpatient diarrhea episodes (1%).
Table 3.
Infectious Diarrheal Pathogens Detected From Stool Specimens Obtained From Pediatric Outpatients, Emergency Department Patients, and Inpatients With the FilmArray Gastrointestinal Panela
| Pathogen | Clinical Setting (n [%]) | ||
|---|---|---|---|
| Outpatient (n = 285) | Emergency (n = 174) | Inpatient (n = 630) | |
| Bacterial pathogens | 75 (26.3) | 49 (28.2) | 166 (26.3) |
| Aeromonas | 3 (1.1) | 3 (1.7) | 2 (0.3) |
| Campylobacter | 4 (1.4) | 5 (2.9)b | 5 (0.8) |
| C difficile | 51 (17.9)c | 19 (10.9)b | 102 (16.2) |
| Plesiomonas shigelloides | 0 (0) | 0 (0) | 3 (0.5) |
| Salmonella | 2 (0.7)c | 7 (4.0) | 12 (1.9) |
| Y enterocolitica | 0 (0) | 0 (0) | 1 (0.2) |
| Vibrio | 0 (0) | 0 (0) | 0 (0) |
| V cholerae | 0 (0) | 0 (0) | 1 (0.2) |
| Shigella/EIEC | 6 (2.1) | 7 (4.0) | 9 (1.4) |
| Diarrheagenic E coli | 39 (13.7) | 32 (18.4)b | 73 (11.6) |
| EAEC | 5 (1.8)c | 9 (5.2) | 16 (2.5) |
| EPEC | 27 (9.5) | 23 (13.2)b | 39 (6.2) |
| ETEC | 3 (1.1) | 3 (1.7) | 5 (0.8) |
| All STEC | 8 (2.8) | 10 (5.7) | 25 (4.0) |
| E coli O157 | 3 (1.1) | 4 (2.3) | 14 (2.2) |
| Non-O157 | 5 (1.8) | 6 (3.4) | 11 (1.7) |
| Viral pathogens | 62 (21.8)c | 78 (44.8)b | 146 (23.2) |
| Adenovirus F 40/41 | 13 (4.6)c | 18 (10.3) | 40 (6.3) |
| Astrovirus | 6 (2.1) | 6 (3.4) | 13 (2.1) |
| Norovirus GI/GII | 29 (10.2)c | 37 (21.3)b | 58 (9.2) |
| Rotavirus A | 1 (0.4)c,d | 11 (6.3) | 28 (4.4) |
| Sapovirus | 19 (6.7) | 20 (11.5)b | 25 (4.0) |
| Protozoal pathogens | 18 (6.3)d | 9 (5.2)b | 9 (1.4) |
| Cryptosporidium | 5 (1.8) | 1 (0.6) | 3 (0.5) |
| C cayetanensis | 0 (0) | 0 (0) | 0 (0) |
| E histolytica | 0 (0) | 0 (0) | 0 (0) |
| G lamblia | 14 (4.9)d | 8 (4.6)b | 6 (1.0) |
| Codetection | 39 (13.7)c | 41 (23.6)b | 80 (12.7) |
| Negative | 147 (51.6)c | 56 (32.2)b | 334 (53.0) |
Abbreviations: EAEC, enteroaggregative E coli; EIEC, enteroinvasive E coli; EPEC, enteropathogenic E coli; ETEC, enterotoxigenic E coli; STEC, shigatoxigenic E coli.
aIn each cell are the number of positive specimens (percentage for diarrheal pathogen). Because multiple pathogens were detected in some children, numbers add up to >100%.
bStatistically significant difference in the frequency of detection between emergency department and inpatient diarrheal episodes (P < .05).
cStatistically significant difference in the frequency of detection between outpatient and emergency department diarrheal episodes (P < .05).
dStatistically significant difference in the frequency of detection between outpatient and inpatient diarrheal episodes (P < .05).
Diarrheal episodes among children with 1 or more underlying chronic medical conditions were less likely to have a pathogen identified than those with no underlying chronic conditions (45% vs 60%, respectively; P < .01) (Supplemental Table 2). When the analysis was restricted to inpatient episodes only, 44% of the children with 1 or more underlying chronic medical conditions had a pathogen detected compared with 55% of inpatients without an underlying chronic medical condition (P = .02). Salmonella, EPEC, STEC, Shigella/EIEC, rotavirus A, and G lamblia were significantly more common among episodes in previously healthy children.
More than 1 pathogen was detected in 160 (15%) diarrheal episodes (Supplemental Table 3). Two pathogens were identified in 118 (11%) of the episodes, and 3 or more pathogens were identified in 42 (4%) episodes. A virus and a bacterium were detected in 7% of the episodes. Viral–viral and bacterial–bacterial codetections were identified at similar rates (4% and 3%, respectively). Mixed viral–protozoal and bacterial–protozoal detections were identified infrequently (1% each).
Seasonality
Diarrheal episodes occurred throughout both years of the study (Figure 2). Bacterial detections were more common in the summer months (April through September), with distinct peaks in EPEC, STEC, and Shigella/EIEC. Viral detections were more common during the winter months (October through March) (P < .01). Toxigenic C difficile and norovirus GI/GII had less distinct seasonality, and cases were identified in every month of the study period.
Figure 2.
Seasonality of pediatric diarrhea according to pathogen, 2010–2012. (A) Numbers of stool specimens tested according to month are presented as the overall height of the bars. Stacked within each of these bars are the numbers of bacterial, viral, protozoal, and mixed (bacterial + viral, bacterial + protozoal, or viral + protozoal) detections and negative test results (white). (B) Detection of enteric bacterial pathogens. (C) Detection of viral pathogens. (D) Detection of protozoal pathogens. The number of diarrheal episodes in which each pathogen was identified are presented as stacked bar graphs for each month of the 2-year study period. Abbreviations: EAEC, enteroaggregative E coli; EIEC, enteroinvasive E coli; EPEC, enteropathogenic E coli; ETEC, enterotoxigenic E coli; STEC, shigatoxigenic E coli.
DISCUSSION
This large study using a testing platform that detects 23 diarrheal pathogens provides insights into the clinical epidemiology of diarrhea among children in an industrialized country. We identified a potential pathogen in 52% of the specimens tested. The proportion of patients with an identified pathogen was higher among younger children, but this result was driven in part by detection of toxigenic C difficile, which likely represented asymptomatic carriage of the organism among infants and young children. Toxigenic C difficile (16%), diarrheagenic E coli (15%), norovirus GI/GII (11%), and adenovirus F 40/41 (7%) were the most commonly detected pathogens. Excluding the detection of toxigenic C difficile in children <1 year old, which is unlikely to represent a true infection, a potential pathogen was detected in 552 (51%) of 1089 children. Despite the marked reduction in rotavirus compared with that found in earlier studies [9, 19], viral pathogens continued to be detected frequently and accounted for more than half of all pathogens identified in children <5 years old. It is striking that norovirus GI/GII and adenovirus F 40/41 infections were more common than rotavirus A, even among children <1 year of age.
A bacterial pathogen was identified in slightly more than 1 in 4 specimens tested, but only 59 (5%) of 1089 episodes were due to “classic” bacterial enteric pathogens that are sought in conventional stool cultures, including Campylobacter spp., Salmonella spp., Shigella spp., Yersinia enterocolitica, Plesiomonas spp., and Vibrio spp. This result is similar to those of recent prospective studies in the United States [8–10]. STEC was detected by molecular methods in 4% of children with diarrhea, which was more frequent than the detection of Salmonella spp., Shigella spp., and Campylobacter spp.; just more than half of the detections were non-E coli O157 STEC. The rate of STEC detection in our study is slightly higher than but consistent with those in other recent studies [7, 9, 10], which may reflect the regional epidemiology of STEC or increased sensitivity of molecular detection methods [20]. Classic bacterial enteric pathogens and STEC were rarely detected (3%) in infants <1 year of age.
The most commonly detected bacterial pathogen was toxigenic C difficile; it was detected in 16% of the specimens tested. The highest isolation rates were found in infants <1 year of age (21%) and in children 1 to 4 years of age (19%). Children in these age groups have high rates of asymptomatic colonization with C difficile. Detection of C difficile in children younger than 2 years is rarely significant, and the clinical significance of detection in children 3 to 4 years old is unclear [21, 22]. In a case-control study for which a cytotoxicity assay was used, Denno et al [10] detected C difficile in 5% of children <3 years of age who presented to the emergency department with diarrhea but in 9% of the controls. Bruijnesteijn et al [23] detected C difficile by PCR in similar proportions of children <5 years of age with and without diarrhea. We did not enroll controls, but in 64% of the infants <1 year of age with toxigenic C difficile, we found a second pathogen, which suggests that most, if not all, toxigenic C difficile detections in this age group represent colonization. Among children <5 years of age with toxigenic C difficile detected, a second pathogen was identified in 47 (47%) of 101 episodes, most commonly norovirus, adenovirus F 40/41, and sapovirus (Supplemental Table 4). However, there is increasing evidence to support the importance of C difficile as a cause of both hospital- and community-onset diarrhea in older children [24, 25]. Among children ≥3 years old, Denno et al [10] detected C difficile in 6% of the cases but only 1% of the controls. C difficile has been detected in 2% to 7% of children in studies of diarrheal illness who presented to an emergency department [8–10]. We identified toxigenic C difficile in 16% of children ≥1 year of age in the outpatient setting, 9% of children who presented to the emergency department, and 16% of inpatients. Additional data on the detection of toxigenic C difficile by PCR in symptomatic and asymptomatic children in different settings are needed to clarify its clinical significance and avoid overtreatment [26, 27].
A significant observation in our study was the detection of diarrheagenic E coli other than STEC in 9% of the diarrheal episodes tested, similar to the rates reported in studies of children in New Haven, Connecticut, Baltimore, Maryland, Cincinnati, Ohio, and Seattle, Washington [7, 8, 10]. We detected EAEC, defined by the presence of aggR and aatA, in 3% of the children ≤5 years of age with diarrhea. Cohen et al [7] found EAEC in 9% of children ≤5 years old in the emergency department with diarrhea and in 3% of the controls. Denno et al [10] identified EAEC in 3% of stool specimens obtained from the Seattle Children’s Hospital emergency department and in 1% of their controls. Nataro et al [8] detected EAEC, defined by the presence of aatA, in 6% of children <18 years old with diarrhea in Baltimore and New Haven and in 3% of their controls. Altogether, these data suggest that EAEC is an important diarrheal pathogen among children in the United States. ETEC was detected in 11 patients in our study. Although we did not collect travel histories, several studies suggest that ETEC infection can be acquired in the United States [28]. We also detected EPEC, defined as the detection of the intimin gene eae in the absence of Shiga toxin genes, which thus includes typical and atypical EPEC, in 8% of the children. However, given the complex genetics of EPEC pathogenicity and the fact that eae-positive EPEC has been detected in similar numbers of cases and controls in some studies, the significance of EPEC detection with the FilmArray requires further delineation through case-control studies [8].
A viral pathogen was identified in more than half of the children in whom a pathogen was identified, and viruses accounted for 62% (194 of 315) of the pathogens identified among children <5 years old. Similar to other data from the United States, norovirus GI/GII was the second most common pathogen in all age groups in our population. In prospective studies in the New Vaccine Surveillance Network, 21% of children <5 years of age who required medical attention had norovirus detected [14, 29]. In our study, we identified norovirus GI/GII in 17% of the children <5 years of age. Norovirus GI/GII was the most common pathogen identified in emergency department encounters (detected in 21% of the children). It is notable that norovirus GI/GII was present in 9% of the hospitalized children, almost as frequently as toxigenic C difficile, which emphasizes the emerging role of norovirus GI/GII in hospital-associated diarrheal illness [30, 31].
Adenovirus F 40/41 was the second most commonly detected virus in this study. Chhabra et al [14] identified adenovirus in 12% of children <5 years of age, similar to the 9% identified among children <5 years of age in the current study. We detected adenovirus in 6% of hospitalized children, in 5% of those in an outpatient setting, and in 10% of those in the emergency department, which is comparable to the results of other studies [8–10, 32]. In marked contrast to the results of studies conducted before the era of childhood rotavirus A vaccination in which rotavirus was found in 10% to 43% of children [8–10, 33], we identified rotavirus in only 4%. Because the FilmArray GI panel detects both wild-type and vaccine-type rotaviruses, some of the 9 detections in children <1 year of age may represent asymptomatic shedding of attenuated vaccine strains.
Protozoal pathogens have not been sought consistently in diagnostic studies of sporadic diarrhea in children in the United States. G lamblia was identified in 28 (3%) of 1089 stool specimens, predominantly among children who presented to the emergency department or an outpatient clinic, which compares with the 1% reported from studies in Seattle [9, 10].
Our study had a number of strengths. Each specimen was comprehensively tested for 23 pathogens by using a method that is equally or more sensitive and specific than standard clinical testing methods [4, 34]. It is one of the largest studies among children in an industrialized country, including 1089 episodes across 2 full calendar years among outpatients, emergency department patients, and inpatients. However, there are a number of important limitations. Pathogens can be detected in asymptomatic children. We did not enroll controls without diarrhea, which makes it difficult to determine whether the identification of a pathogen indicated that the pathogen was the cause of the child’s diarrhea. Case-control studies that use new, highly sensitive multiplex assays are clearly needed to better understand the predictive value of detection. The increased sensitivity of PCR makes comparisons to other case-control studies challenging. We tested samples only from patients for whom a clinician had ordered stool testing, which resulted in potential bias. The study was conducted at 1 institution in Utah, and the distribution of diarrheal pathogens can be influenced by geography, demographics, season, and the occurrence of outbreaks. Thus, our results cannot be generalized. We did not collect comprehensive data on risk factors, symptoms, treatments, or outcomes.
This study provides a detailed picture of the pathogens identified in children who presented with diarrhea in the era of broad rotavirus vaccine use. The results highlight the utility of multipathogen testing, demonstrates the important role of viral agents of gastroenteritis, particularly norovirus, confirms the increasing prevalence of toxigenic C difficile infection among older children, and confirms that EAEC may be an important and underrecognized cause of diarrhea in US children. Additional studies are warranted to determine whether timely and sensitive detection of common viral pathogens reduces unnecessary or potentially dangerous antibiotic use.
Supplementary Data
Supplementary materials are available at Open Forum Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Supplementary Material
Notes
Financial support. This study and the development of the FilmArray GI panel were supported by National Institutes of Health Grant 5R01AI089489.
Potential conflicts of interest. M. R., B. G., M. V., R. C., M. A. P., and S. T. are employees of BioFire Diagnostics, Inc., the maker and manufacturer of the FilmArray GI panel. C. S. and A. T. P. receive support from National Institutes of Health Grant 5R01AI089489 through a subcontract with BioFire Diagnostics, and J. D. and A. T. P. have served as consultants to BioFire Diagnostics. E. K. K. and T. B. have no reported conflicts. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Black RE, Cousens S, Johnson HL et al. . Global, regional, and national causes of child mortality in 2008: a systematic analysis. Lancet 2010; 375 : 1969–87. [DOI] [PubMed] [Google Scholar]
- 2. Lozano R, Naghavi M, Foreman K et al. . Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet 2012; 380 : 2095–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Scallan E, Griffin PM, Angulo FJ, Tauxe RV, Hoekstra RM. Foodborne illness acquired in the United States—unspecified agents. Emerg Infect Dis 2011; 17 : 16–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Buss SN, Leber A, Chapin K et al. . Multicenter evaluation of the BioFire FilmArray gastrointestinal panel for etiologic diagnosis of infectious gastroenteritis. J Clin Microbiol 2015; 53 : 915–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cortes JE, Curns AT, Tate JE et al. . Rotavirus vaccine and health care utilization for diarrhea in U.S. children. N Engl J Med 2011; 365 : 1108–17. [DOI] [PubMed] [Google Scholar]
- 6. Curns AT, Steiner CA, Barrett M, Hunter K, Wilson E, Parashar UD. Reduction in acute gastroenteritis hospitalizations among US children after introduction of rotavirus vaccine: analysis of hospital discharge data from 18 US states. J Infect Dis 2010; 201 : 1617–24. [DOI] [PubMed] [Google Scholar]
- 7. Cohen MB, Nataro JP, Bernstein DI, Hawkins J, Roberts N, Staat MA. Prevalence of diarrheagenic Escherichia coli in acute childhood enteritis: a prospective controlled study. J Pediatr 2005; 146 : 54–61. [DOI] [PubMed] [Google Scholar]
- 8. Nataro JP, Mai V, Johnson J et al. . Diarrheagenic Escherichia coli infection in Baltimore, Maryland, and New Haven, Connecticut. Clin Infect Dis 2006; 43 : 402–7. [DOI] [PubMed] [Google Scholar]
- 9. Klein EJ, Boster DR, Stapp JR et al. . Diarrhea etiology in a children’s hospital emergency department: a prospective cohort study. Clin Infect Dis 2006; 43 : 807–13. [DOI] [PubMed] [Google Scholar]
- 10. Denno DM, Shaikh N, Stapp JR et al. . Diarrhea etiology in a pediatric emergency department: a case control study. Clin Infect Dis 2012; 55 : 897–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Koplan JP, Fineberg HV, Ferraro MJ, Rosenberg ML. Value of stool cultures. Lancet 1980; 2 : 413–6. [DOI] [PubMed] [Google Scholar]
- 12. Guerrant RL, Wanke CA, Barrett LJ, Schwartzman JD. A cost effective and effective approach to the diagnosis and management of acute infectious diarrhea. Bull N Y Acad Med 1987; 63 : 484–99. [PMC free article] [PubMed] [Google Scholar]
- 13. Guerrant RL, Shields DS, Thorson SM, Schorling JB, Groschel DH. Evaluation and diagnosis of acute infectious diarrhea. Am J Med 1985; 78 : 91–8. [DOI] [PubMed] [Google Scholar]
- 14. Chhabra P, Payne DC, Szilagyi PG et al. . Etiology of viral gastroenteritis in children <5 years of age in the United States, 2008–2009. J Infect Dis 2013; 208 : 790–800. [DOI] [PubMed] [Google Scholar]
- 15. Platts-Mills JA, Operario DJ, Houpt ER. Molecular diagnosis of diarrhea: current status and future potential. Curr Infect Dis Rep 2012; 14 : 41–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Feudtner C, Silveira MJ, Christakis DA. Where do children with complex chronic conditions die? Patterns in Washington State, 1980–1998. Pediatrics 2002; 109 : 656–60. [DOI] [PubMed] [Google Scholar]
- 17. Poritz MA, Blaschke AJ, Byington CL et al. . FilmArray, an automated nested multiplex PCR system for multi-pathogen detection: development and application to respiratory tract infection. PloS One 2011; 6 : e26047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Blaschke AJ, Heyrend C, Byington CL et al. . Rapid identification of pathogens from positive blood cultures by multiplex polymerase chain reaction using the FilmArray system. Diagn Microbiol Infect Dis 2012; 74 : 349–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Payne DC, Staat MA, Edwards KM et al. . Active, population-based surveillance for severe rotavirus gastroenteritis in children in the United States. Pediatrics 2008; 122 : 1235–43. [DOI] [PubMed] [Google Scholar]
- 20. Siegler RL, Pavia AT, Christofferson RD, Milligan MK. A 20-year population-based study of postdiarrheal hemolytic uremic syndrome in Utah. Pediatrics 1994; 94 : 35–40. [PubMed] [Google Scholar]
- 21. Rousseau C, Poilane I, De Pontual L, Maherault AC, Le Monnier A, Collignon A. Clostridium difficile carriage in healthy infants in the community: a potential reservoir for pathogenic strains. Clin Infect Dis 2012; 55 : 1209–15. [DOI] [PubMed] [Google Scholar]
- 22. Schutze GE, Willoughby RE. Clostridium difficile infection in infants and children. Pediatrics 2013; 131 : 196–200. [DOI] [PubMed] [Google Scholar]
- 23. Bruijnesteijn van Coppenraet LE, Dullaert-de Boer M, Ruijs GJ et al. . Case-control comparison of bacterial and protozoan microorganisms associated with gastroenteritis: application of molecular detection. Clin Microbiol Infect 2015; 21 : 592e9–19. [DOI] [PubMed] [Google Scholar]
- 24. Sammons JS, Localio R, Xiao R, Coffin SE, Zaoutis T. Clostridium difficile infection is associated with increased risk of death and prolonged hospitalization in children. Clin Infect Dis 2013; 57 : 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sandora TJ, Fung M, Flaherty K et al. . Epidemiology and risk factors for Clostridium difficile infection in children. Pediatr Infect Dis J 2011; 30 : 580–4. [DOI] [PubMed] [Google Scholar]
- 26. Leibowitz J, Soma VL, Rosen L, Ginocchio CC, Rubin LG. Similar proportions of stool specimens from hospitalized children with and without diarrhea test positive for Clostridium difficile. Pediatr Infect Dis J 2015; 34 : 261–6. [DOI] [PubMed] [Google Scholar]
- 27. Dominguez SR, Dolan SA, West K et al. . High colonization rate and prolonged shedding of Clostridium difficile in pediatric oncology patients. Clin Infect Dis 2014; 59 : 401–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Daniels NA. Enterotoxigenic Escherichia coli: traveler’s diarrhea comes home. Clin Infect Dis 2006; 42 : 335–6. [DOI] [PubMed] [Google Scholar]
- 29. Payne DC, Vinje J, Szilagyi PG et al. . Norovirus and medically attended gastroenteritis in U.S. children. N Engl J Med 2013; 368 : 1121–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Koo HL, Ajami NJ, Jiang ZD et al. . A nosocomial outbreak of norovirus infection masquerading as Clostridium difficile infection. Clin Infect Dis 2009; 48 : e75–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Johnston CP, Qiu H, Ticehurst JR et al. . Outbreak management and implications of a nosocomial norovirus outbreak. Clin Infect Dis 2007; 45 : 534–40. [DOI] [PubMed] [Google Scholar]
- 32. Tran A, Talmud D, Lejeune B et al. . Prevalence of rotavirus, adenovirus, norovirus, and astrovirus infections and coinfections among hospitalized children in northern France. J Clin Microbiol 2010; 48 : 1943–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Cardine AM, Gourlain K, Mouterde O et al. . Epidemiology of acute viral gastroenteritis in children hospitalized in Rouen, France. Clin Infect Dis 2002; 34 : 1170–8. [DOI] [PubMed] [Google Scholar]
- 34. Khare R, Espy MJ, Cebelinski E et al. . Comparative evaluation of two commercial multiplex panels for detection of gastrointestinal pathogens by use of clinical stool specimens. J Clin Microbiol 2014; 52 : 3667–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.