Abstract
Background.
Biomarkers that identify critically ill children with systemic inflammatory response syndrome (SIRS) at low risk for bacterial infection may help clinicians reduce unnecessary antibiotic use.
Methods.
We conducted a prospective cohort study of children with SIRS and suspected infection admitted to a pediatric intensive care unit from January 5, 2012 to March 7, 2014. We enrolled patients upon initiation of new antibiotics (Time 0) and measured a panel of 8 serum biomarkers daily over 72 hours. Microbiology, imaging, and clinical data were reviewed to classify bacterial infections using Centers for Disease Control and Prevention definitions. We identified cut points of biomarker combinations to maximize the negative predictive value (NPV) and specificity for bacterial infection. Excess antibiotics were calculated as days of therapy beyond day 2 after SIRS onset in patients without bacterial infection.
Results.
Infections were identified in 46 of 85 patients: bacterial (n = 22) and viral (24), whereas 39 patients had no infection identified. At Time 0, C-reactive protein (CRP) <5 mg/dL plus serum amyloid A <15.0 µg/mL had an NPV of 0.92 (95% confidence interval [CI], 0.79–1.0) and specificity of 0.54 (95% CI, 0.42–0.66) to identify patients without bacterial infection, whereas CRP <4 mg/dL plus procalcitonin <1.75 ng/mL had an NPV of 0.90 (95% CI, 0.79–1.0) and specificity of 0.43 (95% CI, 0.30–0.55). Patients without bacterial infection received a mean of 3.8 excess days of therapy.
Conclusions.
Early measurement of select biomarkers can identify children with SIRS in whom antibiotics might be safely discontinued when there is no other objective evidence of infection at 48 hours.
Keywords: antimicrobial stewardship, biomarkers, critical care, pediatrics.
Sepsis is associated with significant morbidity and mortality in children [1–4], and as many as 1 in 4 critically ill children with severe sepsis die during hospitalization [5, 6]. Although prompt initiation of antimicrobial therapy is paramount to achieving favorable outcomes in patients with suspected sepsis [7, 8], timely discontinuation of antibiotics in children with viral or noninfectious causes of systemic inflammatory response syndrome (SIRS) is challenging. Many critically ill children with no evidence of bacterial infection receive antibiotic therapy, and overuse can lead to patient harm, development of antibiotic resistance, and unnecessary healthcare costs [9]. With up to 70% of children in US pediatric intensive care units (PICU) receiving antibiotics at any given time [10], reliable approaches are needed to ensure the safe discontinuation of antibiotics.
Biomarkers such as procalcitonin (PCT) and C-reactive protein (CRP) have been correlated with severity of illness [11]; they can guide antibiotic initiation, de-escalation, or discontinuation [12, 13]; and they distinguish pathogen type (bacterial vs nonbacterial) in patients with suspected sepsis [14]. In adults, biomarker-driven algorithms have reduced overall antibiotic use in patients with severe sepsis or septic shock [13, 15–19]. However, data on the role of these biomarkers in children are limited [12, 20], and few studies have assessed the performance of biomarkers in combination. We assessed the performance of a panel of serum biomarkers (individually and in combination) in identifying critically ill children with SIRS at low risk of bacterial infection. A secondary objective was to maximize the real-world applicability of the algorithm by focusing on the most parsimonious combination of laboratory markers that are currently available for rapid clinical decision making.
METHODS
Study Design and Population
We conducted a prospective, observational cohort study of children with new-onset SIRS admitted to the PICU at The Children's Hospital of Philadelphia from January 5, 2012 to March 7, 2014. Patients were approached for enrollment in the Emergency Department and PICU. Patients were eligible for enrollment if they were <18 years of age and met criteria for presumed sepsis: (1) presence of ≥2 age-related criteria for SIRS [21] and (2) a blood culture was drawn within 6 hours of SIRS onset; patients had to receive new intravenous antibiotic therapy within 4 hours of blood culture collection to be eligible. Patients with a positive blood culture within 24 hours before SIRS onset (including cultures drawn at outside institutions), fungal infection identified on culture, history of allogeneic bone marrow or solid organ transplant, absolute neutrophil count <500 cells/µL, advanced directives limiting care, or who were not admitted to the PICU after enrollment were excluded. Diagnostic testing for source of infection was performed at the discretion of the clinical team.
The hospital's institutional review board approved the study protocol. Informed consent or assent was obtained from patients and their parents or guardians, as appropriate. A waiver of consent was granted for the initial blood draw so that a baseline blood sample could be collected at the time of blood culture collection; patients' parents or guardians were approached for consent as soon as possible after the baseline blood sample was collected.
Biomarker Measurements
Blood samples were collected during routine clinical blood draws at the following time points for measurement of biomarkers: Time 0 (<4 hours after initiation or expansion of antibiotics), and 24 hours (±12 hours), 48 hours (±12 hours), and 72 hours (±12 hours) following Time 0. Samples were collected in a red top tube, allowed to clot for 30–60 minutes, and centrifuged at 3500 rpm for 10 minutes. Serum was stored at −70°C for batched analysis, and, thus, results were not shared with the clinical team.
Eight serum biomarkers were measured at each time point: α2-macroglobulin (A2 M), CRP, ferritin (FER), haptoglobin (HAP), PCT, serum amyloid A (SAA), serum amyloid P (SAP), and tissue plasminogen activator (TPA). These biomarkers were selected based on their role as acute-phase reactants and diagnostic markers of infection in prior studies [22–27]. Procalcitonin was measured using the VIDAS B.R.A.H.M.S. PCT assay (bioMérieux, Durham, North Carolina), which is a 1-step immunoassay sandwich method using fluorescent detection. The remaining 7 biomarkers were measured using the Bio-Plex Pro Human Acute Phase Multiplex Assay (Bio-Rad Laboratories, Hercules, California). This platform uses bead-based multiplex assays that allow for the measurement of multiple acute-phase biomarkers simultaneously. Measurements were performed according to the manufacturer's instructions with a Luminex 200 reader (Luminex Corporation, Austin, Texas). The range of detection for each biomarker assay was as follows: A2M, 0.5–1.875 ng/mL; CRP, 0.01–50 ng/mL; FER, 3.06–50 000 pg/mL; HAP, 0.1–500 ng/mL; PCT, 0.05–200 ng/mL; SAA, 1–700 ng/mL; SAP, 0.1–250 ng/mL; TPA, 28–5000 pg/mL.
Data Collection
After enrollment, we recorded information from the patient's electronic health record (EHR) including demographic information (eg, age, gender, race), hospital admission diagnosis, presence and category of complex chronic conditions [28], vital signs, and length of hospital and PICU stay before enrollment. Severity of illness on PICU admission was assessed using the Pediatric Risk of Mortality III Score (PRISM III) [29] and the Pediatric Index of Mortality score (PIM-2 ROM) [30]. All medications administered and procedures performed from 48 hours before and through 10 days after enrollment were abstracted from the EHR. Results of all microbiologic cultures and imaging evaluations within 10 days of study enrollment were also collected.
Definitions
The day of enrollment was considered day 0, and patients were observed through day 10. The presence of bacterial infection within 48 hours of enrollment was assessed by independent review of medical records (microbiology, laboratory, imaging, and clinical data) by 2 investigators (J. S. G. and S. E. C.) who were blinded to biomarker values. Bacterial infections were classified as present if Centers for Disease Control and Prevention (CDC) criteria for proven or probable bacterial infection were met [31]. A viral infection was defined as a laboratory-confirmed pathogen by polymerase chain reaction from a clinically obtained specimen. Antibiotic exposure was defined as days of therapy (DOT) and length of therapy (LOT) over the study. Days of therapy tabulates all antibiotics administered on each day separately, whereas LOT describes calendar days on which any antibiotic was administered [32]. Because a typical sepsis “rule-out” involves ≤48 hours of antibiotics, excess antibiotics were defined as those administered on days 3 through 10 in patients without bacterial infections.
Statistical Analysis
Comparisons were made between subjects with bacterial infections and those with either viral or no infection (no bacterial infection detected); patients with fungal infections were excluded. Clinical characteristics and biomarkers at each time point were compared between the 2 groups; the Wilcoxon rank-sum test was used for comparisons of continuous variables, whereas categorical variables were compared using the χ2 test. The area under the receiver operating characteristic (ROC) curve was determined using nonparametric ROC analyses for biomarkers at each time point. The mean (±standard deviation [SD]) number of excess antibiotic days was calculated for patients without bacterial infection.
We derived a biomarker-driven algorithm to identify patients at low risk for bacterial infection. Only biomarkers at Time 0 or 24 hours, which on univariate analysis had P values <.1 for association with infection (by Wilcoxon rank-sum test), were examined. We tested various biomarker combinations to maximize the negative predictive value (NPV) and specificity for bacterial infection. In our study, the NPV represents the probability that patients with both biomarker values below (or above if low values signaled infection) the defined cut points do not have a bacterial infection, whereas the specificity refers to the percentage of patients without bacterial infections detected are designated as test negative by the algorithm. Supplemental Figure 1 displays the 2×2 table used to calculate the test characteristics of biomarker combinations. The consensus of our study team was to only evaluate combinations with NPV of at least 88% because algorithms with a NPV below this would be less clinically applicable in the ICU setting. Of the biomarkers evaluated in this study, only CRP and PCT were routinely performed in our institution's laboratory in a timeframe necessary for clinical decision making about antimicrobial therapy. Hence, we also specifically tested various cut points of CRP and PCT combinations. Statistical analyses were performed using Stata 13.1 (StataCorp LP, College Station, Texas) and SAS version 9.3 (SAS Institute Inc., Cary, North Carolina).
RESULTS
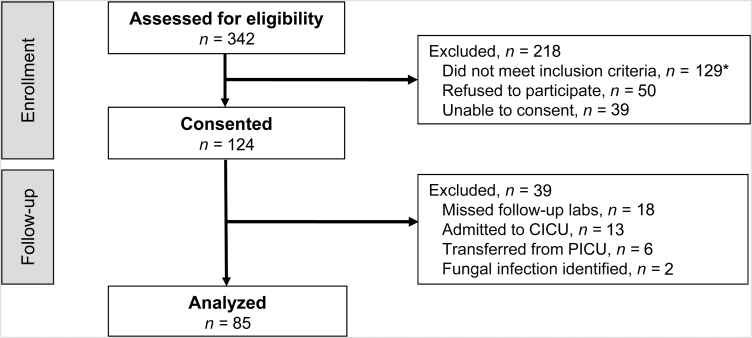
A total of 342 patients were screened and 124 were enrolled (Figure 1). Thirty-seven patients were excluded after enrollment because they did not have biomarkers drawn at 24 or 48 hours, were admitted to the cardiac ICU, or were transferred out of the PICU within 72 hours. Two additional patients were excluded due to presence of fungal infection. The final study cohort comprised 85 patients. Median age was 4.2 years (interquartile range, 1.1–11.6) and 37 (44%) were female (Table 1). Twenty-two patients (26%) had bacterial infections detected; 17 (77%) were definite and 5 (23%) probable infections. One patient with a bacterial infection had surgery 4 days before SIRS onset (zero in the other group). Table 2 displays the characteristics of the infections detected among the cohort.
Figure 1.
Flow chart of entry into the Optimizing Antibiotic Strategies in Sepsis (OASIS) cohort. *Systemic inflammatory response syndrome criteria not met (n = 24); previously enrolled (n = 19); not admitted to the pediatric intensive care unit (PICU) (n = 18); no new antibiotic (n = 16); Time 0 biomarker obtained >4 hours after antibiotic initiated (n = 16); positive blood culture in previous 24 hours (n = 8); do not resuscitate orders in place (n = 7); neutropenic (n = 7); no blood culture (n = 4); transplant (n = 3); no scheduled clinical laboratory tests (n = 5); age >18 years (n = 2). Abbreviation: CICU, cardiac intensive care unit.
Table 1.
Patient and Clinical Characteristics of the Study Population
| Characteristic | Bacterial Infection (n = 22, 26%) |
No Bacterial Infection Detected (n = 63, 74%) |
P Value |
|---|---|---|---|
| Age in years, median (range) | 10.5 (5.4–14.6) | 2.7 (0.8–10.1) | .01 |
| Gender, n (%) | .83 | ||
| Male | 12 (55) | 36 (57) | |
| Female | 10 (45) | 27 (43) | |
| Race, n (%) | .92 | ||
| Asian | 0 | 1 (2) | |
| Black | 9 (41) | 22 (23) | |
| White | 10 (45) | 29 (34) | |
| Unknown | 3 (14) | 11 (17) | |
| Ethnicity, n (%) | .02 | ||
| Hispanic/Latino | 0 | 14 (22) | |
| Non-Hispanic/Latino | 21 (95) | 44 (70) | |
| Unknown | 1 (5) | 5 (8) | |
| Presence of any CCC, n (%) | 13 (59) | 42 (67) | .52 |
| Pre-enrollment PICU length of stay in days, median (range) | 0 (0–2) | 0 (0–4) | .45 |
| Pediatric Risk of Mortality Score (PRISM III),a median (range) | 4 (2–12) | 5 (0–12) | .73 |
| Pediatric Index of Mortality (PIM-2 ROM),a median (range) | 1.4 (0.9–3.7) | 1.2 (0.8–4.4) | .54 |
| Mechanical ventilation,b n (%) | 11 (50) | 29 (46) | .75 |
| Endotracheal intubation | 9 (41) | 24 (38) | .82 |
| Tracheostomy | 2 (9) | 5 (8) | 1.0 |
| Medical device(s) in place,b n (%) | 18 (82) | 60 (95) | .07 |
| Central line | 17 (77) | 59 (94) | .05 |
| Urinary catheter | 14 (64) | 14 (58) | .78 |
| Chest tube | 2 (9) | 1 (2) | .16 |
| Receipt of vasopressor infusion,b n (%) | 7 (32) | 19 (30) | .88 |
| Vasopressor use in days,b median (range) | 0 (0–1) | 0 (0–2) | .86 |
| Underwent surgery,b n (%) | 2 (9) | 5 (8) | 1.0 |
| Final disposition from hospital, n (%) | .88 | ||
| Discharged | 18 (82) | 53 (84) | |
| Transferred to another facility | 2 (9) | 6 (10) | |
| Transferred to rehabilitation unit | 1 (5) | 1 (2) | |
| Died | 1 (5) | 3 (5) |
Abbreviations: CCC, complex chronic condition; PICU, pediatric intensive care unit.
aAt PICU admission.
bAt any time during 10 days after enrollment.
Table 2.
Infections Among the Cohort
| Type of infection | Number |
|---|---|
| Bacterial infections | 22 |
| Bloodstream infection | 7 |
| Staphylococcus aureus | 4 |
| Pseudomonas aeruginosa | 1 |
| Salmonella group B | 1 |
| Streptococcus pneumoniae | 1 |
| Lower respiratory tract infectiona | 8 |
| Wound infection | 2 |
| Proteus species | 1 |
| Streptococcus anginosis group | 1 |
| Otherb | 5 |
| Viral infectionsc | 24 |
| Respiratory tract infections | 22 |
| Blood | 1 |
| Stool | 1 |
aInfiltrate identified on chest radiography; no positive cultures.
bOne patient each had toxic shock syndrome, sinusitis with an epidural abscess, intra-abdominal abscess, Enterobacter cloacae urinary tract infection, and Clostridium difficile colitis.
cViral infections included the following: rhinovirus (7), adenovirus (7), influenza (4), respiratory syncytial virus (4), human metapneumovirus (1), and rotavirus (1).
Biomarkers were measured at Time 0 for all 85 patients, except for PCT, which was measured in 80 because of limited availability of sample. Median CRP was significantly higher at Time 0 and 24 hours in patients with bacterial infections detected (Table 3). Median TPA and SAA were higher at Time 0 in bacterial infection patients, and SAA was also higher at 72 hours in those with bacterial infections. Procalcitonin was higher in patients with bacterial infections at Time 0. Although this difference was not statistically significant, it met our a priori criteria for further evaluation in the combination biomarker algorithm derivation. α2-macroglobulin was lower in patients with bacterial infections at 24 and 48 hours. White blood cell count did not discriminate patients with and without bacterial infections detected (10.6 vs 13.8 cells/mm3, P = .29).
Table 3.
Comparison of Biomarkers Among Individuals With and Without Bacterial Infection Detected
| Biomarkera | Bacterial Infection (n = 22), Median (IQR) | No Bacterial Infection Detected (n = 63), Median (IQR) | Rank Sum P Value |
Area Under the ROC Curve |
|---|---|---|---|---|
| Procalcitonin (ng/mL) | ||||
| Time 0 | 5.95 (0.16–36.31) | 0.65 (0.20–5.04) | .06 | .641 |
| 24 hours | 6.67 (0.76–33.61) | 1.61 (0.31–17.89) | .16 | .614 |
| 48 hours | 3.81 (0.29–39.27) | 1.25 (0.21–12.63) | .40 | .568 |
| 72 hours | 0.97 (0.22–11.39) | 1.61 (0.11–7.17) | .78 | .526 |
| C-reactive protein (mg/dL) | ||||
| Time 0 | 8.7 (3.5–15.3) | 2.2 (0.4–5.8) | <.001 | .766 |
| 24 hours | 9.2 (5.5–16.8) | 3.7 (1.2–7.9) | .002 | .745 |
| 48 hours | 6.1 (2.7–11.8) | 2.8 (0.9–7.8) | .05 | .658 |
| 72 hours | 3.0 (1.8–5.4) | 1.8 (0.5–4.5) | .18 | .620 |
| α2-macroglobulin (mg/dL) | ||||
| Time 0 | 137.3 (109.3–178.1) | 161.5 (123.6–199.7) | .11 | .384 |
| 24 hours | 120.3 (92.5–154.8) | 157.0 (120.7–171.7) | .04 | .337 |
| 48 hours | 116.4 (104.1–127.5) | 150.6 (112.7–185.7) | .004 | .267 |
| 72 hours | 143.1 (80.7–161.4) | 153.1 (122.7–191.6) | .09 | .351 |
| Ferritin (ng/mL) | ||||
| Time 0 | 62.9 (30.0–176.2) | 45.7 (19.0–98.4) | .23 | .587 |
| 24 hours | 79.1 (49.6–228.0) | 53.0 (32.9–117.9) | .09 | .637 |
| 48 hours | 63.8 (42.2–194.6) | 46.6 (27.4–85.0) | .17 | .611 |
| 72 hours | 43.4 (34.2–80.4) | 46.8 (23.3–92.4) | .80 | .523 |
| Haptoglobin (mg/dL) | ||||
| Time 0 | 219.3 (46.0–452.5) | 103.4 (39.2–372.7) | .14 | .606 |
| 24 hours | 369.6 (94.3–498.0) | 106.7 (79.2–402.7) | .09 | .635 |
| 48 hours | 402.6 (144.5–498.0) | 294.0 (38.7–409.6) | .14 | .618 |
| 72 hours | 387.6 (68.3–498.0) | 227.0 (49.7–402.7) | .26 | .602 |
| Serum amyloid A (µg/mL) | ||||
| Time 0 | 18.9 (12.3–26.2) | 11.1 (4.3–22.7) | .009 | .688 |
| 24 hours | 17.8 (12.4–31.5) | 11.4 (7.0–20.5) | .09 | .638 |
| 48 hours | 14.2 (12.7–19.1) | 11.6 (6.1–21.9) | .21 | .602 |
| 72 hours | 11.7 (8.2–18.9) | 7.3 (1.2–11.9) | .03 | .698 |
| Serum amyloid P (ng/mL) | ||||
| Time 0 | 29.2 (22.5–44.3) | 26.7 (19.6–37.3) | .27 | .579 |
| 24 hours | 36.0 (22.5–44.8) | 26.1 (19.1–41.3) | .18 | .608 |
| 48 hours | 36.6 (33.0–43.6) | 30.3 (24.8–36.3) | .05 | .656 |
| 72 hours | 38.6 (28.5–50.3) | 28.4 (21.1–40.7) | .05 | .673 |
| Tissue plasminogen activator (ng/mL) | ||||
| Time 0 | 4.2 (2.9–8.0) | 2.7 (1.6–4.8) | .02 | .671 |
| 24 hours | 3.6 (1.7–7.1) | 2.3 (1.5–3.8) | .07 | .646 |
| 48 hours | 3.2 (2.0–4.1) | 2.4 (1.4–3.3) | .18 | .610 |
| 72 hours | 2.9 (1.5–4.1) | 1.9 (1.1–3.7) | .15 | .630 |
Abbreviations: IQR, interquartile range; ROC, receiver operating characteristic.
aBiomarkers performed at each time point: Time 0 = 85 (80 for procalcitonin), 24 hours = 64 (61 for procalcitonin), 48 hours = 71 (70 for procalcitonin), 72 hours = 58 (57 for procalcitonin). Procalcitonin was performed less often due to limited availability of sample.
Based on univariate analyses, we tested the utility of various combinations of CRP, PCT, SAA, and TPA at Time 0 to capture patients with SIRS without bacterial infection detected. Table 4 displays the optimal cut points identified for each combination of biomarkers at Time 0. Combinations of CRP, FER, A2 M, HAP, SAA, and TPA at 24 hours were also tested, but performance (NPV plus specificity) did not exceed Time 0 analyses (data not shown). Final combinations were limited to Time 0 because biomarkers best discriminated between patients with and without bacterial infection detected at this time point; missing data also limited analytical power at later time points. The biomarker combination that demonstrated the highest NPV plus specificity was a CRP cut point of 5.0 mg/dL plus SAA cut point of 15.0 µg/mL, which yielded a NPV of 0.92 (95% CI, 0.79–1.0) and specificity of 0.54 (95% CI, 0.42–0.66). This combination misclassified 3 infected patients (false negative) and captured 34 of 63 patients without infection. All 3 subjects with false- negative results had readily identifiable infections on clinical evaluation (cultures, imaging).
Table 4.
Optimal Cut Points for Biomarker Combinations at Time 0 to Determine Low Risk of Bacterial Infection
| Biomarkers | Cut Points | Negative Predictive Value (95% CI) | Specificity (95% CI) | Number of Patients Below Both Biomarker Cut Points | |
|---|---|---|---|---|---|
| Viral or No Infectiona (n = 63) | Bacterial Infectionb (n = 22) | ||||
| PCTc and SAA | PCT = 1.00 ng/mL SAA = 14.0 µg/mL |
0.96 (0.88–1.00) | 0.37 (0.24–0.49) | 23 | 1 |
| CRP and SAA | CRP = 5.0 mg/dL SAA = 15.0 µg/mL |
0.92 (0.83–1.00) | 0.54 (0.42–0.66) | 34 | 3 |
| CRP and PCTc | CRP = 4.0 mg/dL PCT = 1.75 ng/mL |
0.90 (0.79–1.00) | 0.43 (0.30–0.55) | 27 | 3 |
| CRP and TPA | CRP = 5.0 mg/dL TPA = 3.75 ng/mL |
0.89 (0.78–0.99) | 0.49 (0.37–0.62) | 31 | 4 |
| PCTc and TPA | PCT = 1.00 ng/mL TPA = 3.0 ng/mL |
0.88 (0.75–1.00) | 0.35 (0.23–0.47) | 22 | 3 |
| SAA and TPA | SAA = 14.0 µg/mL TPA = 3.25 ng/mL |
0.88 (0.77–0.99) | 0.48 (0.35–0.60) | 30 | 4 |
Abbreviations: CI, confidence interval; CRP, C-reactive protein; PCT, procalcitonin; SAA, serum amyloid A; TPA, tissue plasminogen activator.
aNumber of patients without bacterial infection detected who were test negative (true negative).
bNumber of patients who were test negative but had a bacterial infection detected (false negative).
cEighty of 85 patients had PCT performed at Time 0.
Because SAA is not currently available for rapid clinical decision making at most institutions, we next focused on CRP and PCT. When using CRP and PCT, a CRP cut point of 4 mg/dL combined with a PCT cut point of 1.75 ng/mL yielded a NPV of 0.90 (95% CI, 0.79–1.0); this combination identified 24 of 63 patients without bacterial infection detected (specificity 0.43; 95% CI, 0.30–0.55). All 3 patients with bacterial infections who were misclassified as low risk using these PCT and CRP cut points had identifiable bacterial infections diagnosed by standard radiographic studies performed within the initial 48 hours after SIRS onset, including 1 each with community-acquired pneumonia, sinusitis, and hospital-acquired pneumonia. Both the SAA-CRP and PCT-CRP algorithms accurately captured all patients with bacteremia.
The most commonly prescribed antibiotics included vancomycin (83%), cefepime (47%), and cefotaxime (18%). Twenty-five patients (29%) received 1 or more antimicrobials on the day before enrollment and qualified for inclusion in the study due to an addition of antibiotics: 5 of 22 (23%) with and 20 of 63 (32%; P = .42) without bacterial infections detected. Sixty percent of patients without bacterial infections detected received antibiotics beyond day 2. There were a total of 152 days of excess antimicrobials in these patients. Patients without bacterial infection detected had a mean excess LOT and DOT of 2.4 (SD = 2.7) and 3.8 (SD = 4.9) days per patient, respectively. Excess antimicrobial exposure was shorter for patients with viral infections compared with those without any infection detected: mean excess LOT was 1.5 vs 3.0 days per patient (P = .03), and excess DOT was 2.3 vs 4.7 days per patient (P = .05). Application of the CRP-PCT biomarker algorithm might have reduced the total number of DOT by 73 days: 11% of all DOT in patients without bacterial infections detected. Application of the CRP-SAA biomarker algorithm might have decreased DOT by 115 days: 18% of all DOT in patients without bacterial infections detected.
DISCUSSION
We prospectively evaluated the performance of a panel of serum biomarkers to identify a group of critically ill children with SIRS for whom antibiotics may be safely discontinued in the absence of a clinically identifiable bacterial infection. Although the absence of infection might seem a sufficient basis on which to discontinue antibiotics, approximately half of all antibiotics administered over the study period were for patients in whom there was no evidence for bacterial infection, which is consistent with prior studies [33]. This practice resulted in an excess LOT and DOT of 2.4 and 3.8 days per patient, respectively. Application of the CRP-SAA algorithm could have reduced excess DOT by approximately 50% in patients without bacterial infections detected. The few patients with bacterial infections misclassified as low risk by the CRP-PCT and CRP-SAA algorithms were appropriately identified using standard diagnostic testing, reinforcing that any biomarker algorithm is most useful when applied as an adjunctive measure to support decision making in the face of uncertainty.
The best combination of biomarkers to identify patients at low risk of bacterial infection was CRP ≤5.0 mg/dL plus SAA ≤15 µg/mL, which had the highest specificity and NPV combination. Studies are needed to validate these cut points and test their ability to influence antibiotic prescribing in the PICU. Final combinations were limited to Time 0 because this time point (1) showed the greatest capacity to distinguish patients with and without bacterial infection detected, (2) was when blood sampling was universally performed as part of clinical care, and (3) ensured biomarker data were available at the time culture data were being interpreted and clinical decisions about prolonged antimicrobial therapy were commonly made.
We also derived an algorithm using only CRP plus PCT, which are more routinely available laboratory biomarkers that can be rapidly analyzed using standard equipment in most clinical laboratories. Studies support the use of CRP and PCT for the timing of antibiotic discontinuation among adults being treated for known or presumed bacterial sepsis [34]. Our algorithm differs in that we focused on decisions made at 48 hours during a sepsis “rule-out,” designed to determine whether the etiology of SIRS was due to bacterial infections (as opposed to tailoring the duration of therapy). Although this algorithm misclassified 3 children as low risk, each had a readily identifiable infection based on clinical and radiographic evaluations. Therefore, as an adjunct to standard clinical care, we believe that early measurement of CRP and PCT can help identify critically ill children with SIRS at low risk of bacterial infection and promote discontinuation of antibiotics at 48 hours.
Many patients without bacterial infections detected continued on antibiotics beyond 48 hours, even in the setting of laboratory-confirmed viral infection. Most viral infections were respiratory, and it is likely that clinicians felt uncomfortable attributing SIRS to viral respiratory tract infections, consistent with previous reports [35]. Nonetheless, children with viral infections received fewer excess antibiotics than patients with no infections detected, highlighting the benefit of diagnosing viral infections in the PICU. Adjunctive tests, such as biomarkers, can further assist clinicians in antibiotic decision making.
This was an observational study, and results of biomarkers were not made available to the patients' clinical decision makers; however, CRP and PCT could be separately ordered during routine care by the treating clinician. In addition, our institution has an antimicrobial stewardship program (ASP) that requires prior authorization for antibiotic use beyond 48 hours. However, it is unlikely that this influenced antibiotic prescribing specifically for patients in our study, because ASP team members were not made aware of enrolled patients. In addition, because we measured antibiotic use during an ASP-regulated window (beyond 48 hours), (1) the potential impact of this biomarker algorithm on reducing antimicrobial use in centers without ASPs could be even more profound, and (2) ASP practices were insufficient to completely limit unnecessary antibiotic use, suggesting that use of biomarkers may enhance current stewardship efforts.
Our study has several limitations. The performance of the biomarkers in our study was modest because the areas under each of the ROC curves were less than 0.75, likely influenced by our relatively small sample size. However, biomarker values may be impacted differently based on the severity and site (ie, bloodstream, urinary tract, lower respiratory tract, etc) of bacterial infection. This heterogeneity among the bacterial infection group might have biased results towards the null for certain biomarkers. Biomarkers were drawn at times of clinical blood draws, and, thus, several patients had missing values at time points beyond Time 0. Although there were no differences between groups in the proportion of patients with missing biomarkers, the ability of biomarkers to identify patients with bacterial infections after Time 0 might have been impacted by sample availability. We did not exclude patients receiving antibiotics before enrollment as long as they had an expansion or change in antibiotic therapy at SIRS onset; however, the number who received antibiotics before enrollment were similar between study groups. In addition, we found no significant differences in any of the biomarkers based on receipt of antibiotics before enrollment (data not shown). Finally, several factors contributed to a risk of misclassification of infection. Because this was an observational study, patients did not undergo a standardized workup for infection, raising the risk of underdetection of potential infections. In addition, we used CDC surveillance definitions for infection, which provide a structured schema for infection identification and classification but do not always align perfectly with clinicians' diagnoses of infection.
CONCLUSIONS
In conclusion, biomarker measurement at SIRS onset can help identify critically ill children at low risk of bacterial infection in whom antibiotics may be safely discontinued. As an adjunct to standard diagnostic tests and routine clinical evaluations, measurement of CRP in combination with SAA or PCT may help to optimize antibiotic utilization for critically ill children with SIRS by reducing unnecessary therapy in a subset of low-risk patients. Future studies are needed to confirm the utility of our defined cut points.
Supplementary Data
Supplementary materials are available at Journal of the Pediatric Infectious Diseases Society online.
Supplementary Material
Notes
Disclaimer. The funding agencies had no role in the design and conduct of the study, collection, management, analysis, and interpretation of the data, or preparation, review, or approval of the manuscript.
Financial support. This work was supported by the Centers for Disease Control and Prevention Cooperative Agreement under FOA CK11-001, Epicenters for the Prevention of Healthcare Associated Infections (to E. L.). F. B. received career development support from the National Institutes of Health National Heart, Lung, and Blood Institute grant K12-HL109009 and Eunice Kennedy Shriver National Institute of Child Health and Human Development grant 1K23HD082368.
Potential conflicts of interest.All authors: No reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Schlapbach LJ , Straney L , Alexander J , et al. Mortality related to invasive infections, sepsis, and septic shock in critically ill children in Australia and New Zealand, 2002–13: a multicentre retrospective cohort study. Lancet Infect Dis 2015; 15:46–54. [DOI] [PubMed] [Google Scholar]
- 2. Aspesberro F , Mangione-Smith R , Zimmerman JJ. Health-related quality of life following pediatric critical illness. Intensive Care Med 2015; 41:1235–46. [DOI] [PubMed] [Google Scholar]
- 3. Hartman ME , Linde-Zwirble WT , Angus DC , Watson RS. Trends in the epidemiology of pediatric severe sepsis. Pediatr Crit Care Med 2013; 14:686–93. [DOI] [PubMed] [Google Scholar]
- 4. Ruth A , McCracken CE , Fortenberry JD , et al. Pediatric severe sepsis: current trends and outcomes from the Pediatric Health Information Systems database. Pediatr Crit Care Med 2014; 15:828–38. [DOI] [PubMed] [Google Scholar]
- 5. Weiss SL , Fitzgerald JC , Pappachan J , et al. Global epidemiology of pediatric severe sepsis: the sepsis prevalence, outcomes, and therapies study. Am J Respir Crit Care Med 2015; 191:1147–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Balamuth F , Weiss SL , Neuman MI , et al. Pediatric severe sepsis in U.S. children's hospitals. Pediatr Crit Care Med 2014; 15:798–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Weiss SL , Fitzgerald JC , Balamuth F , et al. Delayed antimicrobial therapy increases mortality and organ dysfunction duration in pediatric sepsis. Crit Care Med 2014; 42:2409–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kumar A , Roberts D , Wood KE , et al. Duration of hypotension before initiation of effective antimicrobial therapy is the critical determinant of survival in human septic shock. Crit Care Med 2006; 34:1589–96. [DOI] [PubMed] [Google Scholar]
- 9. Centers for Disease Control and Prevention (CDC). Antibiotic resistance threats in the United States, 2013. Atlanta: CDC , 2013. Available at: http://www.cdc.gov/drugresistance/threat-report-2013/pdf/ar-threats-2013-508.pdf. [Google Scholar]
- 10. Grohskopf LA , Huskins WC , Sinkowitz-Cochran RL , et al. Use of antimicrobial agents in United States neonatal and pediatric intensive care patients. Pediatr Infect Dis J 2005; 24:766–73. [DOI] [PubMed] [Google Scholar]
- 11. Park IH , Lee SH , Yu ST , Oh YK. Serum procalcitonin as a diagnostic marker of neonatal sepsis. Korean J Pediatr 2014; 57:451–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Stocker M , Fontana M , El Helou S , et al. Use of procalcitonin-guided decision-making to shorten antibiotic therapy in suspected neonatal early-onset sepsis: prospective randomized intervention trial. Neonatology 2010; 97:165–74. [DOI] [PubMed] [Google Scholar]
- 13. Bouadma L , Luyt CE , Tubach F , et al. Use of procalcitonin to reduce patients' exposure to antibiotics in intensive care units (PRORATA trial): a multicentre randomised controlled trial. Lancet 2010; 375:463–74. [DOI] [PubMed] [Google Scholar]
- 14. Kofoed K , Andersen O , Kronborg G , et al. Use of plasma C-reactive protein, procalcitonin, neutrophils, macrophage migration inhibitory factor, soluble urokinase-type plasminogen activator receptor, and soluble triggering receptor expressed on myeloid cells-1 in combination to diagnose infections: a prospective study. Crit Care 2007; 11:R38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hohn A , Schroeder S , Gehrt A , et al. Procalcitonin-guided algorithm to reduce length of antibiotic therapy in patients with severe sepsis and septic shock. BMC Infect Dis 2013; 13:158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wilke MH , Grube RF , Bodmann KF. The use of a standardized PCT-algorithm reduces costs in intensive care in septic patients - a DRG-based simulation model. Eur J Med Res 2011; 16:543–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Schroeder S , Hochreiter M , Koehler T , et al. Procalcitonin (PCT)-guided algorithm reduces length of antibiotic treatment in surgical intensive care patients with severe sepsis: results of a prospective randomized study. Langenbecks Arch Surg 2009; 394:221–6. [DOI] [PubMed] [Google Scholar]
- 18. Kopterides P , Siempos II , Tsangaris I , et al. Procalcitonin-guided algorithms of antibiotic therapy in the intensive care unit: a systematic review and meta-analysis of randomized controlled trials. Crit Care Med 2010; 38:2229–41. [DOI] [PubMed] [Google Scholar]
- 19. Albrich WC , Harbarth S. Pros and cons of using biomarkers versus clinical decisions in start and stop decisions for antibiotics in the critical care setting. Intensive Care Med 2015; 41:1739–51. [DOI] [PubMed] [Google Scholar]
- 20. Baer G , Baumann P , Buettcher M , et al. Procalcitonin guidance to reduce antibiotic treatment of lower respiratory tract infection in children and adolescents (ProPAED): a randomized controlled trial. PLoS One 2013; 8:e68419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Goldstein B , Giroir B , Randolph A. International pediatric sepsis consensus conference: definitions for sepsis and organ dysfunction in pediatrics. Pediatr Crit Care Med 2005; 6:2–8. [DOI] [PubMed] [Google Scholar]
- 22. Enguix A , Rey C , Concha A , et al. Comparison of procalcitonin with C-reactive protein and serum amyloid for the early diagnosis of bacterial sepsis in critically ill neonates and children. Intensive Care Med 2001; 27:211–5. [DOI] [PubMed] [Google Scholar]
- 23. Chavez-Bueno S , Beasley JA , Goldbeck JM , et al. ‘Haptoglobin concentrations in preterm and term newborns’. J Perinatol 2011; 31:500–3. [DOI] [PubMed] [Google Scholar]
- 24. Garcia PC , Longhi F , Branco RG , et al. Ferritin levels in children with severe sepsis and septic shock. Acta Paediatr 2007; 96:1829–31. [DOI] [PubMed] [Google Scholar]
- 25. de Boer JP , Creasey AA , Chang A , et al. Alpha-2-macroglobulin functions as an inhibitor of fibrinolytic, clotting, and neutrophilic proteinases in sepsis: studies using a baboon model. Infect Immun 1993; 61:5035–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Noursadeghi M , Bickerstaff MC , Gallimore JR , et al. Role of serum amyloid P component in bacterial infection: protection of the host or protection of the pathogen. Proc Natl Acad Sci U S A 2000; 97:14584–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Winkler F , Kastenbauer S , Koedel U , Pfister HW. Increased serum concentrations of tissue plasminogen activator correlate with an adverse clinical outcome in patients with bacterial meningitis. J Neurol Neurosurg Psychiatry 2002; 73:456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Feudtner C , Hays RM , Haynes G , et al. Deaths attributed to pediatric complex chronic conditions: national trends and implications for supportive care services. Pediatrics 2001; 107:E99. [DOI] [PubMed] [Google Scholar]
- 29. Pollack MM , Patel KM , Ruttimann UE. PRISM III: an updated Pediatric Risk of Mortality score. Crit Care Med 1996; 24:743–52. [DOI] [PubMed] [Google Scholar]
- 30. Slater A , Shann F , Pearson G. PIM2: a revised version of the Paediatric Index of Mortality. Intensive Care Med 2003; 29:278–85. [DOI] [PubMed] [Google Scholar]
- 31. Horan TC , Andrus M , Dudeck MA. CDC/NHSN surveillance definition of health care-associated infection and criteria for specific types of infections in the acute care setting. Am J Infect Control 2008; 36:309–32. [DOI] [PubMed] [Google Scholar]
- 32. Morris AM. Antimicrobial Stewardship Programs: appropriate measures and metrics to study their impact. Curr Treat Options Infect Dis 2014; 6:101–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Blinova E , Lau E , Bitnun A , et al. Point prevalence survey of antimicrobial utilization in the cardiac and pediatric critical care unit. Pediatr Crit Care Med 2013; 14:e280–8. [DOI] [PubMed] [Google Scholar]
- 34. Oliveira CF , Botoni FA , Oliveira CR , et al. Procalcitonin versus C-reactive protein for guiding antibiotic therapy in sepsis: a randomized trial. Crit Care Med 2013; 41:2336–43. [DOI] [PubMed] [Google Scholar]
- 35. Mulpuru S , Aaron SD , Ronksley PE , et al. Hospital resource utilization and patient outcomes associated with respiratory viral testing in hospitalized patients. Emerg Infect Dis 2015; 21:1366–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.