In multivariable analysis, prolonged antimicrobial treatment was a predictor of infant colonization with antimicrobial-resistant Gram-negative bacilli within 7 days of discharge from a neonatal intensive care unit.
Keywords: antimicrobial resistance, cephalosporin agents, gentamicin, meropenem, risk factors
Abstract
Background
The epidemiology of the colonization of infants with antimicrobial-resistant Gram-negative bacilli (GNB) at discharge from the neonatal intensive care unit (NICU) is not well understood.
Methods
A multicenter study in which rectal surveillance samples for culture were obtained at NICU discharge from infants hospitalized ≥14 days was performed. Factors associated with colonization with GNB resistant to gentamicin, third/fourth-generation cephalosporin agents, or carbapenem agents were assessed by using a fixed-effects model.
Results
Of these infants, 9% (119 of 1320) were colonized with ≥1 antimicrobial-resistant GNB. Prolonged treatment (≥10 days) with meropenem or third/fourth-generation cephalosporin agents or treatment for ≥5 days with a β-lactam/β-lactamase combination agent were associated with an increased risk of colonization with GNB resistant to gentamicin. Surgery and ≥5 days of treatment with third/fourth-generation cephalosporin agents, a β-lactam/β-lactamase combination agent, or metronidazole were associated with an increased risk of colonization with GNB resistant to third/fourth-generation cephalosporin agents. Female sex and prolonged treatment (≥10 days) with meropenem were associated with colonization with GNB resistant to carbapenem agents.
Conclusions
Prolonged treatment with broad-spectrum antibiotics was associated with the colonization of infants with antimicrobial-resistant GNB within 7 days of NICU discharge. These findings suggest the potential for dissemination of resistant GNB from colonized infants to other NICUs, the community, or pediatric long-term care facilities. Antimicrobial stewardship efforts aimed at improving appropriate antibiotic use could have a beneficial effect on the emergence of antimicrobial-resistant GNB in the NICU population.
INTRODUCTION
Infants in the neonatal intensive care unit (NICU) frequently receive antimicrobial agents for suspected or confirmed infections and as perioperative prophylaxis. Overall antimicrobial use and the use of broad-spectrum cephalosporin and carbapenem agents have been shown to be associated with colonization and/or infections caused by antimicrobial-resistant Gram-negative bacilli (GNB) [1, 2]. When compared with healthy term infants, colonization of the gastrointestinal (GI) tract of infants in the NICU is delayed, and pathogenic GNB are more common. These differences are likely results of multiple factors, including prematurity, contact with healthcare personnel, use of parenteral nutrition, and exposure to broad-spectrum antimicrobial agents [3–5]. Furthermore, GNB that colonize the GI tract have been identified as the source of subsequent bloodstream infections [6, 7]. However, little is known about the epidemiology of the colonization of infants with antimicrobial-resistant GNB at NICU discharge [8–10].
The objectives of this multicenter study were to examine the rates of GI tract colonization with GNB resistant to selected agents, to determine risk factors associated with colonization, and to assess the concordance of previous healthcare-associated infections (HAIs) caused by antimicrobial-resistant GNB with colonization at NICU discharge. We hypothesized that selective pressure caused by prolonged antimicrobial treatment with broad-spectrum agents would be associated with colonization at NICU discharge with GNB resistant to gentamicin, third/fourth-generation cephalosporin agents, and/or the carbapenem agent meropenem.
MATERIALS AND METHODS
Study Design, Study Sites, and Eligible Infants
This substudy was part of a larger prospective multicenter research study, “Improving Antimicrobial Prescribing Practices in the Neonatal Intensive Care Unit,” performed from May 2009 to April 2012. The study sites included 4 academically affiliated level III NICUs with 247 beds (50–75 per site) and approximately 4500 annual discharges (830–1400 per site). The NICUs were a combination of 2- to 3-bed pods, open units of 8 to 12 beds, and single isolation rooms. Three of the study sites (sites 1, 2, and 4) used vancomycin and gentamicin for empiric therapy for late-onset sepsis, and 1 site (site 3) used vancomycin and cefepime. Site 3 routinely performed chlorhexidine gluconate (CHG) bathing, and sites 1, 2, and 4 used CHG bathing selectively on infants who weighed >1500 g to decolonize them of methicillin-resistant Staphylococcus aureus. Information on the use of CHG was not collected for individual infants in the study. The institutional review board at each participating center approved this study and waived the requirement for documentation of written informed consent. The parents of eligible infants were provided with an information sheet that described the purpose of the study and were given the option to contact the study team to opt out of the substudy.
For this substudy, eligible infants (1) were admitted to a study NICU at <7 days of age, (2) were hospitalized for ≥14 days, and (3) had 3 surveillance cultures performed within 7 days before NICU discharge. These surveillance cultures were performed on a single perirectal swab analyzed for the presence of GNB and enterococci and on 2 swabs for S aureus (anterior nares and skin sites).
Microbiology Methods
Each surveillance swab was obtained and placed in a liquid Amies swab-transport system (bioMérieux, Durham, North Carolina, and Becton Dickinson, Franklin Lakes, New Jersey). The swabs were shipped to the core laboratory at Columbia University Medical Center. For this substudy, rectal swabs were cultured on MacConkey agar (Becton Dickinson) to selectively detect GNB (1–2 predominant morphotypes). The Vitek 2 system (bioMérieux) was used for identification and susceptibility testing of GNB isolates as detailed below. Susceptibility results from the Vitek 2 system were unavailable for Pantoea spp., so Kirby–Bauer antimicrobial discs (Becton Dickinson) were used to test this species. Results were interpreted using 2012 Clinical Laboratory and Standards Institute break points for minimum inhibitory concentration (MIC) interpretive standards and zone-diameter break points [11].
Definition of Antimicrobial-Resistant GNB
GNB isolates were defined as resistant to a selected agent if the MIC or zone diameter was interpreted as intermediate or resistant. Gentamicin-resistant GNB were not susceptible to gentamicin (MIC > 4 μg/ml). For Pseudomonas aeruginosa, third/fourth-generation cephalosporin-resistant P. aeruginosa isolates were not susceptible to ceftazidime (MIC > 8 μg/ml) and/or cefepime (MIC > 16 μg/ml). For non-P. aeruginosa GNB, third/fourth-generation cephalosporin-resistant isolates were not susceptible to cefotaxime, ceftazidime, ceftriaxone, and/or cefepime (Enterobacteriaceae MICs, >1 μg/ml for cefotaxime/ceftriaxone, >8 μg/ml for ceftazidime, and >16 μg/ml for cefepime; Acinetobacter species and other non-Enterobacteriaceae MICs, >8 μg/ml for cefotaxime/ceftazidime/ceftriaxone and >16 μg/ml for cefepime). Carbapenem-resistant GNB were not susceptible to imipenem and/or meropenem (P. aeruginosa MIC, >2 μg/ml; Enterobacteriaceae MIC, >1 μg/ml; Acinetobacter species and other non-Enterobacteriaceae MIC, >4 μg/ml). Potential clusters were identified; clusters were defined as ≥3 antimicrobial-resistant GNB (of any species) collected within 2 weeks of another GNB resistant to an antimicrobial agent in the same category.
Risk Factors for Colonization With Antimicrobial-Resistant GNB
The following risk factors for colonization with antimicrobial-resistant GNB were assessed: demographic characteristics (eg, sex, race), clinical interventions (eg, surgery, mechanical ventilation), and antimicrobial treatment. We evaluated selected agents and selected classes of agents, including (1) the selected individual agents cefazolin, meropenem, metronidazole, and vancomycin; (2) penicillin agents (ie, ampicillin and/or penicillin); (3) aminoglycoside agents (ie, amikacin, gentamicin, and/or tobramycin); (4) β-lactam/β-lactamase combination agents (ie, ampicillin-sulbactam, piperacillin-tazobactam, and/or ticarcillin-clavulanate); and (5) third/fourth-generation cephalosporin agents (ie, cefepime, cefotaxime, ceftazidime, and/or ceftriaxone). Agents or classes of agents (eg, rifampin) used to treat <5 infants across the 4 study sites were not evaluated.
HAIs With Antimicrobial-Resistant GNB
Clinical samples from the infants were obtained for culture at the discretion of the treating clinicians and processed at the study sites per standard care. Each site's research personnel reviewed the medical records of the enrolled infants for HAIs, which were defined as infections diagnosed by an attending neonatologist at ≥4 days of age and treated with intravenous antibiotics for ≥1 day. The antimicrobial susceptibilities of the GNB associated with HAIs were documented to identify antimicrobial-resistant GNB.
Statistical Analysis
Demographic and clinical variables were assessed as predictors of colonization with antimicrobial-resistant GNB. Treatment regimens of individual agents or antimicrobial classes were explored as predictors of colonization by using continuous (eg, number of days of treatment) and categorical variables. After preliminary modeling of antibiotic utilization parameters, the treatment variables of ≥5 continuous antibiotic-days or ≥10 continuous antibiotic-days showed the strongest associations with the study outcomes compared with other measures of antibiotic treatment (eg, number of days of treatment [data not shown]). Furthermore, treatment courses of ≥5 and ≥10 days are commonly used in clinical practice. We performed the χ2 test for categorical variables and the Wilcoxon rank-sum test for continuous variables. Variables found to be significant (P < .2) in unadjusted bivariate analysis were selected as predictors for multivariable models. To avoid a problem of colinearity in the multivariable models, if both ≥5 and ≥10 antibiotic-days of treatment with individual agents or antimicrobial classes were significantly associated with colonization in the bivariate analysis, then the duration with the smaller P value was used in multivariable analyses; if both durations had the same P value, ≥10 days was used. Variance inflation factors were calculated for all the models to ensure that the models did not have other problems with colinearity.
In constructing the multivariable models, we did not use traditional regression approaches because each study site had inherent unmeasured differences, and the outcomes of the subjects at the same site might have been correlated and violated independence assumptions made by traditional regression procedures. We attempted to use a random-effects model (conditional model), but the models did not converge (data not shown). Therefore, we used a fixed-effects model, which is a conditional model that can adjust for differences between sites, and we treated study site as a fixed effect [12, 13]. The final models included significant (P < .05) variables. Statistical analysis was performed by using SAS 9.3 for Windows (SAS Institute, Inc., Cary, North Carolina).
RESULTS
Characteristics of Study Infants
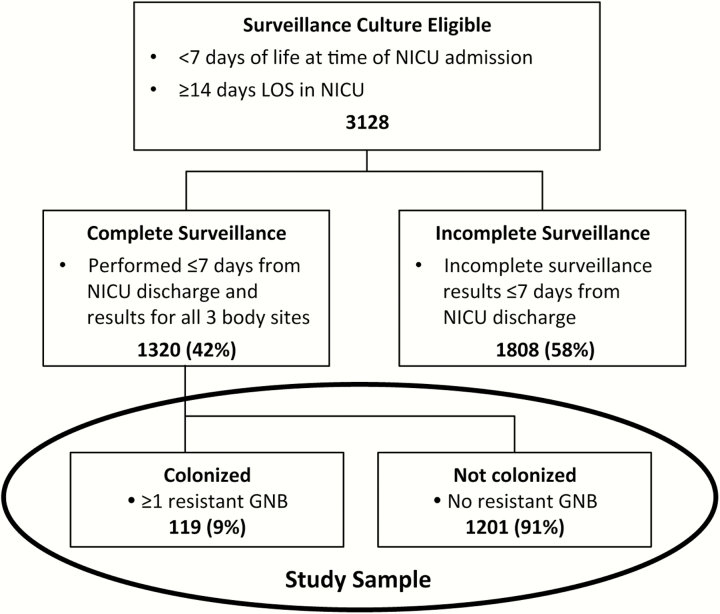
During the study period, 3128 infants met the substudy eligibility criteria for surveillance cultures: admitted to the NICU at <7 days of age and hospitalized for ≥14 days (Figure 1). Of these infants, surveillance culture samples were not obtained for 1808: 506 (16%) infants died or had a prohibitive medical condition, social service issue, language barrier, or parental refusal, and 1302 (42%) infants were excluded because incomplete surveillance cultures. Thus, 1320 (42%) infants had complete surveillance cultures and were included in this substudy sample. Infants without surveillance cultures had a shorter length of NICU stay and higher mean birth weight when compared with infants with surveillance cultures from the same study site (data not shown).
Figure 1.
Numbers of subjects from the multicenter prospective study used in the analysis of infant colonization with antimicrobial-resistant GNB within 7 days of NICU discharge. The study sample included 1320 infants with a complete set of 3 surveillance swabs: 1 swab of the anterior nares for S aureus, 1 swab of the skin for S aureus, and 1 perirectal swab for GNB and enterococci (results of testing for S aureus and enterococci are not provided). Abbreviations: GNB, Gram-negative bacilli; LOS, length of stay; NICU, neonatal intensive care unit.
The demographic characteristics and length of stay of infants from each site are shown in Table 1. Differences in demographic characteristics reflected the referral patterns and local catchment areas of the study sites. The proportions of infants (range, 15%–34%) and patient-days (range, 17%–34%) contributed by each site were similar. Surgery, mostly cardiac or GI procedures, was performed at 3 sites (sites 1, 2, and 3). There were no in-hospital deaths among the infants included in this substudy.
Table 1.
Demographic and Clinical Characteristics of Infants According to Study Site
| Characteristic | Study Sitea | P Valuesb | |||
|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | ||
| Subjects | 443 | 341 | 204 | 332 | .98 |
| Gestational age (mean [SD]) (wk) | 32 (5) | 32 (3) | 36 (3) | 31 (4) | <.01 |
| Birth weight (mean [SD]) (g)c | 1814 (996) | 1692 (658) | 2575 (779) | 1587 (728) | <.01 |
| Birth weight strata(n [row %])c | <.01 | ||||
| ≥2500 g | 129 (29.1) | 32 (9.4) | 116 (56.9) | 31 (9.3) | |
| 1500–2499 g | 94 (21.2) | 169 (49.6) | 67 (32.8) | 132 (39.8) | |
| 1000–1499 g | 104 (23.5) | 98 (28.7) | 15 (7.4) | 103 (31.0) | |
| <1000 g | 116 (26.2) | 42 (12.3) | 6 (2.9) | 66 (19.9) | |
| Sex | .06 | ||||
| Male | 253 (57.1) | 169 (49.6) | 98 (48.0) | 185 (55.7) | |
| Female | 190 (42.9) | 172 (50.4) | 106 (52.0) | 147 (44.3) | |
| Race | <.01 | ||||
| White | 210 (47.4) | 118 (34.6) | 106 (52.0) | 200 (60.3) | |
| Black | 41 (9.3) | 23 (6.7) | 16 (7.8) | 114 (34.3) | |
| Otherd | 182 (41.1) | 44 (12.9) | 81 (39.7) | 10 (3.0) | |
| Unknown | 10 (2.2) | 156 (45.8) | 1 (0.5) | 8 (2.4) | |
| Ethnicity | <.01 | ||||
| Hispanic | 105 (23.7) | 14 (4.1) | 11 (5.4) | 15 (4.5) | |
| Non-Hispanic | 166 (37.5) | 179 (52.5) | 130 (63.7) | 310 (93.4) | |
| Unknown | 172 (38.8) | 148 (43.4) | 63 (30.9) | 7 (2.1) | |
| Length of stay (mean [SD]) (days) | 47 (36) | 45 (38) | 51 (47) | 44 (30) | .50 |
| Patient-days | 20 695 | 15 229 | 10 505 | 14 644 | .98 |
Abbreviation: SD, standard deviation.
aData are numbers (column percent) unless otherwise indicated.
b P values are reported for differences according to site using the χ2 test for categorical variables and the Wilcoxon rank-sum test for continuous variables.
cBirth weight was unavailable for 1 infant.
dIncludes Asian, Native Hawaiian, other Pacific Islander, and other.
GNB Colonization Within 7 Days of Discharge
The GNB isolated from surveillance cultures of samples obtained within 7 days of discharge are shown in Table 2. Overall, 9% (n = 119) of the infants harbored ≥1 antimicrobial-resistant GNB within 7 days of discharge, and 3.5% (n = 46), 4.5% (n = 59), and 1.7% (n = 23) were colonized with GNB resistant to gentamicin, third/fourth-generation cephalosporin agents, and/or meropenem, respectively. The proportions of infants colonized with GNB resistant to gentamicin differed among the sites (P < .01). Most antimicrobial-resistant GNB were Enterobacteriaceae, although 10% of the GNB resistant to third/fourth-generation cephalosporin agents were Acinetobacter baumannii or Pseudomonas spp. Among the 119 colonized infants, 7% (n = 8) harbored GNB resistant to >1 antimicrobial category, including gentamicin and cephalosporin agents (n = 4) or cephalosporin and carbapenem agents (n = 3). One infant was colonized with a Klebsiella pneumoniae carbapenemase-producing strain that was resistant to all 3 antimicrobial categories. Of 20 infants who received antibiotic treatment when the samples for surveillance cultures were obtained, 3 were colonized with antimicrobial-resistant GNB.
Table 2.
Antimicrobial-Resistant GNB That Colonized Infants at NICU Discharge
| Resistance Category and Species | Colonized Infants and Isolatesa,b | Study Sitea | |||
|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | ||
| Gentamicin resistant | 46 (3.5) | 28 (6.3) | 6 (1.8) | 6 (2.9) | 6 (1.8) |
| Citrobacter spp. | 7 | 5 | 0 | 1 | 1 |
| Escherichia coli | 25 | 15 | 3 | 3 | 4 |
| Klebsiella spp. | 15 | 10 | 3 | 2 | 0 |
| Otherc | 2 | 0 | 0 | 1 | 1 |
| Third/fourth-generation cephalosporin resistant | 59 (4.5) | 19 (4.3) | 21 (6.2) | 9 (4.4) | 10 (3.0) |
| Acinetobacter baumannii | 4 | 3 | 1 | 0 | 0 |
| Citrobacter spp. | 13 | 0 | 11 | 1 | 1 |
| Enterobacter spp. | 17 | 5 | 4 | 4 | 4 |
| Escherichia coli | 13 | 7 | 2 | 0 | 4 |
| Klebsiella spp. | 8 | 2 | 3 | 3 | 0 |
| Otherd | 5 | 2 | 1 | 1 | 1 |
| Carbapenem resistant | 23 (1.7) | 7 (1.6) | 8 (2.3) | 1 (0.5) | 7 (2.1) |
| Enterobacter spp. | 8 | 1 | 4 | 0 | 3 |
| Proteus mirabilis | 5 | 1 | 2 | 1 | 1 |
| Serratia marcescens | 3 | 3 | 0 | 0 | 0 |
| Klebsiella pneumoniae e | 1 | 1 | 0 | 0 | 0 |
| Otherf | 76 | 21 | 2 | 0 | 3 |
Abbreviations: GNB, gram-negative bacilli; NICU, neonatal intensive care unit.
aData are presented as number (percent) of colonized infants or number of isolates (infants could be colonized by more than 1 resistant GNB species).
bOne infant was colonized with 2 GNB species, each resistant to a different antimicrobial category, and 8 infants were colonized with 1 GNB species resistant to ≥2 antimicrobial categories.
cOther: 1 Enterobacter spp. and 1 Pseudomonas aeruginosa.
dOther: 2 Morganella morganii, 2 Pseudomonas spp., and 1 Serratia marcescens.
eOne isolate harbored K. pneumoniae carbapenemase and was also resistant to gentamicin and third/fourth-generation cephalosporin.
fOther: 2 Citrobacter spp., 1 Morganella morganii, 2 Pantoea spp., and 1 Escherichia coli.
The largest cluster of antimicrobial-resistant GNB occurred at site 1, spanned 2 months, and involved 10 infants colonized with 11 gentamicin-resistant Enterobacteriaceae species (6 Escherichia coli, 4 Klebsiella spp., and 1 Citrobacter spp.). This cluster represented 37% of gentamicin-resistant GNB at this site. A smaller cluster occurred at site 2 and included 3 infants colonized with Citrobacter spp. resistant to third/fourth-generation cephalosporin agents.
Factors Associated With Colonization With Antimicrobial-Resistant GNB
Bivariate Analysis
As shown in Table 3, several factors were associated (P ≤ .05) with colonization with antimicrobial-resistant GNB in the bivariate analysis. Factors associated with colonization with GNB resistant to gentamicin included demographic characteristics (gestational age and birth weight), clinical characteristics (surgical procedures, mechanical ventilation, and length of stay), and antibiotic treatment (≥10 days of third/fourth-generation cephalosporin agents, meropenem, penicillin agents, or vancomycin and ≥5 days of meropenem, β-lactam/β-lactamase combination agents, or vancomycin). Prolonged treatment with aminoglycoside agents was not associated with colonization with gentamicin-resistant GNB. Factors associated with colonization with GNB resistant to third/fourth-generation cephalosporin agents included clinical characteristics (surgical procedures and mechanical ventilation) and several types of antibiotic treatment, including ≥5 and ≥10 days of treatment with third/fourth-generation cephalosporin agents. Factors associated with colonization with GNB resistant to carbapenem agents included female sex and ≥5 and ≥10 days of treatment with meropenem.
Table 3.
Bivariate Analysis of Risk Factors for Infant Colonization With Resistant GNB at NICU Discharge
| Factor/Variable | Gentamicin-Resistant GNB | Third/Fourth-Generation Cephalosporin-Resistant GNB | Carbapenem-Resistant GNB | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Colonized (n = 46) | Not Colonized (n = 1274) | P Valuesa | Colonized (n = 59) | Not Colonized (n = 1261) | P Valuesa | Colonized (n = 23) | Not Colonized (n = 1297) | P Valuesa | |
| Demographic characteristics | |||||||||
| Gestational age (mean [SD]) (wk) | 33 (5) | 32 (4) | .03 | 33 (5) | 33 (4) | .94 | 32 (4) | 33 (4) | .90 |
| Birth weight (mean [SD]) (g) | 2004 (999) | 1837 (876) | .04 | 1892 (976) | 1840 (877) | .51 | 1670 (837) | 1845 (882) | .42 |
| Female | 24 (52.2) | 591 (46.4) | .27 | 33 (55.9) | 582 (46.2) | .13 | 14 (60.9) | 601 (46.3) | .02 |
| Clinical characteristics | |||||||||
| Surgical procedureb | 16 (34.8) | 248 (19.5) | <.01 | 19 (32.2) | 245 (19.4) | <.01 | 2 (8.7) | 262 (20.2) | .20 |
| Central venous catheter | 33 (71.7) | 758 (59.5) | .51 | 40 (67.8) | 751 (59.6) | .15 | 12 (52.2) | 779 (60.1) | .46 |
| Mechanical ventilation | 35 (76.1) | 734 (57.6) | .01 | 39 (66.1) | 730 (57.9) | <.01 | 11 (47.8) | 758 (58.4) | .12 |
| Length of stay (mean [SD]) (days) | 53 (42) | 46 (37) | .01 | 54 (38) | 46 (37) | .06 | 41 (18) | 46 (38) | .15 |
| Antimicrobial treatment ≥10 days | |||||||||
| Aminoglycoside(s) | 2 (4.3) | 58 (4.6) | .64 | 5 (8.5) | 55 (4.4) | .04 | 1 (4.3) | 59 (4.5) | .99 |
| Third/fourth-generation cephalosporin(s) | 2 (4.3) | 18 (1.4) | .01 | 2 (3.4) | 18 (1.4) | .01 | 0 (0) | 20 (1.5) | NA |
| Meropenem | 2 (4.3) | 11 (0.9) | <.01 | 1 (1.7) | 12 (1.0) | <.01 | 1 (4.3) | 12 (0.9) | <.01 |
| β-Lactam/β-lactamase combination | 3 (6.5) | 33 (2.6) | .06 | 5 (8.5) | 31 (2.5) | .01 | 0 (0) | 36 (2.8) | NA |
| Cefazolin | 1 (2.2) | 21 (1.6) | .06 | 0 (0) | 22 (1.7) | NA | 0 (0) | 22 (1.7) | NA |
| Penicillin | 1 (2.2) | 61 (4.8) | .02 | 5 (8.5) | 57 (4.5) | .16 | 0 (0) | 62 (4.8) | NA |
| Vancomycin | 2 (4.3) | 75 (5.9) | .04 | 3 (5.1) | 74 (5.9) | .57 | 1 (4.3) | 76 (5.9) | .81 |
| Antimicrobial treatment ≥5 days | |||||||||
| Third/fourth-generation cephalosporin(s) | 4 (8.7) | 50 (3.9) | .78 | 6 (10.2) | 48 (3.8) | <.01 | 1 (4.3) | 53 (4.1) | .69 |
| Meropenem | 4 (8.7) | 15 (1.2) | <.01 | 1 (1.7) | 18 (1.4) | .16 | 1 (4.3) | 18 (1.4) | <.01 |
| β-Lactam/β-lactamase combination | 7 (15.2) | 84 (6.6) | .01 | 12 (20.3) | 79 (6.3) | <.01 | 1 (4.3) | 90 (6.9) | .62 |
| Cefazolin | 4 (8.7) | 66 (5.2) | .21 | 2 (3.4) | 68 (5.4) | .09 | 1 (4.3) | 69 (5.3) | .62 |
| Penicillin and/or ampicillin | 9 (19.6) | 269 (21.1) | .35 | 17 (28.8) | 261 (20.7) | .09 | 3 (13.0) | 275 (21.2) | .27 |
| Metronidazole | 1 (2.2) | 18 (1.4) | .75 | 3 (5.1) | 16 (1.3) | <.01 | 0 (0) | 19 (1.5) | NA |
| Vancomycin | 13 (28.3) | 196 (15.4) | <.01 | 11 (18.6) | 198 (15.7) | .60 | 4 (17.4) | 205 (15.8) | .81 |
Abbreviations: GNB, gram-negative bacilli; NICU, neonatal intensive care unit; SD, standard deviation.
a P values for differences according to colonization are from the χ2 test for categorical variables and the Wilcoxon rank-sum test for continuous variables. P is indicated as NA if the methods were not applicable because of small cell sizes.
bSurgical procedures were performed at sites 1, 2, and 3.
Multivariable Analysis
Factors significantly associated with antimicrobial-resistant GNB as determined by multivariable analysis are shown in Table 4. For colonization with gentamicin-resistant GNB, older gestational age was associated with a decreased risk of colonization, whereas increased length of stay and several types of antibiotic treatment were associated with an increased risk of colonization. For colonization with third/fourth-generation cephalosporin-resistant GNB, surgical procedures and several types of antibiotic treatment for ≥5 days (including treatment with third/fourth-generation cephalosporin agents) were associated with an increased risk of colonization, whereas treatment with cefazolin was associated with a decreased risk of colonization. For colonization with carbapenem-resistant GNB, female sex, increased length of stay, and ≥10 days of meropenem treatment were associated with an increased risk of colonization.
Table 4.
Multivariable Analysis of Factors Significantly Associated With Infant Colonization With Antimicrobial-Resistant GNB at NICU Discharge
| Resistance Category | Odds Ratio | 95% Confidence Interval | P Valuesa |
|---|---|---|---|
| Gentamicin-resistant GNB | |||
| Gestational age (wk) | 0.95 | 0.92–0.97 | <.01 |
| Length of stay (days) | 1.00 | 0.99–1.00 | <.01 |
| Meropenem treatment (≥10 days) | 2.68 | 2.25–3.21 | <.01 |
| Third/fourth-generation cephalosporin treatment (≥10 days) | 2.50 | 1.22–5.15 | .01 |
| Penicillin and/or ampicillin treatment (≥10 days) | 0.23 | 0.11–0.52 | <.01 |
| β-Lactam/β-lactamase combination treatment (≥5 days) | 2.30 | 1.18–4.48 | .01 |
| Third/fourth-generation cephalosporin-resistant GNB | |||
| Surgical procedureb | 1.97 | 1.54–2.50 | <.01 |
| Third/fourth-generation cephalosporin treatment (≥5 days) | 2.13 | 1.72–2.66 | <.01 |
| β-Lactam/β-lactamase combination treatment (≥5 days) | 3.07 | 1.45–6.50 | <.01 |
| Metronidazole treatment (≥5 days) | 2.55 | 1.67–3.87 | <.01 |
| Cefazolin treatment (≥5 days) | 0.36 | 0.16–0.82 | .01 |
| Carbapenem-resistant GNB | |||
| Female | 1.92 | 1.12–3.30 | .02 |
| Length of stay (day) | 1.01 | 1.00–1.01 | .03 |
| Meropenem treatment (≥10 days) | 8.12 | 5.69–11.61 | <.01 |
Abbreviations: GNB, gram-negative bacilli; NICU, neonatal intensive care unit.
a P values are reported from the final fixed-effect model for each outcome. Only significant variables (P < .05) in the final models are shown.
bSurgical procedures were performed at sites 1, 2, and 3.
HAIs Caused by Antimicrobial-Resistant GNB
Of the infants in this substudy, 2% (25 of 1320) were diagnosed with an HAI caused by an antimicrobial-resistant GNB; these infections included bloodstream infections (56% [n = 14]), urinary tract infections (40% [n = 10)], respiratory tract infections (16% [n = 4]), and/or other infections (20% [n = 5]). A gentamicin-resistant GNB infected 15 of the infants, 7 (47%) of whom were colonized with the same gentamicin-resistant species within 7 days of discharge. A third/fourth-generation cephalosporin-resistant GNB infected 10 of the infants, 3 (30%) of whom were colonized with the same cephalosporin-resistant species within 7 days of discharge. One infant was infected with carbapenem-resistant GNB and was not colonized with a carbapenem-resistant GNB within 7 days of discharge.
Among the 87 infants who received ≥10 days of gentamicin, third/fourth-generation cephalosporin agents, and/or carbapenem agents, 14 (16%) were diagnosed with an HAI caused by a GNB resistant to ≥1 of these antimicrobial categories. The remaining infants received prolonged treatment but did not have clinical culture results that were positive for GNB.
DISCUSSION
We prospectively assessed infant colonization at NICU discharge with GNB resistant to selected antimicrobial agents. Overall, 9% (119 of 1320) of the infants were colonized with antimicrobial-resistant GNB at discharge from the NICU; 3.5%, 4.5%, and 1.7% were colonized with GNB resistant to gentamicin, third/fourth-generation cephalosporin agents, or carbapenem agents, respectively, and <1% were colonized with GNB resistant to ≥1 antimicrobial category. We confirmed our hypotheses that prolonged treatment with broad-spectrum agents was associated with colonization with antimicrobial-resistant GNB; ≥10 days of treatment with third/fourth-generation cephalosporin agents or meropenem was associated with colonization with GNB resistant to gentamicin, ≥5 days of treatment with third/fourth-generation cephalosporin agents was associated with colonization with GNB resistant to these agents, and ≥10 days of treatment with meropenem was associated with colonization with GNB resistant to carbapenem agents. It is notable that most prolonged treatment with gentamicin, third/fourth-generation cephalosporin agents, and/or meropenem was not associated with a positive culture result; a minority (16% [n = 14 of 87]) of the infants treated with ≥10 days with these agents had a culture-proven HAI caused by an antimicrobial-resistant GNB.
Others have assessed determinants of colonization with antimicrobial-resistant GNB in the NICU population and found associations similar to those noted in our study. Colonization with GNB resistant to gentamicin, piperacillin-tazobactam, or ceftazidime increased as overall antimicrobial treatment-days or treatment-days with vancomycin, ampicillin, an aminoglycoside, piperacillin-tazobactam, and/or a third-generation cephalosporin agent increased [10]. Among very low birth weight infants, colonization with gentamicin-resistant GNB was associated with treatment with carbapenem agents [7]. Colonization with an extended-spectrum β-lactamase (ESBL)–producing Enterobacteriaceae species was associated with treatment with third-generation cephalosporin agents or with concurrent treatment with cephalosporin and aminoglycoside agents [14, 15] or vancomycin [9, 16]. Thus, there exists convincing evidence that selective pressure from antimicrobial agents leads to the emergence of resistant pathogens in the NICU population.
In our study, surgery in an infant was predictive of colonization with GNB resistant to third/fourth-generation cephalosporin agents. Although no previous study has identified surgery as a predictor of colonization among neonates, several studies conducted in adults hospitalized in intensive care units have found that surgical procedures increase the risk of infection and/or colonization with antimicrobial-resistant GNB [17–19]. In addition, treatment with proton pump inhibitors and/or antacids or impaired intestinal motility among infants who undergo surgery may alter the intestinal flora and potentially promote an overgrowth of pathogenic microorganisms [20]. Complex surgical procedures that involve the GI tract may be associated with other unmeasured confounding such as prolonged periods of intravenous parenteral feeding or exposure to potentially pathogenic microorganisms during surgical procedures. Future studies should evaluate the association of specific types of surgery and colonization with pathogenic microorganisms.
We found that female sex was a risk factor for colonization with GNB resistant to carbapenem agents. Female sex was an independent predictor of colonization with ESBL-producing K. pneumoniae during a NICU outbreak of infections caused by ESBL-producing Enterobacteriaceae [15, 21]. The biologic plausibility of these observations is uncertain, but colonization with K pneumoniae may have been a result of the link between GI and genitourinary flora in females.
The optimal strategy, clinical utility, and cost-effectiveness of performing routine surveillance cultures for antimicrobial-resistant GNB are uncertain. Infants hospitalized in the NICU may be colonized with GNB at multiple body sites (eg, the nasopharynx, GI tract, respiratory tract, and skin [3, 5, 22]). Thus, cultures from a single body site may fail to detect all colonized infants, and point-prevalence cultures do not predict the duration of colonization. In addition, an infant's colonization status may not be detected during antimicrobial therapy or may change after antimicrobial therapy. Finally, surveillance efforts are costly, and it is unclear what interventions should be implemented if colonization with antimicrobial-resistant GNB is detected, because decolonization is not generally feasible. Thus, surveillance cultures for GNB should be limited for use in research, during outbreaks, or in infants who are potentially at high risk (eg, older infants transferred to the NICU) [23].
With our analysis, we detected a very low rate of gentamicin- and cephalosporin-resistant GNB occurring in temporal clusters, which might suggest few episodes of unrecognized transmission between infants. Although our findings suggest the potential for transferrable resistance determinants among different GNB species [7, 17, 24–26), the likelihood that interspecies transfer events had occurred seemed rare, because only 4 infants were colonized with 2 species of antimicrobial-resistant GNB, and only 8 were colonized with 1 GNB species resistant to ≥2 antimicrobial classes. However, molecular typing and identification of resistance determinants of surveillance isolates were not performed, so it is hard to fully interpret our findings.
This study had several strengths. Censoring did not occur among study subjects, because the colonization status was determined at the end of the observation period (ie, at NICU discharge), so there was no differential loss to follow-up based on outcome. Surveillance cultures from the 4 study sites were processed by a core microbiology laboratory, which minimized potential misclassification of the outcomes. Finally, the outcomes assessed in this study were measured at the level of the individual, but because the cohort was drawn from 4 study sites, the findings are likely to be more generalizable than those from a single-center study.
This study also had limitations. More than half of the potentially eligible infants did not have samples for surveillance cultures obtained. The rate of colonization with antimicrobial-resistant GNB during the NICU hospitalizations may be underestimated, because we assessed GNB colonization only within 7 days of discharge; surveillance cultures were not performed at admission or at other time points during the hospitalizations, and the results of clinical cultures consistent with colonization not infection (ie, not treated) were not collected. Only 2 dominant colonial morphotypes per rectal swab were analyzed in the study surveillance samples. Furthermore, we cannot determine if colonization occurred before antibiotic exposure. Some potential predictors of colonization (eg, vaginal vs cesarean-section delivery, breast milk vs formula feeding, duration of parenteral nutrition, treatment with proton pump inhibitors, colonization status of mothers) were not evaluated because these data were not collected in the larger study. Finally, 20 infants were receiving antibiotic treatment when the samples for surveillance cultures were obtained, which may have influenced the detection of antimicrobial-resistant GNB, although 3 of these infants were colonized with antimicrobial-resistant GNB.
In conclusion, the rates of colonization with antimicrobial-resistant GNB at NICU discharge were relatively low but similar among the NICUs and across antimicrobial classes. These findings suggest the potential for dissemination of antimicrobial-resistant GNB from colonized infants when they are discharged to another NICU, the community, or a pediatric long-term care facility [8, 10]. We found potentially modifiable risk factors, the most notable of which was prolonged antimicrobial treatment with broad-spectrum agents used for empiric therapy and not associated with positive culture results. Thus, antimicrobial stewardship efforts that are promoted by the Centers for Disease Control and Prevention and professional societies (ie, limiting broad-spectrum agents, targeting the pathogen, and not treating colonization) could have a beneficial effect on preventing the emergence of antimicrobial-resistant GNB in the NICU population [27, 28].
Notes
Acknowledgments. We thank Kristina Rivera and Dana O'Toole for assistance with data collection and Dr. Susan Whittier for invaluable insights into susceptibility testing.
Financial support. This work was supported by National Institute of Nursing Research grant R01 NR010821.
Potential conflicts of interest. All authors: No reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Singh N, Patel KM, Leger MM, et al. Risk of resistant infections with Enterobacteriaceae in hospitalized neonates. Pediatr Infect Dis J 2002; 21:1029–33. [DOI] [PubMed] [Google Scholar]
- 2. Calil R, Marba ST, von Nowakonski A, Tresoldi AT. Reduction in colonization and nosocomial infection by multiresistant bacteria in a neonatal unit after institution of educational measures and restriction in the use of cephalosporins. Am J Infect Control 2001; 29:133–8. [DOI] [PubMed] [Google Scholar]
- 3. Jarvis WR. The epidemiology of colonization. Infect Control Hosp Epidemiol 1996; 17:47–52. [DOI] [PubMed] [Google Scholar]
- 4. Bettelheim KA, Lennox-King SM. The acquisition of Escherichia coli by new-born babies. Infection 1976; 4:174–9. [DOI] [PubMed] [Google Scholar]
- 5. Long SS, Swenson RM. Development of anaerobic fecal flora in healthy newborn infants. J Pediatr 1977; 91:298–301. [DOI] [PubMed] [Google Scholar]
- 6. Graham PL, 3rd, Della-Latta P, Wu F, Zhou J, Saiman L. The gastrointestinal tract serves as the reservoir for Gram-negative pathogens in very low birth weight infants. Pediatr Infect Dis J 2007; 26:1153–6. [DOI] [PubMed] [Google Scholar]
- 7. Smith A, Saiman L, Zhou J, Della-Latta P, Jia H, Graham PL., 3rd Concordance of gastrointestinal tract colonization and subsequent bloodstream infections with Gram-negative bacilli in very low birth weight infants in the neonatal intensive care unit. Pediatr Infect Dis J 2010; 29:831–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Navarro LR, Pekelharing-Berghuis M, de Waal WJ, Thijsen SF. Bacterial colonization patterns in neonates transferred from neonatal intensive care units. Int J Hyg Environ Health 2011; 214:167–71. [DOI] [PubMed] [Google Scholar]
- 9. Ofek-Shlomai N, Benenson S, Ergaz Z, Peleg O, Braunstein R, Bar-Oz B. Gastrointestinal colonization with ESBL-producing Klebsiella in preterm babies—is vancomycin to blame? Eur J Clin Microbiol Infect Dis 2012; 31:567–70. [DOI] [PubMed] [Google Scholar]
- 10. Toltzis P, Dul MJ, Hoyen C, et al. Molecular epidemiology of antibiotic-resistant Gram-negative bacilli in a neonatal intensive care unit during a nonoutbreak period. Pediatrics 2001; 108:1143–8. [DOI] [PubMed] [Google Scholar]
- 11. Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing: 22nd informational supplement. Wayne, PA: CLSI; 2012. [Google Scholar]
- 12. Gardiner JC, Luo Z, Roman LA. Fixed effects, random effects and GEE: what are the differences? Stat Med 2009; 28:221–39. [DOI] [PubMed] [Google Scholar]
- 13. Kahan BC. Accounting for centre-effects in multicentre trials with a binary outcome—when, why, and how? BMC Med Res Methodol 2014; 14:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Linkin DR, Fishman NO, Patel JB, Merrill JD, Lautenbach E. Risk factors for extended-spectrum beta-lactamase-producing Enterobacteriaceae in a neonatal intensive care unit. Infect Control Hosp Epidemiol 2004; 25:781–3. [DOI] [PubMed] [Google Scholar]
- 15. Pessoa-Silva CL, Meurer Moreira B, Camara Almeida V, et al. Extended-spectrum beta-lactamase-producing Klebsiella pneumoniae in a neonatal intensive care unit: risk factors for infection and colonization. J Hosp Infect 2003; 53:198–206. [DOI] [PubMed] [Google Scholar]
- 16. Martins IS, Pessoa-Silva CL, Nouer SA, et al. Endemic extended- spectrum beta-lactamase-producing Klebsiella pneumoniae at an intensive care unit: risk factors for colonization and infection. Microb Drug Resist 2006; 12:50–8. [DOI] [PubMed] [Google Scholar]
- 17. Shakil S, Akram M, Ali SM, Khan AU. Acquisition of extended-spectrum beta-lactamase producing Escherichia coli strains in male and female infants admitted to a neonatal intensive care unit: molecular epidemiology and analysis of risk factors. J Med Microbiol 2010; 59:948–54. [DOI] [PubMed] [Google Scholar]
- 18. Cisneros JM, Rodriguez-Bano J, Fernandez-Cuenca F, et al. Risk-factors for the acquisition of imipenem-resistant Acinetobacter baumannii in Spain: a nationwide study. Clin Microbiol Infect 2005; 11:874–9. [DOI] [PubMed] [Google Scholar]
- 19. Manchanda V, Sanchaita S, Singh N. Multidrug resistant acinetobacter. J Global Infect Dis 2010; 2:291–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Cothran DS, Borowitz SM, Sutphen JL, Dudley SM, Donowitz LG. Alteration of normal gastric flora in neonates receiving ranitidine. J Perinatol 1997; 17:383–8. [PubMed] [Google Scholar]
- 21. Zaoutis TE, Goyal M, Chu JH, et al. Risk factors for and outcomes of bloodstream infection caused by extended-spectrum beta-lactamase-producing Escherichia coli and Klebsiella species in children. Pediatrics 2005; 115:942–9. [DOI] [PubMed] [Google Scholar]
- 22. Westerbeek EA, van den Berg A, Lafeber HN, Knol J, Fetter WP, van Elburg RM. The intestinal bacterial colonisation in preterm infants: a review of the literature. Clin Nutr 2006; 25:361–8. [DOI] [PubMed] [Google Scholar]
- 23. Macnow T, O'Toole D, DeLaMora P, et al. Utility of surveillance cultures for antimicrobial resistant organisms in infants transferred to the neonatal intensive care unit. Pediatr Infect Dis J 2013; 32:e443–50. [DOI] [PubMed] [Google Scholar]
- 24. Nambiar S, Singh N. Change in epidemiology of health care-associated infections in a neonatal intensive care unit. Pediatr Infect Dis J 2002; 21:839–42. [DOI] [PubMed] [Google Scholar]
- 25. Bizzarro MJ, Gallagher PG. Antibiotic-resistant organisms in the neonatal intensive care unit. Semin Perinatol 2007; 31:26–32. [DOI] [PubMed] [Google Scholar]
- 26. Mammina C, Di Carlo P, Cipolla D, et al. Surveillance of multidrug-resistant Gram-negative bacilli in a neonatal intensive care unit: prominent role of cross transmission. Am J Infect Control 2007; 35:222–30. [DOI] [PubMed] [Google Scholar]
- 27. Dellit TH, Owens RC, McGowan JE., Jr. et al. Infectious Diseases Society of America and the Society for Healthcare Epidemiology of America guidelines for developing an institutional program to enhance antimicrobial stewardship. Clin Infect Dis 2007; 44:159–77. [DOI] [PubMed] [Google Scholar]
- 28. Centers for Disease Control and Prevention. Get smart for healthcare. Available at: http://www.cdc.gov/getsmart/healthcare/. Accessed 5 June 2013.