Abstract
β-Cyano-alanine synthase (CAS; EC 4.4.1.9) plays an important role in cyanide metabolism in plants. Although the enzymatic activity of β-cyano-Ala synthase has been detected in a variety of plants, no cDNA or gene has been identified so far. We hypothesized that the mitochondrial cysteine synthase (CS; EC 4.2.99.8) isoform, Bsas3, could actually be identical to CAS in spinach (Spinacia oleracea) and Arabidopsis. An Arabidopsis expressed sequence tag database was searched for putative Bsas3 homologs and four new CS-like isoforms, ARAth;Bsas1;1, ARAth;Bsas3;1, ARAth;Bsas4;1, and ARAth;Bsas4;2, were identified in the process. ARAth;Bsas3;1 protein was homologous to the mitochondrial SPIol;Bsas3;1 isoform from spinach, whereas ARAth;Bsas4;1 and ARAth;Bsas4;2 proteins defined a new class within the CS-like proteins family. In contrast to spinach SPIol;Bsas1;1 and SPIol;Bsas2;1 recombinant proteins, spinach SPIol;Bsas3;1 and Arabidopsis ARAth;Bsas3;1 recombinant proteins exhibited preferred substrate specificities for the CAS reaction rather than for the CS reaction, which identified these Bsas3 isoforms as CAS. Immunoblot studies supported this conclusion. This is the first report of the identification of CAS synthase-encoding cDNAs in a living organism. A new nomenclature for CS-like proteins in plants is also proposed.
β-Cyano-Ala synthase (CAS; EC 4.4.1.9) catalyzes the formation of the non-protein amino acid β-cyano-Ala from Cys and cyanide (Blumenthal et al., 1968), according to the reaction (Eq. 1):
 |
1 |
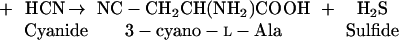 |
This activity has been detected in a broad spectrum of species among bacteria (Dunnill and Fowden, 1965; McAdam and Knowles, 1984), plants (Floss et al., 1965; Hendrickson and Conn, 1969; Miller and Conn, 1980; Ikegami et al., 1988a, 1988b; Maruyama et al., 1998), and insects (Meyers and Ahmad, 1991).
In insects, CAS activity is located principally in mitochondria, which are the main target for cyanide toxicity, and plays a pivotal role in cyanide detoxification (Meyers and Ahmad, 1991). In plants, ethylenesynthesis by 1-aminocyclopropane-1-carboxylic acid oxidase results in the production of cyanide (Peiser et al., 1984). Nevertheless, cyanide does not accumulate in non-cyanogenic plants, even in tissues producing ethylene at a very high rate, because of the involvement of CAS in cyanide fixation (Yip and Yang, 1988). β-Cyano-Ala synthesis also allows the recycling of the reduced nitrogen of cyanide into amino acid synthesis. In blue lupine (Lupinus angustifolius), β-cyano-Ala is metabolized to l-Asn by a β-cyano-Ala hydrolase (Castric et al., 1972). β-Cyano-Ala is also a potent neurotoxin in numerous animals, including monkeys (Ressler et al., 1969), and it is highly abundant as a defense molecule against predators in many species of the genus Vicia, some of which are used for livestock and human consumption as an inexpensive protein source in developing countries (Tate and Enneking, 1992).
In plants, two classes of CAS seem to exist, based on differences in amino acid composition and protein structure (Ikegami et al., 1988a). In blue lupine, CAS is a monomeric enzyme, with a molecular mass of about 52 kD, and contains 1 mol pyridoxal phosphate mol−1 protein, which is essential for the catalytic activity (Akopyan et al., 1975). In spinach (Spinacia oleracea) and Lathyrus latifolius, the enzyme contains two identical subunits of 28 to 30 kD, each containing 1 molecule of pyridoxal phosphate, similar to the CAS of the cyanide-producing eubacterium Chromatium violaceum (McAdam and Knowles, 1984; Ikegami et al., 1988a, 1988b). The structure of this second class of CAS is very close to that of Cys synthase (CS; EC 4.2.99.8), which is a homodimer of about 30- to 35-kD subunits, each containing 1 molecule of pyridoxal phosphate (Masada et al., 1975; Droux et al., 1992). Both CS and CAS belong to the same enzyme family, the β-substituted Ala synthases (Ikegami and Murakoshi, 1994). In fact, purified CAS exhibits detectable CS activity (Hendrickson and Conn, 1969; Ikegami et al., 1988a, 1988b; Maruyama et al., 1998), according to the reaction (Eq. 2):
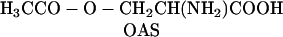 |
2 |
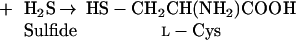 |
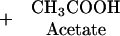 |
On the other hand, purified CSs from various plants species display some CAS activity (Ikegami et al., 1993). In spinach, CAS activity is predominantly present in the mitochondrial fraction but can also be detected in chloroplast and cytosol fractions (Warrilow and Hawkesford, 1998). In cocklebur seed cotyledon, cytosolic CAS activity has been attributed to CS (Maruyama et al., 1998). Compartment-specific CS isoforms have been purified from spinach and cauliflower cytosol, chloroplast, and mitochondrialfractions (Lunn et al., 1990; Rolland et al., 1992). In spinach,cDNAs encoding cytosolic SPIol;Bsas1;1, chloroplastic SPIol;Bsas2;1, and mitochondrial SPIol;Bsas3;1 CS isoforms have been isolated (Saito et al., 1992, 1993, 1994) (for a proposal of a new nomenclature for CS-like genes, Bsas, see Table I). No CAS gene had been identified either in plants or in any other living organism.
Table I.
Gene designation and accession nos.
| Gene Family | Product, EC No. | Gene Designation | Plant Species | Accession No. |
|---|---|---|---|---|
| Bsas1 | OAS acetate-lyase (adding hydrogen sulfide) | ARAth;Bsas1;1 | Arabidopsis | X84097, Z97337, AJ27027 |
| [CS, OAS(thiol)lyase], EC 4.2.99.8 | ARAth;Bsas1;2 | Arabidopsis | X80376 | |
| ARAth;Bsas1;3 | Arabidopsis | X81697 | ||
| ARAth;Bsas1;4 | Arabidopsis | AJ011976 | ||
| BRAju;Bsas1;1 | B. juncea | Y10845 | ||
| BRAju;Bsas1;2 | B. juncea | Y10847 | ||
| CICar;Bsas1;1 | Cicer arietinum | AJ006024 | ||
| CITvu;Bsas1;1 | Citrulus vulgaris | D28777 | ||
| ORYsa;Bsas1;1 | Rice | AF073695 | ||
| ORYsa;Bsas1;2 | Rice | AF073697 | ||
| SOLtu;Bsas1;1 | Solanum tuberosum | AF044172 | ||
| SPIol;Bsas1;1 | Spinach | D10476 | ||
| TRIae;Bsas1;1 | Wheat | D13153 | ||
| ZEAma;Bsas1;1 | Maize | X85803 | ||
| Bsas2 | OAS acetate-lyase (adding hydrogen sulfide) | ARAth;Bsas2;1 | Arabidopsis | X80377, X81698, AC002333 |
| [CS, OAS(thiol)lyase], EC 4.2.99.8 | ARAth;Bsas2;2 | Arabidopsis | X81973 | |
| CAPan;Bsas2;1 | Capsicum annum | X64874 | ||
| SOLtu;Bsas2;1 | S. tuberosum | AF044173 | ||
| SPIol;Bsas2;1 | Spinach | D14722, X66860, E08016, L05184 | ||
| Bsas3 | CAS, EC 4.4.1.9 | ARAth;Bsas3;1 | Arabidopsis | AJ010505, AB024282 |
| SPIol;Bsas3;1 | Spinach | D37963 | ||
| Bsas4 | OAS acetate-lyase (adding hydrogen sulfide) | ARAth;Bsas4;1 | Arabidopsis | AJ011603, AB024284, AC009465 |
| [CS, OAS(thiol)lyase], EC 4.2.99.8 | ARAth;Bsas4;2 | Arabidopsis | AB024283, AJ011044 | |
| BRAju;Bsas4;1 | B. juncea | Y10846 | ||
| Bsas5 | ARAth;Bsas5;1 | Arabidopsis | AB003041, AF082158 | |
| Bsas6 | ORYsa;Bsas6;1 | Rice | AF073696 | |
| ORYsa;Bsas6;2 | Rice | AF073698 |
We have proposed these gene designations in Bsas (β-substituted Ala synthase) family according to the rules recommended by the Commission on Plant Gene Nomenclature (http://mbclserver.rutgers.edu/CPGN/Guide.html).
The extensive structural and functional similarities between CS and CAS prompted us to examine the identity of a CS isoform with CAS. In spinach mitochondria extracts, it was not possible to separate CS from CAS protein (Warrilow and Hawkesford, 1998), raising the possibility that the mitochondrial Bsas3 isoform of CS could actually be a CAS and not a CS, as was thought previously. To test this hypothesis, we searched for SPIol;Bsas3;1 homolog cDNAs in Arabidopsis and determined the kinetic characteristics of CS and CAS activities of spinach and Arabidopsis CS-like recombinant proteins. We conclude that the mitochondrial Bsas3 isoform is the CAS in spinach and Arabidopsis. This is the first report of the identification of CAS-encoding cDNA in living organisms.
RESULTS
Identification of New Members of the CS-Like Protein Family in Arabidopsis
Among the 45,752 expressed sequence tags (ESTs) of Arabidopsis present in the dbEST database (release November 26, 1999), 52 were assigned as encoding a CS. They could be classified into eight families, four of which did not match any previously reported CS sequence. ESTs representative of these families were sequenced and named ARAth;Bsas1;1, ARAth;Bsas3;1, ARAth;Bsas4;1, and ARAth;Bsas4;2. These nucleotide sequences are deposited under the accession numbers AJ011976, AJ010505, AJ011603, and AJ01044, respectively. We proposed a new systematic nomenclature for CS-like genes as Bsas (β-substituted Ala synthase) (Table I).
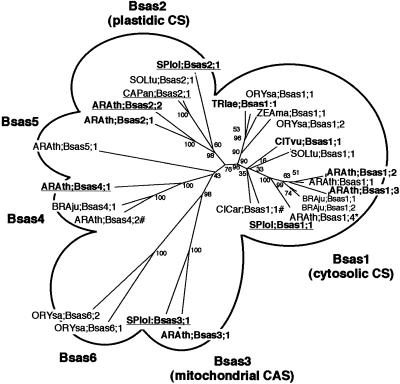
ARAth;Bsas1;1 contained an intron at position 260 to 351 and an in-frame stop codon at position 764 to 766. This imperfectly processed mRNA was corrected manually for the purpose of establishing the phylogeny of plant CS-like proteins. The corrected protein was 325 amino acids long, with a calculated molecular mass of 34 kD, and was predicted to be cytosolic by the PSORT program (Nakai and Kanehisa, 1992). ARAth;Bsas4;1 contained a full-length open reading frame (ORF) encoding a 324-amino acid polypeptide of 34.3 kD, which was predicted to be cytosolic. ARAth;Bsas4;2 contained a partial ORF, comprising only the last 174 amino acids of the protein. ARAth;Bsas3;1 contained a full-length ORF encoding a 368-amino acid polypeptide of 39.9 kD, which was predicted to be mitochondrial. Phylogenetic analysis indicated that plant CS-like proteins could be separated into six groups, defined as Bsas1, Bsas2, Bsas3, Bsas4, Bsas5, and Bsas6 classes (Fig. 1). The ARAth;Bsas1;1 putative protein belonged to the Bsas1 class, whereas ARAth;Bsas4;1 and ARAth;Bsas4;2, together with BRAju;Bsas4;1 from Indian mustard (Brassica juncea), defined the Bsas4 class. The ARAth;Bsas3;1 protein was 74.3% identical and 82.2% similar to the spinach SPIol;Bsas3;1 protein. ARAth;Bsas4;1, ARAth;Bsas4;2, and ARAth;Bsas3;1 appeared to be identical to AtcysD1, AtcysD2, and AtcysC1, respectively, the sequences for which were recently released in the databases.
Figure 1.
Phylogeny of Bsas family proteins in plants. Complete protein sequences were aligned using Clustal X (Thompson et al., 1997). Unrooted phylogenetic tree was calculated by the neighbor-joining method and then drawn using Drawtree from the Phylip package (Phylogeny Inference Package, version 3.57c, Department of Genetics, University of Washington, Seattle). The numbers indicate the bootstrap values for 100 replicates. Accession numbers are summarized in Table I. *, Sequence corrected manually; #, partial sequence only. Sequences whose actual CS or CAS activities have been determined are indicated in bold. Sequences whose actual subcellular localization have been determined are underlined.
Functional Characterization of CS-Like Recombinant Proteins Expressed in a Cys Auxotroph Escherichia coli Mutant
Spinach SPIol;Bsas1;1, SPIol;Bsas2;1, and mature SPIol;Bsas3;1 and Arabidopsis ARAth;Bsas4;1 and mature ARAth;Bsas3;1 recombinant proteins were expressed in the E. coli NK3 mutant, which were deficient in CS activity (Table II). The NK3 mutant transformed with the empty expression vector pTV118N was devoid of any detectable CS and CAS activities (Table III), indicating that CS and CAS are encoded by the same genes in E. coli. Thus, we usedcrude extracts of transformed E. coli for further analysis. SPIol;Bsas1;1 and SPIol;Bsas2;1 recombinant proteins displayed high CS and low CAS activities, with a CS to CAS ratio of 209 and 23, respectively. SPIol;Bsas3;1 and ARAth;Bsas3;1 showed low CS and high CAS activities, with a CS to CAS ratio of 2.6 × 10−3 and 6 × 10−3, respectively. ARAth;Bsas4;1 showed a very low CS activity and no detectable CAS activity.
Table II.
Expression vectors used for this study
| Plant | Isoform Gene | Plasmid | Feature | Ref. |
|---|---|---|---|---|
| Spinach | SPIol;Bsas1;1 | pKM1 | Full length | Saito et al. (1992) |
| SPIol;Bsas2;1 | pCSB2 | Full length | Saito et al. (1993) | |
| SPIol;Bsas3;1 | pCSCMP | 45 N-terminal amino acid deletion | This work | |
| Arabidopsis | ARAth;Bsas4;1 | pOAS3 | Full length | This work |
| ARAth;Bsas3;1 | pOAS5MP | 42 N-terminal amino acid deletion | This work |
SPIol;Bsas3;1 and ARAth;Bsas3;1 were expressed as mature proteins, without their respective mitochondrial transit peptide.
Table III.
CS and CAS activities of recombinant CS-like proteins
| Organism | Isoform Gene | CS Activity | CAS Activity | CS/CAS | CAS/CS |
|---|---|---|---|---|---|
| μmol Cys mg−1 protein min−1 | μmol sulfide mg−1 protein min−1 | ||||
| E. coli NK3 | Empty vector | ND | ND | ||
| Spinach | SPIol;Bsas1;1 | 2,466 | 11.8 | 209 | 4.8 × 10−3 |
| SPIol;Bsas2;1 | 547 | 23.3 | 23 | 4.4 × 10−2 | |
| SPIol;Bsas3;1 | 0.4 | 157 | 2.6 × 10−3 | 382 | |
| Arabidopsis | ARAth;Bsas4;1 | 0.2 | ND | ||
| ARAth;Bsas3;1 | 0.4 | 62.1 | 6 × 10−3 | 168 |
Crude protein extracts of E. coli NK3 transformed with the expression vectors described in Table II were used for the enzyme assays. Empty pTV118N vector was used as a negative control. CS and CAS activities were determined from the same crude protein extracts. ND, Not detected.
The Km values for substrates of the CS and CAS reactions were determined for all recombinant proteins expressed in the NK3 E. coli mutant (Table IV). Km for O-acetyl-l-Ser (OAS) in the CS reaction were in the millimolar range for recombinant SPIol;Bsas1;1, SPIol;Bsas2;1, and ARAth;Bsas4;1. This Km was more than 1 order of magnitude higher in SPIol;Bsas3;1 and ARAth;Bsas3;1. Km values for Na2S in the CS reaction were comparable for all proteins, ranging from 0.99 to 8.24 mm. The Km for Cys in the CAS reaction was very similar for SPIol;Bsas3;1 and ARAth;Bsas3;1 and about 30 times lower in SPIol;Bsas1;1 and SPIol;Bsas2;1. The Km values for KCN in the CAS reaction were in the 100 μm range for SPIol;Bsas3;1 and ARAth;Bsas3;1 and more than 50 times higher in SPIol;Bsas1;1 and SPIol;Bsas2;1. Therefore, the CS reaction is more favored in SPIol;Bsas1;1, SPIol;Bsas2;1, and ARAth;Bsas4;1 than in SPIol;Bsas3;1 and ARAth;Bsas3;1. In contrast, SPIol;Bsas3;1 and ARAth;Bsas3;1 have a far higher affinity for cyanide. Altogether, these results indicate that SPIol;Bsas3;1 and ARAth;Bsas3;1 proteins are actually CAS, whereas SPIol;Bsas1;1, SPIol;Bsas2;1, and ARAth;Bsas4;1 are true CS.
Table IV.
Kinetic characteristics of recombinant CS-like proteins
| Plant | Isoform Gene |
Km for CS Reaction
|
Km for CAS Reaction
|
||
|---|---|---|---|---|---|
| OAS | Na2S | Cys | KCN | ||
| mm | |||||
| Spinach | SPIol;Bsas1;1 | 1.48 | 0.99 | 0.08 | 5.18 |
| SPIol;Bsas2;1 | 1.93 | 1.73 | 0.06 | 5.41 | |
| SPIol;Bsas3;1 | 14.48 | 3.57 | 2.14 | 0.10 | |
| Arabidopsis | ARAth;Bsas4;1 | 0.69 | 4.98 | ND | ND |
| ARAth;Bsas3;1 | 39.88 | 8.24 | 2.54 | 0.06 | |
Crude protein extracts of E. coli NK3 transformed with the expression vectors described in Table II were used for the enzyme assays. All isoforms obeyed the Michaelis-Menten equation. Km values were calculated by linear regression of double-reciprocal plots. ND, Not detected.
Characterization of Anti-CS and Anti-β-CAS Antibodies
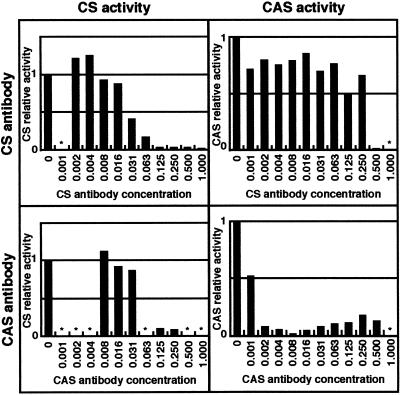
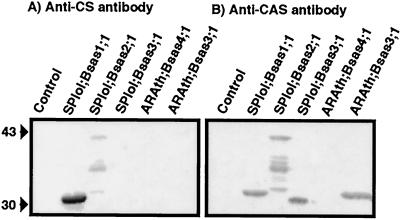
To confirm the CS or CAS nature of the various CS isoforms, we used rabbit polyclonal antibodies directed against biochemically purified spinach CS (isoforms Bsas1 + Bsas2) or CAS. Because CS and CAS are closely related proteins, we investigated the ability of the two antibodies to inhibit the catalytic activities of purified spinach CS and CAS to evaluate their specificity (Fig. 2). The CS antibody could inhibit the CS activity at a concentration of 0.035 dilution, whereas a concentration of 0.5 dilution was necessary to inhibit the CAS activity. In contrast, the CAS antibody could inhibit the CS activity at a concentration of 0.125 dilution, whereas a concentration of 0.002 dilution was necessary to inhibit the CAS activity. We concluded that these anti-CS and anti-CAS antibodies were apparently specific for CS and CAS, respectively. The anti-CS antibody could recognize the SPIol;Bsas1;1 and SPIol;Bsas2;1 isoforms by immunoblotting but could not recognize the SPIol; Bsas3;1, ARAth;Bsas4;1, and ARAth;Bsas3;1 isoforms (Fig. 3). This result indicates that SPIol;Bsas1;1 and SPIol;Bsas2;1 isoforms are different from SPIol;Bsas3;1, ARAth;Bsas3;1, and presumably ARAth;Bsas4;1. In contrast, the anti-CAS antibody could recognize SPIol;Bsas3;1 and ARAth;Bsas3;1 and, to a lesser extent, SPIol;Bsas1;1 and SPIol;Bsas2;1 isoforms, but not ARAth;Bsas4;1.
Figure 2.
Inhibition of purified spinach CS and CAS by anti-CS and anti-CAS antibodies. Specific activity of a mixture of diluted polyclonal rabbit antibodies directed against spinach CS or CAS and purified spinach CAS or CS enzyme were determined. The results are expressed relative to the specific activity of the control reaction without antibody. *, Not determined.
Figure 3.
Immunostaining of crude protein extracts using anti-CS and anti-CAS antibodies. Crude protein extracts were separated on a 12% (w/v) polyacrylamide gel and transferred to a membrane. Immunoblotting was carried out with rabbit polyclonal anti-spinach CS (A) or anti-spinach CAS (B) antibodies. Immunoreactive proteins were visualized using goat anti-rabbit IgG antibodies coupled to phosphatase. Molecular masses in kD are indicated by arrowheads.
DISCUSSION
Identification and Functional Characterization of CAS in Arabidopsis and Spinach
Sequence analysis of the Arabidopsis subset of dbEST database allowed us to isolate four cDNAs encoding CS-like proteins, named ARAth;Bsas1;1, ARAth;Bsas3;1, ARAth;Bsas4;1, and ARAth;Bsas4;2 (Table I). The ARAth;Bsas1;1 cDNA contained an intron and an in-frame stop codon. The protein sequence of this imperfectly processed mRNA was corrected manually for the purpose of establishing the phylogeny of plant CS-like proteins. A low expression of this mRNA was detected by reverse transcriptase-PCR (data not shown), but it remains to be confirmed whether this particular isoform is functional or not. Plant CS-like protein sequences could be separated into six groups: Bsas1, Bsas2, Bsas3, Bsas4, Bsas5, and Bsas6 classes (Fig. 1). ARAth; Bsas3;1 was homologous to spinach SPIol;Bsas3;1, whereas ARAth;Bsas4;1 and ARAth;Bsas4;2, together with BRAju;Bsas4;1 from B. juncea (Indian mustard), belonged to a cytosolic CS-like protein class, named Bsas4 class, which had not been described previously. Among these six classes, only the Bsas1 (Saito et al., 1992; Youssefian et al., 1993; Hell et al., 1994; Noji et al., 1998; Hesse et al., 1999) and Bsas2 (Rolland et al., 1993; Saito et al., 1993; Hesse et al., 1999) classes have been functionally characterized. Because of their sequence homology to the Bsas1 and Bsas2 classes, the function of the other plant CS isoforms, including the SPIol;Bsas3;1 protein, had been assumed to be that of CS but had never been investigated.
The ARAth;Bsas4;1 recombinant protein expressed in the E. coli NK3 mutant strain could not complement the CS mutation (data not shown) and displayed a very low CS activity (Table III). No CAS activity could be detected, indicating that the CAS/CS specific activity ratio of this protein might be low. The ARAth;Bsas4;1 recombinant protein was not recognized either by the anti-CAS or the anti-CS antibody (Fig. 3). Nevertheless, the Km values for the CS reaction were comparable to that of SPIol;Bsas1;1 and CS-B (Table IV). From these observations, we conclude that ARAth;Bsas4;1 is likely to encode a true CS isoform. The low activity in protein extracts, the lack of recognition by the antibodies and the non-complementation of the NK3 mutant are likely due to a problem in the expression of this protein, which has not been investigated.
The catalytic properties of CS-like recombinant proteins (Tables III and IV) show that the CS reaction is favored in SPIol;Bsas1;1 and SPIol;Bsas2;1, compared with SPIol;Bsas3;1 and ARAth;Bsas3;1. In contrast, SPIol;Bsas3;1 and ARAth;Bsas3;1 have a much higher affinity for cyanide than SPIol;Bsas1;1 and SPIol;Bsas2;1. In addition, the E. coli NK3 strain cysK-cysM mutation could not be complemented by expression of SPIol;Bsas3;1 and ARAth;Bsas3;1 proteins (data not shown), indicating that the CS capacities of these proteins may not be sufficient to fulfill the mutant needs for Cys. SPIol;Bsas1;1 and SPIol;Bsas2;1 were able to complement the CS mutation. Immunoblotting experiments with polyclonal antibodies directed against purified CS or CAS proteins showed that SPIol;Bsas1;1 and SPIol;Bsas2;1 proteins were distinct from SPIol;Bsas3;1 and ARAth;Bsas3;1, despite being closely related (Fig. 3). Moreover, the 12 N-terminal residue sequence of purified spinach CAS was identical to residues 28 to 39 of the SPIol;Bsas3;1 deduced protein sequence (Warrilow and Hawkesford, 2000). The partial amino acid sequence of potato CAS was recently shown to be similar to that of SPIol;Bsas3;1 (Maruyama et al., 2000). All of these observations indicate that the mitochondrial Bsas3 class of CS-like proteins encodes CAS in plants, rather than CS as it was assumed previously. This is a good reminder in this genomics era of the limitations of function assignment to genes and proteins relying only on sequence homologies.
Functions of the Various CS-Like Proteins
Significant Cys synthesis is thought to occur in mitochondria, which account for about 14% of the overall CS activity in spinach leaf tissues (Lunn et al., 1990). In the purified mitochondria fraction, the CS to CAS ratio is about 3.4 × 10−1 (Warrilow and Hawkesford, 1998), which is much higher than the value we observed for the recombinant SPIol;Bsas3;1 protein expressed in E. coli (Table III). The low CS capacity of SPIol;Bsas3;1 may not be responsible for the entire observed CS activity in mitochondria, and this observation suggests the existence of a true mitochondrial CS isoform in spinach that has not yet been identified by biochemical methods. The recent functional characterization of a cDNA encoding a mitochondrial CS belonging to the Bsas2 class in Arabidopsis, ARAth;Bsas2;2 (also referred as mtACS1), supports this assumption (Hesse et al., 1999).
Different isoforms of Ser acetyltransferase (SAT), which catalyzes the formation of OAS, are present in cytosol, chloroplast, and mitochondria (Noji et al., 1998). The formation of a SAT-CS multimolecular complex has been demonstrated in plants (Saito et al., 1995; Droux et al., 1998). Plant SATs are also able to form a complex with E. coli CS, despite the genetic distance between plant and bacterial CS (Bogdanova and Hell, 1997; Droux et al., 1998). Therefore, mitochondrial SAT could form a complex with the Bsas3 isoform in mitochondria. SAT is highly unstable in a free form, and efficient OAS synthesis needs such a complex formation, which in turn decreases dramatically the specific activity of the bound CS enzyme (Droux et al., 1998). The occurrence and the effects of such a complex formation on CAS function remain to be investigated.
CS and CAS activities have been detected in cytosol, chloroplast, and mitochondria (Lunn et al., 1990; Maruyama et al., 1998; Warrilow and Hawkesford, 1998). We showed that cytosolic SPIol;Bsas1;1 and chloroplastic SPIol;Bsas2;1 isoforms of spinach are true CS but also display CAS capacities (Tables III and IV). In E. coli, CAS is encoded by one or both of the CS genes, cysK and cysM (Table III), confirming previous speculations (Dunnill and Fowden, 1965). By analogy with bacteria, it is possible that the CAS capacities of CS could account for all of the CAS activity observed in cytosol and chloroplasts, without the involvement of a true CAS lacking CS capacity in these compartments.
1-Aminocyclopropane-1-carboxylic acid oxidase, which catalyzes ethylene formation, is located at the plasma membrane or in the apoplastic space (Zarembinski and Theologis, 1994), and the cyanide resulting from its activity would subsequently diffuse to the various compartments of the cell. In plants, CS catalytic capacity may exceed several hundred-fold the cell needs in Cys synthesis (Schmidt and Jäger, 1992). Therefore, the CS and CAS enzymatic activities of Bsas1 and Bsas2 isoforms are unlikely to compete for the availability of the active site, and the cyanide in cytosol and chloroplasts could be processed by the CAS activity of these isoforms. Nevertheless, the affinity of these CSs for cyanide is rather low (Km approximately 5 mm), and they could not be sufficient to ensure a total cyanide detoxification before this compound could reach the mitochondria. The absolute need of an efficient cyanide detoxification process in mitochondria would be fulfilled by a true CAS with a high affinity to cyanide and dedicated to this function.
Evolution of the CS-Like Protein Family
Plant CS-like proteins are evolutionarily related to bacterial CS and to fungal cystathionine β-synthase (CBS), which catalyzes the formation of cystathionine from Ser and homo-Cys in Cys synthesis (Thomas and Surdin-Kerjan, 1997). The catalytic capacities of the common ancestor enzyme could be that of a general β-substituted Ala synthase and encompass CS, CAS, and CBS functions. During the course of evolution, the kinetic characteristics of some isoforms could have drifted toward the CAS or the CS reactions in plants, to yield enzymes with more narrow capacities, or different substrate specificities in the case of fungal CBS. Such an hypothesis has been proposed to explain the similarities of fungal CBS to plant and bacterial CS (Cherest et al., 1993). In this evolutionary model, we speculate that the Bsas5 and Bsas6 classes may not encode CS or CAS enzymes but still belong to the β-substituted Ala synthases family. The functional characterization of these later isoforms could answer this question.
The kinetic characteristics of CAS for the CS reaction fit well with the proposed function of both enzymes (Table IV): The affinity of CAS for OAS is much lower than that of CS and thus makes the Cys synthesis reaction much less favored in CAS. On the other hand, the kinetic characteristics of CAS and CS for the β-cyano-Ala synthesis reaction are quite puzzling: The CS shows a high affinity to Cys and low affinity to cyanide, whereas the CAS displays the reverse situation. Therefore, none of them is an absolutely efficient or nonefficient CAS. In a functional perspective, this could be explained if a high affinity for cyanide were incompatible with a high affinity for OAS. The selection pressure on CS would maintain a high affinity to OAS, and this enzyme would also have a high affinity to Cys [HS-CH2CH(NH2) COOH], which is structurally similar to OAS [H3CCO-O-CH2CH(NH2) COOH], as a side effect. In contrast, the selection pressure on CAS would maintain a high affinity to cyanide at the expense of the affinity to OAS, and this enzyme would therefore have a low affinity to Cys as a side effect. The affinity for sulfide would be only mildly affected during this process, because of its different nature compared to cyanide. Because Cys synthesis occurs in mitochondria (Rolland et al., 1992), this substrate could be available in concentrations high enough to allow the detoxification of incoming cyanide by CAS, despite the low affinity of this enzyme for Cys. The transformation of CS into CAS and vice versa by mutagenesis should allow us to explore these hypotheses.
MATERIALS AND METHODS
Miscellaneous Techniques
Molecular biology cloning, bacterial media, cultures, and heat shock transformation were performed according to standard procedures (Sambrook et al., 1989).
Isolation of New Members of the CS-Like Protein Family in Arabidopsis
The dbEST database was screened for ESTs encoding putative CS homologs in Arabidopsis using the various BLAST algorithms. The ESTs identified were separated into families based on their sequence homologies with plant CS-encoding cDNAs using the GELMERGE algorithm of the Wisconsin Package version 10.0 (Genetics Computer Group, Madison, WI). ESTs encoding putative new CS-like isoforms were obtained from the Arabidopsis Biological Resource Center (http://aims.cps.msu.edu/aims/) and sequenced by the dideoxynucleotide chain termination method using the Thermo Sequenase kit (Amersham-Pharmacia Biotech, Uppsala) and a DSQ2000 automatic sequencer (Shimazu, Kyoto), following the instructions of the manufacturers.
Expression of CS-Like Isoforms in an E. coli Cys Auxotroph Mutant
Arabidopsis and spinach (Spinacia oleracea) CS isoforms were introduced in-frame at the NcoI site of pTV118N (Takara Shuzo, Kyoto) by engineering a NcoI site containing an in-frame ATG codon in the sequence by PCR, using synthetic oligonucleotide primers (SPIol;Bsas3;1-MP: GTACGCCATGGGGACTAATATTAA AACC; SPIol;Bsas3;1-MP: CTACGCCATGGAGCTATAAGATGCTG; ARAth;Bsas4;1: GGAAGAATTTTGCCATGGAGGAGG; ARAth;Bsas3;1-MP: AGATCTCCCCATGGACTTCCCCTC; M13-20 [for ARAth;Bsas4;1 and ARAth;Bsas3;1-MP]: GTAAAACGACGGCCAGT). The PCR mixture contained 1× buffer, 250 μm each deoxyribonucleotide triphosphates, 0.4 μm primers, 2.5 units of Ex-Taq polymerase (Takara Shuzo), and 10 ng of purified plasmid in a final volume of 50 μL. Amplification conditions were: 95°C, 5 min; 95°C, 1 min to 50°C, 1 min to 72°C, 2 min (30 cycles); 72°C 5 min. The resulting expression vectors (Table II) were introduced into the Escherichia coli CS-deficient strain NK3 (ΔtrpE5 leu-6 thi hsdR hsdM+ cysK cysM).
Determination of CS and CAS Activities
An overnight preculture of transformed E. coli was diluted 100 times in liquid Luria-Bertani broth containing 100 mg/L ampicillin and allowed to grow at 37°C for 3 h under agitation. Isopropyl β-d-thiogalactoside was added to a concentration of 1 mm, and the cultures were grown for 9 h more at 37°C under agitation. Cell-free crude protein extracts were obtained as previously described (Noji et al., 1998). The proteins were assayed by the Bradford method (Bradford, 1976) using a protein assay kit (Bio-Rad Laboratories, Hercules, CA) with bovine serum albumin as a standard.
CS activity of fresh crude protein extracts was determined as reported previously (Saito et al., 1994). The Cys produced was quantified by spectrophotometry using the acid-ninhydrin method (Gaitonde, 1967). One unit was defined as the synthesis of 1 μmol of Cys min−1.
CAS activity of fresh crude protein extracts was determined by quantification of the sulfide produced (Hasegawa et al., 1994). When the l-Cys concentration was modified for kinetic studies, d-Cys was added after the incubation to equilibrate the total dl-Cys concentration to 5 mm prior to sulfide quantification. One unit was defined as the production of 1 μmol of sulfide min−1.
Immunoblotting
Ten micrograms of total proteins from crude E. coli protein extracts was separated by SDS-PAGE (Laemmli, 1970) in a 12% (w/v) acrylamide gel. Immunoblotting was carried-out as published previously (Saito et al., 1991). Rabbit polyclonal antibodies raised against purified spinach CAS or purified spinach CS (Schmidt, 1990) were used at a 1:10,000 dilution. Goat anti-rabbit IgG antibodies conjugated with phosphatase (Kirkegaard & Perry Laboratories, Madison, WI) were used at a 1:4,000 dilution.
Inhibition of Purified Spinach CS and CAS Activities by Anti-CS and Anti-CAS Antibodies
Fifty microliters of biochemically purified spinach CS or CAS (Schmidt, 1990) was incubated with 50 μL of antibody for 15 min at 4°C and then centrifuged at 15,000g for 10 min at 4°C. CS or CAS activity was determined on 50 μL of the supernatant.
ACKNOWLEDGMENTS
We are grateful to the Institut National de la Recherche Agronomique (France) for access to the Genetics Computer Group, version 10.0, package. We also thank Dr. Anthony J. Michael for kindly correcting the grammar.
Footnotes
This work was supported, in part, by Grants-in-Aid for Scientific Research and the Japan Society for Promotion of Science (JSPS) fellows from the Ministry of Education, Science, Sports and Culture, Japan (Monbusho), and by the Research for the Future Program (grant no. 96I00302) from JSPS. Y.H. is supported by a postdoctoral fellowship from JSPS (no. P97.158). A.M. is supported by a research fellowship for young scientists from JSPS (no. 1839).
LITERATURE CITED
- Akopyan T, Braunstein A, Goryachenkova E. β-Cyanoalanine synthase: purification and characterization. Proc Natl Acad Sci USA. 1975;72:1617–1621. doi: 10.1073/pnas.72.4.1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumenthal S, Hendrickson H, Abrol Y, Conn E. Cyanide metabolism in higher plants: III. The biosynthesis of β-cyanoalanine. J Biol Chem. 1968;243:5302–5307. [PubMed] [Google Scholar]
- Bogdanova N, Hell R. Cysteine synthesis in plants: protein-protein interactions of serine acetyltransferase from Arabidopsis thaliana. Plant J. 1997;11:251–262. doi: 10.1046/j.1365-313x.1997.11020251.x. [DOI] [PubMed] [Google Scholar]
- Bradford M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Castric P, Farnden K, Conn E. Cyanide metabolism in higher plants: V. The formation of asparagine from β-cyanoalanine. Arch Biochem Biophys. 1972;152:62–69. doi: 10.1016/0003-9861(72)90193-2. [DOI] [PubMed] [Google Scholar]
- Cherest H, Thomas D, Surdin-Kerjan Y. Cysteine biosynthesis in Saccharomyces cerevisiae occurs through the transsulfuration pathway which has been built up by enzyme recruitment. J Bacteriol. 1993;175:5366–5374. doi: 10.1128/jb.175.17.5366-5374.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Droux M, Martins J, Sajus P, Douce R. Purification and characterization of O-acetylserine(thiol) lyase from spinach chloroplasts. Arch Biochem Biophys. 1992;295:379–390. doi: 10.1016/0003-9861(92)90531-z. [DOI] [PubMed] [Google Scholar]
- Droux M, Ruffet M, Douce R, Job D. Interactions between serine acetyltransferase and O-acetylserine(thiol) lyase in higher plants: structural and kinetic properties of the free and bound enzymes. Eur J Biochem. 1998;255:235–245. doi: 10.1046/j.1432-1327.1998.2550235.x. [DOI] [PubMed] [Google Scholar]
- Dunnill P, Fowden L. Enzymatic formation of β-cyanoalanine from cyanide by Escherichia coli extracts. Nature. 1965;208:1206–1207. doi: 10.1038/2081206a0. [DOI] [PubMed] [Google Scholar]
- Floss H, Hadwiger L, Conn E. Enzymatic formation of β-cyanoalanine from cyanide. Nature. 1965;208:1207–1208. doi: 10.1038/2081207a0. [DOI] [PubMed] [Google Scholar]
- Gaitonde M. A spectrophotometric method for the direct determination of cysteine in the presence of other naturally occurring amino acids. Biochem J. 1967;104:627–633. doi: 10.1042/bj1040627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa R, Tada T, Torii Y, Esashi Y. Presence of β-cyanoalanine synthase in unimbibed dry seeds and its activation by ethylene during pre-germination. Physiol Plant. 1994;91:141–146. [Google Scholar]
- Hell R, Bork C, Bogdanova N, Frolov I, Hauschild R. Isolation and characterization of two cDNAs encoding for compartment specific isoforms of O-acetylserine(thiol) lyase from Arabidopsis thaliana. FEBS Lett. 1994;351:257–262. doi: 10.1016/0014-5793(94)00872-8. [DOI] [PubMed] [Google Scholar]
- Hendrickson H, Conn E. Cyanide metabolism in higher plants: IV. Purification and properties of the β-cyanoalanine synthase of blue lupin. J Biol Chem. 1969;244:2632–2640. [PubMed] [Google Scholar]
- Hesse H, Lipke J, Altmann T, Hofgen R. Molecular cloning and expression analyses of mitochondrial and plastidic isoforms of cysteine synthase (O-acetylserine(thiol) lyase) from Arabidopsis thaliana. Amino Acids. 1999;16:113–131. doi: 10.1007/BF01321531. [DOI] [PubMed] [Google Scholar]
- Ikegami F, Itagaki S, Murakoshi I. Purification and characterization of two forms of cysteine synthase from Allium tuberosum. Phytochemistry. 1993;32:31–34. [Google Scholar]
- Ikegami F, Murakoshi I. Enzymic synthesis of non-protein β-substituted alanines and some higher homologues in plants. Phytochemistry. 1994;35:1089–1104. [Google Scholar]
- Ikegami F, Takayama K, Murakoshi I. Purification and properties of β-cyano-l-alanine synthase from Lathyrus latifolius. Phytochemistry. 1988a;27:3385–3389. [Google Scholar]
- Ikegami F, Takayama K, Tajima C, Murakoshi I. Purification and properties of β-cyano-l-alanine synthase from Spinacia oleracea. Phytochemistry. 1988b;27:2011–2016. [Google Scholar]
- Laemmli U. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lunn J, Droux M, Martin J, Douce R. Localization of ATP sulfurylase and O-acetylserine(thiol) lyase in spinach leaves. Plant Physiol. 1990;94:1345–1352. doi: 10.1104/pp.94.3.1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama A, Ishizawa K, Takagi T. Purification and characterization of β-cyanoalanine synthase and cysteine synthases from potato tubers: are β-cyanoalanine synthase and mitochondrial cysteine synthase the same enzyme? Plant Cell Physiol. 2000;41:200–208. doi: 10.1093/pcp/41.2.200. [DOI] [PubMed] [Google Scholar]
- Maruyama A, Ishizawa K, Takagi T, Esashi Y. Cytosolic β-cyanoalanine synthase activity attributed to cysteine synthases in cocklebur seeds: purification and characterization of cytosolic cysteine synthases. Plant Cell Physiol. 1998;39:671–680. doi: 10.1093/oxfordjournals.pcp.a029421. [DOI] [PubMed] [Google Scholar]
- Masada M, Fukushima K, Tamura G. Cysteine synthase from rape leaves. J Biochem. 1975;77:1107–1115. doi: 10.1093/oxfordjournals.jbchem.a130811. [DOI] [PubMed] [Google Scholar]
- McAdam A, Knowles C. Purification and properties of β-cyanoalanine synthase from the cyanide-producing bacterium Chromobacterium violaceum. Biochem Biophys Acta. 1984;786:123–132. [Google Scholar]
- Meyers D, Ahmad S. Link between l-3-cyanoalanine synthase activity and differential cyanide sensitivity of insects. Biochim Biophys Acta. 1991;1075:195–197. doi: 10.1016/0304-4165(91)90252-c. [DOI] [PubMed] [Google Scholar]
- Miller J, Conn E. Metabolism of hydrogen cyanaide by higher plants. Plant Physiol. 1980;65:1199–1202. doi: 10.1104/pp.65.6.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai K, Kanehisa M. A knowledge base for predicting protein localization sites in eukaryotic cells. Genomics. 1992;14:897–911. doi: 10.1016/S0888-7543(05)80111-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noji M, Inoue K, Kimura N, Gouda A, Saito K. Isoform-dependant differences in feedback regulation and subcellular localization of serine acetyltransferase involved in cysteine biosynthesis from Arabidopsis thaliana. J Biol Chem. 1998;273:32739–32745. doi: 10.1074/jbc.273.49.32739. [DOI] [PubMed] [Google Scholar]
- Peiser G, Wang T-T, Hoffman N, Yang S-F, Liu H-W, Walsh C. Formation of cyanide from carbon 1 of 1-aminocyclopropane-1-carboxylic acid during its conversion to ethylene. Proc Natl Acad Sci USA. 1984;81:3059–3063. doi: 10.1073/pnas.81.10.3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ressler C, Nigam S, Giza Y. Toxic principle in vetch: isolation and identification of γ-l-glutamyl-l-β-cyanoalanine from common vetch seeds: distribution in some legumes. J Am Chem Soc. 1969;91:2758–2765. doi: 10.1021/ja01038a058. [DOI] [PubMed] [Google Scholar]
- Rolland N, Droux M, Douce R. Subcellular distribution of O-acetylserine(thiol) lyase in cauliflower (Brassica oleracea L.) inflorescence. Plant Physiol. 1992;98:927–935. doi: 10.1104/pp.98.3.927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolland N, Droux M, Lebrun M, Douce R. O-acetylserine(thiol) lyase from spinach (Spinacia oleracea L.) leaf: cDNA cloning, characterization, and overexpression in Escherichia coli of the chloroplast isoform. Arch Biochem Biophys. 1993;300:213–222. doi: 10.1006/abbi.1993.1030. [DOI] [PubMed] [Google Scholar]
- Saito K, Miura N, Yamazaki M, Hirano H, Murakoshi I. Molecular cloning and bacterial expression of cDNA encoding a plant cysteine synthase. Proc Natl Acad Sci USA. 1992;89:8078–8082. doi: 10.1073/pnas.89.17.8078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K, Noji M, Ohmori S, Imai Y, Murakoshi I. Integration and expression of a rabbit liver cytochrome P-450 gene in transgenic Nicotiana tabacum. Proc Natl Acad Sci USA. 1991;88:7041–7045. doi: 10.1073/pnas.88.16.7041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K, Tatsuguchi K, Murakoshi I, Hirano H. cDNA cloning and expression of cysteine synthase B localized in chloroplasts of Spinacia oleracea. FEBS Lett. 1993;324:247–252. doi: 10.1016/0014-5793(93)80127-g. [DOI] [PubMed] [Google Scholar]
- Saito K, Tatsuguchi K, Takagi Y, Murakoshi I. Isolation and characterization of cDNA that encodes a putative mitochondrion-localizing isoform of cysteine synthase (O-acetylserine(thiol) lyase) from Spinacia oleracea. J Biol Chem. 1994;269:28187–28192. [PubMed] [Google Scholar]
- Saito K, Yokoyama H, Noji M, Murakoshi I. Molecular cloning and characterization of a plant serine acetyltransferase playing a regulatory role in cysteine biosynthesis from watermelon. J Biol Chem. 1995;270:16321–16326. doi: 10.1074/jbc.270.27.16321. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Ed 2. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Schmidt A. Cysteine synthase. In: Lea P, Dey P, Harborne J, editors. Methods in Plant Biochemistry. Vol. 3. New York: Academic Press; 1990. pp. 349–354. [Google Scholar]
- Schmidt A, Jäger K. Open questions about sulfur metabolism in plants. Annu Rev Plant Physiol Plant Mol Biol. 1992;43:325–349. [Google Scholar]
- Tate M, Enneking D. A mess of red pottage. Nature. 1992;359:357–358. doi: 10.1038/359357a0. [DOI] [PubMed] [Google Scholar]
- Thomas D, Surdin-Kerjan Y. Metabolism of sulfur amino-acids in Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 1997;61:503–532. doi: 10.1128/mmbr.61.4.503-532.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;24:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warrilow A, Hawkesford M. Separation, subcellular location and influence of sulphur nutrition on isoforms of cysteine synthase in spinach. J Exp Bot. 1998;49:1625–1636. [Google Scholar]
- Warrilow A, Hawkesford M (2000) Cysteine synthase (O-acetylserine(thiol) lyase) substrate specificities classify the mitochondrial isoform as a cyanoalanine synthase. J Exp Bot (in press) [DOI] [PubMed]
- Yip W-K, Yang S. Cyanide metabolism in relation to ethylene production in plant tissues. Plant Physiol. 1988;88:473–476. doi: 10.1104/pp.88.2.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youssefian S, Nakamura M, Sano H. Tobacco plants transformed with the O-acetylserine(thiol) lyase gene of wheat are resistant to toxic levels of hydrogen sulphide gas. Plant J. 1993;4:759–769. doi: 10.1046/j.1365-313x.1993.04050759.x. [DOI] [PubMed] [Google Scholar]
- Zarembinski T, Theologis A. Ethylene biosynthesis and action: a case of conservation. Plant Mol Biol. 1994;26:1579–1597. doi: 10.1007/BF00016491. [DOI] [PubMed] [Google Scholar]