Abstract
We present the molecular landscape of pediatric acute myeloid leukemia (AML) and characterize nearly 1,000 participants in Children’s Oncology Group (COG) AML trials. The COG–National Cancer Institute (NCI) TARGET AML initiative assessed cases by whole-genome, targeted DNA, mRNA and microRNA sequencing and CpG methylation profiling. Validated DNA variants corresponded to diverse, infrequent mutations, with fewer than 40 genes mutated in >2% of cases. In contrast, somatic structural variants, including new gene fusions and focal deletions of MBNL1, ZEB2 and ELF1, were disproportionately prevalent in young individuals as compared to adults. Conversely, mutations in DNMT3A and TP53, which were common in adults, were conspicuously absent from virtually all pediatric cases. New mutations in GATA2, FLT3 and CBL and recurrent mutations in MYC-ITD, NRAS, KRAS and WT1 were frequent in pediatric AML. Deletions, mutations and promoter DNA hypermethylation convergently impacted Wnt signaling, Polycomb repression, innate immune cell interactions and a cluster of zinc finger–encoding genes associated with KMT2A rearrangements. These results highlight the need for and facilitate the development of age-tailored targeted therapies for the treatment of pediatric AML.
Acute leukemia is the most common form of childhood cancer1, and its incidence is increasing. Despite constituting only 20% of pediatric acute leukemias, AML is overtaking acute lymphoblastic leukemia (ALL) as the leading cause of childhood leukemic mortality, in part because current prognostic schemas classify many children who will ultimately succumb to their disease as being at low or intermediate risk. Additionally, aside from investigational tyrosine kinase inhibitors for FLT3-activated AML, targeted therapies are not used in pediatric AML. Both problems stem from an inadequate understanding of the biology of childhood AML.
AML is a molecularly heterogeneous group of diseases affecting individuals of all ages2. Recent genome-scale studies have revealed new potentially targetable mutations prevalent in adult de novo AML3–5. However, the relevance of these findings to childhood AML remains unclear, as several of the most common mutations in adults are far less prevalent in pediatric AML6,7.
To date, no comprehensive characterization of pediatric AML has been described. Here we report the initial results of the Therapeutically Applicable Research to Generate Effective Treatments (TARGET) AML initiative, a collaborative COG–NCI project with the aim of comprehensively characterizing the mutational, transcriptional and epigenetic landscapes of a large, well-annotated cohort of pediatric AML. Through comparison of AML molecular profiles across age groups, we show that stark differences in mutated genes, structural variants and DNA methylation patterns distinguish AML in infants, children, adolescents and adults.
RESULTS
Overview of cohort characteristics
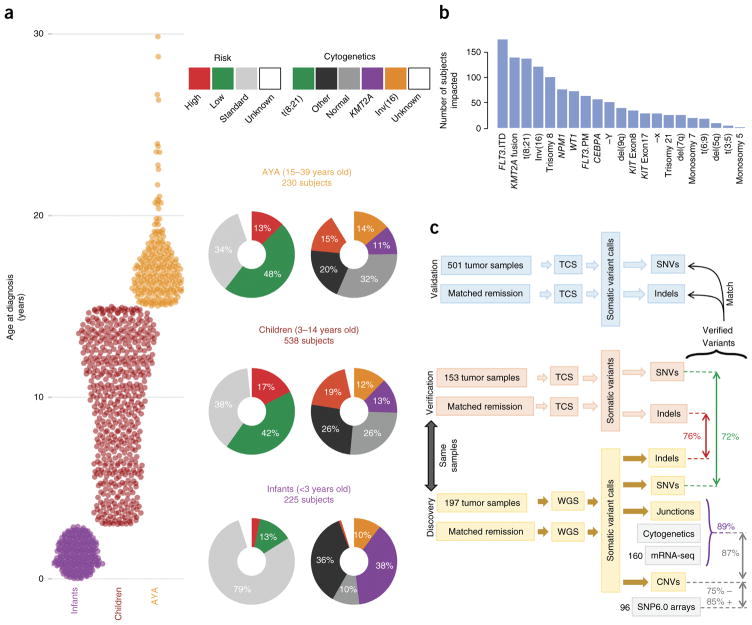
A total of 1,023 children enrolled in COG studies are included in the TARGET AML data set. Comprehensive clinical data, including clinical outcomes and test results for common sequence aberrations (Supplementary Table 1), are available for 993 subjects. Of these, 815 subjects were profiled for somatic mutations at presentation: 197 by whole-genome sequencing (WGS) and 800 by targeted capture sequencing (TCS) at read depths averaging 500× for validation of mutations identified by WGS. The WGS discovery cohort of diagnostic and remission (used as germline comparison) specimens were selected from subjects treated on recent COG studies who achieved an initial remission to induction chemotherapy. In these studies, the type or timing of induction therapy was randomized (CCG-2961)8 or gemtuzumab ozogamicin was administered in a single-arm pilot (AAML03P1)9 or a randomized fashion (AAML0531)10. Specimens for TCS validation were selected from 800 subjects, including 182 from the WGS discovery cohort (153 of whom had matched remission samples). A complete listing of cases and their characterizations is available in the TARGET sample matrix (see URLs). The age at presentation for the TARGET AML participants ranged from 8 d to 29 years (median, 10 years; Fig. 1a). Infants (<3 years old), children (3–14 years old), and adolescents and young adults (AYA; 15–39 years old) differed broadly by cytogenetic and clinical risk group classifications (Fig. 1a; multivariate chi-squared P < 10−22). This is consistent with observed differences in clinically evaluated structural abnormalities and mutations (Fig. 1b). Notably, among these clinically detected abnormalities, only five mutations and five structural aberrations occurred in more than 5% of subjects (mutations in FLT3, NPM1, WT1, CEBPA and KIT; fusions involving RUNX1, CBFB and KMT2A; trisomy 8 and loss of the Y chromosome).
Figure 1.
An overview of the TARGET AML study. (a) The distribution of subjects by clinical risk category and cytogenetic classification is shown adjacent to each age group analyzed (infant, <3 years old; child, 3–14 years old; AYA, 15–39 years old). KMT2A indicates cytogenetic KMT2A abnormality (t(11q23)). (b) Summary of the clinically established molecular aberrations in the cohort (n = 993 subjects). FLT3.ITD, FLT3 internal tandem duplications; FLT3.PM, FLT3 Asp835 point mutations; KIT Exon8 and KIT Exon17, KIT mutations impacting the named exons. (c) Overview of the genomic variant discovery, verification and validation process. We characterized diagnostic and remission (taken as germline) samples from 197 subjects using WGS and verified 153 diagnostic–remission case pairs using TCS of genes recurrently impacted in the WGS samples (an additional 29 WGS cases were verified by TCS of diagnostic cases only; Supplementary Fig. 1). Seventy-two percent of WGS SNVs and 76% of WGS indels were confirmed through TCS (red and green text, respectively). Purple text indicates the percentage of confirmed DNA junctions. For focal copy number (CN) alterations spanning fewer than seven genes, 75% of recurrent WGS deletion or loss and 85% of gain or amplification calls matched recurrent alterations discovered by SNP6 array in 96 matching samples. For chromosomal junctions, we integrated WGS, clinical karyotyping and RNA-seq data by majority vote, confirming 89% of WGS junction calls.
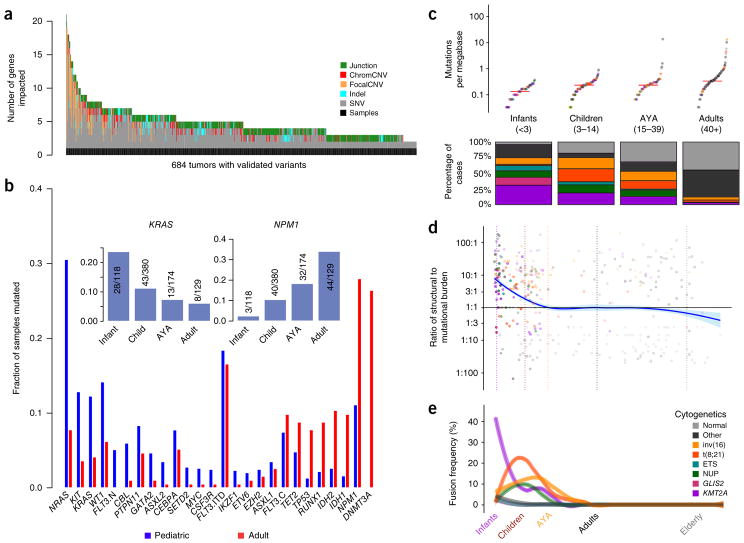
We validated each class of somatic DNA sequence alteration discovered by WGS through secondary assays (Fig. 1c and Supplementary Fig. 1). Single-nucleotide variants (SNVs) and short insertions and deletions (indels) were confirmed through TCS of coding sequences of the genes identified as recurrently altered in the WGS studies. WGS-detected copy number alterations were confirmed using GISTIC2.0 scores from SNP arrays; WGS-detected structural changes (such as translocations and inversions) were confirmed through RNA-seq and clinical leukemia karyotyping data. Across variant types, we found >70% concordance between at least two assays. These variants are henceforth referred to as verified variants. An overview of the multi-platform-verified somatic DNA variants in 684 subjects is presented in Figure 2a. Roughly one-quarter of the subjects possessed a normal karyotype, yet nearly all had at least 1 recurrent verified somatic DNA alteration, and at least 12 common cancer-associated cellular processes were recurrently impacted among subjects (Supplementary Fig. 2 and Supplementary Table 2a,b).
Figure 2.
Age-related differences in mutational and structural alterations in AML. (a) Distribution of variants per sample. At least one variant impacting a gene recurrently altered in pediatric AML was identified using multiplatform-validated variants in 684 subjects. Junction, protein fusion (Online Methods); chromCNV, chromosome-arm- or band-level copy variant; focalCNV, gene-level copy variant. (b) Age-dependent differences in the prevalence of mutations. FLT3 mutations are plotted in three categories: ITD (FLT3.ITD), activation loop domain (FLT3.C) and new, childhood-specific changes (FLT3.N). The inset shows a pattern of waxing or waning mutation rates across age groups that is evident in selected genes (KRAS and NPM1 are illustrated). (c) Top, childhood AML, like adult AML, has a low somatic mutation burden (Supplementary Fig. 5). Bottom, childhood AML is more frequently impacted by common cytogenetic alterations than adult AML. For the color key for c–e, see the legend in e. Midlines represent medians. (d) The ratio of the structural variation burden to that of SNVs and indels is high in infancy and early childhood and declines with age. Vertical dashed lines demarcate age groups, and plot points are represented in the same colors as in e. (e) Using a sliding-window approach to account for uneven sampling by age, the incidence of common translocations in AML is shown to follow age-specific patterns (multivariate chi-squared P < 10−30) and was greatest in infants as compared to all other ages (chi-squared P < 10−22). KMT2A fusions were most common in infants (chi-squared P < 10−20), and CBF fusions tended to affect older children (chi-squared P < 10−7). ETS and NUP refer to mutations in these gene families. GLIS2 and KMT2A refer to fusions involving these genes.
We carried out analyses of microRNAs (miRNAs), mRNAs and/or DNA methylation in 412 subjects. A summary of the assays performed and case–assay overlap is presented in Supplementary Figure 3. We compared our verified variants to those of 177 as adults (40+ years old) with AML cases from The Cancer Genome Atlas (TCGA) project3 stratified by the age groupings defined in Figure 1a. The TARGET and TCGA discovery cohorts both contained numerous AYA subjects (Supplementary Table 3). Notably, our conclusions regarding the molecular characteristics of this age group were identical when analyzing either cohort individually or both cohorts together (Supplementary Fig. 4).
Somatic gene mutations in pediatric AML
Like adult AML, pediatric AML has one of the lowest rates of mutation among cancers that are molecularly well characterized (Supplementary Fig. 5), with <1 somatic change in a protein-coding region per megabase in most cases. However, the landscape of somatic variants in pediatric AML was markedly different from that reported in adults3,4 (Fig. 2b, Supplementary Figs. 6 and 7, and Supplementary Table 4). Alterations in RAS genes, KIT and FLT3, including new, pediatric-specific FLT3 mutations (FLT3.N), were more common in children. Mutational burden increased with age, yet older individuals had relatively fewer recurrent cytogenetic alterations. Indeed, the number of SNVs in protein-coding regions within and across cohorts was best predicted by age (Fig. 2c; P <10−15) and by cytogenetic subgroup. In contradistinction to the higher prevalence of small sequence variants in older subjects, recurrent structural alterations, fusions and focal copy number aberrations were more common in younger subjects (Fig. 2d,e; P < 10−3). Subjects with CBFA2T3-GLIS2, KMT2A or NUP98 fusions tended to have fewer mutations than subjects without these fusions (P < 10−9), and these subgroups demonstrated inferior clinical outcome (Supplementary Fig. 8). Subjects with core binding factor (CBF) rearrangements tended to have more mutations than expected for their age (P < 10−15), yet they had more favorable outcomes than subjects without these rearrangements. The mutational spectrum of SNVs in protein-coding regions (Supplementary Fig. 5) accumulated C→T transitions with age (P < 10−3), with additional C→A transversions in subjects harboring tumors with t(8;21) (P < 10−2) or an aberrant karyotype (P < 10−2).
After adjustment for cytogenetic group and multiple comparisons, we found that NRAS (P < 10−3) and WT1 (P < 10−3) were mutated significantly more often in younger subjects, whereas DNMT3A (P < 10−23), IDH1 or IDH2 (P < 10−4), RUNX1 (P < 10−4), TP53 (P < 10−4) and NPM1 (P < 0.03) were mutated significantly more frequently in older subjects. Mutations in KRAS, CBL, GATA2, SETD2 and PTPN11 appeared to be more common in younger subjects (0.05 < P < 0.1, adjusted; Supplementary Figs. 7 and 9). We identified a prominent hotspot for MYC alterations11 and previously unreported internal tandem duplications (ITDs) that appeared exclusively in children (Supplementary Fig. 7). These observations were replicated in an independent Eastern Cooperative Oncology Group (ECOG) cohort (Supplementary Fig. 10a) of 384 adult subjects with AML5. As gene fusions have characteristic cooperating mutations12, we devised a weighted resampling scheme for comparison of mutation frequencies among 584 TARGET and 131 TCGA AML cases while controlling for karyotypic associations. The results confirm the generality of the pediatric– adult differences described above (Supplementary Fig. 10b).
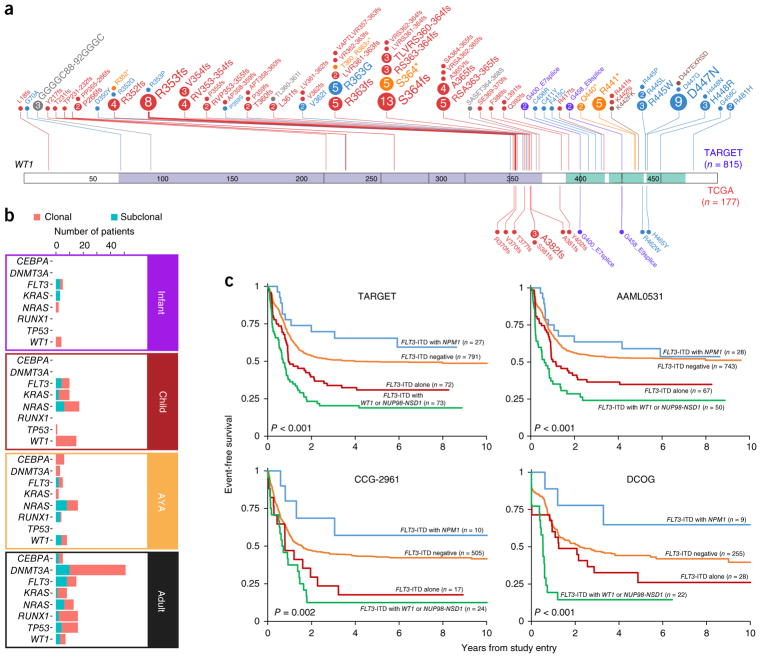
For genes such as CBL, GATA2, WT1, MYC and FLT3, both the frequency and sites of mutation often differed between children and adults (Fig. 3a and Supplementary Fig. 7), with multiple frequently recurrent alterations in pediatric AML distinct from those identified in adult AML. RAS-related mutations (in KRAS, NRAS, PTPN11 or NF1) were common, particularly with KMT2A fusions (Supplementary Fig. 11 and Supplementary Tables 4–6). In addition to being more common and varied, WT1 mutations were more likely to be of clonal origin in younger subjects (Fig. 3b), despite the majority of pediatric subjects presenting with multiple detectable clones (Supplementary Fig. 12).
Figure 3.
Biological and prognostic interactions between alterations of WT1 and NPM1, FLT3-ITD and NUP98-NSD1 fusions. (a) WT1 mutations appear more frequently and impact new sites in childhood AML (for the TARGET cohort, which is expanded above the WT1 representation: 150 alterations among 815 subjects (18.4%); for the TCGA cohort, which is expanded beneath the WT1 representation: 13 alterations among 177 subjects (7.3%); Fisher’s exact test P = 0.0002). Circles indicate sites of mutation with circle size proportional to the number of recurrently detected alterations. Colors indicate type of mutation: red, frameshifting; blue, missense; yellow, nonsense; purple, splice site; gray, in-frame deletion; brown, in-frame insertion. The WT1 domain (purple) and zinc finger domains (green) are shaded on the band. (b) Inference of the clonal origin of selected mutations in 197 TARGET AML (infant, child and AYA) cases with WGS and 177 TCGA AML (adult) cases (Online Methods). (c) The clinical impact of FLT3-ITD is modulated by other sequence aberrations. In the TARGET cohort, 963 subjects had complete data for FLT3-ITD, NPM1 and WT1 mutation status and NUP98-NSD1 fusion status. Subjects with FLT3-ITD plus WT1 mutation and/or NUP98-NSD1 fusion (n = 73) exhibited markedly inferior event-free (multivariate P < 0.001) and overall survival (Supplementary Fig. 13), whereas co-occurrence of NPM1 mutation with FLT3-ITD was associated with improved survival. These findings are confirmed by two separate studies from which TARGET cases were selected (AAML0531 and CCG-2961) as well as an independent cohort of subjects treated on European cooperative group trials (Dutch Childhood Oncology Group; Online Methods).
These differences are of clinical relevance: we have previously shown that newly identified FLT3 mutations were functional and resulted in poor responses to standard therapy13. The established adverse impact of FLT3-ITDs on survival was substantially modulated by cooccurring variants, including WT1 and NPM1 mutations and NUP98 translocations. As shown in Figure 3c and Supplementary Figures 13 and 14, three independent, large-scale studies demonstrated that FLT3-ITDs accompanied by NPM1 mutations were associated with relatively favorable outcomes in pediatric subjects, whereas FLT3-ITDs with WT1 mutations and/or NUP98-NSD1 fusions yielded poorer outcomes than FLT3-ITDs alone.
We found no mutations in protein-coding regions of DNMT3A in pediatric AML despite the high frequency of these mutations in adults. Spontaneous deamination of 5-methylcytosine is strongly associated with aging, and DNMT3A contains a CpG dinucleotide hotspot for Arg882 alteration by C→T deamination14. DNMT3A also directly interacts with TP53 (ref. 15), which is impacted far more frequently in adults. Mutations in DNMT3A or TP53 drive clonal hematopoiesis in many apparently healthy adults16 but are rare in children, as are the IDH1 and IDH2 mutations with which they often co-occur.
The spectrum of somatic structural DNA changes in pediatric AML
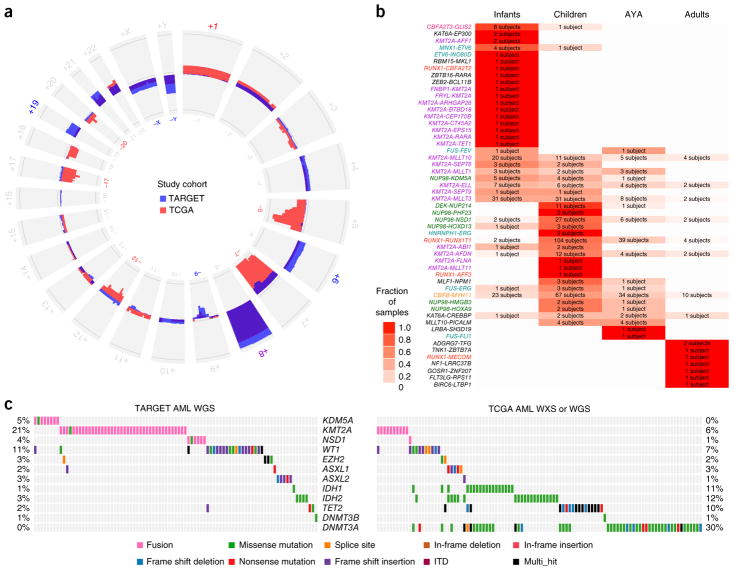
Many pediatric AML cases harbored chromosomal copy number changes distinct from those reported in adults (Fig. 4a). Among the 197 cases assayed by WGS, we identified 14 new focal deletions involving MBNL1, a splicing regulator, or ZEB2, a key regulator of normal17 and leukemic18 hematopoiesis (Supplementary Fig. 15). Despite occurring on separate chromosomes in regions devoid of other deletions, co-deletions of MBNL1 and ZEB2 occurred far more often than expected (P < 10−13). Half of these co-deletions accompanied KMT2A-MLLT3 fusions (P = 0.035; Supplementary Fig. 11 and Supplementary Tables 5 and 6). Samples with MBNL1–ZEB2 co-deletions had a larger number of recurrent mutations than samples without these co-deletions (P = 0.015), and KMT2A fusion samples with deletion of MBNL1 or ZEB2 had a larger number of additional cytogenetic abnormalities (P < 0.0005). Another 15 newly identified, validated focal deletions specifically impacted ELF1, an ETS family transcriptional regulator of MEIS1 (ref. 19). A statistically significant difference in ELF1 mRNA expression existed between ELF1-intact samples and those with ELF1 deletion (P < 0.01), with 63 genes differentially expressed between the two groups (P < 0.01; Supplementary Fig. 16). Among other new recurrent copy losses, we noted five heterozygous deletions of a region containing the IL9R gene (Supplementary Table 5) co-occurring with KIT mutations and t(8;21).
Figure 4.
Chromosomal alterations in pediatric and adult subjects with AML. (a) Patterns of regional and chromosomal gain (outward projection) and loss (inward projection) in the TARGET (blue) and TCGA (red) AML cohorts. Purple indicates the gain occurred in both cohorts. Losses of chromosomes 5q, 7 and 17 predominate in adults, whereas gains of chromosomes 4, 6 and 19 and losses of 9, X and Y are more common in younger subjects. Chromosome numbers are printed on the outside and inside of the circle plot and colored where there are large pediatric–adult differences. (b) Age-specific distributions of validated gene fusions. The fraction of events within an age group for each fusion pair is indicated by white–red shading, and the color of the fusion labels indicates the primary cytogenetic group (colors are the same as in Fig. 1a; see also Supplementary Figs. 17 and 18). The number in each box indicates the number of subjects carrying the indicated translocation (labels at left). (c) Structural and mutational aberrations affecting epigenetic regulators in subjects in the TARGET (WGS) and TCGA AML cohorts. WXS, whole-exome sequencing; Multi_hit, multiple nonsynonymous abberations affecting the same gene.
In accordance with our previous findings regarding NUP98-NSD1 fusions20, we identified an expansive catalog of gene fusions, many of which were observed primarily or exclusively in pediatric cases, underscoring the disproportionate impact of structural variants in younger subjects (Fig. 4b and Supplementary Figs. 17 and 18). But patterns of mutual exclusivity and cooperation were not limited to individuals with recurrent structural alterations: mutated GATA2 was frequently seen in children with normal karyotype (NK) AML, and both GATA2 (P < 10−9) and CSF3R (P < 10−6; Supplementary Fig. 19) mutations co-occurred with mutations of CEBPA21. GATA2 and CEBPA are key regulators of hematopoiesis22,23; the proteins encoded by these genes both interact with RUNX1 in normal hematopoiesis and leukemogenesis24. As with interactions among FLT3-ITD, NUP98-NSD1 and WT1, these findings show that prognostic interactions influence pediatric AML outcome (Supplementary Fig. 19b). RUNX1 mutations and RUNX1-RUNX1T1 gene fusions were significantly mutually exclusive from GATA2 and CEBPA mutations (P = 0.006; Supplementary Fig. 20 and Supplementary Table 7). All four of these alterations occurred in a significantly mutually exclusive manner with KMT2A rearrangements (P < 10−15), CBFB-MYH11 gene fusions (P < 10−11) and ETV6 aberrations (P = 0.01).
DNA methylation subtypes in pediatric AML
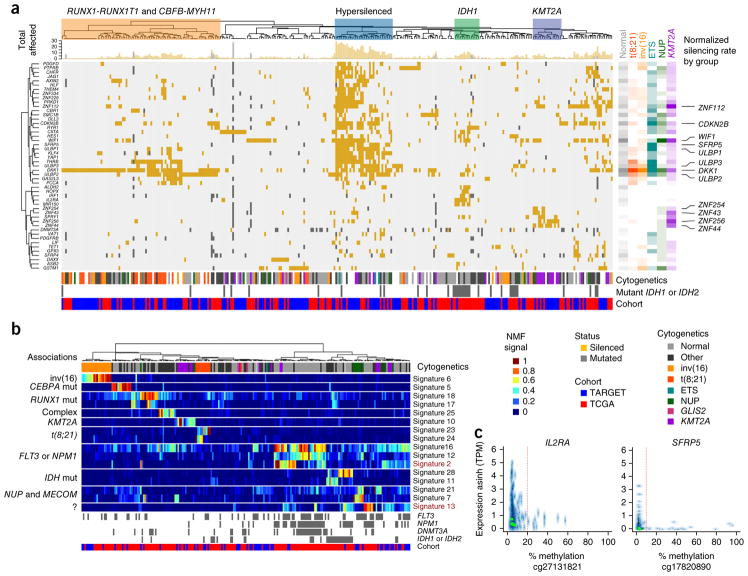
As summarized in Figure 4c, aberrations affecting epigenetic regulators are widespread and rarely overlap in AML, but their origin (structural versus mutational) and frequency differ between children and adults. By combining DNA methylation and mRNA expression results for 56 TARGET and TCGA AML cases, we identified dozens of genes with recurrent transcriptional silencing via promoter hypermethylation across these cases (Fig. 5a,c, Supplementary Tables 8 and 9, and Supplementary Figs. 21 and 22). A number of samples exhibited widespread silencing of genes by aberrant promoter hypermethylation, and this group was enriched for younger subjects with WT1 mutations (P = 0.0012; Fig. 5a, hypersilenced group). Aberrant Wnt–β-catenin signaling is required for the development of leukemic stem cells25, and one or more of the genes encoding Wnt pathway regulators—DKK1, SMAD1, SMAD5, SFRP4, SFRP5, AXIN2, WIF1, FZD3, HES1 and TLE1—were deleted or aberrantly methylated in most AML cases26. Repression of genes encoding activating ligands for natural killer (NK) cells (particularly ULBP1, ULBP2 and ULBP3) appeared to be common in pediatric subjects, and these genes may represent a therapeutic target27. In subjects with rearranged KMT2A, a cluster of poorly characterized zinc finger–encoding genes on chromosome 19 was recurrently silenced.
Figure 5.
Aberrant DNA methylation in adult and pediatric AML. (a) Integrative analysis of genes with recurrent mutations, deletions or transcriptional silencing by promoter DNA hypermethylation (rows) in TARGET and TCGA AML cases (columns). Cluster associations are labeled at the top, including a prominent group enriched for younger subjects with WT1 mutations (P = 0.0012) that shows extensive transcriptional silencing across dozens of genes (blue boxed region). The cytogenetic group, IDH1 or IDH2 mutation status (gray, mutated; white, wild type or unknown) and TARGET or TCGA cohort membership (blue and red, respectively) for each sample are indicated below the main figure. The top marginal histogram indicates the total number of genes impacted for each subject. Right, gene and cytogenetic associations; rate of involvement by cytogenetic class per gene is indicated by color and shading (unfilled, no involvement; full shading, maximum observed involvement of any gene within subjects of the indicated cytogenetic grouping). Wnt regulators and activating NK cell ligands (for example, DKK1 and WIF1; and ULBP1, ULBP2 and ULBP3, respectively) are silenced across cytogenetic subtypes (labeled at far right). Distinct groups of silenced genes are also associated with subjects with mutated IDH1 or IDH2 and with subjects with rearranged KMT2A. A subset of genes (56 of 119) altered in >3 subjects and in subjects (n = 310; 168 TARGET, 142 TCGA) with one or more genes silenced by promoter methylation is illustrated (Supplementary Figs. 21 and 22, and Supplementary Tables 8 and 9, enumeration of all 119 genes in all 456 evaluable subjects.). (b) A subset (16 of 31) of DNA methylation signatures derived by NMF and in silico purification with samples ordered by hierarchical clustering of signatures (labeled at right). Genomic associations are indicated to the left of the main panel. Signature 13 does not correspond directly to any known recurrent alterations; however, it displays potential prognostic relevance, as does signature 2 (Supplementary Fig. 24). The subject-specific score matrix and display of all 31 signatures are provided in Supplementary Table 10 and Supplementary Figure 23. ?, unknown associations. (c) Examples of expression–promoter DNA methylation relationships are shown for IL2RA and SFRP5; these two genes were identified as recurrently silenced (in a) and also contribute to NMF signatures (in b). y axis, transformed expression (asinh(TPM)); x axis, promoter CpG methylation numbers below x-axis, methylation array probe identifiers; TPM, transcripts per million. The vertical red line indicates the empirically established silencing threshold.
We applied non-negative matrix factorization (NMF) to CpG methylation data from 284 TARGET and TCGA subjects with AML for whom DNA methylation data were available. Through cross-validation, we identified 31 signatures (Supplementary Table 10) that best captured DNA methylation differences across samples after controlling for differences in cellularity via in silico purification. Unsupervised clustering of the resulting DNA methylation signatures largely separated subjects by age and karyotypic subtype (Fig. 5b and Supplementary Fig. 23) but also revealed a signature that did not associate strongly with age or established prognostic factors (signature 13; Fig. 5b). Two signatures (signatures 2 and 13) predicted significantly poorer event-free survival (P < 0.05) in both pediatric and adult subjects with above-median scores after stratifying by cohort and adjusting for TP53 mutation status and white blood cell count (Supplementary Fig. 24). Larger sample sizes are needed to evaluate the clinical value of these findings.
The pediatric AML transcriptome is shaped by diverse miRNAs
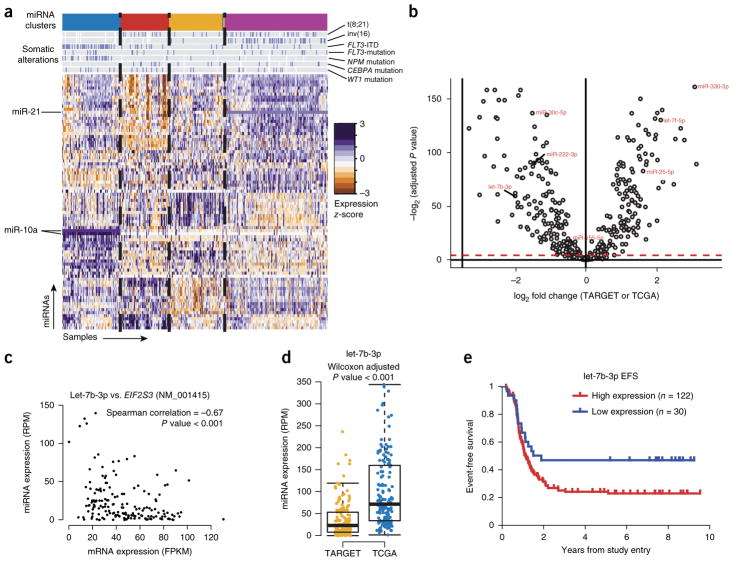
We performed miRNA-seq for 152 subjects to characterize miRNA expression patterns in pediatric AML. Unsupervised clustering of the data revealed four discrete subgroups that were correlated with specific genomic alterations (Fig. 6a and Supplementary Fig. 25), including high miR-10a expression in samples with NPM1 mutations, which is consistent with previous reports28. Further, Cox proportional-hazards analyses identified multiple miRNAs associated with clinical outcome (Supplementary Figs. 26–28 and Supplementary Table 11), including miR-155, which we previously reported to predict poor survival29.
Figure 6.
miRNAs differentially regulate distinct molecular and age subgroups in AML. (a) Unsupervised clustering of miRNA expression patterns in 152 childhood AML cases identifies four subgroups of subjects (colored bands at top) with correlations to somatic alterations as indicated (blue bars on gray background) and subgroup-specific miRNA expression (miR-10 and miR-21 are highlighted as examples). (b) Age-related differences in miRNA expression are evident between adult (n = 162) and pediatric (n = 152) AML. The volcano plot shows differentially expressed miRNAs between adult and pediatric cases (Wilcoxon test, Benjamini–Hochberg adjusted P < 0.05; significance threshold indicated by dashed red line). (c,d) A predicted miRNA:mRNA target relationship involving let-7b (c), which was less abundant in most pediatric cases than in adult cases (d). RPM, reads per million; FPKM, fragments per kilobase of transcript per million mapped reads. In boxplots, the midline represents the median; the upper and lower perimeters mark the interquartile range (IQR); and tails mark the upper and lower bounds of 1.5 times the IQR. (e) High expression of let-7b occurred in a minority of pediatric AML cases and was associated with shorter time to relapse.
Differential expression analyses using Wilcoxon tests identified miRNAs that were differentially expressed between pediatric and adult AML (Fig. 6b). Of note, miR-330 was the most overexpressed miRNA in pediatric samples and has previously been shown to have oncogenic potential in AML30.
Several miRNAs with age-associated expression have binding sites within and expression levels anticorrelated with putative target genes that may be involved in RNA and protein processing, suggesting that these miRNAs could contribute to leukemogenesis through the dysregulation of transcripts and proteins31. Of note, let-7b, which is a potential regulator of protein synthesis via EIF2S3 (Fig. 6c), was typically less abundantly expressed in pediatric AML (Fig. 6d). However, high let-7b expression in pediatric AML was associated with shorter time to relapse (log-rank P < 0.05; Fig. 6e).
DISCUSSION
Using a large cohort of subjects, this study establishes the prevalence of and coincident relationships among recurrent somatic genetic and epigenetic abnormalities in pediatric AML. We observed several features in common between pediatric and adult AML: a low overall mutation rate in comparison to other cancers, a long tail of infrequently affected genes and overlap among recurrently impacted genes. However, pediatric AML exhibits distinctive and critically important characteristics. We and others have previously reported on the presence and clinical impact of new fusion genes in pediatric AML20,32. As this study illustrates, the impact of fusion transcripts in AML is both broad and age dependent. Recognition and comprehensive testing for these alterations are key first steps in the development of new modes of targeted therapy33.
Recurrent focal deletions represent a unique aspect of pediatric AML. Regional (for example, chromosome-arm- and band-level) copy loss differs substantially by age, but surprisingly, focal areas of copy loss are also more common in children, specifically impacting ZEB2, MBNL1 and ELF1. MBNL1 is upregulated by the KMT2A-AF9 fusion protein34, and genes encoding products involved in post-transcriptional processing (SETD2, U2AF1 and DICER1) harbored the sole recurrent mutation in several cases with rearrangements involving KMT2A, suggesting a functional role for altered splicing in pediatric leukemogenesis. Alterations in Zeb2 have been identified as cooperating events in murine CALM-AF10 leukemia models35, and Zeb2-knockout mice develop myelofibrosis36, suggesting a fundamental role for this gene in the pathogenesis of AML.
Many of the genes characteristically mutated in AML are altered at widely variable frequencies across age groups; several (including FLT3 and WT1) are impacted by pediatric-specific variants and hotspots. Clinical tests for a handful of genomic alterations are widely used to stratify patients by risk and to determine treatment regimens. However, the current practice of considering the effect of each somatic alteration in isolation is inadequate. As we illustrate for FLT3-ITD, interactions among sequence variants can have dramatic clinical consequences. Moreover, some interactions appear to be age specific. In pediatric AML, FLT3-ITD and NPM1 mutations co-occur in the absence of DNMT3A mutations in a group of subjects with superior outcomes (Fig. 3c and Supplementary Figs. 13 and 14) in contrast to the inferior outcomes reported in adults in whom FLT3-ITD and NPM1 mutations frequently co-occur with mutations in DNMT3A4. In the TCGA adult AML cohort, over half of the subjects with somatic FLT3 and NPM1 mutations also possessed somatic DNMT3A mutations3. Subsequent studies have established the generality of this result4 and have revealed that DNMT3A mutations are early clonal events37 that often cooperate with later NPM1 and FLT3 mutations to promote chemoresistance, mutagenesis38 and inferior outcomes39. Similarly, the co-occurrence of FLT3-ITDs with WT1 mutations or NUP98-NSD1 fusions accompanies frequent induction failure and dismal outcomes in children with AML (multivariate P < 10−4; Fig. 3c and Supplementary Figs. 13 and 14).
In TARGET, TCGA and ECOG AML cases, WT1 mutations were mutually exclusive with those in ASXL1 and EZH2 (P < 10−3). WT1 recruits EZH2 to specific targets40, and WT1 mutations have been linked to promoter DNA hypermethylation of EZH2 target genes41. Mutations in ASXL1 abolish EZH2-mediated silencing of HOX genes42. The EZH2 gene resides on a recurrently deleted region of chromosome 7, and decreased EZH2 activity is associated with treatment- resistant AML43. In pediatric AML, mutations of WT1 and EZH2 appear to be of exclusively clonal or near-clonal origin, with nearly one-quarter of TARGET cases harboring mutations affecting one or the other. Aberrant WT1, EZH2 or ASXL1 predicted induction failure in TARGET AML cases (multivariate P < 0.05, adjusted for interactions with FLT3 alterations and NUP98-NSD1 and KMT2A fusions) and were largely mutually exclusive of KMT2A rearrangements (P < 10−5). Many of these subjects presented without apparent chromosomal abnormalities at diagnosis, yet less than 20% achieved long-term remission with standard treatment, highlighting the importance of molecular stratification to achieve better outcomes. It is possible that early events in children, such as WT1 mutations and NUP98-NSD1 fusions, may play a similar role to that observed for DNMT3A mutations14 in adults, with considerable implications for risk stratification in AML across age groups.
Our data also demonstrate that DNA methylation and miRNA expression profiles both accompany and complement DNA alterations and can stratify pediatric subjects with AML in terms of both overall and progression-free survival. These findings suggest a need to update pediatric AML clinical risk categories beyond current classifications, with important implications for clinical practice.
Despite incremental improvements with increasingly intensified regimens, contemporary outcomes in pediatric AML have plateaued, with only ~60% of patients achieving long-term survival. As many as 10% of children with AML will die from direct complications of treatment. Survivors suffer unacceptably high rates of long-term morbidity resulting from anthracycline exposure or sequelae of hematopoietic stem cell transplantation. As illustrated herein, pediatric AML is a collection of molecularly diverse diseases with similar phenotypes. No single treatment strategy is likely to be effective for all pediatric AML subtypes, which may explain the repeated failures of randomized clinical trials to improve outcomes in recent years. In keeping with the shift toward comprehensive, molecularly based classification schemas in AML4, the time has come to develop targeted therapies that address specific vulnerabilities of pediatric subtypes. The TARGET AML data set will serve as a foundation for development of pediatric-specific classification schemas and the development of personalized treatment strategies.
URLs
TARGET sample matrix, https://ocg.cancer.gov/programs/target/data-matrix/.
ONLINE METHODS
Sample selection and preparation
All subject samples were obtained by member COG institutions after written consent was obtained from the parents/guardians of minors upon enrolling in the trial. The study was overseen by the Institutional Review Board at Fred Hutchinson Cancer Research Center (protocol 1642, IR file no. 5236). Data on selected clinical (for example, age, presenting hematological indices and cytogenetic classification) and molecular (for example, KIT, RAS genes, NPM, WT1, CEBPA and IDH1 mutations and FLT3-ITD allelic ratios) features were clinically available before genomic analyses and are included in the clinical data file available at the TARGET data matrix. 177 cases from the adult de novo AML TCGA data set3 were selected for analysis after exclusion of those with French-American-British (FAB) system M3 morphology (n = 20) or BCR-ABL1 gene fusion (n = 3), as these subtypes are not represented in the COG–TARGET AML cohort. The age distributions for the TARGET WGS discovery group and the TCGA cohort are outlined in Supplementary Table 3. DNA and RNA were extracted from Ficoll-enriched, viably cryopreserved samples from the COG biorepository using the AllPrep Extraction Kit (Qiagen). Nucleic acids were quantified by NanoDrop (Thermo Scientific). RNA samples were tested for quality and integrity using the Agilent 2100 Bioanalyzer (Agilent Technologies). The integrity of DNA samples was confirmed by visualization on a 0.8% agarose gel.
Whole-genome sequencing
Sequencing libraries were constructed for WGS cases from genomic DNA and sequenced using combinatorial probe anchor ligation by Complete Genomics (CGI)44. Reads were mapped to the GRCh37 reference human genome assembly by the CGI Cancer Sequencing service using software version 2.1 of the CGI cancer analysis pipeline (http://www.completegenomics.com/customer-support/documentation/). Somatic SNVs in protein-coding regions and indels were extracted from the MAF files and filtered in three steps to remove (1) germline variants; (2) low-confidence variants; and (3) paralogs. For step 1, the germline variants used for filtering included those from NLHBI Exome Sequencing Project (http://evs.gs.washington.edu/EVS/), dbSNP 132 (https://www.ncbi.nlm.nih.gov/projects/SNP/), St. Jude–Washington University Pediatric Cancer Genome Project (PCGP) and CGI WGS from the TARGET project. For step 2, a mutation was considered of low confidence if it did not meet one of the following criteria: (i) read count ≥3 more in the tumor than in the matched remission sample; (ii) read count in the tumor significantly higher than that in the normal sample (P < 0.01 by Fisher’s exact test); and (iii) allele fraction in the normal sample of <0.05. For step 3, we ran a BLAT search using a template sequence that included the mutant allele and its 20-bp flanking region to determining the uniqueness of mapping of the mutation. To avoid overfiltering, we implemented a rescue pipeline that retains all ‘gold’ variants that match known somatic mutation hotspots based on our variant classification program Medal Ceremony45. In addition to small variant calls (SNV, indel), the CGI cancer analysis pipeline delivered flat files containing potentially new DNA junctions and segmented copy number ratios derived from normalized read counts from paired tumor–normal specimens. Circos summary plots of the unfiltered CGI data are available through the data matrix. To reduce potentially spurious calls, final copy number variants (CNVs) used for analysis were trimmed after empirical tuning to previously available Affymetrix SNP6 microarray calls in matched samples by requiring a CGI average normalized coverage (avgNormCvg) in the CNV region of ≥20 reads, an s.d. for the lesser allele fraction of ≤0.22 and a CGI ploidyScore of <30 and trimming calls on the mitochondrial chromosome or in centromeric or telomeric regions; adjacent CNVs within 10 kb of each other for each called direction were merged. With these filters, 75% and 85% (loss and gain, respectively) of filtered CNV calls matched CNVs previously called by Affymetrix SNP6 microarray, and 87% of chromosome-arm-level calls matched karyotype abnormalities reported in the clinical data. Putative CNVs underwent further secondary confirmation using the nanoString nCounter assay (Nanostring Technologies). New DNA junctions discovered by WGS were included in cases for which at least one additional level of support was available, from either cytogenetic analysis or RNA-seq studies.
Targeted capture sequencing
Candidate genes identified by WGS analysis were selected for independent verification in 182 samples from the WGS discovery cohort and 618 additional subjects treated on COG AAML0531. Capture baits were designed and ordered using Agilent’s SureDesign (https://earray.chem.agilent.com/suredesign/) for selected genes along with target regions identified in concurrent TARGET studies, targeting coding regions and UTRs with a 10-bp pad. This design (TARGET AML + TARGET other) resulted in an overall target space of 2.376 Mb, with 98.7% of target regions covered by a probe. Probe density was specified at 2×, with moderately stringent repeat masking and balanced boosting options selected.
Genomic DNA libraries, from which gene regions of interest were captured, were constructed according to British Columbia Cancer Agency Genome Sciences Centre (BCGSC) plate-based and paired-end library protocols on a Biomek FX liquid-handling robot (Beckman-Coulter, USA). Briefly, 1 μg of high-molecular-weight genomic DNA was sonicated (Covaris E210) in a 60-μl volume to 200–300 bp. Sonicated DNA was purified with magnetic beads (Agencourt, Ampure). The DNA fragments were end-repaired, phosphorylated and bead-purified in preparation for A-tailing. Illumina sequencing adaptors were ligated overnight at 20 °C, and adaptor-ligated products were bead-purified and enriched with four cycles of PCR using primers containing a hexamer index that enables library pooling. 94-ng aliquots from each of 19 to 24 different libraries were pooled before custom capture using Agilent SureSelect XT Custom 0.5- to 2.9-Mb probes. The pooled libraries were hybridized to the RNA probes at 65 °C for 24 h. Following hybridization, streptavidin-coated magnetic beads (Dynal, MyOne) were used for custom capture. Post-capture material was purified on MinElute columns (Qiagen), followed by post-capture enrichment with ten cycles of PCR using primers that maintained the library-specific indices. Paired-end 100-bp reads were sequenced per pool in a single lane of an Illumina HiSeq 2500 instrument. Illumina paired-end sequencing reads were aligned to the GRCh37-lite reference using Burrows–Wheeler aligner (BWA) version 0.5.7. This reference contains chromosomes 1–22, X, Y and MT together with 20 unlocalized scaffolds and 39 unplaced scaffolds. Multiple lanes of sequences were merged, and duplicated reads were marked with Picard Tools. Small variants (SNVs and indels) from TCS data were identified by parallel methods, integrated and subsequently filtered as follows.
Mpileup
SNVs were analyzed with SAMtools mpileup version 0.1.17 on paired libraries46. Each chromosome was analyzed separately using the -C50- DSBuf parameters. The resulting vcf files were merged and filtered to remove low-quality variants by using SAMtools varFilter (with default parameters) as well as to remove variants with a quality (QUAL) score of less than 20 (vcf column 6). Finally, variants were annotated with gene annotations from Ensembl version 66 using snpEff47, and dbSNP build 137 membership of variants was assigned using snpSift48.
Strelka
Samples were analyzed pairwise with the default settings of Strelka49 version 0.4.7 with primary tumor samples against matched remission samples. Somatic variants called by either mpileup or Strelka were combined and filtered out if they met any of the following criteria: <10 reads in the remission sample, <10 reads in the tumor sample, number of reads called for the altered base (tumor alt base) = 0, adjusted tumor allele frequency = 0, global minor allele frequency (GMAF) > 0.009 and >60 subjects with exactly the same SNVs. For subjects established as being in morphological remission, additional filters included removing variants with an allele fraction of >0.10 in the remission sample and a Fisher’s exact test score of >0.05. For refractory subjects, variants were excluded if they had a >0.35 allele fraction in the post-diagnostic sample. These filtered variants could be ‘rescued’ if a variant was a known Catalogue of Somatic Mutations in Cancer (COSMIC) mutation associated with hematological cancers. The filtering criteria for indel calls were similar. Tandem duplications were identified with Pindel using default parameters50. In addition, results from clinical molecular testing for specific genes (FLT3-ITD and FLT3 codons 835 and 836, CEBPA basic leucine zipper domain (bZIP) and N-terminal domain (NTD) regions, KIT exons 8 and 17, CBL exons 8 and 9, and WT1 exon 7) were merged into the variant calls for final analysis.
DNA variants from discovery and TCS studies were merged to construct the mutation profile for each gene using the web-based program ProteinPaint51. Genome-wide mutational burden was compared to published data from Lawrence et al.52, using the method reported therein.
CBL transcript variant screening by cDNA PCR
Total RNA isolated from subject leukemic cells using the AllPrep DNA/RNA Mini Kit (Qiagen, Germany) was reverse transcribed to cDNA with oligo(dT) primer and additional reagents following the Maxima H Minus First Strand cDNA Synthesis Kit instructions (Thermo Fisher Scientific, Grand Island, NY).
Synthesis of the second-strand cDNA and following PCR were performed using the following primers: forward primer for Genemap: (5′-FAM-TTCCAAGCACTGATTGATGG-3′), forward primer for sequencing (5′-TTCCAAGCACTGATTGATGG-3′), reverse primer: (5′-AACAGAATAT GGCCGGTCTG-3′). PCR was performed in 25-μl volumes containing 12.5 μl of FailSafe PreMix C buffer (2×) (Epicentre Technologies, Madison, WI), 0.5 μl (10 μM) of each primer, 0.25 μl of Platinum Taq Polymerase (Thermo Fisher Scientific–Invitrogen, Grand Island, NY), 1 μl of cDNA and 10.25 μl of nuclease- free water (USB Corporation, Cleveland, OH). The thermocycling program consisted of 5 min of denaturation at 95 °C followed by 35 cycles at 95 °C for 30 s, 60 °C for 30 s and 72 °C for 45 s and a final extension of 7 min at 72 °C in a 96-well Biometra TProfessional Thermocycler (Biometra, Germany).
PCR products were diluted in nuclease-free water (USB Corporation, Cleveland, OH), mixed with deionized formamide and GeneScan-400HD (ROX) size markers (Applied Biosystems, Foster City, CA), and subjected to electrophoresis on an ABI 3730 Genetic Analyzer (Applied Biosystems). After electrophoresis, the fluorescence signals were analyzed using GeneMapper 5.0 software (Applied Biosystems). GeneMapper screening revealed products of the size expected for wild-type CBL transcript (685 bp) and additional products of various sizes corresponding to complete deletion of exon 8 (563 bp), complete deletion of exon 9 (485 bp), and deletion involving both exon 8 and exon 9 (354 bp).
Samples from subjects exhibiting deletions as determined by GeneMapper were then sent for sequence verification. PCR products were treated with ExoSAP-IT PCR Product Cleanup Reagent (USB Corporation, Cleveland, OH). Sequencing was done by Eurofins MWG Operon, LLC. (Huntsville, AL), in accordance with their DNA sequencing process guidelines and methods.
Generalized linear mixed model for coding mutation counts
In order to account for both fixed and random effects that might be present with age and cytogenetic subgroups, we employed a generalized linear mixed model (glmm, Knudson 2016, R package version 1.1.1; https://CRAN.R-project.org/package=glmm/) to model the discrete counts of SNVs in protein-coding regions in each TARGET and TCGA subject having WGS data with a Poisson error distribution (log link). Marginal likelihood-ratio tests for age (as a continuous predictor) and cytogenetic subgroup (as a categorical predictor) were uniformly and highly significant, as reported in the text, and the per-cytogenetic-group random effects accounted for a small (<0.003%) fraction of the variance observed. The model converged in 208 steps; 10,000 Markov chain Monte Carlo (MCMC) iterations were employed to estimate the mixed-effects component of the model, fitted per cytogenetic group assuming a random slopes model.
Generalized Dirichlet-multinomial regression for mutational spectra
To accommodate the possibility of either negative or positive correlation between the counts of each type of mutation (C→T, C→A, C→G, T→C, T→A and T→G) in each subject, we employed a generalized Dirichlet-multinomial model (mglm53, R package version 0.0.7; https://CRAN.R-project.org/package=MGLM/) with age and cytogenetic group as predictors and mutational spectrum (a matrix of counts for each type of mutation) as response. At convergence, the significant predictors of mutational spectrum differences were age (most significant), t(8;21) status and aberrant karyotype (mutually exclusive with t(8;21) and other common recurrent chromosomal abnormalities). C→T transitions are known to increase with age, particularly for methylated cytosines; however, an inflation of C→A transversions was particularly apparent in t(8;21) and aberrant karyotype cases (both t(8;21) and inv(16) affect CBF subunits and both are associated with higher mutational burdens at a given age, but only t(8;21) cases show additional C→A transversions beyond those expected from counts).
Weighted resampling scheme to compare TARGET and TCGA mutation frequencies
Common chromosomal aberrations often co-occur with specific types of additional DNA sequence abnormalities. To account for this observation when determining differences in mutation frequency between TARGET AML and TCGA AML, we first divided each cohort into the following categories: KMT2A fusions, t(8;21), inv(16), del(7), +8, +21, −Y and normal karyotype (NK). A total of 131 unique TCGA and 548 unique TARGET samples fell into one of the above categories. We then sampled equal numbers of specimens from each category and calculated the fraction of samples with mutations in a given gene. To account for sampling variations, we repeated our sampling procedure 5,000 times and calculated the mean and s.d. of the fraction of samples with mutations in each gene of interest.
Variant pairwise mutual exclusivity and co-occurrence
Pairwise mutually exclusive sequence alterations (Supplementary Fig. 20a) were identified using CoMEt54 with the ‘exhaustive’ option (http://compbio.cs.brown.edu/projects/comet/). Pairwise co-occurrence P values (Supplementary Fig. 20b) were calculated directly using a hypergeometric distribution (equivalent to Fisher’s exact test). Statistically significant exclusion and co-occurrence patterns were visualized using Cytoscape55 (http://cytoscape.org/), with edge thickness representing −log10 (P value).
Orthogonal evaluation of mutual exclusivity and co-occurrence via penalized Ising model
A slightly different approach to reconstructing a binary-valued undirected graph (a discrete Markov random field) employs penalized logistic regression of all candidate nodes upon each possible target and selects the most probable graph structure based on extensions of the Bayes information criterion (EBIC). This approach is implemented by Epskamp (https://cran.r-project.org/package=IsingFit/)56 and employs a hyperparameter (γ) for the penalty weight, which eventually determines the density of the estimated network. Adjustment for multiple comparisons was applied to the marginal significance of each gene– gene Fisher’s exact test; this value is not unbiased owing to post-selection inference and is only intended as a guide. The resulting network of correlated and anticorrelated binary indicators (gene- and chromosome-level aberrant or wild type, pediatric or adult) recovered known and CoMEt-detected relationships, but also identified several new and marginally significant (by Fisher’s exact test; see above) relationships, as summarized in Supplementary Table 6.
Hypothesis testing
Except where described by the methods above, P values were calculated using Fisher’s exact test; where an exact binomial test was impractical, we approximated with a chi-squared P value.
Regression fits for structural and sequence variant burden and age-associated recurrent abnormalities
To fit the ratio of structural to sequence variant impact in each subject, we added a smoothing factor of 0.333 to the counts for each clonal event of each type, using all recurrently mutated, fused or silenced genes that were identified in either cohort as candidates for ‘impact’ by structural variants. The transparency of each data point represents the observed over expected mutational burden given the subject’s age but has no impact on the loess regression fit (Fig. 2e). The loess curve was fit by ggplot2 (http://ggplot2.org/) on a log10 scale. To estimate the relative contribution of each of the recurrent fusion neighborhoods across ages (rather than age groups), we used the ‘zoo’ time series package57 to fit a rolling median with expanding time steps (1, 3, 5, 8 and 17 years) across all subjects for whom we had data on fusions. The (smoothed) contribution of each family of fusions to the total number of subjects in a given age window (expanding with advancing age) is plotted in Figure 2d.
Clonality estimation
Several packages (including MAFtools58 (https://github.com/PoisonAlien/maftools/) Gaussian mixture, SciClone’s59 beta mixture model and a weighted penalized logistic mixture model) were compared to validate the results obtained in addition to manual review of all results. Although the proportions of mutations assigned to various clones differed in some cases (especially with and without read support weighting), the primary mutational clones were consistently identified by all methods, and an overall tendency for childhood and AYA subjects to present with greater diagnostic mutational clonality, at the read depths available in the TARGET WGS and TCGA data, was confirmed by all methods. Among AYA subjects (present in both the TCGA and TARGET AML cohorts), no difference in estimated clonality or monoclonal versus polyclonal balance was observed between cohorts (P = 0.7 and P = 0.65, respectively, using Fisher’s exact test), and although a trend toward decreased mutational clonality with increasing age among AYA subjects was observed, it was not statistically significant (P = 0.2). It is important to note that, owing to variable sequencing depths, we do not have the statistical power to reliably detect clones present in less than 5% of the total sample material, although inclusion of variant allele frequencies as low as 0.1% did not change our results or conclusions regarding mutational clonality. Karyotypic clonality was assessed by parsing International System for Human Cytogenetic Nomenclature (ISCN) karyotypes of all TARGET and TCGA subjects with AML and using stemline karyotype to identify the most likely ancestral aberrations for subjects with an abnormal karyotype. Subjects with a normal karyotype were assigned a karyotypic clonality of 1, as were subjects with all metaphases bearing identical aberrations.
Aberrations predicting induction failure
A logistic model with terms for NUP98-NSD1 fusions; FLT3 mutations; interactions between NUP98-NSD1 and FLT3 mutations; and (any one of) WT1, EZH2 or ASXL1 mutation (mutually exclusive); deletion of WT1, EZH2 or ASXL1 (nearly mutually exclusive); or KMT2A rearrangements (also mutually exclusive of WT1, EZH2 or ASXL1 mutation) best fit the data for subjects for whom the first recorded event was induction failure (1) or any other outcome (0). All possible nested models with the same terms, and all other models arrived at by penalized logistic regression (using an elastic net penalty with the glmnet package60, with any observed recurrent lesion eligible for inclusion as an independent predictor), yielded inferior fits both in terms of classification error and Akaike information criterion (AIC). We report the marginal P values for WT1, ASXL1 and EZH2 aberrations as predictors of induction failure in the test based on this model fit.
mRNA sequencing
Total RNA quality was verified on an Agilent Bioanalyzer RNA nanochip or Caliper GX HT RNA LabChip, with samples passing quality control arrayed into a 96-well plate. Poly(A)+ RNA was purified using the 96- well MultiMACS mRNA Isolation Kit on the MultiMACS M96thermo separator (Miltenyi Biotec) using 2 μg total RNA with on-column DNase I treatment per the manufacturer’s instructions. The eluted poly(A)+ RNA was ethanol precipitated and resuspended in 10 μl of SUPERaseIN (Life Technologies) diluted 1:20 in diethyl pyrocarbonate (DEPC)-treated water. First-strand cDNA was synthesized from the purified poly(A)+ RNA using the SuperScript cDNA synthesis kit (Life Technologies) and random hexamer primers at a concentration of 5 μM along with a final concentration of 1 μg/μl actinomycin D, followed by cleanup on AMPure XP SPRI beads on a Biomek FXp robot (Beckman-Coulter). Second-strand cDNA was synthesized following the Superscript cDNA Synthesis protocol by replacing the dTTP with dUTP in dNTP mix, allowing the second strand to be digested using uracil N-glycosylase (UNG, Life Technologies, USA) after adaptor ligation and thus achieving strand specificity. The cDNA was quantified by PicoGreen (Life Technologies) and VICTOR3V Fluorometer (PerkinElmer). The cDNA was fragmented by Covaris E210 sonication for 55 s at a ‘duty cycle’ of 20% and ‘intensity’ of 5. The paired-end sequencing library was prepared following the British Columbia Cancer Agency Genome Sciences Centre strand-specific, plate-based paired-end library construction protocol using a Biomek FX robot (Beckman-Coulter, USA). Briefly, the cDNA was purified in 96-well format using Ampure XP SPRI beads and was subjected to end-repair using T4 DNA polymerase and Klenow DNA polymerase and subsequent phosphorylation using T4 polynucleotide kinase, followed by cleanup using Ampure XP SPRI beads and 3′ A-tailing by Klenow fragment (3′→5′ exo–). After purification using Ampure XP SPRI beads, PicoGreen quantification was performed to determine the amount of Illumina PE adaptors to be used in the next step of the adaptor ligation reaction. The adaptor-ligated products were purified using Ampure XP SPRI beads and digested with UNG (1 U/μl) at 37 °C for 30 min followed by deactivation at 95 °C for 15 min. The digested cDNA was purified using Ampure XP SPRI beads and then PCR amplified with Phusion DNA Polymerase (Thermo Fisher) using Illumina’s PE primer set, with cycle conditions of 98 °C for 30 sec followed by 10–13 cycles of 98 °C for 10 s, 65 °C for 30 s and 72 °C for 30 s and then 1 cycle at 72 °C for 5 min. The PCR products were purified using Ampure XP SPRI beads and checked with Caliper LabChip GX for DNA samples using the High Sensitivity Assay (PerkinElmer, Inc., USA). PCR product of the desired size range was purified using 8% PAGE, the DNA quality and quantity were assessed using an Agilent DNA 1000 series II assay and Quant-iT dsDNA HS Assay Kit on a Qubit fluorometer (Invitrogen), and the DNA was then diluted to 8 nM. The final library concentration was double-checked and determined by Quant-iT dsDNA HS Assay for Illumina Sequencing.
mRNA quantification
Illumina paired-end RNA sequencing reads were aligned to the GRCh37-lite genome-plus-junctions reference using BWA version 0.5.7. This reference combines genomic sequences in the GRCh37-lite assembly and exon–exon junction sequences whose corresponding coordinates were defined based on annotations of any transcripts in Ensembl (version 69), RefSeq and known genes from the UCSC genome browser, which were downloaded on 19 August 2010, 8 August 2010 and 19 August 2010, respectively. Reads that mapped to junction regions were then repositioned back to the genome and were marked with ‘ZJ:Z’ tags. BWA was run using default parameters except that the option (–s) was included to disable Smith–Waterman alignment. Finally, reads failing the Illumina chastity filter were flagged with a custom script, and duplicated reads were flagged with Picard Tools. Gene-, isoform- and exon-level quantifications were performed as previously described61.
Fusion mRNA transcript detection
Transcriptomic data were de novo assembled using ABySS (version 1.3.2) and trans-ABySS (version 1.4.6)62. For RNA-seq assembly, alternate k-mer values from k = 50–96 were aligned using positive-strand and ambiguous-strand reads as well as negative-strand and ambiguous-strand reads. The positive- and negative-strand assemblies were extended where possible, merged and then concatenated together to produce a meta-assembly contig data set. Large-scale rearrangements and gene fusions from RNA-seq libraries were identified from contigs that had high-confidence Genomic Mapping and Alignment Program (GMAP; version 2012-12-20) alignments to two distinct genomic regions. Evidence for the alignments was provided through aligning reads back to the contigs and through aligning reads to genomic coordinates. Events were then filtered on read thresholds. Insertions and deletions were identified by gapped alignment of contigs to the human reference using GMAP. The events were then screened against dbSNP and other variation databases to identify putative new events.
miRNA sequencing
miRNA-containing small RNAs in the flow-through material following mRNA purification on a MultiMACS separator (Miltenyi Biotec) were recovered using ethanol precipitation. miRNA-seq libraries were constructed using a 96-well plate–based protocol developed at the BC Cancer Agency Genome Sciences Centre. Briefly, an adenylated single-stranded DNA 3′ adaptor was selectively ligated to miRNAs using a truncated T4 RNA Ligase 2 (New England Biolabs). An RNA 5′ adaptor was then added using a T4 RNA Ligase (Ambion) and ATP. Next, first-strand cDNA was synthesized using SuperScript II Reverse Transcriptase (Invitrogen); it served as the template for PCR. Index sequences (six nucleotides) were introduced at this PCR step to enable multiplexed pooling of miRNA libraries. PCR products were pooled and then size-selected on an in-house-developed 96-channel robot to enrich the miRNA-containing fraction and remove adaptor contaminants. Each size-selected indexed pool was ethanol precipitated, quality-checked on an Agilent Bioanalyzer DNA 1000 chip and quantified using a Qubit fluorometer (Invitrogen, cat. Q32854). Each pool was then diluted to a target concentration for cluster generation and loaded into a single lane of a HiSeq 2000 flow cell for sequencing with a 31-bp main read (for the insert) and a 7-bp read for the index.
Sequence data were separated into individual samples using the index read sequences, and the reads underwent an initial quality control assessment. The adaptor sequence was then trimmed off, and the trimmed reads for each sample were aligned to the National Center for Biotechnology Information (NCBI) GRCh37-lite reference genome.
Routine quality control was used to assess a subset of raw sequences from each pooled lane for the abundance of reads from each indexed sample in the pool, the proportion of reads that possibly originated from adaptor dimers (i.e., a 5′ adaptor joined to a 3′ adaptor with no intervening biological sequence) and the proportion of reads that mapped to human miRNAs. Sequencing error was estimated by a method originally developed for serial analysis of gene expression (SAGE).
Libraries that passed this quality control stage were preprocessed for alignment. Although the size-selected miRNAs vary somewhat in length, typically they are ~21 bp long and so are shorter than the 31-bp read length. Considering this, each read sequence extends some distance into the 3′ sequencing adaptor. Because this nonbiological sequence can interfere with aligning the read to the reference genome, the 3′ adaptor sequence was identified and removed (trimmed) from the reads. The adaptor-trimming algorithm identified an adaptor sequence that was as long as possible, allowing for a number of mismatches according to the detected adaptor length. A typical sequencing run yields several million reads; using only the first (5′) 15 bases of the 3′ adaptor in trimming made processing efficient and minimized the chance that a miRNA read would match the adaptor sequence.
After each read was processed, a summary report was generated containing the number of reads at each read length. Any trimmed read that was shorter than 15 bp was discarded; remaining reads were submitted for alignment to the reference genome. BWA63 alignment(s) for each read were checked with a series of three filters. A read with more than three alignments was discarded as too ambiguous. Only perfect alignments with no mismatches were used. Reads that failed the Illumina base calling chastity filter were retained, and reads that had soft-clipped Concise Idiosyncratic Gapped Alignment Report (CIGAR) strings were discarded.
For reads retained after filtering, each coordinate for each read alignment was annotated using a reference database, requiring a minimum 3-bp overlap between the alignment and an annotation. If a read had more than one alignment location and the annotations for these were different, we used a priority list to assign a single annotation to the read, as long as only one alignment was to a miRNA. When there were multiple alignments to different miRNAs, the read was flagged as cross-mapped64, and all of its miRNA annotations were preserved while all of its non-miRNA annotations were discarded. This ensured that all annotation information about ambiguously mapped miRNAs was retained, allowing annotation ambiguity to be addressed in downstream analyses. Note that we considered miRNAs to be cross-mapped only if they mapped to different miRNAs, not functionally identical miRNAs that are expressed from different locations in the genome. Such cases are indicated by miRNA miRBase names, which can have up to four separate sections separated by ‘−’, for example, hsa-mir-26a-1. A difference in the final section (for example, ‘−1’) denotes functionally equivalent miRNAs expressed from different regions of the genome, and we considered only the first three sections (for example, ‘hsa-mir-26a’) when comparing names. As long as a read mapped to multiple miRNAs for which the first three sections of the name were identical (for example, hsa-mir-26a-1 and hsa-mir-26a-2), it was treated as if it mapped to only one miRNA and was not flagged as cross-mapped.
The minimum depth of sequencing required to detect the miRNAs expressed in one sample was 1,000,000 reads per library mapped to miRBase (version 21) annotations. Finally, for each sample, the reads that corresponded to particular miRNAs were summed and normalized to 1 million miRNA-aligned reads to generate the quantification files. TARGET and TCGA miRNA quantifications were normalized with the permuted SVA (pSVA) function, preserving known subtype-specific miRNA expression patterns, before comparison65.
Differentially expressed miRNAs and mRNAs were determined using Wilcoxon tests; significantly differentially expressed miRNAs were those with Benjamini–Hochberg multiple test–corrected P value < 0.05. Correlation between miRNA and mRNA expression was determined using Spearman correlation.
DNA methylation analysis
Bisulfite conversion of genomic DNA was performed with EZ DNA Methylation Kit (Zymo Research, Irvine, CA) following the manufacturer’s protocol with modifications for the Infinium Methylation Assay. Briefly, 1 μg genomic DNA was mixed with 5 μl of dilution buffer, incubated at 37 °C for 15 min and then mixed with 100 μl of conversion reagent prepared as instructed in the protocol. Mixtures were incubated in a thermocycler for 16 cycles at 95 °C for 30 s and 50 °C for 60 min. Bisulfite-converted DNA samples were loaded onto the provided 96-column plates for desulfonation, washing and elution. The concentration of bisulfite-converted, eluted DNA was measured by UV absorbance using a NanoDrop ND-1000 (Thermo Fisher Scientific, Waltham, MA). Bisulfite-converted genomic DNA was analyzed using the Infinium HumanMethylation27 BeadChip Kit (Illumina, San Diego, CA, cat. WG-311-1202). DNA amplification, fragmentation, array hybridization, extension and staining were performed with reagents provided in the kit according to the manufacturer’s protocol (Illumina Infinium II Methylation Assay, cat. WG-901-2701). Briefly, 4 μl of bisulfite-converted genomic DNA at a minimum concentration of 20 ng/μl was added to a 0.8-ml 96-well storage plate (Thermo Fisher Scientific), denatured in 0.014 M sodium hydroxide, neutralized and then amplified for 20–24 h at 37 °C. Samples were fragmented at 37 °C for 60 min and precipitated in isopropanol. Resuspended samples were denatured in a 96-well plate heat block at 95 °C for 20 min. Fifteen microliters of each sample was loaded onto a 12-sample BeadChip, assembled in the hybridization chamber as instructed by the manufacturer and incubated at 48 °C for 16–20 h. Following hybridization, the BeadChips were washed and assembled in a fluid flow-through station for primer-extension reaction and staining with the reagents and buffers provided. Polymer-coated BeadChips were scanned in an iScan scanner (Illumina) using Inf Methylation mode. For both HumanMethylation27 and HumanMethylation450 arrays, methylated and unmethylated signal intensity and detection P values were extracted after background correction and (in the case of HumanMethylation450 arrays) dye-bias equalization by normal-exponential convolution (noob66), as implemented in the minfi package67. Data from HumanMethylation450 arrays were additionally normalized using functional normalization (funnorm68), as implemented in the minfi package, and then summarized as beta values (M/(M + U, where M means methylated and U means unmethylated). Probes with an annotated SNV within the CpG or single-base extension site were masked as Not Available (NA) across all samples. Probes with nondetection probability > 0.01 were masked as ‘NA’ for individual samples.
Transcriptional silencing evaluation and tabulation
Transcription is influenced by a large number of features. One such feature is methylation of genomic CpG dinucleotides, which often leads to methyl-binding domain proteins excluding transcriptional activators when the methylation occurs near a transcription start site. Not all gene promoters are influenced by differences in DNA methylation, and not all promoters that are influenced are relevant in a given cell type. Thus, we sought to identify bundles of transcripts (genes) whose expression appeared to be influenced by promoter CpG methylation and whose expression potential was perturbed in a subset of AML cases.
To establish a uniform criterion for ‘calling’ such events, we evaluated over 50,000 loci from the Illumina HumanMethylation450 (‘450k’) microarray near the transcription start sites of over 20,000 transcripts. We retained the locus and gene symbol for further evaluation where any variance in transcript abundance was explained by variation in DNA methylation levels at the locus. With this set of several thousand potential marker pairs, we iteratively sought ‘silencing’ cutoff points such that the maximum expression of a gene in any sample with methylation above the cutoff was less than or equal to the median expression in samples with methylation below the cutoff. The relative levels of DNA methylation and expression appeared to differ systematically between subjects with AML in the TCGA cohort and those in the TARGET cohort. Therefore, we retained the most conservative (highest) cut-point from the two cohorts. A large number of TARGET subjects with AML were previously assayed on the promoter-centric Illumina HumanMethylation27 (‘27k’) microarray; to maximize the sample size for silencing calls, we performed the same conservative procedure as described above with 27k loci. Whenever a locus could be found with a suitable cut-point on both the 27k and 450k arrays, we used the two loci, one from each array, to cross-validate transcriptional silencing behavior between the two (largely disjoint) sets of samples (TCGA subjects with AML were assayed on both the 27k and 450k arrays, so we used the appropriate complementary assay to cross-validate each cutoff in TCGA). The resulting set of ‘tag CpGs’ (loci with satisfactory cutoff values for a given gene) for each platform, along with the results of applying these cutoffs to dichotomize subject samples as silenced or not, is provided in Supplementary Table 9. Selected loci and genes affected across multiple subjects are plotted in Figure 5a, annotated within each major cytogenetic group by the fraction of subjects with silencing.
Non-negative matrix factorization, DNA methylation signature derivation and hierarchical clustering
NMF decomposes a strictly positive data matrix X (with N rows and M columns) into a lower-dimensional N × K weight matrix W and a corresponding K × M score matrix H69. The crux of the decomposition is finding coefficients for W and H that when multiplied most closely recover the original high-dimensional data matrix X, as there is no guarantee that a global optimum exists in the absence of further constraints. This can be approached as an optimization problem: given an estimate of the underlying rank K for the weight matrix W, what coefficients minimize the squared reconstruction error (X − WH)2? When this is formulated as a non-negative least-squares fit, alternating between fits for W and H at each iteration, a fast sequential coordinate descent procedure implemented by E.X. Lin (https://cran.r-project.org/web/packages/NNLM/vignettes/Fast-And-Versatile-NMF.html/) is useful for the large matrix we generate for the input X. To decrease the size of X without discarding information, the HumanMethylation450 data were further collapsed by aggregating signals at adjacent CpG sites (separated by up to 50 bp) using the cpgCollapse function in the minfi package, yielding 221,406 discrete clustered methylation measurements, of which approximately half (118,586) showed nonnegligible variation across diagnostic tumor samples and/or matched remission samples. The underlying identifiable rank K of the low-dimensional weight matrix W was estimated by fivefold cross-validation, using random row × column knockouts (set to NA) in 20% of the matrix entries for each fold, followed by minimization of reconstruction error and maximization of inferred rank. On the basis of this procedure, the optimal rank K (with mean absolute error of 0.02793436) for W was estimated as 31. By masking with W and H matrices derived from normal bone marrow populations (for which K was chosen as 13, again on the basis of reconstruction error as above), we subtracted ‘normal’ hematopoietic cell signals and simultaneously estimated the purity (cellularity) of each tumor sample, which allowed us to amplify disease-specific signals, correcting in silico for estimated purity on a logit scale, and we finally transformed values back to the original proportional 0–1 scale for presentation in Figure 5b. The 31-row by 284-column subject score matrix H is provided in Supplementary Table 10; selected signatures of particular interest are plotted in Figure 5b. Ward’s method was employed to cluster columns (subjects) in the figure panel by Manhattan distance.
Survival analysis
We tested an additional cohort of pediatric subjects with AML for outcome measures associated with alterations of FLT3-ITD, NPM1 and WT1 mutations and NUP98-NSD1 translocations (Fig. 3c, lower right, and Supplementary Fig. 13, ‘DCOG’). Subject data for this cohort were provided by DCOG, the AML ‘Berlin-Frankfurt-Münster’ Study Group (AML-BFM-SG), the Czech Pediatric Hematology (CPH) group, the St. Louis Hospital in Paris, France, the UK Medical Research Council (MRC) and the Italian Association for Pediatric Hematology and Oncology (AIEOP). Subjects were treated on LAME 86, DCOG/AML-BFM 87, DCOG 92-94/AML-BFM 93, AML-BFM 98, AEIOP-2002/01, ELAM02, AML-BFM 04 and MRC-12/15 protocols70–77. These protocols consisted of 4–5 blocks of intensive chemotherapy, using a standard cytarabine and anthracycline backbone. All subjects in this cohort were previously published by Balgobind et al.78 and were extensively screened by RT–PCR or fluorescent in situ hybridization (FISH) for recurrent aberrations, such as KMT2A rearrangements, RUNX1-RUNX1T1, CBFB-MYH11, PML-RARA, NUP98 rearrangements, FLT3-ITD and mutations in NPM1, CEBPA, WT1, NRAS, KRAS and KIT (c-KIT)78–81; this cohort also included 326 subjects with data available on NUP98-NSD1, NPM1, FLT3-ITD and WT1 status. Complete remission was obtained in 74.8% of the subjects. A total of 114 subjects (35.0%) received a hematopoietic stem cell transplant (HSCT); 35 of these subjects (10.7%) received HSCT at first complete remission. The median follow-up time of survivors was 4.5 years (range, 0.3–28 years), and the cohortwide overall survival (OS) and event-free survival (EFS) rates were 59.5% and 41.9%, respectively.
The Kaplan–Meier method was used to estimate OS and EFS. OS is defined as the time from study entry until death. EFS is defined as the time from study entry until death, induction failure or relapse. Subjects lost to follow-up were censored at their date of last known contact. Comparisons of OS and EFS were made using the log-rank test.
TARGET and TCGA subjects were combined in Cox proportional-hazards fits for association of DNA methylation signatures with survival outcome, and model parameters for well-established risk factors (TP53 mutation and white blood cell count at diagnosis) were also estimated. Owing to the nonlinear association of age with survival in pediatric individuals with AML and the difficulty of properly evaluating this relationship, we instead stratified the Cox proportional-hazards fits by cohort.
For miRNA-associated survival analyses, the expression (RPM) cutoff between high- and low-expression groups for each miRNA was defined using the X-tile method82: all separation points between subjects were considered, and the selected cutoff point was the one that provided the optimal (lowest)2 EFS log-rank P value.
Life Sciences Reporting Summary
Further information on experimental design is available in the Life Sciences Reporting Summary.
Data availability
Data used for this analysis are available under database of Genotypes and Phenotypes (dbGaP) accession numbers phs000465 and phs000178. Complete details of sample preparation protocols, clinical annotations and all primary data are available through the TARGET Data Matrix (https://ocg.cancer.gov/programs/target/data-matrix/). Sequence data are also accessible through the NCI Genomic Data Commons (https://portal.gdc.cancer.gov/legacy-archive/search/f) or NCBI dbGaP (https://www.ncbi.nlm.nih.gov/gap/) under accession number phs000218.
Supplementary Material
Acknowledgments
Dedicated to the memory of our colleague, mentor and friend, Dr. Robert Arceci, whose vision and perseverance set this effort in motion: “I may not have gone where I intended to go, but I think I have ended up where I needed to be” (Douglas Adams, The Long Dark Tea-Time of the Soul). The results published here are based on data generated by the TARGET initiative and TCGA. The TARGET initiative is supported by NCI Grant U10CA98543. Work performed under contracts from the NCI, US National Institutes of Health within HHSN261200800001E includes specimen processing (Children’s Oncology Group Biopathology Center), WGS (Complete Genomics), and RNA-seq and TCS (British Columbia Cancer Agency). The content of this publication does not necessarily reflect the views or policies of the US Department of Health and Human Services, nor does mention of trade names, commercial products or organizations imply endorsement by the US government. Computation for the work described in this paper was supported in part by Fred Hutchinson Scientific Computing, University of Southern California’s Center for High-Performance Computing and National Science Foundation (NSF) award ACI-1341935. This work was additionally supported by COG Chairs U10CA180886 and U10CA98543; COG Statistics and Data Center U10CA098413 and U10CA180899; COG Specimen Banking U24CA114766; R01CA114563 (S.M.); St. Baldrick’s Foundation (J.E.F., T.T. and S.M.); Alex’s Lemonade Stand (S.M.); Target Pediatric AML (TpAML), P20GM121293 (J.E.F.); the Arkansas Biosciences Institute (J.E.F.); and the Jane Anne Nohl Hematology Research Fund (T.T.).
Footnotes
AUTHOR CONTRIBUTIONS
M.A.S., D.S.G., S.M. and R.A. conceived and led the project. R.E.R., M.A.M., J.M.G.A., T.M.D., P.G., L.C.H., D.S.G. and S.M. managed the project. H.B., J.E.F., T.T., R.E.R., E.L.L., T.A.A., Y.M., R.M., A.J.M., M.A.M., J.Z., X.M., Yu Liu, Yanling Liu, T.M.D., A.C.H., B.S. and S.R.P. generated, processed and analyzed the data. S.C., G.R., C.M.Z., S.N., E.A.K. and A.S.G. shared critical data and reagents. H.B., J.E.F., T.T., R.E.R., E.L.L. and S.M. drafted the manuscript. All authors edited and approved the manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Note: Any Supplementary Information and Source Data files are available in the online version of the paper.
References
- 1.Steliarova-Foucher E, et al. International incidence of childhood cancer, 2001–10: a population-based registry study. Lancet Oncol. 2017;18:719–731. doi: 10.1016/S1470-2045(17)30186-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li S, et al. Distinct evolution and dynamics of epigenetic and genetic heterogeneity in acute myeloid leukemia. Nat Med. 2016;22:792–799. doi: 10.1038/nm.4125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cancer Genome Atlas Research Network. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 2013;368:2059–2074. doi: 10.1056/NEJMoa1301689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Papaemmanuil E, et al. Genomic classification and prognosis in acute myeloid leukemia. N Engl J Med. 2016;374:2209–2221. doi: 10.1056/NEJMoa1516192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Patel JP, et al. Prognostic relevance of integrated genetic profiling in acute myeloid leukemia. N Engl J Med. 2012;366:1079–1089. doi: 10.1056/NEJMoa1112304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ho PA, et al. Leukemic mutations in the methylation-associated genes DNMT3A and IDH2 are rare events in pediatric AML: a report from the Children’s Oncology Group. Pediatr Blood Cancer. 2011;57:204–209. doi: 10.1002/pbc.23179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Farrar JE, et al. Genomic profiling of pediatric acute myeloid leukemia reveals a changing mutational landscape from disease diagnosis to relapse. Cancer Res. 2016;76:2197–2205. doi: 10.1158/0008-5472.CAN-15-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lange BJ, et al. Outcomes in CCG-2961, a Children’s Oncology Group phase 3 trial for untreated pediatric acute myeloid leukemia: a report from the Children’s Oncology Group. Blood. 2008;111:1044–1053. doi: 10.1182/blood-2007-04-084293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cooper TM, et al. AAML03P1, a pilot study of the safety of gemtuzumab ozogamicin in combination with chemotherapy for newly diagnosed childhood acute myeloid leukemia: a report from the Children’s Oncology Group. Cancer. 2012;118:761–769. doi: 10.1002/cncr.26190. [DOI] [PubMed] [Google Scholar]
- 10.Gamis AS, et al. Gemtuzumab ozogamicin in children and adolescents with denovo acute myeloid leukemia improves event-free survival by reducing relapse risk: results from the randomized phase III Children’s Oncology Group trial AAML0531. J Clin Oncol. 2014;32:3021–3032. doi: 10.1200/JCO.2014.55.3628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lavallée VP, et al. Identification of MYC mutations in acute myeloid leukemias with NUP98-NSD1 translocations. Leukemia. 2016;30:1621–1624. doi: 10.1038/leu.2016.19. [DOI] [PubMed] [Google Scholar]
- 12.Faber ZJ, et al. The genomic landscape of core-binding factor acute myeloid leukemias. Nat Genet. 2016;48:1551–1556. doi: 10.1038/ng.3709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tarlock K, et al. Discovery and functional validation of novel pediatric specific FLT3 activating mutations in acute myeloid leukemia: results from the COG/NCI target initiative. Blood. 2015;126:87. [Google Scholar]
- 14.Ley TJ, et al. DNMT3A mutations in acute myeloid leukemia. N Engl J Med. 2010;363:2424–2433. doi: 10.1056/NEJMoa1005143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang YA, et al. DNA methyltransferase-3a interacts with p53 and represses p53- mediated gene expression. Cancer Biol Ther. 2005;4:1138–1143. doi: 10.4161/cbt.4.10.2073. [DOI] [PubMed] [Google Scholar]
- 16.Genovese G, et al. Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. N Engl J Med. 2014;371:2477–2487. doi: 10.1056/NEJMoa1409405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goossens S, et al. The EMT regulator Zeb2/Sip1 is essential for murine embryonic hematopoietic stem/progenitor cell differentiation and mobilization. Blood. 2011;117:5620–5630. doi: 10.1182/blood-2010-08-300236. [DOI] [PubMed] [Google Scholar]
- 18.Goossens S, et al. ZEB2 drives immature T-cell lymphoblastic leukaemia development via enhanced tumour-initiating potential and IL-7 receptor signalling. Nat Commun. 2015;6:5794. doi: 10.1038/ncomms6794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xiang P, et al. Identification of E74-like factor 1 (ELF1) as a transcriptional regulator of the Hox cofactor MEIS1. Exp Hematol. 2010;38:798–798. doi: 10.1016/j.exphem.2010.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ostronoff F, et al. NUP98/NSD1 and FLT3/ITD coexpression is more prevalent in younger AML patients and leads to induction failure: a COG and SWOG report. Blood. 2014;124:2400–2407. doi: 10.1182/blood-2014-04-570929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maxson JE, et al. CSF3R mutations have a high degree of overlap with CEBPA mutations in pediatric AML. Blood. 2016;127:3094–3098. doi: 10.1182/blood-2016-04-709899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Quintana-Bustamante O, et al. Overexpression of wild-type or mutants forms of CEBPA alter normal human hematopoiesis. Leukemia. 2012;26:1537–1546. doi: 10.1038/leu.2012.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vicente C, Conchillo A, García-Sánchez MA, Odero MD. The role of the GATA2 transcription factor in normal and malignant hematopoiesis. Crit Rev Oncol Hematol. 2012;82:1–17. doi: 10.1016/j.critrevonc.2011.04.007. [DOI] [PubMed] [Google Scholar]
- 24.Ng KP, et al. Runx1 deficiency permits granulocyte lineage commitment but impairs subsequent maturation. Oncogenesis. 2013;2:e78. doi: 10.1038/oncsis.2013.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang Y, et al. The Wnt/β-catenin pathway is required for the development of leukemia stem cells in AML. Science. 2010;327:1650–1653. doi: 10.1126/science.1186624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Valencia A, et al. Wnt signaling pathway is epigenetically regulated by methylation of Wnt antagonists in acute myeloid leukemia. Leukemia. 2009;23:1658–1666. doi: 10.1038/leu.2009.86. [DOI] [PubMed] [Google Scholar]
- 27.Nanbakhsh A, et al. c-Myc regulates expression of NKG2D ligands ULBP1/2/3 in AML and modulates their susceptibility to NK-mediated lysis. Blood. 2014;123:3585–3595. doi: 10.1182/blood-2013-11-536219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marcucci G, et al. MicroRNA expression in cytogenetically normal acute myeloid leukemia. N Engl J Med. 2008;358:1919–1928. doi: 10.1056/NEJMoa074256. [DOI] [PubMed] [Google Scholar]
- 29.Ramamurthy R, et al. miR-155 expression and correlation with clinical outcome in pediatric AML: a report from Children’s Oncology Group. Pediatr Blood Cancer. 2016;63:2096–2103. doi: 10.1002/pbc.26157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fooladinezhad H, Khanahmad H, Ganjalikhani-Hakemi M, Doosti A. Negative regulation of TIM-3 expression in AML cell line (HL-60) using miR-330-5p. Br J Biomed Sci. 2016;73:129–133. doi: 10.1080/09674845.2016.1194564. [DOI] [PubMed] [Google Scholar]
- 31.Lim EL, et al. Comprehensive sequence analysis of relapse and refractory pediatric acute myeloid leukemia identifies miRNA and mRNA transcripts associated with treatment resistance—a report from the COG/NCI-target AML initiative. Blood. 2015;126:687. [Google Scholar]
- 32.Gruber TA, et al. An Inv(16)(p13.3q24.3)-encoded CBFA2T3-GLIS2 fusion protein defines an aggressive subtype of pediatric acute megakaryoblastic leukemia. Cancer Cell. 2012;22:683–697. doi: 10.1016/j.ccr.2012.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liang K, et al. Therapeutic targeting of MLL degradation pathways in MLL-rearranged leukemia. Cell. 2017;168:59–72. doi: 10.1016/j.cell.2016.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Itskovich SS, et al. MBNL1 as a new therapeutic target in MLL-fusion gene leukemia. Blood. 2015;126:462. [Google Scholar]
- 35.Caudell D, et al. Retroviral insertional mutagenesis identifies Zeb2 activation as a novel leukemogenic collaborating event in CALM-AF10 transgenic mice. Blood. 2010;115:1194–1203. doi: 10.1182/blood-2009-04-216184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li J, et al. The EMT transcription factor Zeb2 controls adult murine hematopoietic differentiation by regulating cytokine signaling. Blood. 2017;129:460–472. doi: 10.1182/blood-2016-05-714659. [DOI] [PubMed] [Google Scholar]
- 37.Shlush LI, et al. Identification of pre-leukaemic haematopoietic stem cells in acute leukaemia. Nature. 2014;506:328–333. doi: 10.1038/nature13038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guryanova OA, et al. DNMT3A mutations promote anthracycline resistance in acute myeloid leukemia via impaired nucleosome remodeling. Nat Med. 2016;22:1488–1495. doi: 10.1038/nm.4210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Loghavi S, et al. Clinical features of de novo acute myeloid leukemia with concurrent DNMT3A, FLT3 and NPM1 mutations. J Hematol Oncol. 2014;7:74. doi: 10.1186/s13045-014-0074-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xu B, et al. Tumor suppressor menin represses paired box gene 2 expression via Wilms tumor suppressor protein–Polycomb group complex. J Biol Chem. 2011;286:13937–13944. doi: 10.1074/jbc.M110.197830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sinha S, et al. Mutant WT1 is associated with DNA hypermethylation of PRC2 targets in AML and responds to EZH2 inhibition. Blood. 2015;125:316–326. doi: 10.1182/blood-2014-03-566018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Abdel-Wahab O, et al. ASXL1 mutations promote myeloid transformation through loss of PRC2-mediated gene repression. Cancer Cell. 2012;22:180–193. doi: 10.1016/j.ccr.2012.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Göllner S, et al. Loss of the histone methyltransferase EZH2 induces resistance to multiple drugs in acute myeloid leukemia. Nat Med. 2017;23:69–78. doi: 10.1038/nm.4247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Drmanac R, et al. Human genome sequencing using unchained base reads on self-assembling DNA nanoarrays. Science. 2010;327:78–81. doi: 10.1126/science.1181498. [DOI] [PubMed] [Google Scholar]
- 45.Zhang J, et al. Germline mutations in predisposition genes in pediatric cancer. N Engl J Med. 2015;373:2336–2346. doi: 10.1056/NEJMoa1508054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cingolani P, et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 2012;6:80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cingolani P, et al. Using Drosophila melanogaster as a model for genotoxic chemical mutational studies with a new program, SnpSift. Front Genet. 2012;3:35. doi: 10.3389/fgene.2012.00035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Saunders CT, et al. Strelka: accurate somatic small-variant calling from sequenced tumor–normal sample pairs. Bioinformatics. 2012;28:1811–1817. doi: 10.1093/bioinformatics/bts271. [DOI] [PubMed] [Google Scholar]
- 50.Ye K, Schulz MH, Long Q, Apweiler R, Ning Z. Pindel: a pattern growth approach to detect break points of large deletions and medium sized insertions from paired-end short reads. Bioinformatics. 2009;25:2865–2871. doi: 10.1093/bioinformatics/btp394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhou X, et al. Exploring genomic alteration in pediatric cancer using ProteinPaint. Nat Genet. 2016;48:4–6. doi: 10.1038/ng.3466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lawrence MS, et al. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature. 2014;505:495–501. doi: 10.1038/nature12912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang Y, Zhou H, Zhou J, Sun W. Regression models for multivariate count data. J Comput Graph Stat. 2017;26:1–13. doi: 10.1080/10618600.2016.1154063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Leiserson MD, Wu HT, Vandin F, Raphael BJ. CoMEt: a statistical approach to identify combinations of mutually exclusive alterations in cancer. Genome Biol. 2015;16:160. doi: 10.1186/s13059-015-0700-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shannon P, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.van Borkulo CD, et al. A new method for constructing networks from binary data. Sci Rep. 2014;4:5918. doi: 10.1038/srep05918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zeileis A, Grothendieck G. zoo: S3 infrastructure for regular and irregular time series. J Stat Softw. 2005;14:1–27. [Google Scholar]
- 58.Mayakonda A, Koeffler HP. Maftools: Efficient analysis, visualization and summarization of MAF files from large-scale cohort based cancer studies. 2016 Preprint at https://www.biorxiv.org/content/early/2016/05/11/052662/
- 59.Miller CA, et al. SciClone: inferring clonal architecture and tracking the spatial and temporal patterns of tumor evolution. PLOS Comput Biol. 2014;10:e1003665. doi: 10.1371/journal.pcbi.1003665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Friedman J, Hastie T, Tibshirani R. Regularization paths for generalized linear models via coordinate descent. J Stat Softw. 2010;33:1–22. [PMC free article] [PubMed] [Google Scholar]
- 61.Chun HJ, et al. Genome-wide profiles of extra-cranial malignant rhabdoid tumors reveal heterogeneity and dysregulated developmental pathways. Cancer Cell. 2016;29:394–406. doi: 10.1016/j.ccell.2016.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Robertson G, et al. De novo assembly and analysis of RNA-seq data. Nat Methods. 2010;7:909–912. doi: 10.1038/nmeth.1517. [DOI] [PubMed] [Google Scholar]
- 63.Li H, Durbin R. Fast and accurate long-read alignment with Burrows—Wheeler transform. Bioinformatics. 2010;26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.de Hoon MJ, et al. Cross-mapping and the identification of editing sites in mature microRNAs in high-throughput sequencing libraries. Genome res. 2010;20:257–264. doi: 10.1101/gr.095273.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Leek JT, Johnson WE, Parker HS, Jaffe AE, Storey JD. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics. 2012;28:882–883. doi: 10.1093/bioinformatics/bts034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Triche TJ, Jr, Weisenberger DJ, Van Den Berg D, Laird PW, Siegmund KD. Low-level processing of Illumina Infinium DNA methylation BeadArrays. Nucleic Acids Res. 2013;41:e90. doi: 10.1093/nar/gkt090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Aryee MJ, et al. Minfi: a flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics. 2014;30:1363–1369. doi: 10.1093/bioinformatics/btu049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fortin JP, et al. Functional normalization of 450k methylation array data improves replication in large cancer studies. Genome Biol. 2014;15:503. doi: 10.1186/s13059-014-0503-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lee DD, Seung HS. Learning the parts of objects by non-negative matrix factorization. Nature. 1999;401:788–791. doi: 10.1038/44565. [DOI] [PubMed] [Google Scholar]
- 70.Abrahamsson J, et al. Response-guided induction therapy in pediatric acute myeloid leukemia with excellent remission rate. J Clin Oncol. 2011;29:310–315. doi: 10.1200/JCO.2010.30.6829. [DOI] [PubMed] [Google Scholar]
- 71.Burnett AK, et al. Identification of patients with acute myeloblastic leukemia who benefit from the addition of gemtuzumab ozogamicin: results of the MRC AML15 trial. J Clin Oncol. 2011;29:369–377. doi: 10.1200/JCO.2010.31.4310. [DOI] [PubMed] [Google Scholar]
- 72.Creutzig U, et al. Less toxicity by optimizing chemotherapy, but not by addition of granulocyte colony-stimulating factor in children and adolescents with acute myeloid leukemia: results of AML-BFM 98. J Clin Oncol. 2006;24:4499–4506. doi: 10.1200/JCO.2006.06.5037. [DOI] [PubMed] [Google Scholar]
- 73.Creutzig U, et al. Treatment strategies and long-term results in paediatric patients treated in four consecutive AML-BFM trials. Leukemia. 2005;19:2030–2042. doi: 10.1038/sj.leu.2403920. [DOI] [PubMed] [Google Scholar]
- 74.Gibson BE, et al. Treatment strategy and long-term results in paediatric patients treated in consecutive UK AML trials. Leukemia. 2005;19:2130–2138. doi: 10.1038/sj.leu.2403924. [DOI] [PubMed] [Google Scholar]
- 75.Kardos G, et al. Treatment strategy and results in children treated on three Dutch Childhood Oncology Group acute myeloid leukemia trials. Leukemia. 2005;19:2063–2071. doi: 10.1038/sj.leu.2403873. [DOI] [PubMed] [Google Scholar]
- 76.Perel Y, et al. Impact of addition of maintenance therapy to intensive induction and consolidation chemotherapy for childhood acute myeloblastic leukemia: results of a prospective randomized trial, LAME 89/91. Leucámie Aiqüe Myéloïde Enfant. J Clin Oncol. 2002;20:2774–2782. doi: 10.1200/JCO.2002.07.300. [DOI] [PubMed] [Google Scholar]
- 77.Pession A, et al. Results of the AIEOP AML 2002/01 multicenter prospective trial for the treatment of children with acute myeloid leukemia. Blood. 2013;122:170–178. doi: 10.1182/blood-2013-03-491621. [DOI] [PubMed] [Google Scholar]
- 78.Balgobind BV, et al. Integrative analysis of type-I and type-II aberrations underscores the genetic heterogeneity of pediatric acute myeloid leukemia. Haematologica. 2011;96:1478–1487. doi: 10.3324/haematol.2010.038976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hollink IH, et al. NUP98/NSD1 characterizes a novel poor prognostic group in acute myeloid leukemia with a distinct HOX gene expression pattern. Blood. 2011;118:3645–3656. doi: 10.1182/blood-2011-04-346643. [DOI] [PubMed] [Google Scholar]
- 80.Hollink IH, et al. Clinical relevance of Wilms tumor 1 gene mutations in childhood acute myeloid leukemia. Blood. 2009;113:5951–5960. doi: 10.1182/blood-2008-09-177949. [DOI] [PubMed] [Google Scholar]
- 81.Hollink IH, et al. Favorable prognostic impact of NPM1 gene mutations in childhood acute myeloid leukemia, with emphasis on cytogenetically normal AML. Leukemia. 2009;23:262–270. doi: 10.1038/leu.2008.313. [DOI] [PubMed] [Google Scholar]
- 82.Camp RL, Dolled-Filhart M, Rimm DL. X-tile: a new bio-informatics tool for biomarker assessment and outcome-based cut-point optimization. Clin Cancer Res. 2004;10:7252–7259. doi: 10.1158/1078-0432.CCR-04-0713. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data used for this analysis are available under database of Genotypes and Phenotypes (dbGaP) accession numbers phs000465 and phs000178. Complete details of sample preparation protocols, clinical annotations and all primary data are available through the TARGET Data Matrix (https://ocg.cancer.gov/programs/target/data-matrix/). Sequence data are also accessible through the NCI Genomic Data Commons (https://portal.gdc.cancer.gov/legacy-archive/search/f) or NCBI dbGaP (https://www.ncbi.nlm.nih.gov/gap/) under accession number phs000218.