Abstract
During our search for a cDNA encoding β-galactosidase II, a β-galactosidase/exogalactanase (EC 3.2.1.23) present during tomato (Lycopersicon esculentum Mill.) fruit ripening, a family of seven tomato β-galactosidase (TBG) cDNAs was identified. The shared amino acid sequence identity among the seven TBG clones ranged from 33% to 79%. All contained the putative active site-containing consensus sequence pattern G-G-P-[LIVM]-x-Q-x-E-N-E-[FY] belonging to glycosyl hydrolase family 35. Six of the seven single-copy genes were mapped using restriction fragment length polymorphisms of recombinant inbred lines. RNA gel-blot analysis was used to evaluate TBG mRNA levels throughout fruit development, in different fruit tissues, and in various plant tissues. RNA gel-blot analysis was also used to reveal TBG mRNA levels in fruit of the rin, nor, and Nr tomato mutants. The TBG4-encoded protein, known to correspond to β-galactosidase II, was expressed in yeast and exo-galactanase activity was confirmed via a quantified release of galactosyl residues from cell wall fractions containing β(1→4)-d-galactan purified from tomato fruit.
β-Galactosidases (EC 3.2.1.23), a widespread family of glycosyl hydrolases, are characterized by their ability to hydrolyze terminal, non-reducing β-d-galactosyl residues from β-d-galactosides. β-Galactosidases are commonly associated with the hydrolysis of the milk sugar lactose into Gal and Glc by mammals and Escherichia coli. Due to the hardy and easily assayed nature of some β-galactosidases, E. coli LacZ has become one of the most conspicuous reporter gene systems in use today (Bayer and Campos-Ortega, 1992; Young and Hope, 1993; Cui et al., 1994; Timmons et al., 1997; Lewis et al., 1998). However, LacZ has not been used for plant-based reporter gene systems due to high endogenous β-galactosidase activity in most plant tissues at neutral pH (Jefferson et al., 1987). Although the LacZ product can be easily assayed, β-galactosidases have been shown to function under a variety of reaction conditions, to have numerous substrate specificities, and to be located in various subcellular and extracellular locations in eukaryotes (Gossrau, 1976; Dey and del Campillo, 1984; Singh and Knox, 1984; Kulikova et al., 1990). It has become clear that β-galactosidases play numerous roles in the metabolism of a multitude of galactosyl-containing substrates.
Numerous studies have shown that β-galactosidases catalyze the hydrolysis of terminal galactosyl residues from carbohydrates, glycoproteins, and galactolipids. β-Galactosidase action has been proposed to release stored energy for rapid growth (lactose hydrolysis in mammals and bacteria, xyloglucan mobilization in cotyledons), release free Gal during normal metabolic recycling of galactolipids, glycoproteins, and cell wall components, and degrade cell wall components during senescence (Lo et al., 1979; Bhalla and Dalling, 1984; Maley et al., 1989; Raghothama et al., 1991; De Veau et al., 1993; Ross et al., 1993; Buckeridge and Reid, 1994; Hall, 1998). Furthermore, many β-galactosidases have specific biosynthetic activities by both transglycosylation and reverse hydrolysis under favorable thermodynamic in vitro conditions (Bonnin et al., 1995; Yoon and Ajisaka, 1996).
The study of the role of β-galactosidases in tomato fruit has resulted from physiological and biochemical data showing that Gal is the most dynamic sugar residue of the cell wall during tomato fruit development. In particular, these physiological studies showed that there was a significant net loss of galactosyl residues from the wall throughout fruit development and the rate of galactosyl residue loss increased during ripening. Also shown was that free Gal levels, although stable throughout the preripening stages of fruit development, increased rapidly during ripening (Kim et al., 1991). Physiological studies also showed that when free Gal was infiltrated into mature green fruit at a concentration equivalent to the red ripe stage of fruit development, ripening was hastened (Gross, 1985). Furthermore, the un-conjugated N-glycans, Man5GlcNAc and Man3(Xyl)GlcNAc, were able to predictably promote or delay ripening of excised pericarp discs, but only in the presence of physiologically relevant levels of free Gal (Yunovitz and Gross, 1994).
The purification of three tomato β-galactosidases (TBGs), of which only β-galactosidase II had exo-galactanase activity, was first reported by Pressey (1983). It was proposed that β-galactosidase II might be important in the softening of tomato fruit during ripening. This contention was further supported by demonstrating that, although total β-galactosidase activity was unchanged during ripening of wild-type and mutant fruit, β-galactosidase II activity increased 4-fold in wild-type fruit, but did not change during the equivalent chronological period in the ripening mutants, ripening inhibitor (rin) and non-ripening (nor) (Carey et al., 1995). The cDNA corresponding to β-galactosidase II was subsequently cloned (Smith et al., 1998). In the process of cloning the β-galactosidase II cDNA, six additional cDNAs were identified that had significant shared amino acid sequence identity. We report on the characterization of this family of β-galactosidase genes, which may be involved in Gal metabolism during cell wall turnover, conversion of chloroplasts into chromoplasts, fruit growth, and senescence.
RESULTS
There is some contradiction in the literature concerning the nomenclature of β-galactosidases versus cell wall (or other native substrates) galactosyl-specific hydrolases. To clarify the work presented here, the following terms are used: β-galactosidase, an enzyme that can hydrolyze a β-galactosyl residue linked to a variety of aglycones (e.g. lactose, p-nitrophenyl-β-d-galactopyranoside [PNP-gal], etc.); exogalactanase, an enzyme that is specific for the non-reducing end of galactan and has no action on PNP-gal or lactose and whose affinity for the substrate increases the depolymerization of the substrate; and galactanase, an enzyme that cleaves internal bonds in galactan. Moreover, an enzyme that can hydrolyze galactose from PNP-gal and the non-reducing end of galactan is referred to as a β-galactosidase/exogalactanase (B. Henrissat, personal communication; Henrissat, 1998).
Sequence Analysis
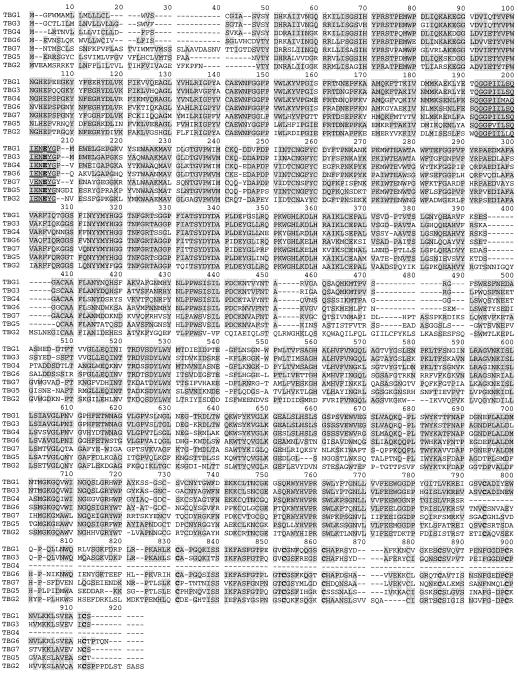
The open reading frames (ORFs) of seven cDNA clones that had a significant level of shared amino acid sequence identity to each other and other published plant β-galactosidases were identified (Fig. 1). All of the TBGs contain the putative active site-containing consensus sequence pattern G-G-P-[LIVM]-x-Q-x-E-N-E-[FY] belonging to glycosyl hydrolase family 35; where amino acids separated by dashes are conserved, amino acids [LIVM] and [FY] are conserved substitutions and x is any amino acid (Fig. 2; Henrissat, 1998). Due to a number of short amino acid insertions and deletions throughout their sequences (e.g. Fig. 2, positions 472–491), TBG2, TBG5, and TBG7 shared less than 41% amino acid sequence identity to each other and to the other β-galactosidases. The deduced amino acid sequence in the TBG1 ORF is nearly identical to that of a previously described cDNA (accession no. P48980) from tomato cv Ailsa Craig (Carey et al., 1995). Also, the ORFs of TBG1, TBG3, and TBG4 are identical to the cDNA clones (accession nos. CAA10174, CAA10173, and CAA10175, respectively) isolated from tomato cv Money Maker. To our knowledge, no data on these clones has been published. These clones are likely derived from the same gene, but differ in their non-coding region and nucleotide sequences due to their varietal origins. The nucleotide sequence accession numbers for the TBGs are: TBG1, AF023847; TBG2, AF154420; TBG3, AF154421; TBG4, AF020390; TBG5, AF154423; TBG6, AF154424; and TBG7, AF154422.
Figure 1.
β-Galactosidase phylogenetic tree based on shared amino acid sequence identity. A, TBG cDNAs. B, Plant β-galactosidases. Performed using the Higgins-Sharp algorithm (unweighted pair group method with arithmetic means).
Figure 2.
Deduced amino acid sequence alignment of TBG ORFs. Regions of identity are highlighted. Numbers correspond to positions within the alignment. Underlined is the consensus sequence for the putative active site of the glycosyl hydrolase family 35, and the Glu (E) shown in bold is the putative active site (proton donor) residue. Performed using the Higgins-Sharp algorithm (unweighted pair group method with arithmetic means).
BLAST searches of the non-redundant database at GenBank also indicated significant shared amino acid sequence identity among domains of the TBGs and mammalian and fungal β-galactosidases, however, little shared sequence identity was detected with most bacterial β-galactosidases (data not shown).
All the TBG ORFs were predicted to have a signal sequence (Table I). However, cellular targeting was not fully confirmed using the two prediction programs SignalP and PSORT. Only TBG4, TBG5, and TBG6 were predicted to be secreted by both programs. TBG7 was predicted to be targeted to the chloroplast by PSORT. The deduced amino acid sequences of the seven clones were also subjected to analysis using the program DNASIS (Hitachi, Tokyo) for predicting molecular mass, pI, and potential N-linked glycosylation sites (Table I).
Table I.
TBG cDNA nucleotide and deduced amino acid sequence data
| Clone | Length | kD | pI | N-Link | Target |
|---|---|---|---|---|---|
| kb | |||||
| TBG1 | 3.2 | 90.8 | 6.2 | 2 | er/out |
| TBG2 | 3 | 97.0 | 6.2 | 6 | pm |
| TBG3 | 3 | 90.5 | 8.2 | 1 | er/out |
| TBG4 | 2.5 | 77.9 | 8.9 | 3 | out |
| TBG5 | 2.8 | 89.8 | 6.6 | 6 | out |
| TBG6 | 3 | 91 | 6.9 | 1 | out |
| TBG7 | 3 | 93.3 | 8.0 | 6 | chlor |
N-link, Possible N-linked glycosylation sites; er, endoplasmic reticulum; out, secreted; pm, tethered to plasma membrane; chlor, chloroplast.
Sequence analysis of the full-length TBG and plant β-galactosidases clones showed that the β-galactosidases could be divided into two distinct groups based on mass and other characteristics. The first group of proteins (apple, carnation, lupin, papaya, and TBG4) had a molecular mass range of 80.5 to 82.8 kD and was comprised of 721 to 731 amino acids. The second group (asparagus, TBG1, TBG2, TBG3, TBG5, TBG6, and TBG7) had a molecular mass range of 89.8 to 99.9 kD and was comprised of 832 to 888 residues. The larger size of the proteins in the second group was due primarily to an addition of approximately 100 amino acids at their carboxyl termini. The addition at the carboxyl terminus was remarkable due to the presence of a number of highly conserved residues, and in particular, seven conserved cystines (Fig. 2, shown in bold). It is unknown if the similarity within or differences between these two groups of β-galactosidases has any functional significance.
DNA Gel-Blot Analysis and Mapping
DNA gel-blot analysis was done using the 3′-untranslated regions of TBG1, TBG2, TBG3, TBG4, and TBG7 and nucleotides 286 to 1,041 and 254 to 1,003 of TBG5 and TBG6, respectively, as probes against restriction enzyme-digested genomic DNA. The genes corresponding to the clones appeared to be present as single copies (data not shown). The same probes were used to map six of the seven genes (Table II) using RFLPs of recombinant inbred lines (J. Giovannoni, personal communication).
Table II.
TBG gene map positions
| Gene | Chromosome | R1 Line |
|---|---|---|
| TBG1 | 12a | Overlap of IL 12-2, IL 12-3 |
| TBG2 | 9 | IL 9-3 |
| TBG3 | 3 | IL 3-5 |
| TBG4 | 12a | Overlap of IL 12-2, IL 12-3 |
| TBG5 | 11 | IL 11-3 |
| TBG6 | 2 | Overlap of IL 2-4, IL 2-5 |
| TBG7 | No RFLP |
Genes were mapped by DNA gel-blot analysis using RFLPs of recombinant inbred lines described in Eshed and Zamir (1994). Data provided by J. Giovannoni and S. Miller (personal communication).
TBG1 and TBG4 did not hybridize to restriction fragments of the same size.
RNA Gel-Blot Analysis
RNA gel-blot analysis was performed so that the relative TBG hybridization signals within each experiment were approximately comparable. This was accomplished by doing preliminary RNA gel-blot analysis to estimate the signal intensity that each TBG probe produced among all RNA samples (data not presented). Based on these preliminary results, RNA gel-blot analysis was repeated to confirm the previous results and present the data in a comparative manner (Figs. 3–6). Further care was taken on this point by ensuring that probes were synthesized to similar specific activities using TBG cDNA regions that did not cross-hybridize (see “Materials and Methods” for details).
Figure 3.
RNA gel-blot analysis of TBG temporal expression in fruit tissues. Twenty micrograms of total RNA extracted from all fruit tissues except seeds was loaded in each lane. Fruit was harvested at 10, 20, 30, 35, and 40 dpp and at the breaker (Br), turning (Tr), pink (Pk), red (Rd), and over-ripe (OR) stages. Blots were hybridized using the probes indicated to the right. PG was used as a fruit-ripening-specific control. A 26S ribosomal gene clone was used as a loading control for each blot and one example is shown.
Figure 6.
RNA gel-blot analysis of TBG expression in normal and mutant fruit tissues. Twenty micrograms of total RNA extracted from peel and outer pericarp tissue at various dpp was hybridized to specified probes. PG was also used as fruit-ripening-specific control. A 26S ribosomal gene clone from soybean was used as a loading control for each blot; one example is shown. WT, Wild type.
Temporal Expression Pattern in Fruit
The temporal expression pattern of the seven genes in fruit tissue was examined using RNA extracted from all fruit tissues except seeds. Transcripts for all seven genes were detected during some stage of fruit development (Fig. 3). TBG1 and TBG3 had similar constitutive expression patterns throughout the breaker to over-ripe stages. TBG2 transcript was detected at a very low level from 30 d post-pollination (dpp) to the over-ripe stage. TBG4 transcript was first detected at the breaker stage, peaked at the turning stage, and declined as fruit ripened fully. Although TBG5 had a similar expression pattern to TBG4 during the ripening stages, TBG5 mRNA was also detected throughout all of the earlier stages of fruit development. TBG6 had a distinctive expression pattern in that the presence of its high abundance transcript was limited to preripening. TBG7 also had a unique expression pattern, being transiently expressed very early and late in fruit development.
Spatial Expression Pattern in Fruit
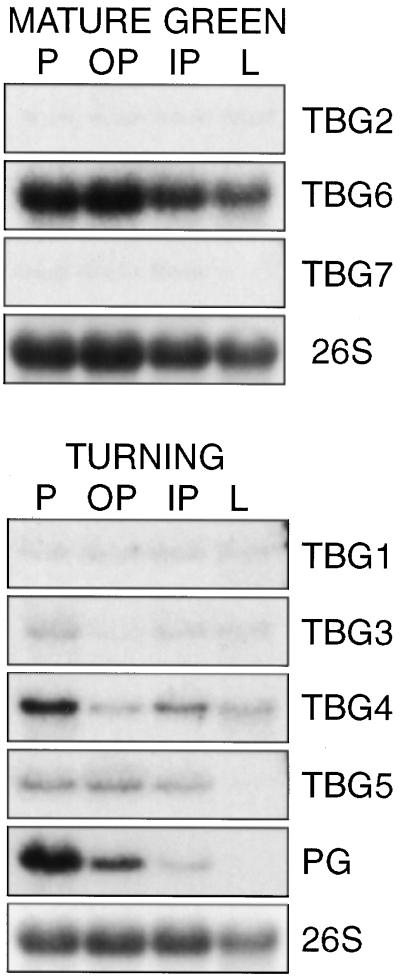
RNA gel-blot analysis was also performed to determine transcript accumulation in specific tissues within immature and maturing fruit (Fig. 4). Within all mature green fruit tissues, both TBG2 and TBG7 transcripts were detected, albeit at a very low level. TBG6 transcript was also detected in all mature green fruit tissues, but at a far more abundant level. TBG1, TBG3, and TBG4 transcripts were detected in all turning stage fruit tissues, although TBG4 transcript, like tomato polygalacturonase (PG), was most abundant in the peel. TBG5 transcript was detected at moderate abundance in the peel and inner and outer pericarp, but similar to PG, was conspicuously absent in locular tissue.
Figure 4.
RNA gel-blot analysis of TBG spatial expression in fruit tissues. Twenty micrograms of total RNA extracted from mature green or turning stage fruit peel (P), outer pericarp (OP), inner pericarp (IP), and locular (L) tissue was loaded in each lane. Blots were hybridized using the probes indicated to the right. A cDNA clone for PG was used as a fruit-ripening-specific control. A 26S ribosomal gene clone from soybean was used as a loading control for each blot; one example is shown.
Tissue Specificity
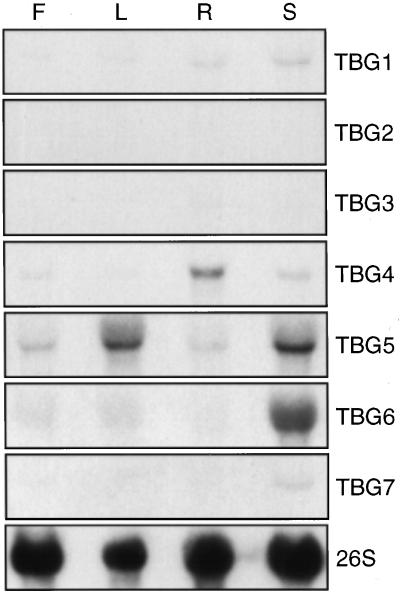
RNA gel-blot analysis was performed to reveal whether any of the TGBs were expressed in non-fruit tissues. With the exception of TBG2, transcripts of all other clones were detected in non-fruit tissues (Fig. 5). Although TBG3 transcript may appear to be absent in Figure 5, after 2-fold-longer film exposures, its transcript was detected at very low levels in root and stem tissues (data not shown). Transcripts of TBG1, TBG4, TBG5, and TBG6 were detected in all tissues tested. However, only transcripts from TBG5 and TBG6 were detected at levels approximately as abundant in vegetative tissues as in growing fruit.
Figure 5.
RNA gel-blot analysis of TBG expression in various plant tissues. Twenty micrograms of total RNA extracted from flowers (F), leaves (L), roots (R), and stems (S) was loaded in each lane. Blots were hybridized using the probes indicated to the right. A 26S ribosomal gene clone from soybean was used as a loading control for each blot; one example is shown.
Expression in Normal versus Mutant Fruit
Gene expression in rin and nor fruit was shown to be impaired for most ripening-related genes via the lack of climacteric ethylene-inducible gene expression (DellaPenna et al., 1989; Knapp et al., 1989; Harriman et al., 1991; Picton et al., 1993). The expression of many developmentally regulated genes (e.g. PG, E8, and 1-aminocyclopropane-1-carboxylic acid synthase) was also shown to be impaired in rin and nor fruit (DellaPenna et al., 1989; Theologis et al., 1993). Although rin and nor fruit cannot be ripened by exogenous ethylene, ethylene-regulated gene expression can be induced by exogenous ethylene. Therefore, rin and nor fruit were shown to be competent to respond to ethylene (Tigchelaar et al., 1978; Lincoln and Fischer 1988; Giovannoni et al., 1989; Gray et al., 1992). The never ripe (Nr)-encoded gene product is believed to be an ethylene receptor and the Nr mutation is characterized by a block in a wide range of ethylene responses (Lanahanet al., 1994; Wilkinson et al., 1995). To suggest the potential roles of the TBG products and transcriptional regulatory mechanisms, RNA gel-blot analysis was performed using fruit tissue from the ripening-impaired mutants rin, nor, and Nr.
The results of RNA gel-blot analysis of ripening-impaired fruit suggested that transcript levels of TBG1, TBG2, TBG3, TBG5, and TBG7 were present in a chronological (dpp) pattern similar to that of wild-type accumulation (Fig. 6). However, the abundance of TBG4 and TBG6 transcripts was different in the mutant fruit. TBG4 transcript accumulation was significantly impaired in all three mutants relative to wild-type accumulation. Interestingly, TBG6 transcript accumulation persisted up to 50 dpp in fruit of all three mutants (Fig. 6), whereas it was not detected in wild-type fruit after 43 dpp (Figs. 3 and 6).
Enzyme Activity
To determine the role of the TBG encoded products, we initiated experiments to express the cDNA encoded enzymes using heterologous expression systems. A yeast expression system, which secretes a mature amino-terminal-(FLAG) fusion protein into the culture medium, was tested. TBG4 expression was attempted first because the corresponding enzyme β-galactosidase II has been purified from tomato fruit and characterized in some detail (Carey et al., 1995; Smith et al., 1998). Therefore, the activity of the heterologous system-expressed protein could be compared to the native enzyme purified from tomato. Expression of a FLAG-TBG4-encoded fusion protein construct resulted in the production of approximately 1 mg of active enzyme per 50 mL of culture (data not shown). Subsequently, the FLAG-TBG4 protein was affinity-purified using an anti-FLAG affinity resin (Fig. 7).
Figure 7.
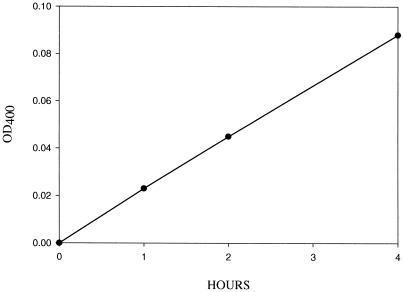
β-Galactosidase assay of the TBG4-encoded protein. The TBG4-encoded protein was expressed in yeast, affinity column-purified, and assayed for β-galactosidase activity using p-nitrophenyl-β-d-galactopyranoside.
The affinity-purified TBG4 enzyme had β(1→4)-d-galactosidase activity by virtue of its ability to hydrolyze the synthetic substrates PNP-gal (Fig. 8) and 5-bromo-4-chloro-3-indoxyl-β-d-galactopyranoside (X-gal; data not shown). The enzyme was also able to hydrolyze lactose, albeit poorly (Table III). Most significantly, the enzyme was able to release galactosyl residues from a variety of galactosyl-containing cell wall substrates, and therefore had exogalactanase activity (Table III). Thus, this enzyme should be termed a β-galactosidase/exo-galactanase.
Figure 8.
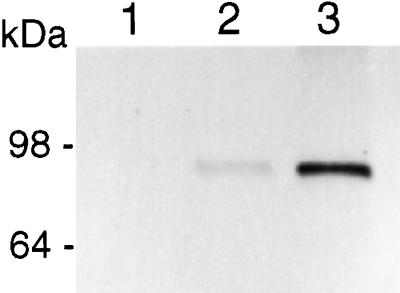
Protein gel-blot analysis of FLAG-TBG4 fusion protein expression by yeast. Lane 1, Concentrated and desalted culture medium of a YEpFLAG1 transformed yeast clone, used as a negative control. Lane 2, Concentrated and desalted culture medium of yeast clone expressing FLAG-TBG4 fusion protein. Lane 3, Affinity-purified FLAG-TBG4 fusion protein.
Table III.
Enzyme activity of TBG4 expressed in yeast
| Substrate | Galactose Released
|
|
|---|---|---|
| Boiled | Active | |
| μg | ||
| Lactose | 0 | 0.1 |
| CSP | 0 | 5 |
| ASP | 0 | 15 |
| HCF | 0 | 4 |
| LG | 0 | 117 |
Galactosyl residue hydrolysis from lactose, chelator soluble pectin (CSP), alkali soluble pectin (ASP), and hemicellulosic (HCF) fractions purified from tomato fruit cell walls and a commercially prepared lupin galactan (LG).
DISCUSSION
During our search for a cDNA coding for β-galactosidase II, a total of seven cDNA clones with a significant level of shared sequence identity were found. All of the genes appear to have a single genomic copy and six of the seven genes were mapped. The shared sequence identity of the seven clones to each other and to other plant β-galactosidases and the presence of the glycosyl hydrolase family 35 consensus sequence (Fig. 2), suggests that they all may have β-galactosidase activity. However, this does not make it possible to predict their in vivo substrate specificities.
The expression patterns of the TBGs varied widely. Only TBG2 mRNA was found to be potentially fruit specific by RNA gel-blot analysis, however, more sensitive mRNA quantification assays are required to conclusively determine TBG2's expression pattern. The wide range of expression patterns among the TBGs suggests that the substrates for most of the TBG-encoded enzymes are distributed throughout the tomato plant. The prediction, by both PSORT and Signal P, that the signal sequences of TBG2, TBG4, TBG5, and TBG6 may target these proteins outside the cell suggests that they may all be involved in cell wall galactosyl modification (Table I). Although PSORT predicted that TBG1 and TBG3 proteins are targeted to the endoplasmic reticulum, the program SignalP predicted that TBG1 and TBG3 are secreted. Therefore, it is conceivable that these enzymes are involved in cell wall galactosyl modification as well (Table I).
The idea that six of the seven TBG proteins may be involved in cell wall galactosyl modifications is plausible based on the increasing number of reports on the substrate specificities of β-galactosidase/exogalactanases involved in xyloglucan mobilization in cotyledons of several species. One recent example of the specificity that various β-galactosidase/exogalactanases can have was reported by de Alcantara et al. (1999). They showed that a β-galactosidase/exogalactanase purified from Copaifera langsdorffii cotyledons could only hydrolyze terminal galactosyl residues if linked to a xylosyl residue adjacent to the non-reducing end glucosyl residue of an endo-β-glucanase-derived xyloglucan oligomer, and that this enzyme was unable to hydrolyze a galactosyl residue linked to a xylosyl residue adjacent to a reducing end glucosyl residue. Moreover, the removal of this terminal galactosyl residue by β-galactosidase/exogalactanase was implicated to be a prerequisite for the hydrolysis of the reducing end glucosyl residue by β-glucosidase.
One interesting finding in the present study was that TBG5 mRNA was present at relatively high levels during early fruit development, dropped throughout the immature and mature green stages, rose at the onset of ripening, and fell again throughout the later stages of ripening (Fig. 3). It is possible that the TBG5-encoded enzyme, if targeted to the cell wall, is needed for rapid expansion and again during the disassembly of the wall during ripening. The pattern of TBG5 mRNA accumulation throughout normal fruit development was similar to that of the tomato endo-β-glucanase genes Cel1 and Cel2 (Gonzalez-Bosch et al., 1996). The overlapping expression pattern of the endo-β-glucanase and exo-β-galactanase genes may indicate that a coordinated effort of these enzymes is necessary for hemicellulosic modifications that occur during cell division, cell growth and fruit ripening.
TBG7 mRNA is present at maximum levels at 10 dpp and in over-ripe fruit and at relatively low levels throughout the other stages of fruit development. The TBG7-encoded enzyme is predicted to be targeted to the chloroplast, and, therefore, it may be involved in galactolipid turnover/degradation, which occurs in chloroplasts and chromoplasts during tomato fruit development (Güçlü et al., 1989; Whitaker, 1992). However, not enough is known about galactolipid metabolism throughout fruit development to predict what, if any, relationship TBG7's expression pattern has with galactolipid metabolism.
The rin, nor, and Nr ripening-impaired mutants are distinguished by multiple aberrant fruit ripening phenotypes (Tigchelaar et al., 1979). Rin and nor fruit are characterized by a lack of climacteric ethylene and cannot be ripened by the application of exogenous ethylene (Tigchelaar et al., 1979). Because ethylene-regulated gene expression is attenuated (DellaPenna et al., 1989; Knapp et al., 1989; Harriman et al., 1991; Picton et al., 1993), ethylene-regulated gene expression can be induced by exogenous ethylene (Lincoln and Fischer, 1988; Giovannoni et al., 1989; Gray et al., 1992), and expression of several developmentally regulated genes (e.g. PG, E8, and 1-aminocyclopropane-1-carboxylic acid synthase) is attenuated (DellaPenna et al., 1989; Theologis et al., 1993), rin and nor are believed to be lesions in a developmentally regulated pathway of fruit ripening. The Nr mutation results in a block of a wide range of ethylene responses (Lanahan et al., 1994). The Nr gene has been cloned and is believed to be involved in ethylene perception and signal transduction (Wilkinson et al., 1995). Of particular interest to this study is that rin, nor, and Nr fruit soften little when compared to ripening wild-type fruit at the equivalent chronological stage (Tigchelaar et al., 1979). Therefore, experiments were done to compare the accumulation of TBG mRNA in wild-type versus ripening-impaired mutant fruit. TBG1, TBG2, TBG3, and TBG5 transcripts are present in rin, nor, and Nr fruit in a chronological (dpp) pattern similar to that of wild-type accumulation (Fig. 6). Therefore, it is unlikely that these TBGs could be solely responsible for cell wall modifications that result in fruit softening during ripening. It is also unlikely that the increase in mRNA of these genes during ripening is regulated solely by ethylene. In contrast, the abundance of TBG4 and TBG6 transcripts differed in the mutant versus wild-type fruit. TBG4 transcript accumulation is significantly impaired in all three mutants relative to wild-type accumulation. This observation implies that the TBG4-encoded β-galactosidase II may be involved in cell wall modifications that lead to fruit softening and TBG4 transcription may be up-regulated by ethylene.
Accumulation of TBG6 mRNA persisted up to 50 dpp in the fruit of all three mutants (Fig. 6), whereas it was not detected in wild-type fruit after 43 dpp (Figs. 3 and 6). It is consequently unlikely that the TBG6-encoded gene product plays a crucial role in cell wall degradation leading to fruit softening during ripening. The activities of most β-galactosidases/exogalactanases are believed to result in degradation of the cell wall, although it is known that numerous β-galactosidases have specific biosynthetic activities by both transglycosylation and reverse hydrolysis under favorable thermodynamic in vitro conditions (Bonnin et al., 1995; Yoon and Ajisaka, 1996). Therefore, it is interesting to speculate that the persistence of the TBG6-encoded gene product in mutant fruit tissues results in increased fruit firmness via some type of biosynthetic (cell wall strengthening) activity. It is unfortunate that we do not know of any example that demonstrates biosynthetic activity by a plant β-galactosidase under in vivo conditions to support this hypothesis.
The significance of TBG expression patterns will be more fully understood after cellular localization and substrate specificities of their encoded enzymes is known. We and others have attempted to make antibodies to various plant β-galactosidases, but have been unable to produce satisfactory results (data not shown; G. Seymour, personal communication). This has unfortunately made cellular localization of the various β-galactosidases difficult, and reporter gene or epitope fusions with the various mature forms of β-galactosidases may be needed to elucidate their cellular localization using transgenic plants.
Cloning and characterization of the TBG4 cDNA was previously described by Smith et al. (1998) and was included here for comparison to the other TBG clones. Here, use of a yeast expression system enabled us to produce enough enzyme to test for exogalactanase activity on a variety of substrates. TBG4 codes for β-galactosidase II, as described by Pressey (1983) and Carey et al. (1995). Our yeast-expressed protein has similar attributes to the fruit-purified enzyme, in that its molecular mass using SDS-PAGE is 75 kD, the pH optimum is 4.0, and it has both β-galactosidase and exo-galactanase activity on a variety of natural substrates containing β-1,4-linked galactans. It is not surprising that the TBG4-encoded enzyme is most active on alkali-soluble pectin (Table III), since there is a correlation between increasing β-galactosidase II activity and decreasing galactosyl content of alkali-soluble, but not chelator-soluble, pectin during tomato fruit ripening (Carrington and Pressey, 1996).
The analysis of an increasing number of cloned plant β-galactosidases and native β-galactosidases is resulting in a much better understanding of their wide range of in vitro substrate specificities. However, the precise role of the individual β-galactosidases in plant development remains speculative. Therefore, we will take a molecular genetic approach to elucidate the consequences of modifying expression of the TBGs. Both antisense and over-expression studies of TBGs in transgenic tomato plants are being conducted to further evaluate the functional consequences of altering TBG gene expression.
MATERIALS AND METHODS
Plant Material
Tomato (Lycopersicon esculentum Mill. cv Rutgers) plants were grown in a greenhouse using standard cultural practices. The ripening mutants rin, nor and Nr (Tigchelaar et al., 1978), were all in the cv Rutgers background. Flowers were tagged at anthesis and fruit was harvested according to the number of dpp, or based on surface color using the ripeness stages previously described by Mitcham et al. (1989). For gene expression studies, only mature green fruit (40–42 dpp) at the MG4 stage were used for RNA extractions. The MG4 stage was confirmed by cutting fruit open and observing entirely liquefied locular gel and no cut seeds. Leaf, flower, and stem tissues were harvested from greenhouse-grown plants. Roots were harvested from seedlings grown in liquid culture medium consisting of the salts and vitamins described by Murashige and Skoog (1962) and 1% (w/v) Suc for 4 weeks.
RNA Extraction
Fruits were processed immediately after harvest by chilling on ice, excising the various tissues, and freezing them in liquid nitrogen. Tissue samples were ground using a mortar and pestle and stored at −80°C. RNA was extracted using the method described by Verwoerd et al. (1989), except that a second chloroform extraction was performed.
cDNA Isolation
Reverse transcriptase-PCR, cDNA library construction, library screening, and sequencing were done exactly as described in Smith et al. (1998). The 5′-cDNA ends of TBG2, TBG5, and TBG6 were isolated using the 5′ system for RACE (Gibco-BRL, Cleveland). The gene-specific primers used for 5′ RACE were DS251 (ggaccgaatgagctttcaaca), DS252 (gagcgactcagatatcataaga), and DS253 (tgagatccgactagctttgc) for TBG2; DS245 (cgagactcaatatcaccattg), DS246 (tgtgaatcgcttcatttctgc), and DS247 (agctctctccaccaacttca) for TBG5; and DS248 (ctccaagtaccttggcttga), DS249 (agcataccctttcattgcgtt), and DS250 (gccctgctttctgaatcgtt) for TBG6. A 3′-RACE kit was used to amplify the 3′-cDNA ends of TBG5 and TBG6. The gene-specific primers used for 3′ RACE were DS262 (agtctcgttatggtcctcgt), DS263 (aacacccaagatgtggactg), and DS264 (gaagacatcgctttcgctgt) for TBG5, and DS265 (tcaagccaaggtacttggag), DS266 (ctgcaatttggactgaagct), and DS267 (aggatttggcatttgctgttg) for TBG6. 5′ and 3′ RACE were performed following the manufacturer's instructions and Platinum Taq (Gibco-BRL) was used for PCR. Full-length TBG5 and TBG6 cDNAs were amplified by PCR using Platinum Pfx DNA polymerase (Gibco/BRL), the 3′-RACE products described above as templates, and the primers DS282 (tttttttctagaagttgatcggaaaattgaaga) and DS283 (tattgaagctagttttcttttatta) for TBG5, and DS277 (aaagagtctagaagggaggtggaatcatggag) and DS273 (gatgcaaattacacttttccattg) for TBG6. The TBG5 and TBG6 PCR products were digested with XbaI and were ligated into XbaI plus SmaI digested pBluescript II KS (Stratagene, La Jolla, CA). Both strands of the cDNAs were sequenced by primer walking.
Sequence Analysis
Nucleotide and deduced amino acid sequence comparisons against the databases were done using BLAST searches (Altschul et al., 1990). Sequence data were analyzed and aligned using MacDNAsis (Hitachi, San Bruno, CA) software. Protein classification was performed using the interactive web site Protomap (Yona et al., 1998). Signal sequence predictions were conducted using the interactive web sites PSORT (Nakai and Kanehisa, 1992) and SignalP (Nielsen et al., 1997).
RNA Gel-Blot Analysis
Total RNA (20 μg per lane) was separated in a formaldehyde/MOPS [3-(N-morpholino)-propanesulfonic acid] agarose gel, transferred to Hybond-N+ nylon membrane (Amersham, Arlington Heights, IL), fixed by incubating for 2 h at 80°C, hybridized overnight in a hybridization incubator using a buffer described by Church and Gilbert (1984), washed to a final stringency of 0.1× SSC with 0.2% (w/v) SDS at 65°C, and autoradiographed. An RNA ladder standard was used to estimate the length of the mRNAs. Probes were synthesized using a Random Primed DNA Labeling Kit (Roche Molecular Biochemicals, Indianapolis) with [32P]dATP (3,000 Ci/mmol) as the label. Probes were synthesized using DNA fragments derived from PCR-amplification of the 3′-untranslated regions of TBG1, TBG2, TBG3, TBG4, and TBG7 and nucleotides 286 to 1,041 and 254 to 1,003 of TBG5 and TBG6, respectively. Probes were used only when the 32P incorporation level was between 70% and 85% and adjusted to 106 dpm/mL for hybridization. For all RNA gel-blot experiments, the TBG blots were exposed to X-Omat (Eastman Kodak, Rochester, NY) film using an intensifying screen for 4 d at −80°C. As a loading control, RNA blots were stripped and reprobed at a reduced hybridization and washing stringency using a soybean 26S rDNA fragment (Turano et al., 1997). Blots hybridized with the rDNA and PG probes were exposed for 4 and 16 h respectively.
Expression in Yeast, Protein-Blot Analysis, and Enzyme Activity
TBG proteins were expressed using a the FLAG Yeast N-Terminal Expression System (Sigma-Aldrich, St. Louis). The ORF of TBG4 was PCR-amplified using the oligonucleotides DS214 (agtgagatctagtgtttcttatgatgacaga) and DS218 (tcgagtgtcgactcttgatctcctgactagaga) so that the signal peptide (aa 1–23) was removed and a BglII and SalI restriction site was created at the 5′ and 3′ end of the ORF, respectively. The 2.183-kb fragment was cloned into a BglII plus SalI digested YEpFLAG1 vector to produce an extracellular N-terminal FLAG fusion protein when expressed. The vector was transformed into the Saccharomyces cerevisiae host strain BJ3505. Yeast cells transformed with the YEpFLAG1 vector were used as a negative control. Cultures were grown and protein harvesting was done as described by the manufacturer, except that the cells were grown at room temperature for maximal expression.
Samples of yeast culture medium or affinity-purified TBG4 protein were subjected to SDS-PAGE and transferred to nitrocellulose. Blots were subjected to protein-blot analysis using M1 anti-FLAG primary antibody, rabbit anti-mouse secondary antibody conjugated to alkaline phosphatase, and colorimetric detection using Sigma-Fast substrate following the manufacturer's recommendations.
The yeast culture medium from YEpFLAG1 and YEpFLAGTBG4 transformed cells was concentrated and desalted using Centriprep30 columns (Millipore, Bedford, MA). Enzyme assays were performed using the desalted medium and were repeated using column-purified TBG4 enzyme to confirm the specificity of TBG4 enzyme's activity. β-Galactosidase activity was assayed as described by Pressey (1983) using 10 μL of affinity-column-purified enzyme per reaction and PNP-gal as substrate; 1 unit of activity was defined as the amount of enzyme that liberated 1 μmol PNP min−1 at 37°C. Exo-galactanase activity was determined by incubating cell wall material with the enzyme samples essentially as described by Carey et al. (1995). One-milliliter assays consisted of 2 mg of substrate, 0.005 unit of enzyme, 0.1% (w/v) bovine serum albumin, and 50 mm sodium acetate buffer at pH 4. Cell wall material was purified from mature green tomato fruit using the methods described by Gross (1984). A lupin galactan, pretreated with α-l-arabinofuranosidase, was obtained from Megazyme (Wicklow, Ireland). Free Gal was identified and quantified by gas chromotography/mass spectrometry-selected ion monitoring of the galactitol acetate derivative prepared from the released Gal product in reaction mixtures using the method of Gross and Acosta (1991).
ACKNOWLEDGMENTS
The authors wish to thank Heather Torlina, Karen Green, and Norman Livsey for excellent technical assistance, Dr. Graham Seymour for helpful comments and critical review of the manuscript, and Dr. James Giovannoni and Stephanie Miller for mapping the TBGs.
LITERATURE CITED
- Altschul SF, Gish W, Miller W, Meyers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Ausubel F, Brent R, Kingston R, Moore D, Seidman J, Smith J, Struhl K. Current Protocols in Molecular Biology. New York: John Wiley & Sons; 1987. [Google Scholar]
- Bayer TA, Campos-Ortega JA. A transgene containing lacZ is expressed in primary sensory neurons in zebrafish. Development. 1992;115:421–426. doi: 10.1242/dev.115.2.421. [DOI] [PubMed] [Google Scholar]
- Bhalla PL, Dalling MJ. Characteristics of a β-galactosidase associated with the stroma of chloroplasts prepared from mesophyll protoplasts of the primary leaf of wheat. Plant Physiol. 1984;76:92–95. doi: 10.1104/pp.76.1.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnin E, Lahaye M, Vigouroux J, Thibault JF. Preliminary characterization of a new exo-β-(1,4)-galac-tanase with transferase activity. Int J Biol Macromol. 1995;17:345–351. doi: 10.1016/0141-8130(96)81844-7. [DOI] [PubMed] [Google Scholar]
- Buckeridge MS, Reid JS. Purification and properties of a novel β-galactosidase or exo-(1→4)-β-d-galactanase from the cotyledons of germinated Lupinus angustifolius L. seeds. Planta. 1994;192:502–511. doi: 10.1007/BF00203588. [DOI] [PubMed] [Google Scholar]
- Carey AT, Holt K, Picard S, Wilde R, Tucker GA, Bird CR, Schuch W, Seymour GB. Tomato exo-β(1→4)-d-galactanase: isolation, changes during ripening in normal and mutant tomato fruit, and characterization of a related cDNA clone. Plant Physiol. 1995;108:1099–1107. doi: 10.1104/pp.108.3.1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrington CMS, Pressey R. β-Galactosidase II activity in relation to changes in cell wall galactosyl composition during tomato ripening. J Am Hortic Soc. 1996;121:132–136. [Google Scholar]
- Church GM, Gilbert W. Genomic sequencing. Proc Natl Acad Sci USA. 1984;81:1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui C, Wani MA, Wight D, Kopchick J, Stambrook PJ. Reporter genes in transgenic mice. Transgenic Res. 1994;3:182–194. doi: 10.1007/BF01973986. [DOI] [PubMed] [Google Scholar]
- de Alcântara PHN, Dietrich SMC, Buckeridge MS. Xyloglucan mobilization and purification of a (XLLG/XLXG) specific β-galactosidase from cotyledons of Copaifera langsdorffii. Plant Physiol Biochem. 1999;37:653–663. [Google Scholar]
- DellaPenna D, Lincoln JE, Fischer RL, Bennett AB. Transcriptional analysis of polygalacturonase and other ripening associated genes in Rutgers, rin, nor, and Nr tomato fruit. Plant Physiol. 1989;90:1372–1377. doi: 10.1104/pp.90.4.1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Veau EJI, Gross KC, Huber DJ, Watada AE. Degradation and solubilization of pectin by β-galacto-sidases purified from avocado mesocarp. Physiol Plant. 1993;87:279–285. [Google Scholar]
- Dey PM, del Campillo E. Biochemistry of the multiple forms of glycosidases in plants. In: Meister A, editor. Advances in Enzymology. Vol. 56. New York: John Wiley & Sons; 1984. pp. 141–249. [DOI] [PubMed] [Google Scholar]
- Eshed Y, Zamir D. A genomic library of Lycopersicon pennellii in L. esculentum: a tool for fine mapping of genes. Euphytica. 1994;79:175–179. [Google Scholar]
- Giovannoni J, DellaPenna D, Bennett A, Fischer R. Expression of a chimeric polygalacturonase gene in transgenic rin (ripening inhibitor) tomato fruit results in polyuronide degradation but not fruit softening. Plant Cell. 1989;1:53–63. doi: 10.1105/tpc.1.1.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Bosch C, Brummell DA, Bennett AB. Differential expression of two endo-1,4-β-glucanase genes in pericarp and locules of wild-type and mutant tomato fruit. Plant Physiol. 1996;111:1313–1319. doi: 10.1104/pp.111.4.1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gossrau R. Localization of glycosidases with naphthyl substrates. Histochem J. 1976;8:271–282. doi: 10.1007/BF01003816. [DOI] [PubMed] [Google Scholar]
- Gray JE, Picton S, Shabbeer J, Schuch W, Grierson D. Molecular biology of fruit ripening and its manipulation with antisense genes. Plant Mol Biol. 1992;19:69–87. doi: 10.1007/BF00015607. [DOI] [PubMed] [Google Scholar]
- Gross K, Acosta PB. Fruits and vegetables are a source of galactose: implications in planning the diets of patients with galactosemia. J Inher Dis. 1991;14:253–258. doi: 10.1007/BF01800599. [DOI] [PubMed] [Google Scholar]
- Gross KC. Fractionation and partial characterization of cell walls from normal and non-ripening mutant tomato fruit. Physiol Plant. 1984;62:25–32. [Google Scholar]
- Gross KC. Promotion of ethylene evolution and ripening of tomato fruit by galactose. Plant Physiol. 1985;79:306–307. doi: 10.1104/pp.79.1.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Güçlü J, Paulin A, Soudain P. Changes in polar lipids during ripening and senescence of cherry tomato (Lycopersicon esculentum): relation to climacteric and ethylene increases. Physiol Plant. 1989;77:413–419. [Google Scholar]
- Hall BG. Determining the evolutionary potential of a gene. Mol Biol Evol. 1998;15:1055–1061. doi: 10.1093/oxfordjournals.molbev.a026004. [DOI] [PubMed] [Google Scholar]
- Harriman R, Tieman D, Handa A. Molecular cloning of tomato pectin methylesterase gene and its expression in Rutgers, ripening inhibitor, nonripening, and Never ripe tomato fruits. Plant Physiol. 1991;97:80–87. doi: 10.1104/pp.97.1.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henrissat B. Glycosidase families. Biochem Soc Trans. 1998;26:153–156. doi: 10.1042/bst0260153. [DOI] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW. GUS fusions: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 1987;6:3901–3907. doi: 10.1002/j.1460-2075.1987.tb02730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Gross KC, Solomos T. Galactose metabolism and ethylene production during development and ripening of tomato fruit. Postharv Biol Technol. 1991;1:67–80. [Google Scholar]
- Knapp J, Moureau P, Schuch W, Grierson D. Organization and expression of polygalacturonase and other ripening related genes in Ailsa Craig “Neverripe” and “Ripening inhibitor” tomato mutants. Plant Mol Biol. 1989;12:105–116. doi: 10.1007/BF00017453. [DOI] [PubMed] [Google Scholar]
- Kulikova AK, Gomarteli MM, Tsereteli AK, Bezborodov AM, Kvesitadze GI, Bilai TI. β-Galactosidases of lower eukaryotes. Appl Biochem Microbiol. 1990;25:621–632. [Google Scholar]
- Lanahan MB, Yen HC, Giovannoni JJ, Klee HJ. The Never ripe mutation blocks ethylene perception in tomato. Plant Cell. 1994;6:521–530. doi: 10.1105/tpc.6.4.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis JC, Feltus A, Ensor CM, Ramanathan S, Daunert S. Applications of reporter genes. Anal Chem. 1998;70:579A–585A. doi: 10.1021/ac9819638. [DOI] [PubMed] [Google Scholar]
- Lincoln J, Fischer R. Regulation of gene expression by ethylene in wild-type and rin tomato (Lycopersicon esculentum) fruit. Plant Physiol. 1988;88:370–374. doi: 10.1104/pp.88.2.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo JT, Mukerji K, Awasthi YC, Hanada E, Suzuki K, Srivastava SK. Purification and properties of sphingolipid β-galactosidase from human placenta. J Biol Chem. 1979;254:6710–6715. [PubMed] [Google Scholar]
- Maley F, Trimble RB, Tarentino AL, Plummer TH. Characterization of glycoproteins and their associated oligosaccharides through the use of endoglycosidases. Anal Biochem. 1989;180:195–204. doi: 10.1016/0003-2697(89)90115-2. [DOI] [PubMed] [Google Scholar]
- Mitcham EJ, Gross KC, Ng TJ. Tomato fruit cell wall synthesis during development and senescence: in vivo radiolabeling of cell wall fractions using [14C]sucrose. Plant Physiol. 1989;89:477–481. doi: 10.1104/pp.89.2.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T, Skoog F. A revised medium for the growth and bioassays with tobacco tissue culture. Physiol Plant. 1962;15:473–497. [Google Scholar]
- Nakai K, Kanehisa M. A knowledge base for predicting protein localization sites in eukaryotic cells. Genomics. 1992;14:897–911. doi: 10.1016/S0888-7543(05)80111-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen H, Engelbrecht J, Brunak S, von Heijne G. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 1997;10:1–6. doi: 10.1093/protein/10.1.1. [DOI] [PubMed] [Google Scholar]
- Picton S, Gray JE, Barton SL, AbuBaker U, Lowe A, Grierson D. cDNA cloning and characterization of novel ripening-related mRNAs with altered patterns of accumulation in the ripening inhibitor (rin) tomato ripening mutant. Plant Mol Biol. 1993;23:193–207. doi: 10.1007/BF00021431. [DOI] [PubMed] [Google Scholar]
- Pressey R. β-Galactosidases in ripening tomatoes. Plant Physiol. 1983;71:132–135. doi: 10.1104/pp.71.1.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghothama KG, Lawton KA, Goldsbrough PB, Woodson WR. Characterization of an ethylene-regulated flower senescence-related gene from carnation. Plant Mol Biol. 1991;17:61–71. doi: 10.1007/BF00036806. [DOI] [PubMed] [Google Scholar]
- Ross GS, Redgwell RJ, MacRae EA. Kiwifruit β-galactosidase: isolation and activity against specific fruit cell-wall polysaccharides. Planta. 1993;189:499–506. [Google Scholar]
- Singh MB, Knox RB. Quantitative cytochemistry of β-galactosidase in normal and enzyme deficient (gal) pollen of Brassica campestris: application of the indogenic method. Histochem J. 1984;16:1273–1296. doi: 10.1007/BF01003726. [DOI] [PubMed] [Google Scholar]
- Smith DL, Starrett DA, Gross KC. A gene coding for tomato fruit β-galactosidase II is expressed during fruit ripening. Plant Physiol. 1998;117:417–423. doi: 10.1104/pp.117.2.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theologis A, Oeller P, Wong L, Rothmann W, Gantz D. Use of a tomato mutant constructed with reverse genetics to study fruit ripening, a complex developmental process. Dev Genetics. 1993;14:282–259. doi: 10.1002/dvg.1020140406. [DOI] [PubMed] [Google Scholar]
- Tigchelaar EC, McGlasson WB, Buescher RW. Genetic regulation of tomato fruit ripening. Hortic Sci. 1978;13:508–513. [Google Scholar]
- Timmons L, Becker J, Barthmaier P, Fyrberg C, Shearn A, Fyrberg E. Green fluorescent protein/β-galactosidase double reporters for visualizing Drosophila gene expression patterns. Dev Genet. 1997;20:338–347. doi: 10.1002/(SICI)1520-6408(1997)20:4<338::AID-DVG5>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- Turano FJ, Thakkar SS, Fang T, Weisemann JM. Characterization and expression of NAD(H) dependent glutamate dehydrogenase genes in Arabidopsis thaliana. Plant Physiol. 1997;113:1329–1341. doi: 10.1104/pp.113.4.1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verwoerd TC, Dekker BMM, Hoekema A. A small-scale procedure for the rapid isolation of plant RNAs. Nucleic Acids Res. 1989;17:2362. doi: 10.1093/nar/17.6.2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitaker BD. Changes in galactolipid and phospholipid levels of tomato fruits stored at chilling and nonchilling temperatures. Phytochemistry. 1992;31:2627–2630. [Google Scholar]
- Wilkinson J, Lanahan M, Yen H, Giovannoni J, Klee H. An ethylene-inducible component of signal transduction encoded by Never-ripe. Science. 1995;270:1807–1809. doi: 10.1126/science.270.5243.1807. [DOI] [PubMed] [Google Scholar]
- Yona G, Linial N, Tishby N, Linial M. A map of the protein space: an automated hierarchical classification of all protein sequences. Intelligent Syst Mol Biol. 1998;6:212–221. [PubMed] [Google Scholar]
- Yoon JH, Ajisaka K. The synthesis of galactopyranosly derivatives with β-galactosidases of different origins. Carbohydr Res. 1996;292:153–163. doi: 10.1016/s0008-6215(96)91041-1. [DOI] [PubMed] [Google Scholar]
- Young JM, Hope IA. Molecular markers of differentiation in Caenorhabditis elegans obtained by promoter trapping. Dev Dyn. 1993;196:124–132. doi: 10.1002/aja.1001960206. [DOI] [PubMed] [Google Scholar]
- Yunovitz H, Gross KC. Delay of tomato fruit ripening by an oligosaccharide N-glycan: interactions with IAA, galactose and lectins. Physiol Plant. 1994;90:152–156. [Google Scholar]