Abstract
The root-hypocotyl of Arabidopsis produces a relatively large amount of secondary vascular tissue when senescence is delayed by the removal of inflorescences, and plants are grown at low population density. Peptidase zymograms prepared from isolated xylem and phloem revealed the existence of distinct proteolytic enzyme profiles within these tissues. cDNA libraries were constructed from isolated xylem and bark of the root-hypocotyl and screened for cDNAs coding for cysteine, serine, and aspartic peptidases. Three cDNAs, two putative papain-type cysteine peptidases (XCP1 and XCP2) and one putative subtilisin-type serine peptidase (XSP1), were identified from the xylem library for further analysis. Using RNA gel blots it was determined that these peptidases were expressed in the xylem and not in the bark. Quantitative reverse transcriptase-polymerase chain reaction confirmed the RNA gel-blot results and revealed high levels of XCP1 and XCP2 mRNA in stems and flowers of the infloresence. A poly-histidine-tagged version of XCP1 was purified from Escherichia coli by denaturing metal-chelate chromatography. Following renaturation, the 40-kD recombinant XCP1 was not proteolytically active. Activation was achieved by incubation of recombinant XCP1 at pH 5.5 and was dependent on proteolytic processing of the 40-kD inactive polypeptide to a 26-kD active peptidase.
In view of the economic significance of wood formation and Suc transport, understanding the physiology of vascular tissues is of obvious importance. Xylem, phloem, and the vascular cambium are also tissues of great fundamental significance, representing opportunities for investigations aimed at understanding how plants regulate differentiation, programmed cell death (pcd), secondary cell wall biosynthesis, and lignification. That xylem formation is inducible by plant growth regulators, Suc, and wounding has greatly facilitated investigations of tracheary element (TE) differentiation. Examples of TE model systems include wound-induced differentiation in Coleus × hybridus stems (Stephenson et al., 1996), cultured explants from Jerusalem artichoke and lettuce pith (Phillips and Dodds, 1977; Wilson et al., 1994), and zinnia mesophyll cell cultures (Kohlenbach and Schmidt, 1975; Fukuda and Komamine, 1980). Wood-forming tissues of economically important tree species loblolly pine (Loopstra and Sederoff, 1995) and poplar (Sterky et al., 1998) have been isolated and used to generate expressed sequence tag (EST) collections. In Arabidopsis a pendant stem phenotype (Zhong et al., 1997) has led to the identification of a gene that affects fiber and vascular differentiation (Zhong and Ye, 1999).
We are interested in the roles played by proteolytic enzymes in the differentiation of TEs. The zinnia system has yielded the most information to date concerning the biochemistry and molecular biology of proteolytic pathways active during this process. The ability of the 26S proteasome inhibitor lactacystin (Omura et al., 1991) to prevent TE differentiation implicates the ubiquitin-proteasome pathway of proteolysis as a regulator of TE differentiation (Woffenden et al., 1998). Increases in TE-specific Cys peptidase activities have been observed (Minami and Fukuda, 1995; Ye and Varner, 1996; Beers and Freeman, 1997), and a cDNA encoding a papain-type Cys endopeptidase has been cloned from differentiating zinnia TEs (Ye and Varner, 1996). Although direct demonstrations of the roles played by Cys peptidases have not been presented, it was recently shown that a peptide-aldehyde inhibitor of papain-type peptidases blocked complete autolysis of TEs (Woffenden et al., 1998). In addition to their probable role as autolytic enzymes during TE differentiation, papain-type peptidases have been implicated in numerous aspects of plant growth and development including seed germination, organ senescence (for recent reviews, see Buchanan-Wollaston, 1997; Granell, 1998), stress and pathogen response (Solomon et al., 1999), and anther dehiscence (Koltunow et al., 1990). Cultured zinnia TEs also exhibit increased activity of Ser peptidases (Ye and Varner, 1996; Beers and Freeman, 1997; Groover and Jones, 1999). Neither protein nor cDNA sequences for zinnia TE Ser peptidases have been reported. The Ser peptidase present in TE extracts is the approximate molecular mass (60 kD) expected for mature subtilisin-type (family S8) Ser endopeptidases. Plant subtilisin-type enzymes are also associated with fruit ripening (Yamagata et al., 1994), microsporogenesis (Taylor et al., 1997), and pathogen response (Tornero et al., 1997).
In this report we describe a method for using Arabidopsis to study gene expression in secondary vascular tissues. Although not typically considered as a model for secondary growth, Arabidopsis is capable of producing relatively large quantities of secondary vascular tissue (Kondratieva-Melville and Vodolaz-sky, 1982; Lev-Yadun, 1994; Dolan and Roberts, 1995; Busse and Evert, 1999) when grown at low-population density and under conditions that prevent senescence, i.e. removal of reproductive structures (Lev-Yadun, 1994). We have taken advantage of the potential for secondary growth by Arabidopsis and developed a simple method for preparing xylem and phloem-enriched samples for biochemical analyses of peptidase activities. We have also isolated xylem and bark for the construction of cDNA libraries. From the xylem cDNA library, we have cloned full-length cDNAs predicted to code for two papain-type peptidases (XCP1 and XCP2) and a subtilisin-type peptidase (XSP1). Quantitative reverse transcriptase (RT)-PCR and RNA gel blots support the conclusion that within the root-hypocotyl, mRNA transcripts encoding these peptidases localize to the xylem. We expressed a poly-His-tagged version of XCP1 and confirmed that this cDNA codes for an active peptidase.
RESULTS
Isolation of Secondary Vascular Tissues from Root-Hypocotyl Sections
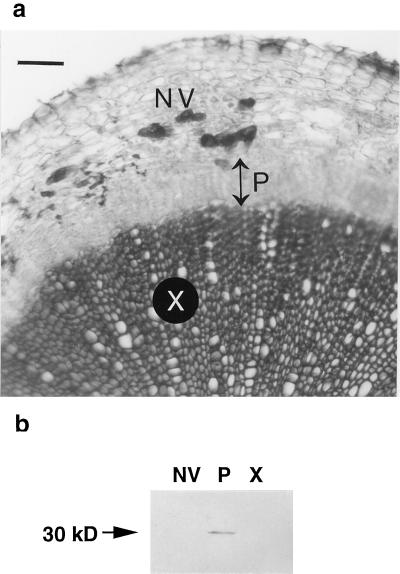
A transverse section through an 8-week-old root-hypocotyl segment shows extensive secondary growth including numerous files of secondary xylem and phloem (Fig. 1a). Separation of root-hypocotyl segments into three components, xylem, phloem-enriched, and non-vascular (cortex plus epidermis) tissues was performed under the dissecting microscope. Alternatively, root-hypocotyl segments were separated into two components, xylem and bark (phloem plus non-vascular), as for cDNA library construction. Lignification, visible as the dark, phloroglucinol-stained central core of the root-hypocotyl section in Figure 1a, was a convenient histochemical marker for confirming the purity of isolated tissues, i.e. lignified secondary xylem did not partition with secondary phloem (data not shown). We do not know whether the vascular cambium remained with the phloem, as would be expected for woody species from which similar xylem and bark samples have been prepared (Loopstra and Sederoff, 1995; Sterky et al., 1998). Secondary phloem was more difficult to separate from non-vascular tissue. Consequently, it is possible that small amounts of non-vascular tissue partitioned with the phloem preparations. For this reason, isolated phloem is referred to as phloem enriched. Immunoblot analysis was used to determine whether phloem cells were partitioning with non-vascular tissue (or with xylem) during tissue isolation. The immunoblot shown in Figure 1b reveals a 30-kD sieve element-specific protein limited to the phloem-enriched fraction, confirming the absence of detectable levels of phloem sieve elements from xylem and non-vascular preparations. The 30-kD phloem protein was recognized by the monoclonal antibody RS32 that recognizes a 32-kD protein (R. Sjölund, personal communication) from sieve element cultures of Streptanthus tortuosus (Brassicacae) (Wang et al., 1995).
Figure 1.
Transverse section through an 8-week-old Arabidopsis root-hypocotyl segment and immunoblot detection of a 30-kD, sieve element-specific protein in the phloem-enriched fraction. Extensive secondary vascular tissue production is observed in the root-hypocotyl following 8 weeks of growth at low population density and with removal of emerging inflorescences (a). Files of secondary xylem (X) appear darker than surrounding tissue due to phloroglucinol staining of lignified TEs. Double-headed arrow indicates the extent of secondary phloem (P). Non-vascular (NV) tissue consists of cortex and epidermis. Bar = 100 μm. Secondary xylem and non-vascular tissue can be separated from secondary phloem in the root-hypocotyl (b). A monoclonal antibody, RS32, recognizes a 30-kD sieve element protein in an extract from the phloem-enriched fraction (P). The 30-kD protein is not detectable in extracts from xylem (X) or non-vascular (NV) tissues.
Root-Hypocotyl Tissue-Specific Peptidase Profiles
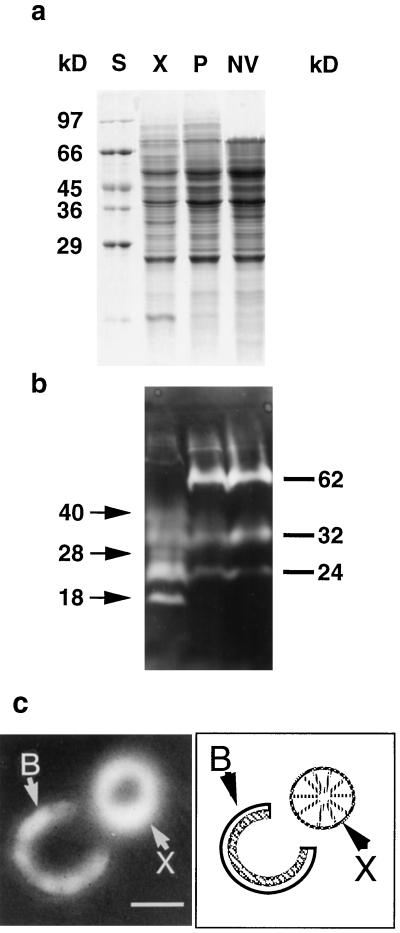
To determine whether xylem and phloem tissues exhibit differential expression of peptidase genes, gelatin-impregnated zymograms were prepared. Coomassie staining of proteins revealed only minor detectable differences between protein profiles from xylem, phloem-enriched, and non-vascular preparations (Fig. 2a). The peptidase profiles for these tissues were distinct, however, and revealed three xylem-specific activities at 40, 28, and 18 kD (Fig. 2b). Other characteristics of the zymogram included a 24-kD species that was detectable in all three extracts but that was most active in xylem, a 32-kD activity present at similar levels in all three tissues, and a 62-kD species not detected in xylem but equally represented in both phloem-enriched and non-vascular tissue extracts. Incubation of gelatin-impregnated gels with inhibitors (E-64 or cystatin) of papain-type Cys endopeptidases resulted in the loss of all activities of 40 kD and smaller (data not shown). The 62-kD enzyme was unaffected by E-64 and cystatin and was only partially inhibited by 10 mm phenylmethylsulfonyl fluoride (data not shown). A tissue print zymogram of a dissected root transverse section localized the xylem peptidase activity to the secondary xylem at the periphery of that tissue (Fig. 2c). Activity was not associated with the oldest central portion of the root.
Figure 2.
SDS-PAGE and tissue print zymogram analyses of total protein and peptidases extracted from xylem, phloem-enriched, and non-vascular preparations from Arabidopsis root-hypocotyl. Protein was extracted from xylem (X), phloem-enriched (P), and non-vascular (NV) preparations and subjected to SDS-PAGE followed by Coomassie staining (a) or incubation with a gelatin-impregnated substrate gel (b) for detection of peptidase activity. The Coomassie-stained gel was loaded with 30 μg of protein in each lane. The SDS-PAGE zymogram was produced using 45 μg of protein in each lane. Molecular mass standards (S) are indicated to the left of the Coomassie-stained gel. Molecular masses of xylem-specific peptidase are indicated by arrows and the molecular masses of peptidases either common to all three tissues (32 and 24 kD) or absent from xylem (62 kD) are indicated by bars to the right of the zymogram. For the tissue print zymogram (c), a thin transverse section was prepared from an 8-week-old primary root, dissected to separate bark from xylem, and incubated on a gelatin-impregnated polyacrylamide gel to visualize peptidase activity. Peptidase activity is visible throughout the bark (B) but is limited to the periphery of the isolated xylem (X). The diagram to the right of the tissue print zymogram shows the relative positions of dissected bark and xylem during tissue printing. In the diagram the hatched inner layer of the bark represents secondary phloem and the clear outer layer of the bark represents non-vascular tissue. Bar = 1 mm.
Screening for Xylem-Specific Peptidases
To identify papain-type peptidases expressed in vascular tissues, we designed degenerate primers for PCR screening of xylem and bark cDNA libraries (Table I). We also designed degenerate primers for subtilisin-type Ser and for Asp peptidases as these enzymes are also reported to be associated with tissues engaged in pcd (Taylor et al., 1997; Runeberg-Roos and Saarma, 1998; Neuteboom et al., 1999; Panavas et al., 1999), and hence, may be associated with xylem. In addition, because vascular tissue is likely to be under-represented in most cDNA libraries derived from whole organs, we used selected gene-specific primers to determine whether papain-type peptidases predicted from genomic sequences but not yet described either experimentally or as ESTs were represented in our cDNA libraries. Finally, primers Cys369 and Cys604 were used because they target genes coding for peptidases sharing 65% identity with a zinnia TE Cys peptidase p48-17 (Ye and Varner, 1996).
Table I.
Results of xylem (X) and bark (B) cDNA library screen using degenerate sense (S) and antisense (A) and gene-specific antisense primers for Ser, Cys, and Asp peptidases
| Peptidase | Primer Name | Primer Sequence | PCR Product (Accession No.) | X | B |
|---|---|---|---|---|---|
| Ser | Ser1 (S) | WTMGCSRYSTACAARGTYTGTTGG | AF069299 | + | − |
| Ser2 (A) | AACRTGAGGRCAKGCCATTGARGT | AL022023 | + | + | |
| Cys | Cys1 (S) | GGWGGWYTYATGGAYTAYGC | D13043 | + | + |
| Cys2 (A) | GGRTAMTCRTGWGGRCARCA | ||||
| Cys132 (A) | CTACAGCCTCCATTAACGCT | AC000132 | + | + | |
| Cys707 (A) | TGCATTTTCCGCCGAATCCA | Z99707 | − | − | |
| Cys604 (A) | CTTGGTCTTGGTAGGATATGAGGC | AL022604 | + | − | |
| Cys369 (A) | TCAAGATCAACCCCGCACCGC | AC007369 | + | − | |
| Asp | Asp1 (S) | TTTTTGAYACYGGRAGCTCTAACC | U51036 | + | + |
| Asp2 (A) | CCAGAATCTGCTATCGCAGAACA |
Gene-specific antisense primers Cys132, Cys707, Cys604, and Cys369 were used in combination with the T3 primer as the sense primer. PCR product was amplified (+) or not amplified (−) from the cDNA libraries as indicated. Identity of PCR products was confirmed by sequence determination for all products except the Cys132/T3 product.
Table I lists primers, accession numbers for Arabidopsis peptidases, and results of the screen. Degenerate primers Cys1 and Cys2 yielded a partial cDNA corresponding to the desiccation-induced Cys peptidase gene rd21A (Koizumi et al., 1993). The Cys707/T3 primer combination yielded no PCR product. Cys132/T3 yielded a product of the expected size from both the xylem and bark libraries. Cys604/T3 and Cys369/T3 products were detected only when xylem cDNA was used as template. Degenerate primers Ser1 and Ser2 yielded two cDNAs coding for predicted subtilisin-type peptidases. One cDNA clone (accession no. AL022023) was amplified from both libraries, whereas the second (accession no. AF069299) was amplified from the xylem library only. Primers Asp1 and Asp2 produced a single PCR product from both libraries encoding a previously reported Asp peptidase (accession no. U51036; D'Hondt et al., 1997). Its expression in vascular tissues was not previously reported. Thus, three potentially xylem-specific cDNAs designated XCP1, XCP2, (xylem Cys peptidases 1 and 2), and XSP1 (xylem Ser peptidase 1) were selected for cloning and further study. No bark-specific peptidase cDNAs were detected in this study.
Analysis of XCP1, XCP2, and XSP1 cDNA Sequences
We were unable to obtain the 5′-untranslated region (UTR) for XCP1 (accession no. AF191027) by the methods used for this study, and an EST for this gene has not been reported. The coding region is 1,068 bp and the 3′-UTR from a partial cDNA clone of XCP1 is 129 bp long. We cloned a version of XCP2 (accession no. AF191028) that included 5 bp upstream of the predicted initiating ATG, a 1,071-bp coding region, and a 125-bp 3′-UTR. When the XCP2 sequence was used in a BLAST search, EST T21368 including 24 bp upstream of the predicted initiator ATG was retrieved. The genes corresponding to XCP1 and XCP2 are structurally similar, both containing three introns and four exons. Intron sizes are very similar, ranging from 83 to 97 bp when both genes are considered. Intron identity between genes is approximately 27% for all three introns. With the exception of exon 1, which at 478 bp is 3 bp longer in XCP2 than in XCP1, the corresponding exons are of identical sizes, i.e. 236, 141, and 216 bp for exons 2, 3, and 4, respectively. Levels of sequence identity between XCP1 and XCP2 are 78%, 73%, 68%, and 58% for exons 1 through 4, respectively. The predicted open reading frames (ORFs) for cDNAs XCP1 and XCP2 share 100% identity with the predicted ORFs for Arabidopsis genes F23E12.90 (accession no. AL022604) and F9H16.17 (accession no. AC007369), respectively.
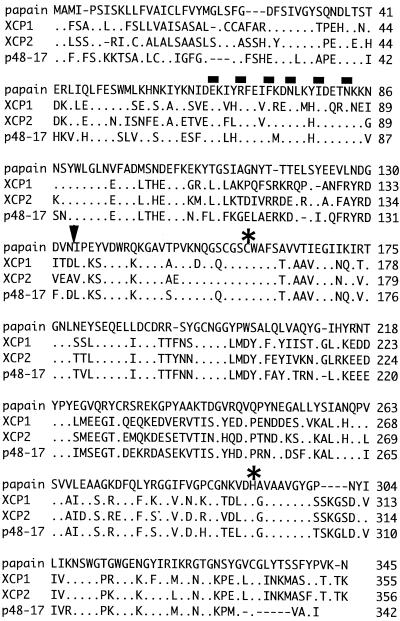
Alignments of the derived amino acid sequences for the 355- and 356-residue proteins XCP1 and XCP2 are shown in Figure 3. Sequences for papain and p48-17 (Ye and Varner, 1996) are also shown. XCP1 and XCP2 share 70% identity with each other, and they are both 65% identical to p48-17. Both XCP1 and XCP2 share approximately 44% sequence identity with papain. Cleavage of putative signal sequences for XCP1 and XCP2 are predicted to occur after residues 28 and 26, respectively (Nakai and Kanehisa, 1992). Prodomains for both peptidases possess the ERFNIN signature present in mammalian cathepsin L but absent from cathepsin B (Karrer et al., 1993). Prodomain cleavage yielding the mature, active enzyme is predicted from that known for papain. Amino acids surrounding the Cys and His residues of the catalytic dyad of XCP1, XCP2, and p48-17 are identical to those of papain with the exception of an Ala to Gly substitution common to XCP1, XCP2, and p48-17, adjacent to the catalytic His (Fig. 3).
Figure 3.
Derived amino acid sequences for XCP1, XCP2, and p48-17 (Ye and Varner, 1996) aligned with the sequence for papain. The prepropeptide is shown for all proteins. The alignment was produced using the megalign program (DNASTAR, Inc., Madison, WI). Dots indicate identity; dashes indicate gaps in the alignment; bars indicate positions of residues in the ERFNIN motif, arrowhead indicates the predicted cleavage site between the prodomain and the mature protein based on the N-terminal residue of mature papain. Asterisks indicate the positions of the active site Cys and His residues.
The cDNA clone XSP1 (accession no. AF190794) consists of a 12-bp 5′-UTR, a 2,247-bp coding region, and a 164-bp 3′-UTR. The amino acid sequence (Fig. 4) consisting of a predicted 28-residue signal sequence (Nakai and Kanehisa, 1992), an 87-residue propeptide, and a 634-residue mature polypeptide is compared with cucumisin with which it shares 40% overall identity. Complete or nearly complete identity is shared between these two sequences in regions that surround the conserved residues of the catalytic triad, Asp, His, and Ser (Fig. 4). Processing of the proprotein is predicted to occur at a pair of conserved Thr residues, Thr116/117 (Neuteboom et al., 1999). There is currently no sequence that matches XSP1 among the Arabidopsis ESTs. A BLAST search of genomic DNA revealed a predicted subtilisin-type peptidase, gene F6N15.3 (accession no. AF069299) on chromosome IV, which shared 100% identity over the majority of XSP1 but appeared to lack the signal sequence and 30 amino acid residues of the prodomain predicted from XSP1 cDNA. In addition a 13-residue insert near the midpoint of the protein, contributed by a misidentified exon, was also predicted for the genomic sequence. As we obtained sequence coding only for the protein shown in Figure 4 from independent cDNA clones, and because the prepropeptide predicted from cDNA XSP1 reflects the typical structure of a subtilisin-type peptidase, we believe that our predicted sequence is correct. With regard to gene structure, therefore, XSP1 consists of 11 exons and 10 introns rather than 10 exons and nine introns predicted in accession number AF069299.
Figure 4.
Alignment of the derived amino acid sequences for XSP1 and cucumisin. Dots indicate identity; dashes indicate gaps in the alignment; arrowheads show predicted cleavage sites after the end of the putatative signal sequence at Ser-28 and after the end of the prodomain at His-115; asterisks indicate positions of the active site Asp, His, and Ser residues.
Expression Patterns of XCP1, XCP2, and XSP1
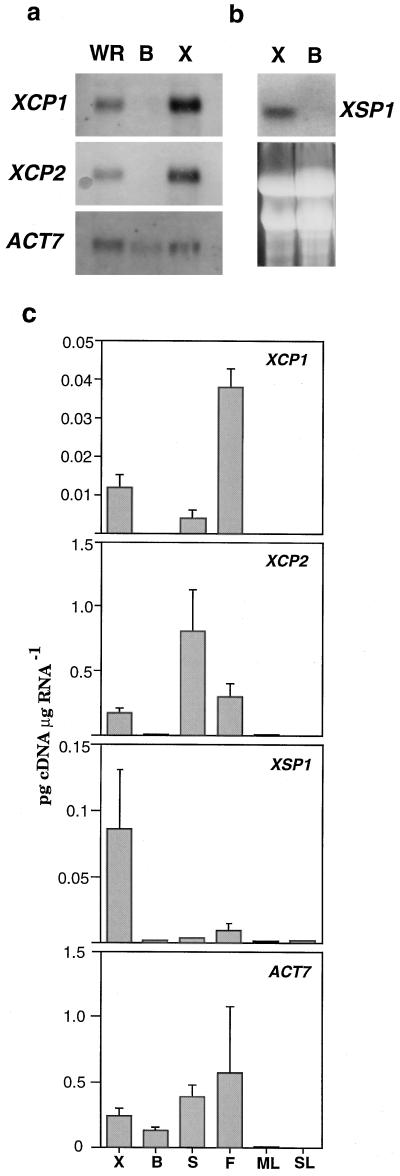
On RNA gel blots, XCP1, XCP2, and XSP1 mRNA was detectable in whole root (data not shown for XSP1) and xylem but not in bark (Fig. 5, a and b). Although XCP1 and XCP2 sequences are 70% identical, probes prepared from XCP1 and XCP2 cDNA do not cross-hybridize at high stringency in Southern blots (data not shown). Nevertheless, it is possible that these probes cross-hybridize with RNA targets. Therefore, we performed quantitative RT-PCR as an independent, gene-specific method of investigating the expression patterns for the Cys peptidase genes and XSP1. Results from three independent quantitative RT-PCR experiments are presented for xylem, bark, inflorescence stem and flower, and mature and senescing leaves (Fig. 5c). Although very low levels of mRNA for all three genes were detectable in bark, this was not surprising as RT-PCR is capable of amplifying transcripts from single cells (e.g. Brandt et al., 1999), and it was possible that differentiating xylem cells were present in small numbers in bark preparations. In contrast, such low levels of contamination would not be detected at the light microscope, zymogram, or RNA gel-blot levels. Quantitative RT-PCR also revealed that in xylem, XCP2 mRNA levels exceeded those of XCP1 by approximately 20-fold. This difference in mRNA levels is not evident in the RNA gel-blots presented in Figure 5, indicating that cross hybridization between XCP1 and XCP2 may have occurred. The actin gene ACT7 used as the loading control in Figure 5a was also subjected to quantitative RT-PCR. The amplification of the ACT7 cDNA from bark to a level that was approximately one-half of that observed for xylem (Fig. 5c) is consistent with the RNA gel-blot results (Fig. 5a). Transcript levels for all three genes were low to non-detectable in both mature and senescing leaves. In contrast, inflorescence stem and flower RNA yielded relatively high levels of Cys peptidase cDNA. For example, the level of XCP1 in flower was 3-fold greater than that observed for root-hypocotyl xylem, and the level of XCP2 in stems was 4-fold greater than that observed for root-hypocotyl xylem (Fig. 5). Tissue localization of the peptidase transcripts is not yet known for stems or flowers.
Figure 5.
RNA gel-blot and quantitative RT-PCR analyses of XCP1, XCP2, and XSP1 expression in organs and tissues of Arabidopsis. For XCP1, XCP2, and loading control ACT7, poly(A+) RNA (1.5 μg) isolated from whole root (WR), bark (B), and xylem (X) was separated by agarose gel electrophoresis, transferred to a nylon membrane, and hybridized with probes directly labeled with alkaline phosphatase, for visualization by chemiluminescence (a). For XSP1, total RNA (20 μg) from xylem and bark was hybridized with 32P-labeled probe (b). EtBr-stained RNA separated by agarose gel electrophoresis is shown to indicate levels of RNA transferred to nylon membrane for XSP1 analysis. For quantitative RT-PCR (c), total RNA was isolated from xylem and bark dissected from the root-hypocotyl, and from influorescence stems (S), flowers (F), mature leaves (ML), and senescing leaves (SL). Following reverse transcription of mRNA, cDNA from the indicated tissues and organs was used in six PCR reactions, each containing competitor cDNA of known concentration (dilution series of 10−1 to 10−6 ng/μL). The resulting PCR products were separated by agarose gel electrophoresis, and the density of EtBr-stained bands was determined. The values shown were determined from the intersection of curves depicting the levels of product from the competitor and levels of product from the target cDNAs. The values shown are means + sd determined from three independent RT-PCR experiments.
In Vitro Functional Assay of XCP1
The cDNAs described here are predicted to code for papain-type and subtilisin-type peptidases with well-known structural and functional characteristics. We have confirmed the prediction for one of the cDNAs, XCP1, by demonstrating that a recombinant, poly-His-tagged version of XCP1 codes for a protein with peptidase activity. From cloning vectors, the final recombinant protein received 16 additional N-terminal amino acid residues and 20 additional C-terminal residues, including the poly-His tag. Poly-His-tagged XCP1 also lacks the signal sequence and 20 residues of the prodomain but retains the ERFNIN-containing domain necessary for prodomain-mature protein interaction (Groves et al., 1998).
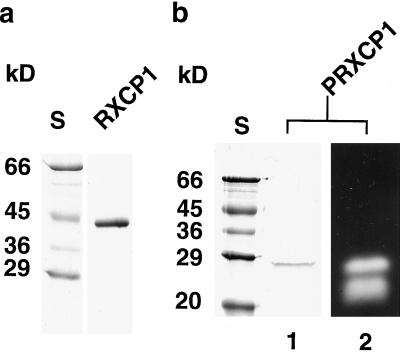
The expressed protein was purified and tested for activity (Fig. 6). The expected 40-kD polypeptide of XCP1 was efficiently purified from inclusion bodies by denaturing metal-chelate chromatography (Fig. 6a). Following renaturation (Smith and Gottesman, 1989), the full-length recombinant protein was not active (data not shown) but could be activated by incubation at pH 5.5. Activation was coincident with the disappearance of the 40-kD form and the appearance of a smaller 26-kD form of XCP1 (Fig. 6b). Activation and processing to the smaller form were prevented by inclusion of the papain-type peptidase inhibitor leupeptin in the activation buffer (data not shown). Processing resulted in the production of two forms, 26 and 22 kD, of active peptidase, although only the 26-kD form was abundant enough to be visible following Coomassie staining. No activity was detected when E-64 was added to the processed protein preparation prior to SDS-PAGE (data not shown). As indicated by the presence of two bands of active peptidase, cleavage at more than one site can yield an active processed enzyme. These experiments also indicate that, at least in vitro, XCP1 does not appear to require additional peptidases for processing to the active forms. Although the 26- and 22-kD processed forms of recombinant XCP1 are similar in mass to the 28- and 18-kD xylem-specific peptidase activities visible in Figure 2b, it would be premature to suggest that specific peptidase activities observed on zymograms from plant extracts are the result of XCP1 or XCP2 gene products.
Figure 6.
SDS-PAGE and zymogram analyses of the purification and activation of poly-His-tagged XCP1. Following Ni2+-chelate purification of poly-His-tagged XCP1, only the 40-kD recombinant propeptide (RXCP1) is detected by Coomassie staining (a). Following proteolytic processing during incubation at pH 5.5, a smaller (26-kD) version (PRXCP1) is detectable by Coomassie staining (lane 1) and zymogram (lane 2) (b). An additional activity at 22 kD is detectable on the zymogram but not on the Coomassie-stained gel. Molecular masses of standards (S) are shown to the left of each Coomassie-stained gel.
DISCUSSION
The overall objective of this research was to develop a method for using Arabidopsis for the study of gene expression in secondary vascular tissue and in particular for investigating the roles played by proteolytic enzymes in the xylem. By exploiting the secondary growth potential of Arabidopsis, we were able to construct xylem and bark cDNA libraries, thereby providing a herbaceous complement to the loblolly pine (Loopstra and Sederoff, 1995; Allona et al., 1998) and poplar (Sterky et al., 1998) cDNA libraries. Three xylem peptidase cDNA clones, named XCP1, XCP2, and XSP1, were initially identified by PCR screening of xylem and bark cDNA libraries. Within the root-hypocotyl, transcripts for XCP1, XCP2, and XSP1 were restricted almost exclusively to the xylem. In addition, transcripts for XCP1 and XCP2 were present in inflorescences at levels that exceeded those observed for the root-hypocotyl. The peptidase genes described here do not appear to be associated with leaf senescence. Although XCP2 was cloned prior to this report (EST T21368), expression of XCP1 and XSP1 was not previously reported. We have confirmed the predicted identity for one of the papain-type peptidases by demonstrating in vitro processing and proteolytic activity of a poly-His-tagged version of XCP1.
It is clear from published expression patterns of papain-type peptidases from several models that these enzymes are expressed under a wide variety of developmental programs and inducible responses. Considering Arabidopsis alone, expression of specific papain-type enzymes is known to increase during senescence (Hensel et al., 1993; Lohman et al., 1994) and in response to drought (Koizumi et al., 1993) and ethylene (Grbic and Bleecker, 1995). In addition, we show that two genes encoding very similar papain-type peptidases exhibit similar tissue and organ distribution but a 20-fold difference in expression levels (Fig. 5). The observed higher level of expression for XCP2 versus XCP1 is consistent with the fact that ESTs identical to XCP2 have been reported whereas those for XCP1 have not.
That XCP1 and XCP2 are detectable primarily in tissues and organs (stems and flowers) where cells are expanding and differentiating and not in mature tissues suggests that they have specialized functions associated with growth and/or differentiation, including, but perhaps not limited to, autolysis of xylem TEs. Although XCP1 and XCP2 are potentially linked to TE suicide, xylem cell-type localization has not yet been established for these transcripts, and hence, they may localize to xylem parenchyma where they may encode enzymes that function in routine protein turnover. Alternatively, XCP1 and XCP2 may serve in the extensive modification of the proteome that is likely to occur in concert with changes in gene expression during differentiation at the vascular cambium. For this study we did not localized peptidase transcripts at the tissue level for the influorescence stem or flowers. For stems, xylem localization is a reasonable hypothesis, however, as extensive vascular tissue development is a characteristic of stems, and they have recently been used for molecular genetic studies of xylem differentiation (Turner and Somerville, 1997; Zhong and Ye, 1999). In flowers, vascularization is not the only process that uses Cys peptidases during pcd. Increased expression of Cys peptidases is associated with several anther cell types that degenerate prior to dehiscence (Koltunow et al., 1990) and with petal senescence (Jones et al., 1995; Guerrero et al., 1998).
In addition to the papain-type enzymes described here, we identified a xylem subtilisin-type peptidase cDNA, XSP1. Subtilisin mRNA levels increase during lateral root emergence (Neuteboom et al., 1999), which may be associated with pcd of adjacent root cortex cells (Kosslak et al., 1997). Flowers exhibited the second highest level of XSP1 mRNA after xylem. A subtilisin-type peptidase has been localized to the apoplast of lily anthers (Taylor et al., 1997). Studies have implicated plant subtilisin-type peptidases in the processing of peptides in the extracellular matrix (Schaller and Ryan, 1994; Kinal et al., 1995; Tornero et al., 1996). Other S8 family members, the yeast and mammalian kexin/furin-type Ser peptidases known as proprotein convertases, are involved in the generation of bioactive peptides by proteolytic processing of inactive precursors (Seidah et al., 1998). Whether XSP1 encodes a xylem peptidase that functions during autolysis of TEs, in the processing of peptide precursors, or has other functions not yet described, is currently unknown.
Expression that is high in flower and stem and low in mature leaves as noted here for the actin gene ACT7 is similar to the expression patterns for ACT7 promoter-GUS fusions reported by McDowell et al. (1996). These authors also reported a reduction of expression as roots matured. However, root-hypocotyl tissues exhibiting extensive secondary growth were not assayed by McDowell et al. (1996), and hence, the ACT7 mRNA we detected in root tissues may reflect a second wave of expression associated with lateral expansion driven by the vascular cambium. Although mRNA for all genes tested was very low in leaf RNA preparations, ethidium bromide (EtBr) staining following electrophoresis of RNA from all sources prior to RT-PCR indicated that all RNA used for quantitative RT-PCR was of high quality. Hence, we do not believe that apparent low cDNA levels were the result of RNA degradation.
In Arabidopsis both papain-type and subtilisin-type peptidases belong to multi-gene families. By establishing a method for studying vascular tissue gene expression in Arabidopsis, we have identified at least some of the family members that may have xylem-specific functions. The findings reported here set the stage for future reverse genetics experiments aimed at identifying functions for these peptidases. It is also expected that the method for vascular tissue isolation and the cDNA libraries described here will be useful for investigations of other proteins with structural, metabolic, or regulatory functions required for vascular tissue development.
MATERIALS AND METHODS
Plant Growth and Isolation of Xylem and Phloem
Arabidopsis ecotype Columbia was grown in Sunsine Mix 1 (Wetsel Seed Co., Harrisonburg, VA) under continuous light, at a planting density of four to six plants per 10-cm pot. It is important to firmly tamp the potting medium before sowing seed. Plants were watered with nutrient solution according to Somerville and Ogren (1982). Under these conditions inflorescences were typically visible 3 weeks after germination. For the next 5 weeks, influorescences were routinely removed as they emerged. Eight-week-old plants were harvested, and potting medium was washed from the roots with a strong stream of cool tap water. Approximately 1 cm of root-hypocotyl was excised from just below the cotyledons, and lateral roots were trimmed from the primary root with a razor blade. Prior to dissection, root-hypocotyl segments were washed with distilled, deionized water, blotted dry, and placed on ice. Root-hypocotyl segments harvested for peptidase zymograms were surface sterilized in 1% (w/v) calcium hypochlorite plus 1% (v/v) Triton X-100 for 10 min before extensive rinsing with distilled, deionized water. Separation of root-hypocotyl segments into xylem, phloem-enriched, and non-vascular fractions or xylem and bark fractions was done under the dissecting microscope. Briefly, using a double-edged razor blade, a longitudinal cut was made along the entire length of the root-hypocotyl segment passing through the non-vascular tissue and secondary phloem but not into the xylem. Using a dissecting probe and forceps, the non-vascular tissue was peeled from the phloem and placed in liquid N2. Xylem and phloem were then separated and placed in liquid N2. When xylem and bark samples were isolated, the non-vascular tissue and phloem were separated from the xylem as a unit.
Protein Extraction
Xylem, phloem-enriched, and non-vascular tissues were powdered in liquid N2 and extracted in 100 mm sodium phosphate, pH 7.2, 7 mm 2-mercaptoethanol, 20 μm leupeptin using a buffer:tissue ratio of 2 mL:60 xylem (approximately 0.6 g), phloem (approximately 0.25 g), or non-vascular segments (approximately 0.5 g). Homogenized tissue extract was clarified by centrifugation at 12,000g (4°C) for 20 min. The supernatant was concentrated approximately 10-fold using YM10 concentrators (Millipore, Bedford, MA). Samples were stored at −80°C for subsequent use in either zymograms or immunoblots. Protein content of concentrates was determined using bicinchoninic acid (Sigma, St. Louis), according to the supplier's instructions.
Peptidase Zymograms and Immunoblots
Gelatinase zymograms were prepared according to Beers and Freeman (1997). Immunoblots were prepared according to Woffenden et al. (1998), using the monoclonal antibody RS32 (gift of R. Sjölund) as a 100:1 dilution in “Blotto” (Johnson et al., 1984) as the primary antibody.
Construction and Screening of cDNA Libraries
Approximately 300 root-hypocotyl segments were dissected into xylem and bark fractions and used to isolate the 5 μg of poly(A+) RNA necessary for each of the two cDNA libraries constructed for this report. Total RNA was isolated by phenol/chloroform extraction and LiCl precipitation (Ecker and Davis, 1987). Poly(A+) RNA was isolated using PolyATract mRNA isolation system III (Promega, Madison, WI). Complementary DNA libraries were constructed using the ZAP-cDNA kit and instructions provided by Stratagene (La Jolla, CA). Libraries were screened by PCR (for primer sequences, see Table I). PCR mix (50 μL) consisted of approximately 106 plaque-forming units from either xylem or phloem cDNA library as template, 0.25 μm each primer, 1× REDtaq reaction buffer (Sigma), 1.5 units of REDtaq DNA polymerase, and 125 μm dNTPs. PCR began with denaturation at 94°C for 5 min. Thereafter, for 32 cycles, conditions were 94°C, 55°C, and 72°C each for 1 min. The cycling ended with a 10-min extension at 72°C.
Cloning cDNAs for XCP1, XCP2, and XSP1
XCP1 cDNA was synthesized from xylem poly(A+) RNA by RT-PCR using sense (ATGGCTTTTTCTGCACCATCACTT) and antisense (CTTGGTCTTGGTAGGATATGAGGC) primers to amplify the ORF predicted for accession number AL022604, gene F23E12.90, and ligating the resulting PCR product into the plasmid vector pCR2.1 (Invitrogen, Carlsbad, CA), yielding pXCP1. Using sense (TGTGACAGGAAACTTGACAACAT) and antisense (Cys369, Table I) primers in combination with T7 or T3 phagemid primers, respectively, the 3′ and 5′ regions of XCP2 were amplified and cloned into pGEM-TEasy vectors (Promega). The full-length clone, pXCP2, was constructed by subcloning the 5′ and 3′ ends together into pBluescript using an internal restriction site common to the overlapping central region of both partial cDNA clones. All sequencing for this report was performed at the Virginia Polytechnic Institute DNA sequencing facility. The full-length cDNA clone for XSP1, pXSP1, was obtained using the approach described for pXCP2 with primers TGCTTCAGCCGAAGATGAAC (sense) and TTCCTCATCGCG-TGAAACGA (antisense).
RNA Gel-Blot Analyses
For XCP1, XCP2, and ACT7, 1.5 μg of poly(A+) RNA isolated from xylem and bark was resolved by formaldehyde gel electrophoresis and transferred to nylon overnight by capillary action. Transferred RNA was UV cross-linked to the membrane. For probe synthesis, 550 bp and the entire 3′-UTR from XCP1 and 670 bp from the coding region of XCP2 were obtained by digesting cDNA clones with the restriction enzymes HindIII/EcoRI. The probe for ACT7 was a 680-bp coding region fragment prepared by restriction enzyme digestion of a partial cDNA clone with EcoRI/BglI. The cDNA fragments were purified using QIAEXII Gel Extraction Kit (Qiagen, Valencia, CA) and directly labeled with alkaline phosphatase using AlkPhos Direct (Amersham Pharmacia Biotech, Uppsala). Hybridization at 55°C, blot washes, and chemiluminescent signal detection with CDP-Star (Roche Molecular Biochemicals, Indianapolis) were conducted without modification according to instructions provided by the supplier. For XSP1, total RNA (20 μg/lane) isolated from xylem and bark was blotted as for poly(A+) RNA. For probe synthesis, PCR product corresponding to approximately 900 bp of the 5′ end of the ORF of pXSP1 was purified using QIAEXII Gel Extraction Kit, and 32P-labeling was performed using RTS RadPrime DNA Labeling System (Life Technologies, Frederick, MD). Hybridization was at 42°C in northern-hybridization buffer (5× SSC, 1% [w/v] SDS, 5× Denhardt's, and 200 μg/mL salmon-sperm DNA) overnight. The membrane was washed with 0.1× SSC and 0.1% (w/v) SDS once at 42°C and then twice at 50°C. The hybridization signal was recorded on x-ray film at −70°C using an intensifying screen.
Quantitative RT-PCR
Total RNA was isolated from xylem, bark, inflorescence stems, flowers, and mature leaves as described for cDNA library construction. Total RNA was isolated from senescing leaves according to Puissant and Houdebine (1990). All RNA was DNase-treated prior to RT-PCR. From each sample, 2 μg of total RNA was used for reverse transcription using RETROscript and oligo(dT) primers (Ambion, Austin, TX). Twenty-five cycles of PCR were performed as described above, using 5% of the RT reaction for each PCR with the following primers: XCP1, ATGGCTTTTTCTGCACCATCACTT, CTTGGTCTTGGTAGGATATGAGGC; XCP2, TGTGACAGGAAACTTGACAACAT, TCAAGATCAACCCCGCACCGC; XSP1, TGCTTCAGCCGAAGATGAAC, TTCCTCATCGCGTGAAACGA; ACT7, GGCC-GATGGTGAGGATATTC, CTGACTCATCGTACTCACTC.Competitors consisted of a 70-bp EcoRV/EcoRV deletion mutant of pXCP1 and for XCP2, XSP1, and ACT7, partial genomic clones approximately 180, 700, and 300 bp, respectively, larger than the target cDNAs. The intron-containing competitors were cloned by PCR using genomic DNA and the primers listed above for competitive PCR. Densitometric analysis of EtBr-stained gels was conducted with Alpha Imager 2000 (Alpha Innotech, San Leandro, CA). The point of equivalency, i.e. where target cDNA amplified was equal to competitor cDNA amplified, was determined from the intersection of the curves plotted for competitor and target.
Construction of Poly-His-Tagged Papain-Type Peptidase XCP1
XCP1 with a C-terminal poly-His tag was cloned using Qiagen type-III vectors, pQE50 (provides initiator Met) and pQE16 (provides C-terminal poly-His tag), according to instructions provided by the supplier (Qiagen). Briefly, a HindIII/BamHI fragment from pXCP1 was ligated with a 1,025-bp HindIII/BglI fragment from pQE50 and a 2,428-bp BglII/BglI fragment from pQE16, creating pXCP1H6. That pXCP1H6 coded for the amino acid sequence predicted from XCP1 cDNA was confirmed by DNA sequencing.
Purification of Poly-His-Tagged XCP1
Escherichia coli cells (100 mL) were cultured according to instructions provided by Qiagen USA. Expression was induced by addition of isopropylthio-β-galactoside to 1 mm. At 4 h postinduction, cells were harvested by centrifugation at 4,000g for 20 min. All purification steps were performed at 4°C. Cells were resuspended in 12 mL of lysis buffer (50 mm NaH2PO4, pH 8.0, 300 mm NaCl, and 1 mg/mL lysozyme) and incubated on ice for 30 min prior to sonication. The sonicated cell lysate was pelleted at 10,000g for 30 min. The supernatant was discarded, and the remaining pellet containing inclusion bodies was solubilized in 1 mL of denaturing buffer (8 m urea, 100 mm NaH2PO4, and 10 mm Tris-HCl, pH 8.0). The solubilized sample was clarified by centrifugation at 14,000g for 10 min.
The resulting supernatant was added to a microfuge tube containing 300-μL bed volume of nickel-nitrilotriacetic acid agarose beads equilibrated in denaturing buffer, and poly-His-tagged protein was allowed to bind during gentle mixing for at least 30 min. After binding, beads were pelleted by centrifugation at 2,000g for 2 min. The supernatant containing unbound proteins was discarded. The following washes were used to remove background proteins from nickel-nitrilotriacetic acid agarose beads: 2 mL of denaturing buffer; 5 mL of denaturing buffer, pH 6.5; 5 mL of lysis buffer, without lysozyme; 2 mL of lysis buffer, pH 4.5. Purified protein was released from beads into elution buffer (8 m urea, 100 mm NaH2PO4, 10 mm Tris-HCl, 500 mm imidazole, and 10 mm 2-mercapto-ethanol, pH 8.0) by adding 1 mL of elution buffer, gently mixing beads, pelleting beads by centrifugation, and removing the supernatant. The elution was repeated twice, and supernatants containing purified protein were pooled for renaturation.
Purified denatured poly-His-tagged XCP1 was renatured essentially as described by Smith and Gottesman (1989). Purified protein in elution buffer was added slowly to 200 volumes of renaturation buffer (50 mm KH2PO4, pH 10.7, 5 mm EDTA, and 10 mm 2-mercaptoethanol) and stirred overnight at 4°C. The pH of the renaturation buffer was then adjusted to 8.0 with HCl followed by concentration using a YM10 membrane.
ACKNOWLEDGMENTS
We thank Drs. R.F. Evert, S. Gan, K. Robinson-Beers, and S. Scheckler for valuable advice and Dr. A. Esen for critical reading of the manuscript. Dr. R.F. Evert kindly provided the English translation from the Russian language publication by Kondratieva-Melville and Vodolazsky (1982). Dr. R. Sjölund kindly provided the monoclonal antibody RS32. Technical assistance was provided by T.B. Freeman, V. Funk, H.E. Petzold, and L. Weigt.
Footnotes
This work was supported by the U.S. Department of Agriculture-National Research Initiative Competitive Grants Program (grant no. 9801401 to E.P.B.) and by the National Science Foundation (grant no. MCB–9418377). B.K. was supported by a scholarship from the Royal Thai Government. C.Z. was supported in part by the Anhui Academy of Agricultural Sciences (Hefei, People's Republic of China).
LITERATURE CITED
- Allona I, Quinn M, Shoop E, Swope K, Cyr SS, Carlis J, Riedl J, Retzel E, Campbell MM, Sederoff R, Whetten RW. Analysis of xylem formation in pine by cDNA sequencing. Proc Natl Acad Sci USA. 1998;95:9693–9698. doi: 10.1073/pnas.95.16.9693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beers EP, Freeman TB. Proteinase activity during tracheary element differentiation in zinnia mesophyll cultures. Plant Physiol. 1997;113:873–880. doi: 10.1104/pp.113.3.873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandt S, Kehr J, Walz C, Imlau A, Willmitzer L, Fisahn J. A rapid method for detection of plant gene transcipts from single epidermal, mesophyll and companion cells of intact leaves. Plant J. 1999;20:245–250. doi: 10.1046/j.1365-313x.1999.00583.x. [DOI] [PubMed] [Google Scholar]
- Buchanan-Wollaston V. The molecular biology of leaf senescence. J Exp Bot. 1997;48:181–199. [Google Scholar]
- Busse JS, Evert RF. Vascular differentiation and transition in the seedling of Arabidopsis thaliana (Brassicaceae) Int J Plant Sci. 1999;160:241–251. [Google Scholar]
- D'Hondt KD, Stack S, Gutteridge S, Vandekerckhove J, Krebbers E, Gal S. Aspartic proteinase genes in the Brassicaceae Arabidopsis thaliana and Brassica napus. Plant Mol Biol. 1997;33:187–192. doi: 10.1023/a:1005794917200. [DOI] [PubMed] [Google Scholar]
- Dolan L, Roberts K. Secondary thickening in roots of Arabidopsis thaliana: anatomy and cell surfaces. New Phytol. 1995;131:121–128. doi: 10.1111/j.1469-8137.1995.tb03061.x. [DOI] [PubMed] [Google Scholar]
- Ecker J, Davis R. Plant defense genes are regulated by ethylene. Proc Natl Acad Sci USA. 1987;84:5202–5206. doi: 10.1073/pnas.84.15.5202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda H, Komamine A. Establishment of an experimental system for the study of tracheary element differentiation from single cells isolated from the mesophyll of Zinnia elegans. Plant Physiol. 1980;65:57–60. doi: 10.1104/pp.65.1.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granell A. Plant cysteine proteinases in germination and senescence. In: Barrett AJ, Rawlings ND, Woessner JF, editors. Handbook of Proteolytic Enzymes, Chapter 199. New York: Academic Press; 1998. [Google Scholar]
- Grbic V, Bleecker AB. Ethylene regulates the timing of leaf senescence in Arabidopsis. Plant J. 1995;8:595–602. [Google Scholar]
- Groover A, Jones AM. Tracheary element differentiation uses a novel mechanism coordinating programmed cell death and secondary cell wall synthesis. Plant Physiol. 1999;119:375–384. doi: 10.1104/pp.119.2.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groves MR, Coulomber R, Jenkins J, Cygler M. Structural basis for specificity of papain-like cysteine protease proregions toward their cognate enzymes. Proteins Struct Funct Genet. 1998;32:504–514. [PubMed] [Google Scholar]
- Guerrero C, de la Calle M, Reid MS, Valpuesta V. Analysis of the expression of two thiolprotease genes from daylily (Hemerocallis spp.) during flower senescence. Plant Mol Biol. 1998;36:565–571. doi: 10.1023/a:1005952005739. [DOI] [PubMed] [Google Scholar]
- Hensel LL, Grbic V, Baumgarten DA, Bleecker AB. Developmental and age-related processes that influence the longevity and senescence of photosynthetic tissues in Arabidopsis. Plant Cell. 1993;5:553–564. doi: 10.1105/tpc.5.5.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson DA, Gautsel JW, Sportsman JR, Elder JH. Improved technique utilizing nonfat dry milk for analysis of proteins and nucleic acids transferred to nitrocellulose. Gene Anal Tech. 1984;1:3–8. [Google Scholar]
- Jones ML, Larsen PB, Woodson WR. Ethylene-regulated expression of a carnation cysteine proteinase during flower petal senescence. Plant Mol Biol. 1995;28:505–512. doi: 10.1007/BF00020397. [DOI] [PubMed] [Google Scholar]
- Karrer KM, Peiffer SL, DiToms ME. Two distinct gene subfamilies within the family of cysteine protease genes. Proc Natl Acad Sci USA. 1993;90:3063–3067. doi: 10.1073/pnas.90.7.3063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinal H, Park C, Berry JO, Koltin Y, Bruenn JA. Processing and secretion of a virally encoded antifungal toxin in transgenic tobacco plants: evidence for a Kex2p pathway in plants. Plant Cell. 1995;7:677–688. doi: 10.1105/tpc.7.6.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohlenbach HW, Schmidt B. Cytodifferenzierung in form einer direkten umwandlung isolierter mesophyllzellen zu tracheiden. Z Pflanzenphysiol. 1975;75:369–374. [Google Scholar]
- Koizumi M, Yamaguchi-Shinozaki K, Tsuji H, Shinozaki K. Structure and expression of two genes that encode distinct drought-inducible cysteine proteinases in Arabidopsis thaliana. Gene. 1993;129:175–182. doi: 10.1016/0378-1119(93)90266-6. [DOI] [PubMed] [Google Scholar]
- Koltunow AM, Truettner J, Cox KH, Wallroth M, Goldberg RB. Different temporal and spacial gene expression patterns occur during anther development. Plant Cell. 1990;2:1201–1224. doi: 10.1105/tpc.2.12.1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondratieva-Melville EA, Vodolazsky LE. Morphological and anatomical structure of Arabidopsis thaliana (Brassicaceae) in ontogenesis. Bot J. 1982;67:1060–1069. [Google Scholar]
- Kosslak RM, Chamberlin MA, Palmer RG, Bowen BA. Programmed cell death in the root cortex of soybean root necrosis mutants. Plant J. 1997;11:729–745. doi: 10.1046/j.1365-313x.1997.11040729.x. [DOI] [PubMed] [Google Scholar]
- Lev-Yadun S. Induction of schlereid differentiation in the pith of Arabidopsis thaliana (L.) Heynh. J Exp Bot. 1994;45:1845–1849. [Google Scholar]
- Lohman KN, Gan S, John MC, Amasino RM. Molecular analysis of natural leaf senescence in Arabidopsis thaliana. Physiol Plant. 1994;92:322–328. [Google Scholar]
- Loopstra CA, Sederoff RR. Xylem-specific gene expression in loblolly pine. Plant Mol Biol. 1995;27:277–291. doi: 10.1007/BF00020183. [DOI] [PubMed] [Google Scholar]
- McDowell JM, An Y, Huang S, McKinney EC, Meagher RB. The Arabidopsis ACT7 actin gene is expressed in rapidly developing tissues and responds to several stimuli. Plant Physiol. 1996;111:699–711. doi: 10.1104/pp.111.3.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minami A, Fukuda H. Transient and specific expression of a cysteine endoproteinase associated with autolysis during differentiation of Zinnia mesophyll cells into tracheary elements. Plant Cell Physiol. 1995;36:1599–1606. [PubMed] [Google Scholar]
- Nakai K, Kanehisa M. A knowledge base for predicting protein localization sites in eukaryotic cells. Genomics. 1992;14:897–911. doi: 10.1016/S0888-7543(05)80111-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuteboom LW, Ng JMY, Kuyper M, Clijesdale OR, Hooykass PJJ, van der Zaal BJ. Isolation and characterization of cDNA clones corresponding with mRNAs that accumulate during auxin-induced lateral root formation. Plant Mol Biol. 1999;39:273–287. doi: 10.1023/a:1006104205959. [DOI] [PubMed] [Google Scholar]
- Omura S, Fujimoto T, Otoguro K, Matsuzaki K, Moriguchi R, Tanaka H, Sasaki Y. Lactacystin, a novel microbial metabolite, induces neuritogenesis of neuroblastoma cells. J Antibiot. 1991;44:113–116. doi: 10.7164/antibiotics.44.113. [DOI] [PubMed] [Google Scholar]
- Panavas T, Pikla A, Reid PD, Rubinstein B, Walker EL. Identification of senescence-associated genes from daylily petals. Plant Mol Biol. 1999;40:237–248. doi: 10.1023/a:1006146230602. [DOI] [PubMed] [Google Scholar]
- Phillips R, Dodds JH. Rapid differentiation of tracheary elements in cultured explants of Jerusalem artichoke. Planta. 1977;135:207–212. doi: 10.1007/BF00384891. [DOI] [PubMed] [Google Scholar]
- Puissant C, Houdebine L-M. An improvement of the single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Biotechniques. 1990;8:148–149. [PubMed] [Google Scholar]
- Runeberg-Roos P, Saarma M. Phytepsin, a barley vascular aspartic proteinase, is highly expressed during autolysis of developing tracheary elements and sieve cells. Plant J. 1998;15:139–145. doi: 10.1046/j.1365-313x.1998.00187.x. [DOI] [PubMed] [Google Scholar]
- Schaller A, Ryan CA. Identification of a 50-kDa systemin-binding protein in tomato plasma membranes having Kex2p-like properties. Proc Natl Acad Sci USA. 1994;91:11802–11806. doi: 10.1073/pnas.91.25.11802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidah NG, Day R, Marcinkiewica M, Chrétien M. Precursor convertase: an evolutionary ancient, cell-specific, combinatorial mechanism yielding diverse bioactive peptides and proteins. Ann NY Acad Sci. 1998;839:9–24. doi: 10.1111/j.1749-6632.1998.tb10727.x. [DOI] [PubMed] [Google Scholar]
- Smith SM, Gottesman MM. Activity and deletion analysis of recombinant human cathepsin L expressed in Escherichia coli. J Biol Chem. 1989;264:20487–20495. [PubMed] [Google Scholar]
- Solomon M, Belenghi B, Delledonne M, Menachem E, Levine A. The involvement of cysteine proteases and protease inhibitor genes in the regulation of programmed cell death in plants. Plant Cell. 1999;11:431–443. doi: 10.1105/tpc.11.3.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somerville CR, Ogren WL. Isolation of photorespiration mutants in Arabidopsis thaliana. In: Edelman M, Hallick RB, Chua N-H, editors. Methods in Chloroplast Molecular Biology. New York: Elsevier Biomedical Press; 1982. pp. 129–139. [Google Scholar]
- Stephenson P, Collins BA, Reid PD, Rubinstein B. Localization of ubiquitin to differentiating vascular tissues. Am J Bot. 1996;83:140–147. [Google Scholar]
- Sterky F, Regan S, Karlsson J, Hertzberg M, Rohde A, Holmberg A, Amini B, Bhalerao R, Larsson M, Villarroel R, Van Montagu M, Sandberg G, Olsson O, Teeri TT, Boerjan W, Gustafsson P, Uhlén M, Sundberg B, Lundeberg J. Gene discovery in the wood-forming tissue of poplar: analysis of 5,692 expressed sequence tags. Proc Natl Acad Sci USA. 1998;95:13330–13335. doi: 10.1073/pnas.95.22.13330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor AA, Horsch A, Rzepczyk A, Hasenkampf CA, Riggs CD. Maturation and secretion of a serine proteinase is associated with events of late microsporogenesis. Plant J. 1997;12:1261–1271. doi: 10.1046/j.1365-313x.1997.12061261.x. [DOI] [PubMed] [Google Scholar]
- Tornero P, Conejer V, Vera P. Identification of a new pathogen-induced member of the subtilisin-like processing protease family from plants. J Biol Chem. 1997;272:14412–14419. doi: 10.1074/jbc.272.22.14412. [DOI] [PubMed] [Google Scholar]
- Tornero P, Mayda E, Gomez MD, Canas L, Conejero V, Vera P. Characterization of LRP, a leucine-rich repeat (LRR) protein from tomato plants that is processed during pathogenesis. Plant J. 1996;10:315–330. doi: 10.1046/j.1365-313x.1996.10020315.x. [DOI] [PubMed] [Google Scholar]
- Turner SR, Somerville CR. Collapsed xylem phenotype of Arabidopsis identifies mutants deficient in cellulose deposition in the secondary cell wall. Plant Cell. 1997;9:689–701. doi: 10.1105/tpc.9.5.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Monroe J, Sjölund RD. Identification and characterization of a phloem-specific β-amylase. Plant Physiol. 1995;109:743–750. doi: 10.1104/pp.109.3.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson JW, Roberts LW, Wilson PMW, Gresshoff PM. Stimulatory and inhibitory effects of sucrose concentration on xylogenesis in lettuce pith explants: possible mediation by ethylene biosynthesis. Ann Bot. 1994;73:65–73. [Google Scholar]
- Woffenden BJ, Freeman TB, Beers EP. Proteasome inhibitors prevent tracheary element differentiation in zinnia mesophyll cell cultures. Plant Physiol. 1998;118:419–430. doi: 10.1104/pp.118.2.419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagata H, Masuzawa T, Nagaoka Y, Ohnishi T, Iwasaki T. Cucumisin, a serine protease from melon fruits, shares structural homology with subtilisin and is generated from a large precursor. J Biol Chem. 1994;269:32725–32731. [PubMed] [Google Scholar]
- Ye Z-H, Varner JE. Induction of cysteine and serine proteinases during xylogenesis in Zinnia elegans. Plant Mol Biol. 1996;30:1233–1246. doi: 10.1007/BF00019555. [DOI] [PubMed] [Google Scholar]
- Zhong R, Taylor JJ, Ye Z-H. Disruption of interfascicular fiber differentiation in an Arabidopsis mutant. Plant Cell. 1997;9:2159–2170. doi: 10.1105/tpc.9.12.2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R, Ye Z-H. IFL1, a gene regulating interfascicular fiber differentiation in Arabidopsis encodes a homeodomain-leucine zipper protein. Plant Cell. 1999;11:2139–2152. doi: 10.1105/tpc.11.11.2139. [DOI] [PMC free article] [PubMed] [Google Scholar]