Abstract
Increasing studies have suggested that dysregulation of RNA‐binding proteins (RBPs) contributes to cancer progression. Neuro‐oncological ventral antigen 1 (NOVA1) is a novel RBP and plays an important role in tumour development. However, the expression and role of NOVA1 in melanoma remain unknown. In this study, we indicated that NOVA1 expression was up‐regulated in melanoma samples and cell lines. Moreover, we demonstrated that knockdown of NOVA1 suppressed melanoma cell proliferation, migration and invasion in both A375 and A875 cell lines. In addition, we showed that suppressed expression of NOVA1 enhanced forkhead box O3a (FOXO3a) expression while inhibited AKT expression in melanoma cell. Furthermore, we demonstrated that inhibited expression of FoxO3A rescued NOVA1‐mediated cell proliferation, migration and invasion in melanoma cell line A375. These results suggested that NOVA1 acted as an oncogene in the development of melanoma partly through regulating FoxO3A expression.
Keywords: FOXO3a, melanoma, NOVA1, RNA‐binding proteins
1. INTRODUCTION
Melanoma is derived from melanocytes and responsible for most of skin tumour‐related deaths in human beings.1, 2, 3, 4, 5 There are about 76 380 new patients of melanoma and approximately 10 130 melanoma‐related deaths in 2016 in the United States.6 Although various treatments such as surgery, gene‐targeted therapy, chemotherapy, radiotherapy and immune therapy have been developed, the prognosis of cases with advanced melanoma is still unsatisfied.2, 7, 8, 9, 10 Unfortunately, the mechanism of the high rate of metastasis in melanoma remains unknown.3, 11, 12 Therefore, it is important to find new biomarkers and potential therapeutic targets for melanoma patients.
Recently, increasing studies have showed that RNA‐binding proteins (RBPs) are important for post‐transcriptional regulation.13, 14, 15, 16 Several studies have investigated the roles of RBPs in the prognosis and progression of melanoma.17, 18, 19 Neuro‐oncological ventral antigen 1 (NOVA1) is one member of RBPs and is involved in the programme of neural splicing.20, 21 The NOVA superfamily has two members such as NOVA1 and NOVA2. NOVA1 is expressed in central nervous system and can bind to the YCAY motif through its KH domain, thereby modulating alternative splicing.22, 23, 24 Growing evidence suggested that NOVA1 was deregulated in several tumours such as gastric cancer (GC), astrocytoma and oligodendroglioma, hepatocellular carcinoma (HCC) and lymphomas.21, 25, 26, 27 For example, Zhang et al28 demonstrated that overexpression of NOVA1 increased the HCC growth and retro‐regulated GABAARγ2 and GABA expression. Shen et al29 showed that ectopic expression of miR‐339 suppressed GC cell growth, invasion, migration and tumorigenicity through regulating NOVA1 expression. However, the expression and role of NOVA1 in melanoma remain unknown.
In this study, we firstly detected the expression of NOVA1 in melanoma tissues and control tissues. Then, we investigated the role of NOVA1 in melanoma cell. Furthermore, we explored the potential mechanism of NOVA1 in melanoma.
2. MATERIALS AND METHODS
2.1. Clinical specimens
Melanoma tissues and the normal adjacent samples were collected in our hospital. These samples were cut into two copies: One was fixed with the formalin for histopathological confirming, and the other was stored in liquid nitrogen. All patients provided written consent for this research experiment. Our study was approved by the Ethics Committee of Peking Union Medical College Hospital. The detailed clinical information was shown as follows: case 1: T3, ulceration, foot; case 2: T3, no ulceration, hand; case 3: T3, ulceration, foot.
2.2. Immunohistochemistry
Tissues were fixed with the 4% paraformaldehyde and then washed with ddH2O and PBS. Next, H2O2 (3%) was added and incubated at RT for a half‐hour and the tissue was washed with PBS for three times. Then, the tissue was incubated with 0.3% Triton X‐100 for 30 minutes, followed by incubation in 12% normal goat serum. The slides were incubated with anti‐NOVA1 primary antibodies (1:500, dilution, Abcam) and then incubated with biotin‐conjugated goat anti‐rabbit IgG. The slide was dehydrated and mounted, and figure was captured by microscopy.
2.3. Cell culture and cell transfection
Human melanoma cell lines (A375, A875 and SK‐MEL‐1) and normal melanocyte cell line (D78) were collected from Cell bank Center of Institute of Chinese Academy of Medical Sciences and Peking Union Medical College (http://www.cellresource.cn/cellsearch.aspx) (Beijing, China). These cell lines were cultured in the Dulbecco's modified Eagle's medium (DMEM) supplemented with FBS (foetal bovine serum) and penicillin and streptomycin and were kept at 37°C in the humidified atmosphere (5% CO2). Cells were transfected with the siRNA‐NOVA1 and siRNA‐control; siRNA‐forkhead box O3a (FOXO3a) and siRNA‐control plasmid using Lipofectamine 2000 (Invitrogen, CA, USA) according to the instruction.
2.4. RNA isolation and qRT‐PCR
Total RNA from cell or samples was extracted using Trizol reagent (Invitrogen, Carlsbad, CA, USA). cDNA was carried out by the M‐MLV reverse transcriptase (Invitrogen, CA, USA) from 1 ug total RNA, and qRT‐PCR was performed to determine the expression of NOVA1 and GAPDH using SYBR Green Kit (Qiagen) on the iQ5 Real‐Time PCR assat System (Bio‐Rad, USA) following the manufacturer's instructions. GAPDG was selected as the internal control. The mRNA expression was detected using 2−DDCT method. The corresponding PCR primer was shown as follows: NOVA1, forward, 5′‐GGTCTCAGCCAAGCAGCAGCAA‐3′ and reverse, 5′‐TTGCAGCAGTAGCAGCAGCCAG‐3′; GAPDH, forward, 5′‐TGGAT CTGACATGCCGCCTGGA‐3′ and reverse, 5′‐AGGTC CACCA CCCTGTTGCTGT‐3′.
2.5. Western blotting
Protein lysates from cells or tissues were prepared using the RIPA kit (Beyotime, China) following the manufacturer's instruction. The protein concentration was measured by BCA protein kit. Protein was separated by 12% SDS‐PAGE and transferred to PVDF membrane. After blocking with non‐fat milk, the membrane was immunoblotted with primary antibodies (anti‐NOVA1, anti‐FoxO3A and anti‐AKT, diluted 1:1000) overnight. After washing for three times, the membrane was incubated with HRP‐linked secondary antibodies for 1 hour. The signal was measured by enhanced chemiluminescence (ECL) (Pierce, USA). GAPDH was used as the internal control.
2.6. Cell proliferation, migration and invasion assay
Cell growth was monitored using a Cell Counting Kit‐8 (CCK‐8, Dojindo, Japan) following the manufacturer's information. Cells (1 x 105 cells) were cultured on the 96‐well plate, and cell proliferation was measured at 0, 1, 2 and 3 days. The number of cells was determined by measurement of OD at 450 nm using microplate reader (TECAN). Scratch wound‐healing assay was used to measure cell migration. When cells were grown to confluence on the 12‐well plate, scratch wound was created using 200 μL pipette tip. Cells were cultured in serum‐free medium; picture was taken at 0, 48 and 60 hours. For cell invasion assay, transwell was used. Cells were seeded on the top side of transwell chamber coated with Matrigel in serum‐free medium; medium with 10% serum was added in the lower chamber. The invaded cells on the lower membrane were washed and stained with crystal violet.
2.7. Statistical analysis
All data were shown as mean ± SD (standard deviation), and statistical assay was performed using the SPSS 19.0 (SPSS Inc., Chicago, IBM). Student's t‐test and one‐way ANOVA were used to calculate the significance between different groups. P < .05 was indicated as significant.
3. RESULTS
3.1. NOVA1 expression was up‐regulated in melanoma tissues and melanoma cell lines
Firstly, we measured the expression of NOVA1 in three patients with melanoma and two cases of cutaneous nevus using immunostaining analysis. Our result showed that the expression of NOVA1 in melanoma was overexpressed in all three patients with melanoma compared to that in two cases of cutaneous nevus (Figure 1). Furthermore, we detected NOVA1 expression in melanoma tissues using qRT‐PCR and Western blot. Our data showed that NOVA1 protein was expressed in three melanoma samples (Figure 2A). In line with this, NOVA1 mRNA was also detected in three melanoma tissues (Figure 2B). In addition, we demonstrated that the expression of NOVA1 was up‐regulated in melanoma cell lines (A875, A375 and SK‐MEL‐1) compared to normal melanocyte cell line (D78) using Western blot (Figure 2C) and qRT‐PCR (Figure 2D).
Figure 1.
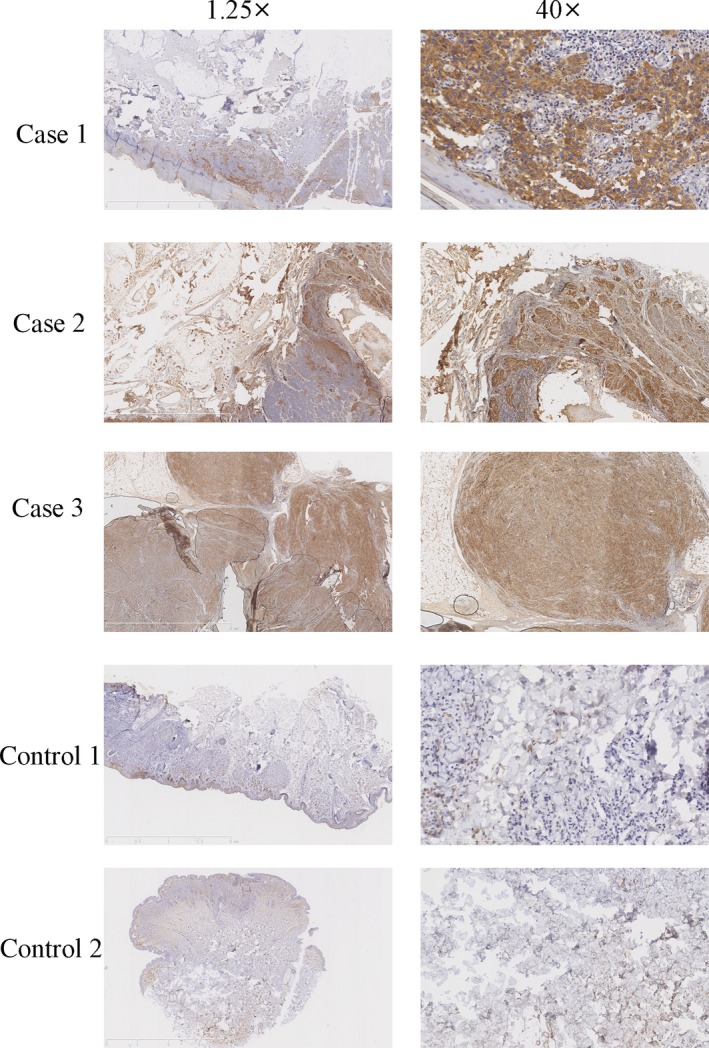
Neuro‐oncological ventral antigen 1 (NOVA1) expression was up‐regulated in the melanoma tissues. The expression of NOVA1 in the three patients with melanoma samples and two patients of cutaneous nevus was measured using immunostaining analysis. Magnification: 1.25× and 40×. The brown was indicated as the positive of NOVA1
Figure 2.

Neuro‐oncological ventral antigen 1 (NOVA1) expression was up‐regulated in the melanoma cell lines. A, The protein expression of NOVA1 in these three cases with melanoma tissues was determined by Western blot. B, The mRNA expression of NOVA1 in these three cases with melanoma tissues was determined by qRT‐PCR. C, The protein expression of NOVA1 in the melanoma cell lines (A875, A375 and SK‐MEL‐1) and normal melanocyte cell line (D78) using Western blot. D, The mRNA expression of NOVA1 was measured by qRT‐PCR
3.2. Knockdown of NOVA1 suppressed melanoma cell proliferation
To explore the functions of FoxO3a in melanoma progression, siRNA‐NOVA1 was transfected into melanoma cell lines (A375 and A875). As shown in Figure 3A,B, the expression of NOVA1 was significantly decreased in A375 and A875 cells after treated with siRNA‐NOVA1. In addition, the mRNA expression of NOVA1 was inhibited in A375 and A875 cells treated with siRNA‐NOVA1 (Figure 3C,D). Moreover, inhibited expression of NOVA1 suppressed A375 (Figure 3E) and A875 (Figure 3F) proliferation using the CCK‐8 assay. We also demonstrated that down‐regulation of NOVA1 suppressed the cyclin D1 expression in the A375 (Figure 3G) and A875 cells (Figure 3H).
Figure 3.
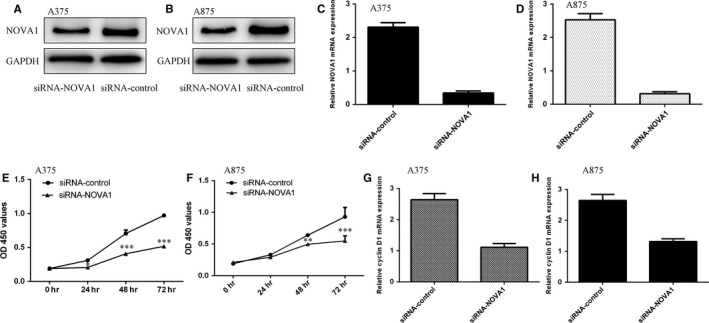
Knockdown expression of NOVA1 suppressed the melanoma cell proliferation. A, The protein expression of NOVA1 in the melanoma cell line A375 was determined by Western blot. B, The protein expression of NOVA1 in the melanoma cell line A875 was determined by Western blot. C, The mRNA expression of NOVA1 in the melanoma cell line A375 was determined by qRT‐PCR. D, The mRNA expression of NOVA1 in the melanoma cell line A875 was determined by qRT‐PCR. E, Inhibition expression of NOVA1 suppressed the A375 cell growth. F, Down‐regulation expression of NOVA1 inhibited A875 cell proliferation. G, The mRNA expression of cyclin D1 in the A375 was measured by qRT‐PCR. H, The mRNA expression of cyclin D1 in the A875 was measured by qRT‐PCR. **P < .01 and ***P < .001
3.3. Inhibited expression of NOVA1 suppressed melanoma cell migration and invasion
Furthermore, we demonstrated that suppressed expression of NOVA1 inhibited A875 cell migration using wound‐healing assay (Figure 4A), and the relative ratio of wound closure was shown in the right. In addition, we also showed that inhibited expression of NOVA1 suppressed A375 cell migration, and the relative ratio of wound closure was shown in the right (Figure 4B). Moreover, down‐regulated expression of NOVA1 suppressed the A375 cell invasion using transwell assays (Figure 4C). In line with this, we also showed that inhibited expression of NOVA1 decreased A875 cell invasion (Figure 4D).
Figure 4.
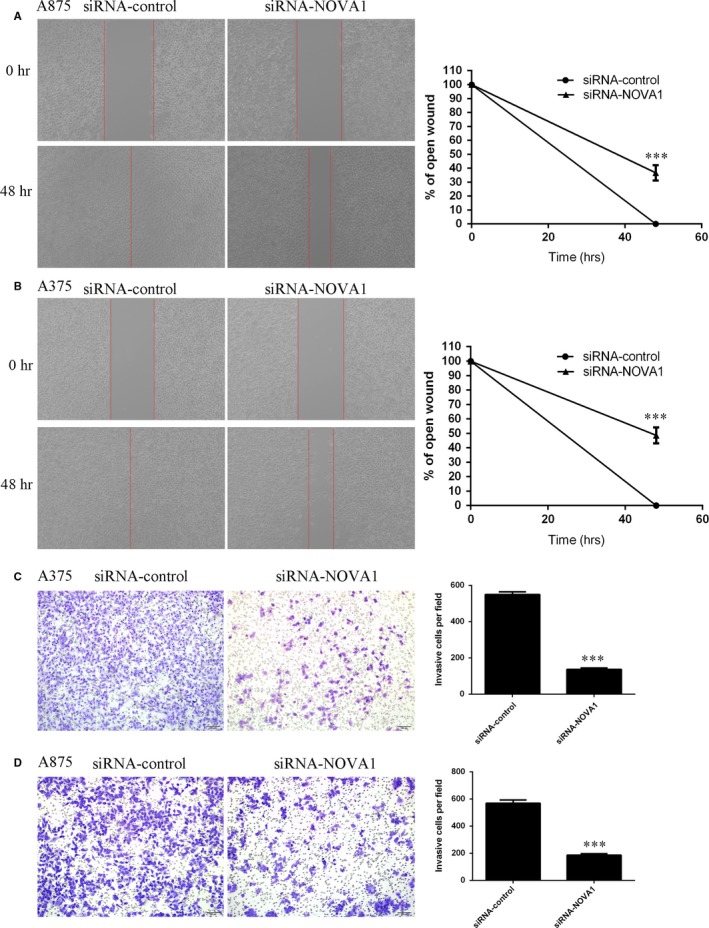
Inhibition expression of NOVA1 inhibited the melanoma cell migration and invasion. A, Scratch wound‐healing assay was performed to determine the cell migration. The relative ratio of wound closure was shown in the right. B, Inhibition expression of NOVA1 inhibited the A375 cell migration, and the relative ratio of wound closure was shown in the right. C, Down‐regulation expression of NOVA1 suppressed the A375 cell invasion using transwell assays. D, Inhibition expression of NOVA1 decreased the A875 cell invasion. ***P < .001
3.4. NOVA1 involvement in FoxO3A and AKT expressions
Next, we studied the mechanism of NOVA1 in melanoma. We showed that knockdown of NOVA1 enhanced FoxO3A expression in melanoma A375 cell (Figure 5A). Moreover, we indicated that down‐regulated expression of FoxO3A inhibited the AKT expression in A375 cell (Figure 5B).
Figure 5.

Knockdown expression of NOVA1 enhanced the FoxO3A expression and decreased AKT expression. A, The protein expression of FoxO3A was determined in the A375 cell by Western blot. B, The protein expression of AKT was determined in the A375 cell by Western blot
3.5. Inhibited expression of FoxO3A rescued NOVA1‐mediated cell proliferation, migration and invasion
To study the role of FoxO3A in NOVA1‐regulated melanoma cell proliferation, migration and invasion, A375 was treated with siRNA‐FoxO3A. We confirmed that the expression of FoxO3A was significantly down‐regulated in the A375 cell after treated with siRNA‐FoxO3A (Figure 6A). Moreover, we showed that inhibited expression of FoxO3A partly rescued NOVA1‐mediated cell proliferation (Figure 6B). In line with this, we showed that suppressed expression of FoxO3A promoted the cyclin D1 expression in the siRNA‐NOVA1‐treated A375 cell (Figure 6C). Furthermore, inhibited expression of FoxO3A partly rescued NOVA1‐mediated cell migration (Figure 6D). We also indicated that suppressed expression of FoxO3A enhanced cell invasion in the siRNA‐NOVA1‐treated A375 cell (Figure 6E).
Figure 6.
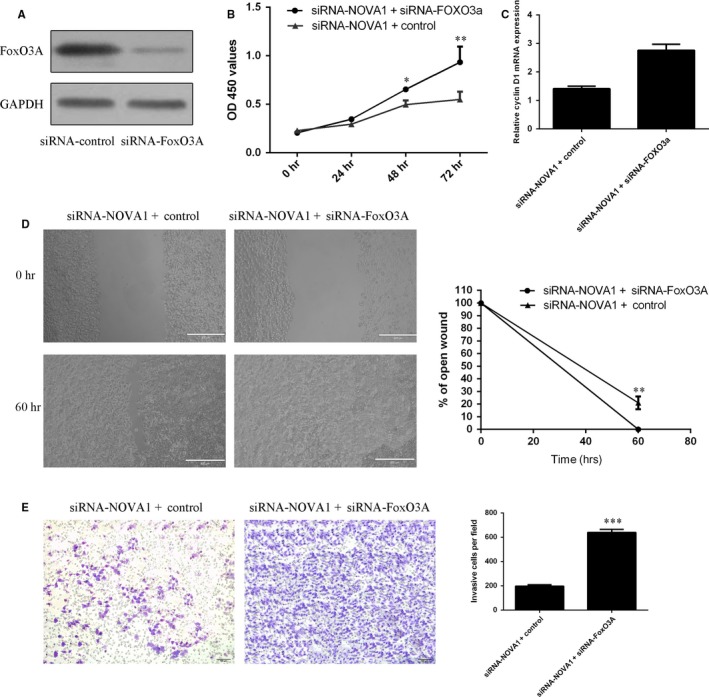
Inhibition expression of FoxO3A rescued NOVA1‐mediated cell proliferation, migration and invasion. A, The protein expression of FoxO3A was determined in the A375 cell by Western blot. B, Cell proliferation was determined using CCK‐8 analysis. C, The mRNA expression of cyclin D1 was measured by qRT‐PCR. D, Inhibition expression of FoxO3A partly rescued NOVA1‐mediated cell migration. E, Cell invasion was determined in the A375 cell using transwell assay. *P < .05, **P < .01 and ***P < .001
4. DISCUSSION
We investigated the expression and functional role of NOVA1 in melanoma. Firstly, we indicated that NOVA1 expression was up‐regulated in melanoma samples and cell lines. Moreover, we demonstrated that knockdown of NOVA1 suppressed melanoma cell proliferation, migration and invasion in both A375 and A875 cells. In addition, we showed that suppressed expression of NOVA1 enhanced FoxO3A expression and suppressed AKT expression in melanoma cell line A375. Furthermore, we demonstrated that inhibited expression of FoxO3A rescued NOVA1‐mediated cell proliferation, migration and invasion in melanoma cell line A375. These results suggested that NOVA1 acted as an oncogene in the development of melanoma partly through regulating FoxO3A expression.
Growing evidence has proved that RBPs play crucial roles in cell biology such as cell growth, apoptosis, migration, differentiation and invasion.30, 31, 32 Several studies suggested that RBPs acted crucial roles in the development of many tumours.32, 33 NOVA1 is one neuron‐specific RBP, which can influence the ligand‐binding, electrophysiological and signal transducing properties.20, 34, 35 Recently, several references have demonstrated that NOVA1 plays an important role in the progression of cancers. For example, Zhang et al36 indicated that NOVA1 enhanced hepatocellular carcinoma growth in vivo partly because of its interaction with the GABAA Receptor‐γ2. Kim et al25 demonstrated that suppressed expression of NOVA1 was found in the gastric cancer (GC) microenvironment, and down‐regulated expression of NOVA1 in GC cells was correlated with GC progression and poor prognosis. Yoon et al37 showed that NOVA1 was one candidate biomarker for prognosis of GC, which was regulated by miR‐146b‐5p. Zhi et al26 demonstrated that miR‐181b‐5p suppressed the astrocytoma growth, invasion and migration and enhanced cell apoptosis partly through inhibiting NOVA1 expression. However, the expression and functional role of NOVA1 in melanoma are still uncovered. Therefore, we firstly detected the expression of NOVA1 in melanoma samples and cell lines. We found that NOVA1 expression was up‐regulated in melanoma tissues and cell lines. In addition, we indicated that inhibited expression of NOVA1 suppressed melanoma cell growth, migration and invasion.
Previous study showed that suppressed expression of NOVA1 enhanced FoxO3A expression and inhibited AKT expression in the pancreatic beta cell.38 As a crucial transcription factor, FOXO3a was a downstream factor which was negatively modulated by PI3K/AKT signal pathway in several human tumours and the p‐FOXO3a catalysed by AKT phosphorylation will significantly inhibit FOXO3a transcriptional activity.39, 40, 41 FoxO3a was shown as a transcription factor which was involved in modulation of the apoptosis, stress response and autophagy.42, 43 Recently, a study showed that inhibited expression of FOXO3a enhanced tumour metastasis and was negatively correlated with the metastasis‐free survival in cases with clear cell renal cell carcinoma.44 Yan et al45 demonstrated that ectopic expression of FoxO3a attenuated basal migration and invasion of uveal melanoma. In our study, we demonstrated that knockdown expression of NOVA1 enhanced the FoxO3A expression and decreased AKT expression. In addition, we showed that inhibited expression of FoxO3A rescued NOVA1‐mediated cell proliferation, migration and invasion. These results suggested that NOVA1 enhanced the melanoma cell growth, migration and invasion partly through suppressing the FoxO3A expression.
In conclusion, we indicated that NOVA1 expression was up‐regulated in melanoma tissues and cell lines. Moreover, we demonstrated that knockdown of NOVA1 suppressed melanoma cell proliferation, migration and invasion partly through regulating FoxO3A expression. NOVA1 acts an oncological role in melanoma, and knockdown may provide a novel therapeutic target for melanoma.
CONFLICT OF INTEREST
The authors declare no competing financial interests.
Yu X, Zheng H, Chan MTV, Wu WKK. Nova1 acts as an oncogene in melanoma via regulating FOXO3a expression. J Cell Mol Med. 2018;22:2622–2630. https://doi.org/10.1111/jcmm.13527
REFERENCES
- 1. Asangani IA, Harms PW, Dodson L, et al. Genetic and epigenetic loss of microRNA‐31 leads to feed‐forward expression of EZH2 in melanoma. Oncotarget. 2012;3:1011‐1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Li J, Martinka M, Li G. Role of ING4 in human melanoma cell migration, invasion and patient survival. Carcinogenesis. 2008;29:1373‐1379. [DOI] [PubMed] [Google Scholar]
- 3. Liu S, Howell PM, Riker AI. Up‐regulation of miR‐182 expression after epigenetic modulation of human melanoma cells. Ann Surg Oncol. 2012;20:1745‐1752. [DOI] [PubMed] [Google Scholar]
- 4. Mazar J, DeBlasio D, Govindarajan SS, Zhang S, Perera RJ. Epigenetic regulation of microRNA‐375 and its role in melanoma development in humans. FEBS Lett. 2011;585:2467‐2476. [DOI] [PubMed] [Google Scholar]
- 5. Mazar J, Khaitan D, DeBlasio D, et al. Epigenetic regulation of microRNA genes and the role of miR‐34b in cell invasion and motility in human melanoma. PLoS ONE. 2011;6:e24922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Siegel R, Miller K, Jemal A. Cancer statistics. CA Cancer J Clin. 2016;66:7‐30. [DOI] [PubMed] [Google Scholar]
- 7. Cantile M, Scognamiglio G, Marra L, et al. HOTAIR role in melanoma progression and its identification in the blood of patients with advanced disease. J Cell Physiol. 2017;232:3422‐3432. [DOI] [PubMed] [Google Scholar]
- 8. Sarkar D, Leung EY, Baguley BC, Finlay GJ, Askarian‐Amiri ME. Epigenetic regulation in human melanoma: past and future. Epigenetics. 2015;10:103‐121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Streicher KL, Zhu W, Lehmann KP, et al. A novel oncogenic role for the miRNA‐506‐514 cluster in initiating melanocyte transformation and promoting melanoma growth. Oncogene. 2012;31:1558‐1570. [DOI] [PubMed] [Google Scholar]
- 10. Sinnamon A, Neuwirth M, Gimotty P, et al. Association of first‐in‐class immune checkpoint inhibition and targeted therapy with survival in patients with stage IV melanoma. JAMA Oncol. 2018;4:126‐128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Jukic DM, Rao UN, Kelly L, et al. Microrna profiling analysis of differences between the melanoma of young adults and older adults. J Transl Med. 2010;8:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kitago M, Martinez SR, Nakamura T, Sim MS, Hoon DS. Regulation of RUNX3 tumor suppressor gene expression in cutaneous melanoma. Clin Cancer Res. 2009;15:2988‐2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Neelamraju Y, Gonzalez‐Perez A, Bhat‐Nakshatri P, Nakshatri H, Janga S. Mutational landscape of RNA‐binding proteins in human cancers. RNA Biol 2018;15:115‐129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Heller D, Krestel R, Ohler U, Vingron M, Marsico A. ssHMM: extracting intuitive sequence‐structure motifs from high‐throughput RNA‐binding protein data. Nucleic Acids Res. 2017;45:11004‐11018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Xu D, Xu S, Maung Kyaw A, et al. RNA binding protein, Ybx2, regulates RNA stability during cold‐induced brown fat activation. Diabetes. 2017;66:2987‐3000. [DOI] [PubMed] [Google Scholar]
- 16. Li Y, Xiao M, Shi B, et al. Identification of high‐confidence RNA regulatory elements by combinatorial classification of RNA‐protein binding sites. Genome Biol. 2017;18:169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wurth L, Papasaikas P, Olmeda D, et al. UNR/CSDE1 drives a post‐transcriptional program to promote melanoma invasion and metastasis. Cancer Cell. 2016;30:694‐707. [DOI] [PubMed] [Google Scholar]
- 18. Schiera G, Di Liegro C, Puleo V, et al. Extracellular vesicles shed by melanoma cells contain a modified form of H1.0 linker histone and H1.0 mRNA‐binding proteins. Int J Oncol. 2016;49:1807‐1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Roesch A, Becker B, Meyer S, et al. Retinoblastoma‐binding protein 2‐homolog 1: a retinoblastoma‐binding protein downregulated in malignant melanomas. Mod Pathol. 2005;18:1249‐1257. [DOI] [PubMed] [Google Scholar]
- 20. Xin Y, Li Z, Zheng H, Ho J, Chan M, Wu W. Neuro‐oncological ventral antigen 1 (NOVA1): Implications in neurological diseases and cancers. Cell Prolif. 2017;50:1‐4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kim E, Yoon S, Kim S, Yang W, Cho Y, Kim S. Upregulated neuro‐oncological ventral antigen 1 (NOVA1) expression is specific to mature and immature T‐ and NK‐cell lymphomas. J Pathol Transl Med. 2016;50:104‐112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Saito Y, Miranda‐Rottmann S, Ruggiu M, et al. NOVA2‐mediated RNA regulation is required for axonal pathfinding during development. Elife. 2016;5:pii:e14371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jelen N, Ule J, Zivin M, Darnell R. Evolution of Nova‐dependent splicing regulation in the brain. PLoS Genet. 2007;3:1838‐1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Fletcher C, Okano H, Gilbert D, et al. Mouse chromosomal locations of nine genes encoding homologs of human paraneoplastic neurologic disorder antigens. Genomics. 1997;45:313‐319. [DOI] [PubMed] [Google Scholar]
- 25. Kim E, Yoon S, Jung W, et al. Implications of NOVA1 suppression within the microenvironment of gastric cancer: association with immune cell dysregulation. Gastric Cancer. 2017;20:438‐447. [DOI] [PubMed] [Google Scholar]
- 26. Zhi F, Wang Q, Deng D, et al. MiR‐181b‐5p downregulates NOVA1 to suppress proliferation, migration and invasion and promote apoptosis in astrocytoma. PLoS ONE. 2014;9:e109124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gimenez M, Marie S, Oba‐Shinjo S, et al. Quantitative proteomic analysis shows differentially expressed HSPB1 in glioblastoma as a discriminating short from long survival factor and NOVA1 as a differentiation factor between low‐grade astrocytoma and oligodendroglioma. BMC Cancer. 2015;15:481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhang Y, Liu H, Zhu J, Zhang D, Shen X, Liu T. RNA binding protein Nova1 promotes tumor growth in vivo and its potential mechanism as an oncogene may due to its interaction with GABAA Receptor‐γ2. J Biomed Sci. 2016;23:71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Shen B, Zhang Y, Yu S, et al. MicroRNA‐339, an epigenetic modulating target is involved in human gastric carcinogenesis through targeting NOVA1. FEBS Lett. 2015;589:3205‐3211. [DOI] [PubMed] [Google Scholar]
- 30. Lucchesi C, Zhang J, Chen X. Modulation of the p53 family network by RNA‐binding proteins. Transl Cancer Res. 2016;5:676‐684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hao J, Duan F, Wang Y. MicroRNAs and RNA binding protein regulators of microRNAs in the control of pluripotency and reprogramming. Curr Opin Genet Dev. 2017;46:95‐103. [DOI] [PubMed] [Google Scholar]
- 32. Pereira B, Billaud M, Almeida R. RNA‐binding proteins in cancer: old players and new actors. Trends Cancer. 2017;3:506‐528. [DOI] [PubMed] [Google Scholar]
- 33. Dang H, Takai A, Forgues M, et al. Oncogenic activation of the RNA binding protein NELFE and MYC signaling in hepatocellular carcinoma. Cancer Cell. 2017;32:101‐114. e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Fryssira H, Tsoutsou E, Psoni S, et al. Partial monosomy14q involving FOXG1 and NOVA1 in an infant with microcephaly, seizures and severe developmental delay. Mol Cytogenet. 2016;9:55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Herve M, el Ibrahim C. MicroRNA screening identifies a link between NOVA1 expression and a low level of IKAP in familial dysautonomia. Dis Model Mech. 2016;9:899‐909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zhang YA, Zhu JM, Yin J, et al. High expression of neuro‐oncological ventral antigen 1 correlates with poor prognosis in hepatocellular carcinoma. PLoS ONE. 2014;9:e90955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Yoon SO, Kim EK, Lee M, et al. NOVA1 inhibition by miR‐146b‐5p in the remnant tissue microenvironment defines occult residual disease after gastric cancer removal. Oncotarget. 2016;7:2475‐2495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Villate O, Turatsinze J, Mascali L, et al. Nova1 is a master regulator of alternative splicing in pancreatic beta cells. Nucleic Acids Res. 2014;42:11818‐11830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ling N, Gu J, Lei Z, et al. microRNA‐155 regulates cell proliferation and invasion by targeting FOXO3a in glioma. Oncol Rep. 2013;30:2111‐2118. [DOI] [PubMed] [Google Scholar]
- 40. Zhang S, Liu L, Wang R, et al. MicroRNA‐217 promotes angiogenesis of human cytomegalovirus‐infected endothelial cells through downregulation of SIRT1 and FOXO3A. PLoS ONE. 2013;8:e83620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Aldonza MB, Hong JY, Alinsug MV, Song J, Lee SK. Multiplicity of acquired cross‐resistance in paclitaxel‐resistant cancer cells is associated with feedback control of TUBB3 via FOXO3a‐mediated ABCB1 regulation. Oncotarget. 2016;7:34395‐34419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Nestal de Moraes G, Bella L, Zona S, Burton M, Lam E. Insights into a critical role of the FOXO3a‐FOXM1 axis in DNA damage response and genotoxic drug resistance. Curr Drug Targets 2016;17:164‐177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Chiacchiera F, Simone C. The AMPK‐FoxO3A axis as a target for cancer treatment. Cell Cycle. 2010;9:1091‐1096. [DOI] [PubMed] [Google Scholar]
- 44. Ni D, Ma X, Li H, et al. Downregulation of FOXO3a promotes tumor metastasis and is associated with metastasis‐free survival of patients with clear cell renal cell carcinoma. Clin Cancer Res. 2014;20:1779‐1790. [DOI] [PubMed] [Google Scholar]
- 45. Yan F, Liao R, Farhan M, et al. Elucidating the role of the FoxO3a transcription factor in the IGF‐1‐induced migration and invasion of uveal melanoma cancer cells. Biomed Pharmacother. 2016;84:1538‐1550. [DOI] [PubMed] [Google Scholar]


