Abstract
New treatments to overcome the obstacles of conventional anti-cancer therapy are a permanent subject of investigation. One promising approach is the application of toxins linked to cell-specific ligands, so-called immunotoxins. Another attractive option is the employment of toxin-encoding plasmids. However, immunotoxins cause hepatoxicity, and DNA therapeutics, among other disadvantages, bear the risk of insertional mutagenesis. As an alternative, this study examined chemically modified mRNAs coding for diphtheria toxin, subtilase cytotoxin, and abrin-a for their ability to reduce cancer cell growth both in vitro and in vivo. The plant toxin abrin-a was the most promising candidate among the three tested toxins and was further investigated. Its expression was demonstrated by western blot. Experiments with firefly luciferase in reticulocyte lysates and co-transfection experiments with EGFP demonstrated the capability of abrin-a to inhibit protein synthesis. Its cytotoxic effect was quantified employing viability assays and propidium iodide staining. By studying caspase-3/7 activation, Annexin V-binding, and chromatin condensation with Hoechst33258 staining, apoptotic cell death could be confirmed. In mice, repeated intratumoral injections of complexed abrin-a mRNA resulted in a significant reduction (89%) of KB tumor size compared to a non-translatable control mRNA.
Keywords: mRNA therapy, cancer, toxins
Introduction
Conventional anti-cancer therapy is often limited by chemoresistance,1 radiation resistance,2 and severe side effects like cardiotoxicity3 or neurocognitive deficits.4 Therefore, the need for innovative cancer therapeutic options is immense. For instance, administration of immunotoxins as anti-cancer therapeutics is being tested in clinical studies,5 with one drug gaining FDA approval.6 Immunotoxins are cell-specific ligands linked either to plant or to bacterial toxins, leading to a selective killing of target cells.7, 8 The advantage of this approach, compared to other tumor treatments, is high tumor specificity and hence less damage of healthy tissue. A further benefit is their applicability to both solid and non-solid tumors.9 Still, despite their cell specific toxicity in vitro, in some cases damage to healthy tissue, particularly to the liver, has been observed.10, 11 Another disadvantage is the comparatively slow protein uptake from blood vessels by tumors due to their abnormal tissue architecture.11
An alternative that might avoid these deficiencies but shares many of the benefits of immunotoxins is the employment of nucleic acids coding for toxic proteins as suicide cancer therapy. Hochberg and co-workers12, 13 have shown the potential of this approach by applying a plasmid coding for diphtheria toxin. Expression of diphtheria toxin is under the control of H19 promoter, which is highly active in a range of human cancers, ensuring that toxin production is confined to the tumor. Clinical trials (phase 2b) with this plasmid-based approach have been successful.12 Analogously, tumor growth was reduced after intratumoral delivery of a recombinant adeno-associated virus containing DNA coding for the toxin trichosanthin.14 Nevertheless, DNA-based gene therapeutics bear some risks and disadvantages, with potential genomic integration followed by mutation being one of the most important safety concerns.15 The employment of messenger RNA instead of DNA is an attractive approach to circumvent several deficiencies of plasmid-based transfection. The most important benefits of mRNA application versus DNA therapeutics are (1) no risk of insertional mutagenesis,15 (2) no need to enter the nucleus, therefore resulting in higher transfection efficiency and earlier onset of protein expression,16 (3) expression being self-limited because of short mRNA half-life,17, 18 and (4) only the sequence of interest being introduced into the cell.16 Also, using the herpes simplex virus-thymidine kinase/ganciclovir (TK/GCV) suicide gene therapy system, Wang et al.19 observed that the reduction in tumor growth was significantly higher for mRNA than for plasmid DNA (pDNA).
In spite of the advantages of mRNA over pDNA as therapeutic agent, its presumably low stability and high immunogenicity prevented its broad application until the last decade. Since Wolff et al.20 showed the first successful mRNA transfection in vivo in 1990, research has identified ways to evade these issues. Several studies have reported increased stability and decreased immunogenicity after incorporating modified nucleotides in in vitro transcribed mRNAs.21, 22, 23, 24, 25 Accordingly, in the present study, chemically modified mRNA (cmRNA) comprising 5′-methylcytosine and 2′-thiouridine, which was shown previously to result in stabilized non-immunogenic mRNA,24 was applied.
In this study, we combine the advantages of immunotoxins and mRNA-based therapeutics. We investigated the cytotoxic potential of mRNA transcripts coding for three toxins that have been previously used as immunotoxins.9, 26, 27, 28 Diphtheria toxin, produced by Corynebacterium diphtheria,29 subtilase cytotoxin, produced by shiga toxigenic Escherichia coli (STEC),30 and the plant-derived abrin-a, isolated from Abrus precatorius,31 all belong to the family of conventional AB-toxins.30, 32, 33 After binding of the B-subunit to the target cell, the catalytic A-subunit of AB-toxins enters the cell34 and mediates the toxic effects via impairment of protein synthesis, resulting in cell death.35, 36, 37 We focused on these three toxins since they represent different signaling pathways of protein synthesis inhibition and have shown promising anti-tumor effects in pre-clinical or clinical studies.26, 28, 38
In a first step, we compared the three cmRNAs regarding their potency to inhibit cell viability in two different tumor cell lines. The best-performer abrin-a, A-chain (AA) cmRNA was then investigated in detail to estimate its potential as new anti-tumor agent. In order to exclude any possible unspecific effects of mRNA transfection, a cmRNA (AAstop) was utilized that shows no translation. This was achieved by scrambling the Kozak element39 and mutating the start as well as all in frame downstream ATGs into TAGs. In in vitro experiments, the expression of AA was verified by western blot, and it was assessed for its capacity to decrease protein synthesis, its cytotoxicity, and the apoptotic characteristics of induced cell death. In vivo, experiments in mice showed that intratumoral injection of AA cmRNA resulted in a significantly reduced tumor growth.
This is a first proof-of-concept study demonstrating the efficacy of “killer RNAs” as promising anti-tumor agents. Further studies with different tumor models will be highly valuable in determining the true potential of such mRNA-based therapeutics.
Results
Comparison of the Decrease in Cell Viability Caused by SubA, DTA, and AA cmRNA in HuH7 and KB Cells
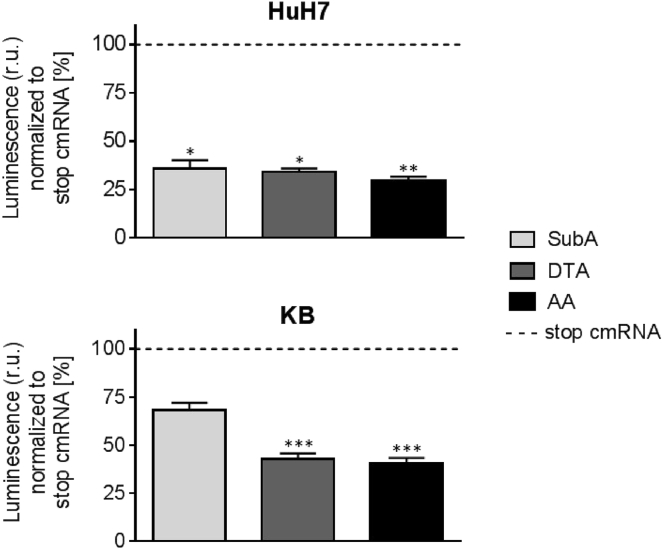
To determine the effectivity of the different cmRNAs, namely subtilase cytotoxin, A-chain (SubA), diphtheria toxin, A chain (DTA), and AA, cell viability measurements using the human cervix carcinoma cell line KB and the human hepatocarcinoma cell line HuH7 were performed. As control, stop cmRNAs were designed by introducing small changes in the sequence of the toxins that prevent translation. Cells were transfected with 100 ng of cmRNA. At 48 hr after transfection, cell viability was assessed by measuring the ATP content using the CellTiter-Glo Luminescence Viability Assay (Figure 1). In HuH7 cells, all three toxins induced a significant reduction in cell viability of 65%–70% (SubA, p = 0.0127; DTA, p = 0.0494; AA, p = 0.0017) in comparison to cells transfected with the respective stop cmRNA. In KB cells, the decrease in cell viability by SubA cmRNA was low with 32%, whereas DTA cmRNA and AA cmRNA decreased cell viability significantly by almost 60% compared to cells transfected with the respective stop cmRNA (SubA, p = 0.2144; DTA, p = 0.0009; AA, p = 0.0002). Considering the different size of the two RNAs (AA, 1.0 kb, versus DTA, 0.8 kb), more DTA molecules were required to induce similar effects on cell viability compared to AA. This led to the conclusion that AA was the most potent toxin and was therefore used in the following experiments with KB cells.
Figure 1.
Comparison of the Decrease in Cell Viability Caused by SubA, DTA, and AA cmRNA in HuH7 and KB Cells
Forty-eight hours post-transfection of KB and HuH7 cells with either of the cmRNAs SubA (subtilase cytotoxin, A-chain), DTA (diphtheria toxin, A-chain), AA (abrin-a, A-chain), or one of their stop cmRNAs, cell viability was assessed by quantifying the ATP content using the CellTiter-Glo Luminescence Viability Assay. Cell viability was proportional to the measured luminescence. 100 ng cmRNA were applied. Data are presented as mean in % ± SEM of cells transfected with the respective stop cmRNA (dotted line). Statistical significance for HuH7 and KB cells versus cells transfected with the respective stop cmRNA was assessed by Kruskal-Wallis test adjusted for multiple comparisons, with *p < 0.05, **p < 0.01, ***p < 0.001, and n = 3.
Detection of Abrin-a Protein after Transfection of KB Cells with AA cmRNA by Western Blot
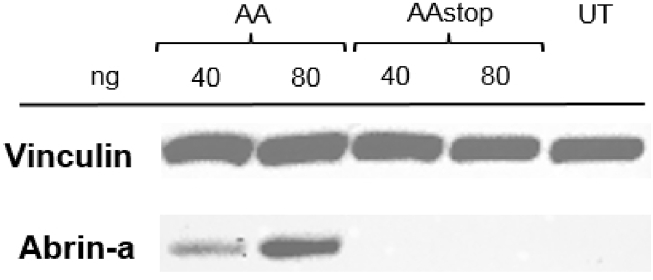
The expression of abrin-a after transfection of KB cells with AA cmRNA was verified by western blot analysis. For this, KB cells were transfected with 40 ng and 80 ng AA or AAstop cmRNA, lysed 24 hr post-transfection, and SDS-PAGE and western blot was performed. Figure 2 clearly demonstrates abrin-a protein production in KB cells after transfection with AA cmRNA in a concentration-dependent manner. After transfection with AAstop cmRNA and in case of untransfected control cells (UTs), no abrin-a protein was detected.
Figure 2.
Detection of Abrin-a Protein after Transfection of KB Cells with AA cmRNA by Western Blot
KB cells were transfected with 40 or 80 ng of AA (abrin-a, A-chain) or AAstop cmRNA. Twenty-four hours post-transfection, cells were lysed and SDS-PAGE and western blot was performed. Anti-vinculin antibody was used as loading control. A representative blot, including untransfected control cells (UT), is shown. n = 2.
Inhibition of Protein Synthesis by AA cmRNA
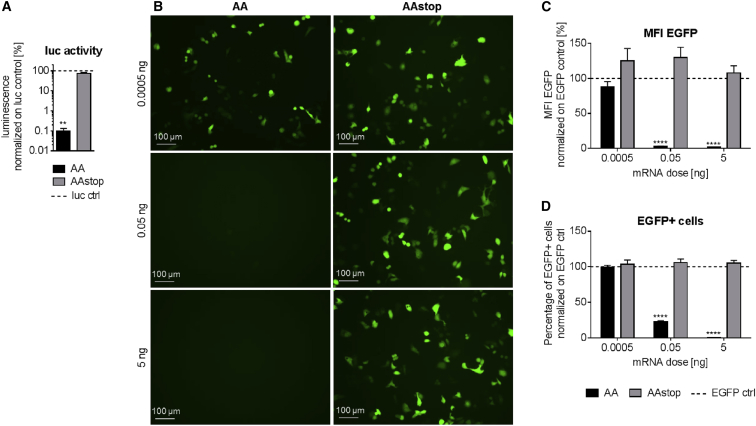
The capability of AA to inhibit protein synthesis was assessed by co-treatment of rabbit reticulocyte lysate with 1 μg firefly luciferase (luc) cmRNA and co-transfection of KB cells with 50 ng EGFP cmRNA. Forty-five minutes after treatment of lysate with 0.1 μg AA cmRNA, luminescence was reduced significantly (p = 0.0079 in comparison to AAstop cmRNA) to 0.1% in comparison to lysate treated with luc cmRNA only (luc control, Figure 3A). Addition of AAstop cmRNA, however, did not result in decreased luc activity. KB cells transfected with AA cmRNA showed considerable EGFP fluorescence at 0.0005 ng, but no EGFP fluorescence was visible at 0.05 or 5 ng after 24 hr (Figure 3B). EGFP fluorescence in cells transfected with AAstop cmRNA was not affected at any of the tested concentrations. These microscopic observations were further confirmed by flow cytometry analysis, showing both the mean fluorescence intensity (MFI) of EGFP (Figure 3C) and the percentage of EGFP-positive cells (Figure 3D). Compared to cells transfected with EGFP cmRNA only (EGFP control), the MFI of EGFP was unchanged at 0.0005 ng AA cmRNA. At 0.05 and 5 ng AA cmRNA, the MFI of EGFP was decreased to 3.4% and 1.7%, respectively, compared to EGFP control cells. Similar to the results obtained with MFI, the number of EGFP-positive cells was unchanged at 0.0005 ng AA cmRNA (compared to EGFP control), but reduced by 77% at 0.05 ng or 100% at 5 ng. The differences between AA and AAstop cmRNA were significant for 0.05 and 5 ng (p < 0.0001). These results demonstrate that production of abrin-a after treatment of reticulocyte lysate or of KB cells with AA cmRNA was active and led to inhibition of protein synthesis of co-applied luc or EGFP cmRNA.
Figure 3.
Inhibition of Protein Synthesis by AA cmRNA
(A) Inhibition of translation of firefly luciferase (luc) cmRNA in a rabbit reticulocyte lysate system by AA cmRNA. Rabbit reticulocyte lysate was treated with 1 μg luc cmRNA and 0.1 μg AA (abrin-a, A-chain) or AAstop cmRNA and luminescence as a measure of luc activity was determined 45 min after start of the reaction. Data are presented as mean in % ± SEM of lysate treated only with luc cmRNA (luc ctrl, dotted line). Statistical significance versus AAstop cmRNA was assessed by Mann-Whitney U test, with **p < 0.01 and n = 3. (B–D) Inhibition of EGFP fluorescence in KB cells by AA cmRNA. KB cells were co-transfected with 50 ng EGFP cmRNA and either AA (abrin-a, A-chain) or AAstop cmRNA. Twenty-four hours post-transfection, inhibition of protein synthesis was assessed by fluorescence microscopy (B) and flow cytometry (C and D). Representative fluorescence images are shown (B). Mean fluorescence intensity (MFI) (C) and percentage of EGFP-positive cells (D) are depicted. Data are presented as mean in % ± SEM of control cells transfected only with EGFP cmRNA (EGFP ctrl, dotted line). Statistical significance versus AAstop cmRNA was assessed by two-way ANOVA adjusted for multiple comparisons, with ****p < 0.0001 and n = 3.
Toxicity of AA cmRNA on KB Cells
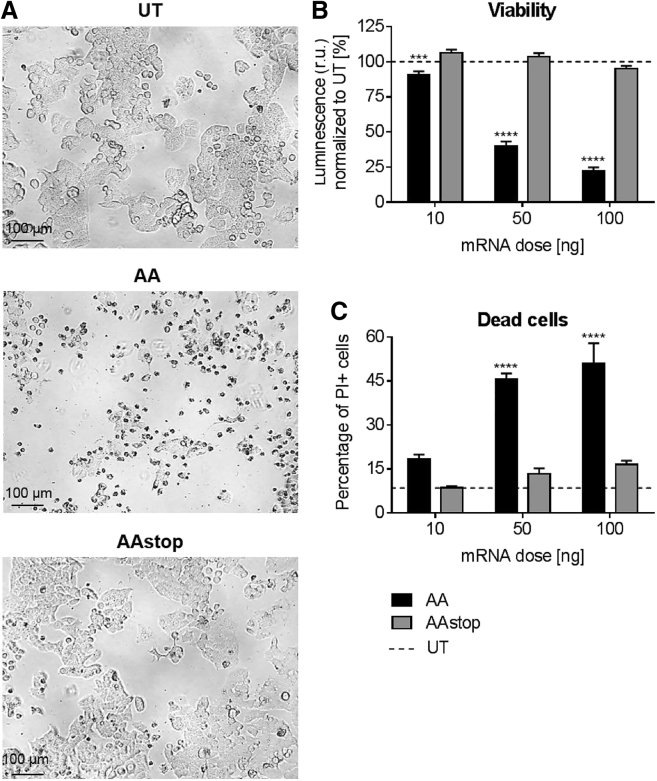
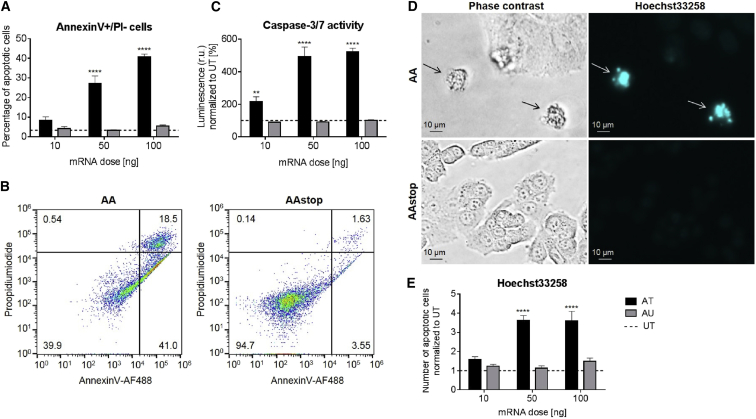
The toxic effect of AA cmRNA on KB cells in comparison to AAstop cmRNA was evident in microscopic images taken 48 hr after transfection (Figure 4A). Cell viability measurements (Figure 4B) showed reduction at all tested concentrations of AA cmRNA compared to UTs. Toxicity at 10 ng was low with 9.2% decrease in viability but increased considerably at 50 ng and 100 ng with 60.2% and 77.6% reduction of cell viability compared to UT cells. The percentage of dead cells, determined by staining with propidium iodide (PI), was in agreement with these findings. At 10 ng AA cmRNA, the number of PI-positive cells was twice as high as for UT. For 50 and 100 ng, the percentage of dead cells increased to 45%–51% of all cells. With the exception of PI staining at 10 ng dose, the differences between AA and AAstop cmRNA were statistically significant (10 ng, p = 0.0002; others, p < 0.0001). It could be shown that AA cmRNA not only distinctly decreases cell viability but also induces cell death.
Figure 4.
Toxicity of AA cmRNA on KB Cells
Forty-eight hours post-transfection of KB cells with AA (abrin-a, A-chain) or AAstop cmRNA. (A) Representative pictures were taken of cells transfected with 100 ng cmRNA or of untransfected cells (UT). (B) Cell viability was determined by measuring ATP content with the CellTiter-Glo Luminescence Viability Assay. Cell viability was proportional to the measured luminescence. Data is presented as mean in % ± SEM of untransfected control cells (UT, dotted line). (C) Number of dead cells was counted using propodium iodide (PI) staining and flow cytometry analysis. Percentage of PI-positive cells was compared to untransfected control cells (UT, dotted line) and is shown as mean in % ± SEM. Statistical significance versus AAstop cmRNA was assessed by two-way ANOVA adjusted for multiple comparisons, with ***p < 0.001, ****p < 0.0001, and n = 3.
Apoptotic Cell Death after Transfecting KB Cells with AA cmRNA
The occurrence of apoptotic cell death after transfection with AA cmRNA was demonstrated by investigating Annexin V-binding, caspases-3 and -7 activity, and chromatin condensation using Hoechst33258 staining. At 24 hr after transfection with 10 ng AA cmRNA, binding of Annexin V to the cell membrane increased compared to UTs (Figure 5A). The fraction of cells that were Annexin V positive but PI negative was 27% for 50 ng and 41% for 100 ng dose group compared to UT with 3.4%. The increase of apoptotic cells after transfection with AA cmRNA was dose dependent. Figure 5B shows a representative dot plot of the Annexin V-PI fluorescence cytometry assay. In comparison to AAstop cmRNA, a clear shift from viable cells (double negative) to apoptotic (Annexin V positive, PI negative) and dead cells (double positive) was detectable for AA cmRNA-transfected cells. Twenty-four hours after transfection, an increase in caspase-3 and -7 activity could be observed at 10 ng (Figure 5C). For 50 and 100 ng doses groups, the activity of caspases-3 and -7 was five times higher for AA cmRNA than for UT. Forty-eight hours post-transfection with AA cmRNA, the apoptotic characteristics “membrane blebbing” and nuclear fragmentation could be observed (Figure 5D) as well as shrinkage and rounding of cells (Figure 4A) In Figure 5E, the Hoechst33258-positive cells were quantified. The increase in apoptotic cells was low compared to UT for 10 ng AA cmRNA. The number of apoptotic cells in case of AA cmRNA was three times higher than for UT at 50 and 100 ng. As observed for caspase-3 and -7 activity, no major difference in degree of apoptosis could be detected for AA cmRNA transfections with 50 or 100 ng. AAstop cmRNA did not show any induction of apoptotic cell death or morphological aberrations under the tested conditions. For all tested doses, concerning 10 ng only for caspase-3 and -7 activity, increase in apoptosis was statistically significant for AA compared to AAstop cmRNA (10 ng, p = 0.0075; others, p < 0.0001). Taken together, the results suggest that transfection of KB cells with AA cmRNA leads to predominantly apoptotic cell death.
Figure 5.
Apoptotic Cell Death after Transfecting KB Cells with AA cmRNA
At 24 hr (A–C) or 48 hr (D and E) post-transfection of KB cells with AA (abrin-a, A-chain) or AAstop cmRNA, cells were examined for apoptosis. (A and B) Annexin V-AF488 and propidium iodide (PI) double staining flow cytometry assay. (A) Percentages of cells in early apoptotic stage (Annexin V-AF488 positive, PI negative). Data is shown as mean in % ± SEM and is compared to untransfected control cells (UT, dotted line). (B) Representative dot plots of cells transfected with 100 ng cmRNA. The percentages of the different populations are given in the respective corners. (C) Caspase-Glo 3/7 assay; activity was proportional to luminescence. Data is represented as mean in % ± SEM of untransfected control cells (UT, dotted line). (D and E) Hoechst33258 staining. (D) Representative phase contrast and fluorescence images of cells transfected with 100 ng cmRNA are displayed. Arrows depict cells showing “membrane blebbing” or nuclear fragmentation. (E) Number of Hoechst33258-positive cells. Data (mean ± SEM) is presented as multiple of untransfected control cells (UT, dotted line). Statistical significance versus AAstop cmRNA was assessed by two-way ANOVA adjusted for multiple comparisons, with **p < 0.01, ****p < 0.0001, and n = 3.
Decrease in Cell Viability by AA-LF132 In Vitro and Inhibition of Tumor Growth In Vivo
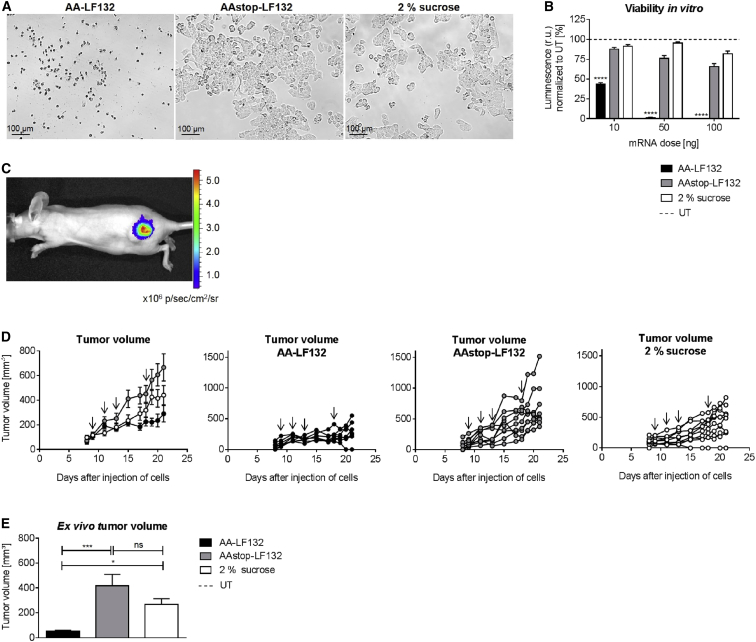
Prior to performing the in vivo experiment, the Ethris’ proprietary cationic lipid formulation LF132 was tested in vitro on KB cells for its effectiveness. Forty-eight hours after transfection, very high toxicity of AA-LF132 but no toxic effect of AAstop-LF132 or 2% sucrose (vehicle control) was observed (Figure 6A). This was further confirmed by assessing cell viability (Figure 6B). For 10, 50, or 100 ng AA cmRNA, cell viability was reduced by 56%, 99%, or 100% compared to UTs, respectively. Accordingly, in comparison to AA-Lipofectamine 2000 (cf. Figure 4B), potency of inducing toxicity of AA-LF132 was considerably higher. AAstop-LF132 also showed some toxicity at higher concentrations, but far less compared to AA-LF132. The reduction in luminescence by AA-LF132 was statistically significant in comparison to AAstop-LF132 for all tested doses (p < 0.0001).
Figure 6.
Decrease in Cell Viability by AA-LF132 In Vitro and Its Influence on Tumor Growth In Vivo
(A and B) In vitro assessment of toxicity on KB cells at 48 hr post-transfection with AA-LF132, AAstop-LF132, or treatment with 2% sucrose (vehicle control). (A) Representative pictures of KB cells transfected with 100 ng cmRNA. (B) CellTiter-Glo Luminescence Viability Assay. Cell viability was proportional to the measured luminescence. Data is presented as mean in % ± SEM of untransfected control cells (UT, dotted line). Statistical significance versus AAstop-LF132 was assessed by two-way ANOVA adjusted for multiple comparisons, with ****p < 0.0001 and n = 3. (C) Luciferase activity. 5 × 106 KB cells were injected into the flank of immuno-deficient NMRI-nu mice. 10 μg of lipid nanoparticle formulated cmRNA coding for firefly luciferase was injected intratumorally on days 9, 11, and 13 after injection of tumor cells. On day 14, bioluminescence was determined. (D and E) In vivo anti-tumor activity of AA-LF132. 5 × 106 KB cells were injected into the flank of immuno-deficient NMRI-nu mice. 10 μg of AA-LF132, 10 μg of AAstop-LF132 or 2% sucrose were injected intratumorally on days 9, 11, 13, and 18 after injection of tumor cells. (D) Tumor volume was measured in vivo throughout the experiment using a caliper. Arrows display days of treatment. Data represent means ± SEM (left) or individual values of each mouse. n = 7 for AA-LF132; n = 10 for AAstop-LF132 and 2% sucrose. (E) Tumor volume was determined ex vivo on day 21 after injection of tumor cells. Data represent means ± SEM. Statistical significance was assessed by Kruskal-Wallis test adjusted for multiple comparisons, with *p < 0.05, ***p < 0.001, n = 7 for AA-LF132, n = 10 for AAstop-LF132 and 2% sucrose.
To test the anti-tumor activity of AA-LF132 in vivo, 5 × 106 KB cells were injected into the flank of immuno-deficient Naval Medical Research Institute-nude (NMRI-nu) mice. In a small pre-experiment, the expression characteristics of intratumorally injected RNA were investigated. Therefore, 10 μg lipid nanoparticle formulated cmRNA coding for firefly luciferase was injected three times. Twenty-four hours after the third application, considerable and locally defined luciferase activity was observed (Figure 6C). In the main experiment, the complexed cmRNAs or 2% sucrose were injected intratumorally as soon as the tumors reached a sufficient size (∼100 mm3). The treatment was repeated three times in 2- to 5-day intervals. Surveillance of general condition and body weight during treatment as well as examination of blood parameters (white blood cells, red blood cells, hemoglobin, hematocrit, and platelets) on day 21 after injection of tumor cells showed no disparities between the three groups (data not shown). However, while the formation of cutaneous lesions up to ulcers was not observed in AAstop-LF132 or 2% sucrose-treated mice, it was present in all but one animal of the AA-LF132 group. Measurement of the tumor volume throughout the treatment demonstrated a marked difference in growth rate between the different groups (Figure 6D). Twelve days after the first injection, tumor volume was determined ex vivo (Figure 6E). It was shown that treatment resulted in a significantly lower tumor size for AA-LF132 than for AAstop-LF132 (p = 0.0010) or for 2% sucrose (p = 0.0350). With a mean volume of 50 mm3, AA-LF132 treated tumors were 89% smaller than tumors injected with AAstop-LF132. The difference in tumor size concerning AAstop-LF132 and 2% sucrose was statistically not significant. This experiment clearly demonstrates the potential of cmRNA coding for toxic proteins to reduce tumor growth in vivo.
Discussion
Employment of mRNA coding for different cellular and viral proteins for anti-cancer therapies has been investigated in previous studies. Van der Jeught et al.,40 for instance, delivered mRNA coding for interferon β fused to the transforming growth factor β (TGF-β) receptor II intratumorally to enhance tumor-specific immunity. Also, an mRNA encoding the TK/GCV suicide system was applied intravenously to suppress tumor growth.19 In both studies, reduction of tumor growth was observed. The present study aimed to investigate whether protein toxins are suitable for employment as mRNA cancer therapeutics. For this purpose, cmRNAs coding for the catalytic A-chain of three AB-toxins, namely subtilase cytotoxin (SubA), diphtheria toxin (DTA) and abrin-a (AA), were employed.
A cell viability assay performed on the cancer cell lines HuH7 and KB revealed AA as the most promising candidate for further investigation. Concerning HuH7 cells, the decline in viability 48 hr after transfection was comparably high for all three toxins. While AA and DTA cmRNA showed a similar effect on KB cells, SubA cmRNA showed reduced toxicity compared to HuH7 cells. Because of differences in cell number and cell size, a direct comparison between the two cell lines regarding their sensitivity to toxin mediated cell death is limited. As DTA inactivates EF-2 by ADP-ribosylation29, 35 and AA blocks the binding of EF-2 to the ribosome by cleaving an adenine from the rRNA,41 the two toxins show a similar and irreversible mode of action. The activation of unfolded protein response (UPR), as cause for cell death by SubA,36 distinguishes it clearly from the other toxins. As the induction of the UPR also increases the expression of GRP78,42 the substrate of SubA,36 it seems possible that UPR-induced apoptosis can be evaded. This might account for the cell-dependent differences in effectiveness observed when comparing SubA with DTA and AA. As the molecular weight of AA compared to DTA cmRNA is higher (AA, 1.0 kb, versus DTA, 0.8 kb), AA was more effective than DTA when considering molecular toxicity. As one challenge of successful mRNA-based therapy is transfection efficiency, high effectivity per mRNA molecule is desirable. Moreover, with high molecular toxicity, comparably lower doses of AA are sufficient, thereby reducing potential toxic side effects of mRNA delivery.
To ensure that the various effects caused by transfection of AA were specific, a control cmRNA (AAstop) was applied in this study. AAstop displays the same sequence as AA but has a scrambled Kozak39 element, and the start as well as all in-frame downstream ATGs mutated to TAGs. Prior to translation, the 40 S ribosome scans the mRNA sequence, starting at the 5′ end, and initiates translation when it reaches the first AUG codon.39 Binding to this first AUG by the ribosome is strongly supported by a consensus sequence (Kozak element) directly upstream. Consequently, as could be asserted in this study, the introduced alterations in AAstop cmRNA prevented its translation.
The successful expression of abrin-a protein after transfection of KB cells with AA cmRNA was demonstrated by western blot analysis. At the lower dose of 40 ng cmRNA, protein production was reduced compared to 80 ng cmRNA.
A rabbit reticulocyte lysate system assessed that AA almost completely inhibits protein synthesis of co-applied luc cmRNA. Moreover, fluorescence inhibition of co-transfected EGFP cmRNA confirmed that transfected AA cmRNA was translated into an active toxin. For 0.05 ng dose, only one-fourth of transfected cells were EGFP positive with a weak EGFP signal (low MFI). In those cells, the comparably low amount of AA presumably was not able to inactivate all ribosomes before enough EGFP molecules for fluorescence detection had been synthesized. In contrast, none of the cells transfected with 5 ng AA cmRNA showed detectable EGFP fluorescence. This dose dependency of translational inhibition by abrin has been shown before.43 The results demonstrate that AA exerts its influence shortly after the start of translation. As AA directly inhibits protein synthesis by cleaving an adenine from the 28 S rRNA,41 thereby immediately blocking translation, such immediate effects on translation are conceivable. Also, Hung et al.44 detected depurination of isolated rat liver ribosomes already 15 min after treatment with the A-chain of the protein abrin-a. Previous studies demonstrated that inhibition of protein synthesis is the main reason for abrin-a-induced cell death.43
At low doses (9,100 molecules AA cmRNA per cell), translation of EGFP cmRNA was inhibited substantially, while cell death was only detected at higher doses. This discrepancy indicates that higher amounts of AA are needed to disturb protein metabolism to such an extent that it results in cell death. Furthermore, it has been assumed that abrin-a can induce cell death independent of inhibition of protein synthesis.45, 46 Potentially, higher concentrations of abrin-a are necessary for this toxicity-enhancing effect. In accordance with what has been reported by different groups,45, 47 this study demonstrates that abrin-a induces apoptotic cell death. Caspase-3 activation has been shown to be a key component of abrin induced apoptosis with peak activation ranging from 18 hr to 48 hr.47, 48 Cells undergoing apoptosis bind to Annexin V but are impermeable to PI.49 Qu and Qing45 showed Annexin V-positive but PI-negative cells at 8–36 hr post-treatment with abrin, while at 40 hr the majority of cells was positive for Annexin V and PI. They also detected DNA fragmentation 15 hr after exposure of cells to abrin, as was likewise shown by other groups at 12 hr and 24 hr.47, 50 After staining with Hoechst, chromatin condensation has been observed in previous studies.45 While it is established that cells treated with abrin-a undergo apoptosis, different pathways have been proposed. Qu and Qing45 suggest that the inhibition of protein synthesis and mitochondrial membrane damage after reactive oxygen species (ROS) production present two independent pathways. The intrinsic mitochondrial pathway has been confirmed by various groups.47, 50 Saxena et al.48 showed the involvement of the Fas ligand and thereby of the extrinsic pathway after exposure to abrin.
In this study, besides toxicity in vitro, tumor growth in mice could be diminished considerably by four intratumoral injections of 10 μg formulated AA cmRNA. Twelve days after start of the treatment, the volume of tumors treated with AA-LF132 was significantly reduced by 89% compared to tumors that had been injected with AAstop-LF132. In comparison, a previous study using plasmid DNA coding for the A-chain of diphtheria toxin under a target-cell-specific promoter could diminish tumor size by 68% compared to the luc-expressing control plasmid group.51 To achieve these effects, three injections, each with 25 μg plasmid, were employed. Ramnath et al.52 showed a reduction in tumor volume in mice up to 62% compared to the control group after five intralesional treatments with the protein abrin-a. Another group could show that by single injection of an immunotoxin containing the A-chain of abrin-a, tumor growth in mice could be delayed by 7 to 10 days.28
For clinical application, a neoadjuvant therapy can be envisaged which comprises intratumoral co-administration of a toxin-encoding mRNA with an immune-stimulatory agent analogous to the employment of armed oncolytic viruses.53 Their anti-tumor potency could be extensively enhanced by inclusion of chemokines, cytokines or tumor-associated antigens.54 Due to its immunological properties, the mRNA molecule itself might serve as adjuvant.55 Conceivably, this approach could be pursued prior to surgical removal of a primary tumor with the aim of inducing tumor shrinkage and a systemic immune response to disseminated tumor cells. Besides direct activation of the immune system, blockage of molecules inhibiting T cell differentiation or function, e.g. PD-1, displays a promising tool for stimulation of the immune system.56
The present study clearly showed that cmRNA encoding for the A-chain of abrin-a toxin displays effective anti-tumor properties. By repeated injections of complexed cmRNA into tumors in mice, tumor growth could be constrained in a manner comparable to previous in vivo studies applying abrin-a or toxin-encoding plasmids. The employment of mRNA is very attractive, as it shows various safety-relevant benefits compared to pDNA and limited toxicity has been associated with immunotoxins. The promising results obtained with AA prompt further studies using different tumor models to fully appreciate the anti-tumor efficacies of toxin encoding cmRNAs.
Materials and Methods
Plasmid Preparation
The toxin (SubA, DTA, AA) and control (SubAstop, DTAstop, AAstop) sequences were cloned at the KpnI site (Thermo Fisher, Waltham, MA) into the backbone pVAX1-A120.24 Toxin and control sequences codon optimized for expression in Homo sapiens were produced by GeneArt in two parts. Sequences were retrieved from NCBI GenBank (SubA, AF399919.3; DTA, K01722.1; AA, AY458627.1). Only the A-chain of the toxins was utilized. Subsequent sub-cloning into pVAX1-A120 was performed using the GeneArt Seamless Cloning and Assembly Enzyme Mix (Invitrogen, Darmstadt, Germany) and One Shot TOP10 Chemically Competent E. coli (Invitrogen, Darmstadt, Germany).
Generation of cmRNA
DNA plasmids were linearized downstream of the poly(A) tail with the restriction enzyme NotI (Thermo Fisher, Waltham, MA), and purification was performed by chloroform extraction and ethanol precipitation. The linearized plasmids were used as template for in vitro transcription (IVT). IVT-mix was as follows: 0.1 μg/μL plasmid, transcription buffer 1 (Ethris, Planegg, Germany), 1 U/μL RiboLock Rnase Inhibitor (Thermo Fisher, Waltham, MA), 0.015 U/μL Inorganic Pyrophosphatase 1 (Thermo Fisher, Waltham, MA), 2 U/μL T7 Polymerase (Thermo Fisher, Waltham, MA), 7.5 mM rATP, 7.5 mM rGTP, 5.6 mM rCTP, 5.6 mM rUTP, 1.9 mM 5′-methyl-rCTP, and 1.9 mM 2′-thio-rUTP. The nucleotides were purchased from Jena Biosciences (Jena, Germany). The complete IVT-mix was incubated at 37°C for 4.5 hr, following which 1 U/μL DNase I (Thermo Fisher, Waltham, MA) was added to remove the plasmid template and the reaction incubated an additional 25 min at 37°C. RNA was precipitated with ammonium acetate at a final concentration of 2.5 mM and washed twice with 70% ethanol before it was re-suspended in aqua ad injectabilia. RNA concentration was measured on a spectrophotometric device (Nanodrop) and its correct size and purity determined with 1% agarose gel and RiboRuler High Range RNA Ladder (Thermo Fisher, Waltham, MA). For subsequent capping, cmRNA was employed at a final concentration of 1 mg/mL and denaturated in advance at 65°C for 15 min. The capping reaction mix contained 1× capping buffer (NEB, Frankfurt, Germany), 0.5 mM GTP (NEB, Frankfurt, Germany), 0.2 mM S-Methyladenosine (NEB, Frankfurt, Germany), 0.5 U/μL Vaccinia Virus Capping Enzyme (NEB, Frankfurt, Germany), 2.5 U/μL mRNA Cap 2′-o-Methyltransferase (NEB, Frankfurt, Germany), and 1 U/μL RiboLock RNase Inhibitor (Thermo Fisher, Waltham, MA). Reaction mix was incubated at 37°C for 60 min before the RNA was precipitated with ammonium acetate at a final concentration of 2.5 mM and washed twice with 70% ethanol before it was re-suspended in aqua ad injectabilia.
Cell-free Protein Synthesis System
In order to determine protein synthesis of luciferase firefly from luc cmRNA (kindly provided by Ethris, Planegg, Germany) in a cell-free system, the Rabbit Reticulocyte Lysate System (Promega, Madison, WI) was applied according to the manufacturer’s instructions. Per reaction of 35 μL lysate, 1 μg luc cmRNA and either 0.1 μg AA or 0.1 μg AAstop cmRNA were employed simultaneously. After an incubation of 45 min at 30°C, luminescence was measured using Tecan InfiniteR 200 PRO (Tecan, Männedorf, Switzerland).
Cell Culture
Cells were maintained in a humidified incubator at 37°C and 5% CO2. HuH7 cells (CSC-C9441L, Creative Bioarray, Shirley, NY) were cultured in DMEM (1×) + GlutaMax supplemented with 10% fetal bovine serum (FBS) and 1% Pen/Strep. For KB cells (ACC-136, German Collection of Microorganisms and Cell Cultures [DSMZ], Braunschweig, Germany), RPMI1640 (1×) + GlutaMax supplemented with 10% FBS and 1% Pen/Strep was used as culture medium. Both cell lines were authenticated by DSMZ.
Transfections in Cell Culture
If not stated otherwise, transfections were performed with Lipofectamine 2000 (Invitrogen, Darmstadt, Germany) according to the manufacturer’s instructions using 2 μL Lipofectamine 2000 per μg mRNA in medium without additives. Transfections were performed in 96-well plates and 100 μL final volume of medium per well, unless specified differently. 1 × 104 KB cells and 3 × 103 Huh7 cells per 100 μL medium were used in all in vitro experiments but for the western blot, where 2 × 104 cells per 100 μL medium were applied. For co-transfections, 50 ng/100 μL of EGFP cmRNA (kindly provided by Ethris, Planegg, Germany) and either 5, 0.05, or 0.0005 ng/100 μL of AA or AAstop cmRNAs were transfected. For western blot detection, 40 and 80 ng/100 μL of AA or AAstop cmRNA were used for transfection. In all other experiments, 10 ng, 50 ng, or 100 ng of mRNA were applied in a final volume of 100 μL. At 4 hr after transfection, medium was discarded, and supplemented cell culture medium was added. For in vivo purposes, cmRNAs (AA and AAstop) were formulated using Ethris’ (Planegg, Germany) proprietary cationic lipid formulation LF132, which was based on Jarzebińska et al.57 In order to examine their effectiveness in vitro, cells were transfected with 10, 50, and 100 ng/100 μL of AA-LF132 or AAstop-LF132. As vehicle control, 2% sucrose (diluted accordingly) was applied. Just like with transfections using Lipofectamine 2000, at 4 hr after transfection medium was discarded and supplemented cell culture medium added.
Cell Culture Assays
For all assays, luminescence was measured using Tecan InfiniteR 200 PRO (Tecan, Männedorf, Switzerland). The Attune NxT (Thermo Fisher, Waltham, MA) was employed for flow cytometery analysis. Microscopic images were taken with Leica DMi8 (Leica, Wetzlar, Germany). Except for the western blot analysis (two independent technical replicates), all cell culture assays were performed in three independent biological replicates, each treatment in triplicates.
SDS-PAGE and Western Blot
For western blot detection, cells were seeded in 6-well plates. The assay was conducted 24 hr post-transfection. After washing the cells with cold PBS, they were detached from the well using cell scrapers and lysed in lysis buffer (25 mM Tris-HCl, 0.1% Triton X-100 [pH: 7.8], 1× cOmplete protease inhibitor [Sigma Aldrich, Steinheim, Germany], 150 U/mL DNase I [Thermo Fisher, Waltham, MA]) for 30 min on ice. Total protein content was determined by the bicinchoninic acid (BCA) assay, following the manufacturer’s instructions (Thermo Fisher, Waltham, MA). Cell lysates (45 μg of total protein per sample) were separated on 4%–12% polyacrylamide gels (Thermo Fisher, Waltham, MA) and transferred to polyvinylidene fluoride (PVDF) membranes (Bio-Rad). Membranes were blocked in Western Breeze blocking solution (Thermo Fisher, Waltham, MA) and probed with antibodies against abrin-a (6.7 μg/mL, Tetracore, Rockville, MD) and vinculin (1:10,000, Abcam, Cambridge, UK). For protein detection, horseradish peroxidase (HRP)-conjugated anti-rabbit (1:10,000, sc2004; Santa Cruz Biotechnology, Dallas, TX) antibodies were added. For signal detection, Luminata Western HRP substrate was applied according to the manufacturer’s protocol (Merck Chemicals, Darmstadt, Germany).
Protein Synthesis Inhibition
To determine EGFP expression 24 hr post-transfection, culture medium was put aside, cells trypsinized with TrypLE Express Enzyme (Thermo Fisher, Waltham, MA), re-suspended in the cell culture medium, and analyzed by flow cytometry. Analysis was performed on singlet cells.
Cell Viability/Toxicity Assays
Assays were conducted 48 hr post-transfection. To determine cell viability, the CellTiter-Glo Luminescent Cell Viability Assay (Promega, Madison, WI) was performed according to the manufacturer’s instruction. Before addition of the substrate, the plate was centrifuged for 5 min at 1,100 rpm, the medium discarded, and fresh supplemented medium added. For dead cell staining, culture medium was put aside, and cells were trypsinized with TrypLE Express Enzyme (Thermo Fisher, Waltham, MA) and re-suspended in the cell culture medium. PBS containing 5 μg/mL PI (Sigma Aldrich, Steinheim, Germany) was added at 1:5 (v/v) to the cell suspension before analysis with flow cytometry. Analysis was performed on singlet cells.
Apoptotic Assays
Annexin V-PI double staining was performed 24 hr post-transfection. Annexin V, Alexa Fluor 488 conjugate (Thermo Fisher, Waltham, MA) was used according to the manufacturer’s instruction, and PI solution (Sigma Aldrich, Steinheim Germany) was applied at 1 μg/mL. Analysis was performed on singlet cells. For this assay, 24-well plates, containing 4 × 104 cells per well in a final volume of 400 μL medium, were used. Activity of caspases-3 and -7 was assessed 24 hr post-transfection with the Caspase-Glo 3/7 Assay (Promega, Madison, WI) according to the manufacturer’s instruction. At 48 hr post-transfection, Hoechst33258 (Thermo Fisher, Waltham, MA) was employed at 10 μg/mL in supplemented cell culture medium and cells incubated for 15 min at room temperature. Before examination, Hoechst33258 was discarded, and fresh supplemented cell culture medium was added. Of each replicate, nine images at 20× magnification were taken, and the number of Hoechst33258-positive cells was counted.
Expression of Firefly Luciferase in the Tumor and Reduction of Tumor Growth in Mice
5 × 106 KB cells were injected into the flank of female, 6-week-old NMRI-nu mice (RjOrl:NMRI-Foxn1nu /Foxn1nu, Janvier Labs, Saint Berthevin Cedex, France). For the pre-experiment, 10 μg cmRNA coding for firefly luciferase (kindly provided by Ethris, Planegg, Germany) in 50 μL 2% sucrose was injected into the formed tumors on days 9, 11, and 13 after injection of tumor cells. Twenty-four hours after the third application, luminescence was measured using an IVIS In Vivo Imaging System with Living Image software 3.2 (Caliper Life Sciences, Mountain View, CA). 100 μl luciferin (60 mg/mL in PBS) were injected intraperitoneally 15 min prior to imaging. For the reduction of tumor growth, the cmRNAs (AA and AAstop) were formulated in a proprietary lipid formulation (LF132) by Ethris (Planegg, Germany) in 2% sucrose (Sigma Aldrich, Steinheim, Germany). The cationic lipid formulation LF132 was based on Jarzebińska et al.57 Treatment was started as soon as tumors had reached a sufficient size (100 mm3, day 9). 50 μl of solution containing either 10 μg of AA-LF132, 10 μg of AAstop-LF132 or 2% sucrose were injected intratumorally on days 9, 11, 13, and 18 post-injection of tumor cells. Throughout the experiment, tumor volume was determined with a caliper using the formula a × b2/2, with a indicating the length of the tumor and b the width. On day 21 after injection of tumor cells, mice were euthanized by cervical dislocation. Tumor was explanted, and tumor size was measured again using the formula a × b × c/2. The protocols for animal experiments were approved by the animal ethics committee and the government of Oberbayern (May 26, 2014; Permit Number, Az. 55.2-1-54-2531-53-09), and experiments were executed in accordance with the German Animal Welfare Law.
Data Analysis
Flow cytometry data was analyzed by FlowJo v.10.0.8 (FlowJo, Ashland, OR). GraphPad Prism 6 (GraphPad Software, La Jolla, CA) was utilized for statistical analysis.
Author Contributions
K.H. performed the experiments and wrote the manuscript. C.D. developed the LF132 formulation and contributed together with A.H.-J. to the planning of the in vivo experiment. A.J. formulated the RNAs and contributed to the cultivation of the cells for the in vivo experiment. E.W. contributed to the planning of the in vivo experiment and supervised the in vivo experiment. E.K. contributed to the planning of the in vivo experiment and performed the in vivo experiment. V.K. supervised the project and together with M.K.A. provided intellectual input and contributed to the manuscript. C.R. together with C.P. supervised the work, provided input on the experimental design, and contributed to and edited the manuscript.
Conflicts of Interest
C.P. and C.R. are founders and shareholders of Ethris GmbH, a company that develops mRNA therapeutics. K.H., C.D., A.H-.J., V.K., and M.K.A. are employees of Ethris GmbH.
Acknowledgments
Financial support by Ethris GmbH and the German Research Foundation, Excellence Cluster “Nanosystems Initiative Munich (NIM),” is gratefully acknowledged. We thank Dr. Daniela Emrich and Sarah Kern for their valuable contribution to the in vivo experiment.
References
- 1.Sharom F.J. ABC multidrug transporters: structure, function and role in chemoresistance. Pharmacogenomics. 2008;9:105–127. doi: 10.2217/14622416.9.1.105. [DOI] [PubMed] [Google Scholar]
- 2.Rich J.N. Cancer stem cells in radiation resistance. Cancer Res. 2007;67:8980–8984. doi: 10.1158/0008-5472.CAN-07-0895. [DOI] [PubMed] [Google Scholar]
- 3.Monsuez J.J., Charniot J.C., Vignat N., Artigou J.Y. Cardiac side-effects of cancer chemotherapy. Int. J. Cardiol. 2010;144:3–15. doi: 10.1016/j.ijcard.2010.03.003. [DOI] [PubMed] [Google Scholar]
- 4.Monje M., Dietrich J. Cognitive side effects of cancer therapy demonstrate a functional role for adult neurogenesis. Behav. Brain Res. 2012;227:376–379. doi: 10.1016/j.bbr.2011.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kreitman R.J., Stetler-Stevenson M., Margulies I., Noel P., Fitzgerald D.J., Wilson W.H., Pastan I. Phase II trial of recombinant immunotoxin RFB4(dsFv)-PE38 (BL22) in patients with hairy cell leukemia. J. Clin. Oncol. 2009;27:2983–2990. doi: 10.1200/JCO.2008.20.2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eklund J.W., Kuzel T.M. Denileukin diftitox: a concise clinical review. Expert Rev. Anticancer Ther. 2005;5:33–38. doi: 10.1586/14737140.5.1.33. [DOI] [PubMed] [Google Scholar]
- 7.Kreitman R.J. Immunotoxins for targeted cancer therapy. AAPS J. 2006;8:E532–E551. doi: 10.1208/aapsj080363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pastan I. Targeted therapy of cancer with recombinant immunotoxins. Biochim. Biophys. Acta. 1997;1333:C1–C6. doi: 10.1016/s0304-419x(97)00021-8. [DOI] [PubMed] [Google Scholar]
- 9.Potala S., Sahoo S.K., Verma R.S. Targeted therapy of cancer using diphtheria toxin-derived immunotoxins. Drug Discov. Today. 2008;13:807–815. doi: 10.1016/j.drudis.2008.06.017. [DOI] [PubMed] [Google Scholar]
- 10.Pai-Scherf L.H., Villa J., Pearson D., Watson T., Liu E., Willingham M.C., Pastan I. Hepatotoxicity in cancer patients receiving erb-38, a recombinant immunotoxin that targets the erbB2 receptor. Clin. Cancer Res. 1999;5:2311–2315. [PubMed] [Google Scholar]
- 11.Pastan I., Hassan R., FitzGerald D.J., Kreitman R.J. Immunotoxin treatment of cancer. Annu. Rev. Med. 2007;58:221–237. doi: 10.1146/annurev.med.58.070605.115320. [DOI] [PubMed] [Google Scholar]
- 12.Gofrit O.N., Benjamin S., Halachmi S., Leibovitch I., Dotan Z., Lamm D.L., Ehrlich N., Yutkin V., Ben-Am M., Hochberg A. DNA based therapy with diphtheria toxin-A BC-819: a phase 2b marker lesion trial in patients with intermediate risk nonmuscle invasive bladder cancer. J. Urol. 2014;191:1697–1702. doi: 10.1016/j.juro.2013.12.011. [DOI] [PubMed] [Google Scholar]
- 13.Mizrahi A., Czerniak A., Levy T., Amiur S., Gallula J., Matouk I., Abu-lail R., Sorin V., Birman T., de Groot N. Development of targeted therapy for ovarian cancer mediated by a plasmid expressing diphtheria toxin under the control of H19 regulatory sequences. J. Transl. Med. 2009;7:69. doi: 10.1186/1479-5876-7-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang Y.H., Wang Y., Yusufali A.H., Ashby F., Zhang D., Yin Z.F., Aslanidi G.V., Srivastava A., Ling C.Q., Ling C. Cytotoxic genes from traditional Chinese medicine inhibit tumor growth both in vitro and in vivo. J. Integr. Med. 2014;12:483–494. doi: 10.1016/s2095-4964(14)60057-1. [DOI] [PubMed] [Google Scholar]
- 15.Hacein-Bey-Abina S., Von Kalle C., Schmidt M., McCormack M.P., Wulffraat N., Leboulch P., Lim A., Osborne C.S., Pawliuk R., Morillon E. LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1. Science. 2003;302:415–419. doi: 10.1126/science.1088547. [DOI] [PubMed] [Google Scholar]
- 16.Rejman J., Tavernier G., Bavarsad N., Demeester J., De Smedt S.C. mRNA transfection of cervical carcinoma and mesenchymal stem cells mediated by cationic carriers. J. Control. Release. 2010;147:385–391. doi: 10.1016/j.jconrel.2010.07.124. [DOI] [PubMed] [Google Scholar]
- 17.Yamamoto A., Kormann M., Rosenecker J., Rudolph C. Current prospects for mRNA gene delivery. Eur. J. Pharm. Biopharm. 2009;71:484–489. doi: 10.1016/j.ejpb.2008.09.016. [DOI] [PubMed] [Google Scholar]
- 18.Sahin U., Karikó K., Türeci Ö. mRNA-based therapeutics—developing a new class of drugs. Nat. Rev. Drug Discov. 2014;13:759–780. doi: 10.1038/nrd4278. [DOI] [PubMed] [Google Scholar]
- 19.Wang Y., Su H.H., Yang Y., Hu Y., Zhang L., Blancafort P., Huang L. Systemic delivery of modified mRNA encoding herpes simplex virus 1 thymidine kinase for targeted cancer gene therapy. Mol. Ther. 2013;21:358–367. doi: 10.1038/mt.2012.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wolff J.A., Malone R.W., Williams P., Chong W., Acsadi G., Jani A., Felgner P.L. Direct gene transfer into mouse muscle in vivo. Science. 1990;247:1465–1468. doi: 10.1126/science.1690918. [DOI] [PubMed] [Google Scholar]
- 21.Kariko K., Muramatsu H., Welsh F.A., Ludwig J., Kato H., Akira S., Weissman D. Incorporation of pseudouridine into mRNA yields superior nonimmunogenic vector with increased translational capacity and biological stability. Mol. Ther. 2008;16:1833–1840. doi: 10.1038/mt.2008.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Naylor R., Ho N.W., Gilham P.T. Selective chemical modifications of uridine and pseudouridine in polynucleotides and their effect on the specificities of ribonuclease and phosphodiesterases. J. Am. Chem. Soc. 1965;87:4209–4210. doi: 10.1021/ja01096a050. [DOI] [PubMed] [Google Scholar]
- 23.Anderson B.R., Muramatsu H., Jha B.K., Silverman R.H., Weissman D., Karikó K. Nucleoside modifications in RNA limit activation of 2′-5′-oligoadenylate synthetase and increase resistance to cleavage by RNase L. Nucleic Acids Res. 2011;39:9329–9338. doi: 10.1093/nar/gkr586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kormann M.S., Hasenpusch G., Aneja M.K., Nica G., Flemmer A.W., Herber-Jonat S., Huppmann M., Mays L.E., Illenyi M., Schams A. Expression of therapeutic proteins after delivery of chemically modified mRNA in mice. Nat. Biotechnol. 2011;29:154–157. doi: 10.1038/nbt.1733. [DOI] [PubMed] [Google Scholar]
- 25.Nallagatla S.R., Bevilacqua P.C. Nucleoside modifications modulate activation of the protein kinase PKR in an RNA structure-specific manner. RNA. 2008;14:1201–1213. doi: 10.1261/rna.1007408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Backer J.M., Krivoshein A.V., Hamby C.V., Pizzonia J., Gilbert K.S., Ray Y.S., Brand H., Paton A.W., Paton J.C., Backer M.V. Chaperone-targeting cytotoxin and endoplasmic reticulum stress-inducing drug synergize to kill cancer cells. Neoplasia. 2009;11:1165–1173. doi: 10.1593/neo.09878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gadadhar S., Karande A.A. Abrin immunotoxin: targeted cytotoxicity and intracellular trafficking pathway. PLoS ONE. 2013;8:e58304. doi: 10.1371/journal.pone.0058304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wawrzynczak E.J., Zangemeister-Wittke U., Waibel R., Henry R.V., Parnell G.D., Cumber A.J., Jones M., Stahel R.A. Molecular and biological properties of an abrin A chain immunotoxin designed for therapy of human small cell lung cancer. Br. J. Cancer. 1992;66:361–366. doi: 10.1038/bjc.1992.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pappenheimer A.M., Jr. Diphtheria toxin. Annu. Rev. Biochem. 1977;46:69–94. doi: 10.1146/annurev.bi.46.070177.000441. [DOI] [PubMed] [Google Scholar]
- 30.Paton A.W., Srimanote P., Talbot U.M., Wang H., Paton J.C. A new family of potent AB(5) cytotoxins produced by Shiga toxigenic Escherichia coli. J. Exp. Med. 2004;200:35–46. doi: 10.1084/jem.20040392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lin J.Y., Lee T.C., Hu S.T., Tung T.C. Isolation of four isotoxic proteins and one agglutinin from jequiriti bean (Abrus precatorius) Toxicon. 1981;19:41–51. doi: 10.1016/0041-0101(81)90116-1. [DOI] [PubMed] [Google Scholar]
- 32.Naglich J.G., Metherall J.E., Russell D.W., Eidels L. Expression cloning of a diphtheria toxin receptor: identity with a heparin-binding EGF-like growth factor precursor. Cell. 1992;69:1051–1061. doi: 10.1016/0092-8674(92)90623-k. [DOI] [PubMed] [Google Scholar]
- 33.Bagaria A., Surendranath K., Ramagopal U.A., Ramakumar S., Karande A.A. Structure-function analysis and insights into the reduced toxicity of Abrus precatorius agglutinin I in relation to abrin. J. Biol. Chem. 2006;281:34465–34474. doi: 10.1074/jbc.M601777200. [DOI] [PubMed] [Google Scholar]
- 34.Gill D.M. Academic Press; 1978. Seven Toxic Peptides That Cross Cell Membranes; pp. 291–322. [Google Scholar]
- 35.Frankel A.E., Rossi P., Kuzel T.M., Foss F. Diphtheria fusion protein therapy of chemoresistant malignancies. Curr. Cancer Drug Targets. 2002;2:19–36. doi: 10.2174/1568009023333944. [DOI] [PubMed] [Google Scholar]
- 36.Paton A.W., Beddoe T., Thorpe C.M., Whisstock J.C., Wilce M.C., Rossjohn J., Talbot U.M., Paton J.C. AB5 subtilase cytotoxin inactivates the endoplasmic reticulum chaperone BiP. Nature. 2006;443:548–552. doi: 10.1038/nature05124. [DOI] [PubMed] [Google Scholar]
- 37.Endo Y., Tsurugi K. RNA N-glycosidase activity of ricin A-chain. Mechanism of action of the toxic lectin ricin on eukaryotic ribosomes. J. Biol. Chem. 1987;262:8128–8130. [PubMed] [Google Scholar]
- 38.Frankel A.E., Powell B.L., Hall P.D., Case L.D., Kreitman R.J. Phase I trial of a novel diphtheria toxin/granulocyte macrophage colony-stimulating factor fusion protein (DT388GMCSF) for refractory or relapsed acute myeloid leukemia. Clin. Cancer Res. 2002;8:1004–1013. [PubMed] [Google Scholar]
- 39.Kozak M. Initiation of translation in prokaryotes and eukaryotes. Gene. 1999;234:187–208. doi: 10.1016/s0378-1119(99)00210-3. [DOI] [PubMed] [Google Scholar]
- 40.Van der Jeught K., Joe P.T., Bialkowski L., Heirman C., Daszkiewicz L., Liechtenstein T., Escors D., Thielemans K., Breckpot K. Intratumoral administration of mRNA encoding a fusokine consisting of IFN-β and the ectodomain of the TGF-β receptor II potentiates antitumor immunity. Oncotarget. 2014;5:10100–10113. doi: 10.18632/oncotarget.2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Endo Y., Glück A., Wool I.G. Ribosomal RNA identity elements for ricin A-chain recognition and catalysis. J. Mol. Biol. 1991;221:193–207. doi: 10.1016/0022-2836(91)80214-f. [DOI] [PubMed] [Google Scholar]
- 42.Ye J., Rawson R.B., Komuro R., Chen X., Davé U.P., Prywes R., Brown M.S., Goldstein J.L. ER stress induces cleavage of membrane-bound ATF6 by the same proteases that process SREBPs. Mol. Cell. 2000;6:1355–1364. doi: 10.1016/s1097-2765(00)00133-7. [DOI] [PubMed] [Google Scholar]
- 43.Mishra R., Kumar M.S., Karande A.A. Inhibition of protein synthesis leading to unfolded protein response is the major event in abrin-mediated apoptosis. Mol. Cell. Biochem. 2015;403:255–265. doi: 10.1007/s11010-015-2355-9. [DOI] [PubMed] [Google Scholar]
- 44.Hung C.H., Lee M.C., Chen J.K., Lin J.Y. Cloning and expression of three abrin A-chains and their mutants derived by site-specific mutagenesis in Escherichia coli. Eur. J. Biochem. 1994;219:83–87. doi: 10.1111/j.1432-1033.1994.tb19917.x. [DOI] [PubMed] [Google Scholar]
- 45.Qu X., Qing L. Abrin induces HeLa cell apoptosis by cytochrome c release and caspase activation. J. Biochem. Mol. Biol. 2004;37:445–453. doi: 10.5483/bmbrep.2004.37.4.445. [DOI] [PubMed] [Google Scholar]
- 46.Narayanan S., Surendranath K., Bora N., Surolia A., Karande A.A. Ribosome inactivating proteins and apoptosis. FEBS Lett. 2005;579:1324–1331. doi: 10.1016/j.febslet.2005.01.038. [DOI] [PubMed] [Google Scholar]
- 47.Narayanan S., Surolia A., Karande A.A. Ribosome-inactivating protein and apoptosis: abrin causes cell death via mitochondrial pathway in Jurkat cells. Biochem. J. 2004;377:233–240. doi: 10.1042/BJ20030797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Saxena N., Yadav P., Kumar O. The Fas/Fas ligand apoptotic pathway is involved in abrin-induced apoptosis. Toxicol. Sci. 2013;135:103–118. doi: 10.1093/toxsci/kft139. [DOI] [PubMed] [Google Scholar]
- 49.Koopman G., Reutelingsperger C.P., Kuijten G.A., Keehnen R.M., Pals S.T., van Oers M.H. Annexin V for flow cytometric detection of phosphatidylserine expression on B cells undergoing apoptosis. Blood. 1994;84:1415–1420. [PubMed] [Google Scholar]
- 50.Bhutia S.K., Mallick S.K., Maiti S., Mishra D., Maiti T.K. Abrus abrin derived peptides induce apoptosis by targeting mitochondria in HeLa cells. Cell Biol. Int. 2009;33:720–727. doi: 10.1016/j.cellbi.2009.04.012. [DOI] [PubMed] [Google Scholar]
- 51.Amit D., Tamir S., Birman T., Gofrit O.N., Hochberg A. Development of targeted therapy for bladder cancer mediated by a double promoter plasmid expressing diphtheria toxin under the control of IGF2-P3 and IGF2-P4 regulatory sequences. Int. J. Clin. Exp. Med. 2011;4:91–102. [PMC free article] [PubMed] [Google Scholar]
- 52.Ramnath V., Kuttan G., Kuttan R. Antitumour effect of abrin on transplanted tumours in mice. Indian J. Physiol. Pharmacol. 2002;46:69–77. [PubMed] [Google Scholar]
- 53.Cattaneo R., Miest T., Shashkova E.V., Barry M.A. Reprogrammed viruses as cancer therapeutics: targeted, armed and shielded. Nat. Rev. Microbiol. 2008;6:529–540. doi: 10.1038/nrmicro1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Guo Z.S., Liu Z., Bartlett D.L. Oncolytic immunotherapy: dying the right way is a key to eliciting potent antitumor immunity. Front. Oncol. 2014;4:74. doi: 10.3389/fonc.2014.00074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tavernier G., Andries O., Demeester J., Sanders N.N., De Smedt S.C., Rejman J. mRNA as gene therapeutic: how to control protein expression. J. Control. Release. 2011;150:238–247. doi: 10.1016/j.jconrel.2010.10.020. [DOI] [PubMed] [Google Scholar]
- 56.Palucka A.K., Coussens L.M. The basis of oncoimmunology. Cell. 2016;164:1233–1247. doi: 10.1016/j.cell.2016.01.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jarzębińska A., Pasewald T., Lambrecht J., Mykhaylyk O., Kümmerling L., Beck P., Hasenpusch G., Rudolph C., Plank C., Dohmen C. A single methylene group in oligoalkylamine-based cationic polymers and lipids promotes enhanced mRNA delivery. Angew. Chem. Int. Ed. Engl. 2016;55:9591–9595. doi: 10.1002/anie.201603648. [DOI] [PubMed] [Google Scholar]