Abstract
Single-cell genome sequencing has proven valuable for the detection of somatic variation, particularly in the context of tumor evolution. Current technologies suffer from high library construction costs which restrict the number of cells that can be assessed and thus impose limitations on the ability to measure heterogeneity within a tissue. Here, we present Single cell Combinatorial Indexed Sequencing (SCI-seq) as a means of simultaneously generating thousands of low-pass single cell libraries for somatic copy number variant detection. We constructed libraries for 16,698 single cells from a combination of cultured cell lines, primate frontal cortex tissue, and two human adenocarcinomas, including a detailed assessment of subclonal variation within a pancreatic tumor.
Introduction
Single cell sequencing has uncovered the breadth of genomic heterogeneity between cells in a variety of contexts, including somatic aneuploidy in the mammalian brain1–4 and intra-tumor heterogeneity5–8. Studies have taken one of two approaches: high depth of sequencing per cell for single nucleotide variant detection2,9, or low-pass sequencing to identify copy number variants (CNVs) and aneuploidy1,10,11. In the latter approach, the lack of an efficient, cost-effective method to produce large numbers of single cell libraries has made it difficult to quantify the frequency of CNV-harboring cells at population scale, or to provide a robust analysis of heterogeneity in the context of cancer12.
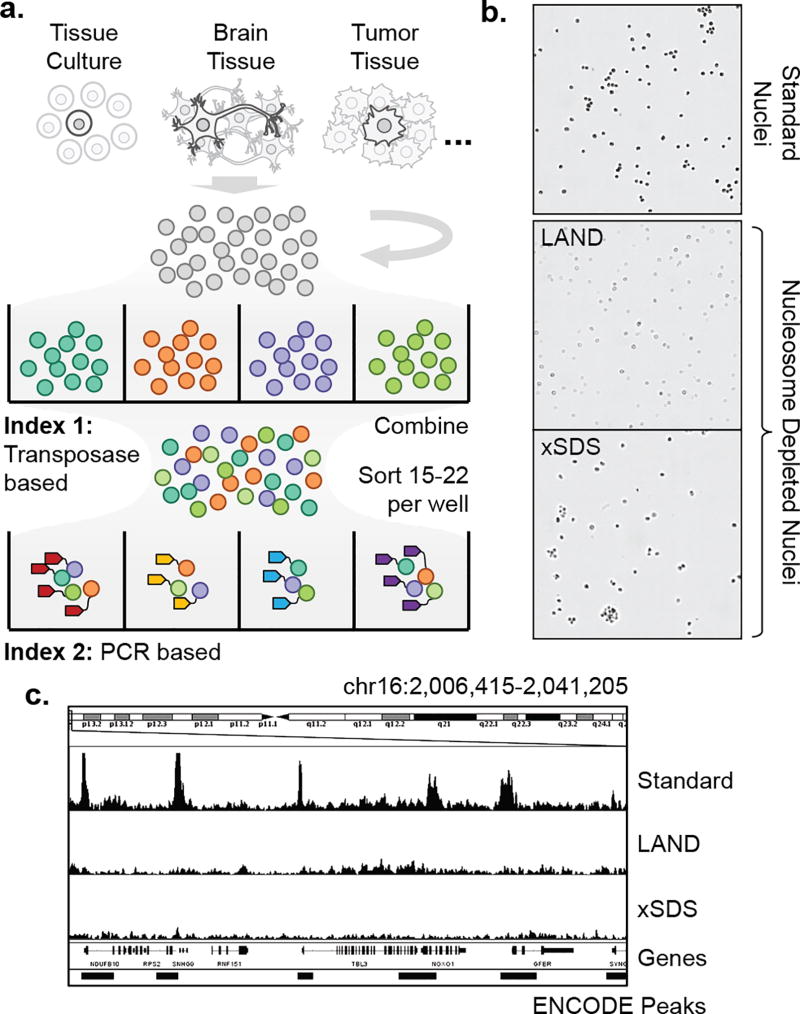
Recently, we established CPT-seq, a method to produce thousands of individually barcoded libraries of linked sequence reads using a transposase-based combinatorial indexing strategy13–15. We applied CPT-seq to the problem of genomic haplotype resolution14 and de novo genome assembly15. This concept was then integrated into the chromatin accessibility assay, ATAC-seq16, to produce profiles of active regulatory elements in thousands of single cells17 (sciATAC-seq, Fig. 1a). In combinatorial indexing, nuclei are first barcoded by the incorporation of one of 96 indexed sequencing adaptors via transposase. The 96 reactions are then combined and 15–25 of these randomly indexed nuclei are deposited into each well of a PCR plate by Fluorescence Activated Nuclei Sorting (FANS, Supplementary Fig. 1). The probability of any two nuclei having the same transposase barcode is therefore low (6–11%)17. Each PCR well is then uniquely barcoded using indexed primers. At the end of this process, each sequence read contains two indexes: Index 1 from the transposase plate, and Index 2 from the PCR plate, which facilitate single cell discrimination. As proof of principle, Cusanovich and colleagues produced over 15,000 sciATAC-seq profiles and used them to separate a mix of two cell types by their accessible chromatin landscapes17. We reasoned that a similar combinatorial indexing strategy could be extended to single cell whole genome sequencing.
Figure 1. Single cell combinatorial indexing with nucleosome depletion.
(a) Single cell combinatorial indexing workflow. (b) Phase contrast images of intact nuclei generated by standard isolation followed by nucleosome depletion using Lithium Assisted Nucleosome Depletion (LAND) or crosslinking and SDS treatment (xSDS). Scale bar: 100 µm. (c) Nucleosome depletion produces genome-wide uniform coverage that is not restricted to sites of chromatin accessibility.
Results
Nucleosome depletion for uniform genome coverage
The key hurdle to adapt combinatorial indexing to produce uniformly distributed sequence reads is the removal of nucleosomes bound to genomic DNA without compromising nuclear integrity. The sciATAC-seq method is carried out on native chromatin, which permits the conversion of DNA into library molecules only within regions of open chromatin (1–4% of the genome)18. This restriction is desirable for epigenetic characterization; however, for CNV detection, it results in biological bias and severely limited read counts (~3,000 per cell)17. We therefore developed two strategies to unbind nucleosomes from genomic DNA while retaining nuclear integrity for SCI-seq library construction. The first, Lithium Assisted Nucleosome Depletion (LAND), utilizes the chaotropic agent, Lithium diiodosalycylate, to disrupt DNA-protein interactions in the cell, therefore releasing DNA from histones. The second, crosslinking with SDS (xSDS), uses the detergent SDS to denature histone proteins and render them unable to bind DNA. However, SDS has a disruptive effect on nuclear integrity, thus necessitating a crosslinking step prior to denaturation in order to maintain intact nuclei.
To test the viability of these strategies, we performed bulk (30,000 nuclei) preparations on the HeLa S3 cell line, for which chromatin accessibility and genome structure has been extensively profiled19,20, and carried out LAND or xSDS treatments along with a standard control. In all three cases, nuclei remained intact – a key requirement for the SCI-seq workflow (Fig. 1b). Prepared nuclei were then carried through standard ATAC-seq library construction16. The library prepared from untreated nuclei produced the expected ATAC-seq signal with a 10.8 fold enrichment of sequence reads aligning to annotated HeLa S3 accessibility sites. Both the LAND and xSDS preparations had substantially lower enrichments of 2.8 and 2.2 fold respectively, close to the 1.4 fold observed for shotgun sequencing (Fig. 1c, Supplementary Table 1). Furthermore, the projected number of unique sequence reads present in the LAND and xSDS preparations were 1.7 billion and 798 million respectively, much greater than for the standard library at 170 million, suggesting a larger proportion of the genome was converted into viable sequencing molecules.
SCI-seq with nucleosome depletion
To assess the performance of nucleosome depletion with our single cell combinatorial indexing workflow, we first focused on the deeply profiled, euploid lymphoblastoid cell line GM1287814,15,19. We produced a total of six SCI-seq libraries with a variety of LAND conditions, each using a single 96-well plate at the PCR indexing stage, and a single xSDS library with 3×96-well PCR plates. To serve as a comparison to existing methods, we prepared 42 single cell libraries using quasi-random priming (QRP, 40 passing QC) and 51 using degenerate oligonucleotide primed PCR (DOP, 45 passing QC). Finally, we karyotyped 50 cells to serve as a non-sequencing means of aneuploidy measurement (Supplementary Table 2).
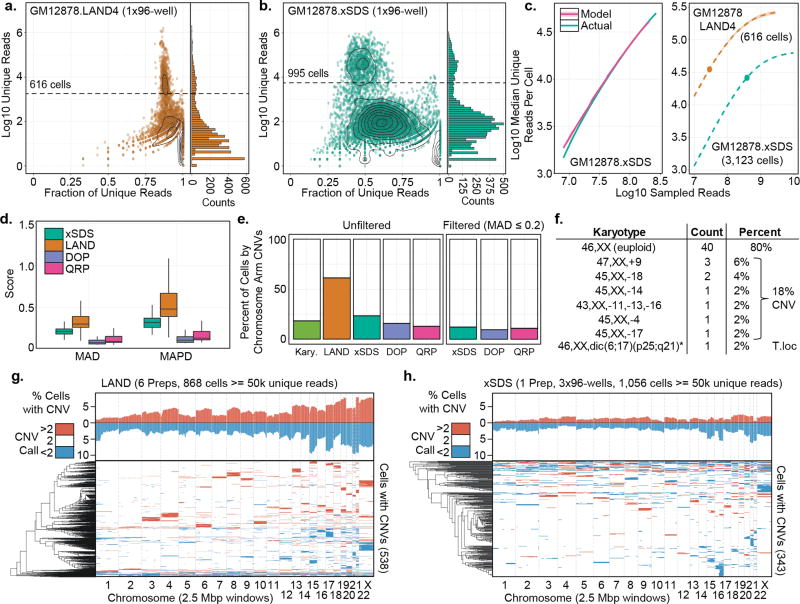
For each SCI-seq preparation, the number of potential index combinations is 96 (transposase indexing) × N (PCR indexing, 96 per plate); however, not all index combinations represent a single cell library, as each PCR well contains only 15–25 transposase-indexed nuclei. To identify non-empty index combinations, we generated a log10 transformed histogram of unique (i.e. non-PCR duplicate), high-quality (MQ ≥ 10) aligned reads for each potential index combination. This resulted in a bimodal distribution comprised of a low-read-count, noise component centered between 50 and 200 reads, and a high-read-count, single cell component centered between 10,000 and 100,000 reads (Fig 2a,b, Supplementary Fig. 2, Supplementary Software). We then used a mixed model to identify indexes that fall in this high-read-count component (Supplementary Fig. 3), which resulted in 4,643 single cell libraries across the six SCI-seq preparations that used LAND for nucleosome depletion and 3,123 for the xSDS preparation.
Figure 2. Comparison of LAND and xSDS nucleosome depletion methods with SCI-seq.
(a) Complexity for one of six LAND SCI-seq preparations on GM12878. Right panel, histogram showing distribution of read counts. Dashed line represents single cell read cutoff. (b) As in (a) but for xSDS nucleosome depletion for one of three PCR plates. (c) Left, model built on downsampled reads for the GM12878 xSDS preparation and used to predict the full depth of coverage. Right, Projections for one of the LAND preparations and the full xSDS preparation. Shading represents standard deviation over multiple models. Points represent actual depth of sequencing. (d) Coverage uniformity scores for SCI-seq using LAND or xSDS and for quasi-random priming (QRP) and degenerate oligonucleotide PCR (DOP). (e) Summary of the percentage of cells showing aneuploidy at the chromosome arm level across all preparations with and without imposing a variance filter. (f) Karyotyping results of 50 GM12878 cells. (g–h) Summary of windowed copy number calls and clustering of GM12878 single cells produced using LAND (g) or xSDS (h). Top represents a chromosome-arm scale summary of gain or loss frequency for all cells, bottom is the clustered profile for cells that contain at least one CNV call.
To confirm that the majority of putative single cell libraries contain true single cells, we carried out four SCI-seq library preparations on a mix of human and mouse cells using LAND (2,369 total cells) with either 22 or 25 nuclei per PCR well, and one preparation using xSDS split between two FANS conditions (1,367 total cells; Supplementary Figure 4). For each experiment we analyzed the proportion of putative single cells with ≥ 90% of their reads that aligned exclusively to the human or mouse genome. The remaining cells represent human-mouse collisions (i.e. doublets) and make up approximately half of the total collision rate (the remaining half being human-human or mouse-mouse). The total collision rates varied between 0–23.6%, and were used to decide upon 22 nuclei per well with restrictive sorting conditions for a target doublet frequency of <10%, comparable to sciATAC-seq17 or high throughout single cell RNA-seq technologies21.
The unique read count produced for each library in a SCI-seq preparation is a function of library complexity and sequencing depth. Due to the inhibitive cost of deeply sequencing every preparation during development, we implemented a model to project the anticipated read count and PCR duplicate percentage that would be achieved with increased sequencing depth (Fig. 2c, Methods). As a means of quality assessment, we identified the depth at which a median of 50% of reads across cells are PCR duplicates (M50), representing the point at which additional sequencing becomes excessive (i.e. greater than 50% of additional reads provide no new information), along with several other metrics (Supplementary Table 3). Model projections from a subset of the sequenced reads accurately predicted the actual median unique read count within a median of 0.02% (maximum 2.25%, mean 0.41%) across all libraries. As further confirmation, additional sequencing of a subset of PCR wells from several preparations produced unique reads counts for each cell that were within a median of 0.13% (maximum 3.56%, mean 0.72%) of what was predicted by our model (Supplementary Fig. 5).
Coverage uniformity was assessed using mean absolute deviation (MAD)22 and mean absolute pairwise deviation (MAPD)2, which indicated substantially better uniformity using xSDS over LAND (MAD: mean 1.57-fold improvement, p = <1×10−15; MAPD: 1.70-fold improvement, p = <1×10−15, Welch’s t-test). The deviation using xSDS is similar to multiple displacement amplification methods, though still greater than for QRP and DOP (Fig. 2d)22. While LAND preparations had higher coverage bias, they also produced higher unique read counts per cell (e.g. M50 of 763,813 for one of three HeLa LAND preparations) when compared to xSDS (e.g. M50 of 63,223 for the GM12878 preparation). For all libraries, we observed the characteristic 9 basepair overlap of adjacent read pairs due to the mechanism of transposition13,23, indicating we are able to sequence molecules on either side of a transposase insertion event (Supplementary Fig. 6).
Copy number variant calling using SCI-seq
For any single cell genome sequencing study, determining how to filter out failed libraries without removing true aneuploid cells is a significant challenge. We initially proceeded with CNV calling on our SCI-seq preparations without any filtering in order to directly compare with other methods. For all preparations, we used cells with a minimum of 50,000 unique, high quality aligned reads (868 across all LAND libraries, 1,056 for the xSDS library), applied Ginkgo22, Circular Binary Segmentation (CBS)24, and a Hidden Markov Model (HMM)25, with variable-sized genomic windows (target median of 2.5 million bp) for CNV calling (Supplementary Fig. 7) and conservatively retained the intersection of all three methods. To compare our sequencing-based calls with karyotyped cells, we focused on chromosome-arm level events (Fig. 2e,f). Consistent with the coverage uniformity differences, our LAND SCI-seq preparations produced a high aneuploidy rate (61.9%), suggesting an abundance of false positives due to lack of coverage uniformity (Fig. 2e,g). However, the xSDS nucleosome depletion strategy with SCI-seq resulted in an aneuploidy frequency of 22.6%, much closer to the karyotyping results (Fig. 2e,h) as well as DOP and QRP (15.0% and 13.5%, respectively) (Supplementary Fig. 8).
We next determined filtering criteria based on MAD and MAPD scores across a variety of resolutions and read count thresholds (Supplementary Fig. 9). This analysis revealed a greater range of variability in the resolution of our SCI-seq preparations, which is largely driven by the wider range of unique reads per cell when compared to standard methods. By applying a MAD variance filter of 0.2 across all methods, aneuploidy rates for xSDS, DOP and QRP dropped to 12.2%, 9.7% and 10.5% respectively, all below the rate determined by karyotyping, yet closer to one another than prior to filtering (Supplementary Fig. 10).
Copy number variation in the Rhesus brain
Estimates of aneuploidy and large-scale CNV frequencies in the mammalian brain vary widely, from <5% to 33%1–4. This uncertainty largely stems from the inability to profile sufficient numbers of single cells to produce quantitative measurements. The Rhesus macaque is an ideal model for quantifying the abundance of aneuploidy in the brain, as human samples are challenging to acquire and are confounded by high variability in lifetime environmental exposures. Furthermore, the Rhesus brain is phylogenetically, structurally and physiologically more similar to humans than rodents26.
To demonstrate the versatility of our platform, we applied LAND and xSDS SCI-seq to archived frontal cortex tissue (Individual 1), along with 38 cells using QRP (35 passing QC), and 35 cells using DOP (30 passing QC). Our low-capacity LAND preparation (16 PCR indexes) produced 340 single cell libraries with a median unique read count of 141,449 (248 cells ≥ 50,000 unique reads), and our xSDS preparation generated 171 single cell libraries with a median unique read count of 55,142 (92 cells ≥ 50,000 unique reads). The number of cells produced in our xSDS preparation was lower than expected, largely due to nuclei aggregates during sorting that may be remedied by additional cell dis-aggregation steps.
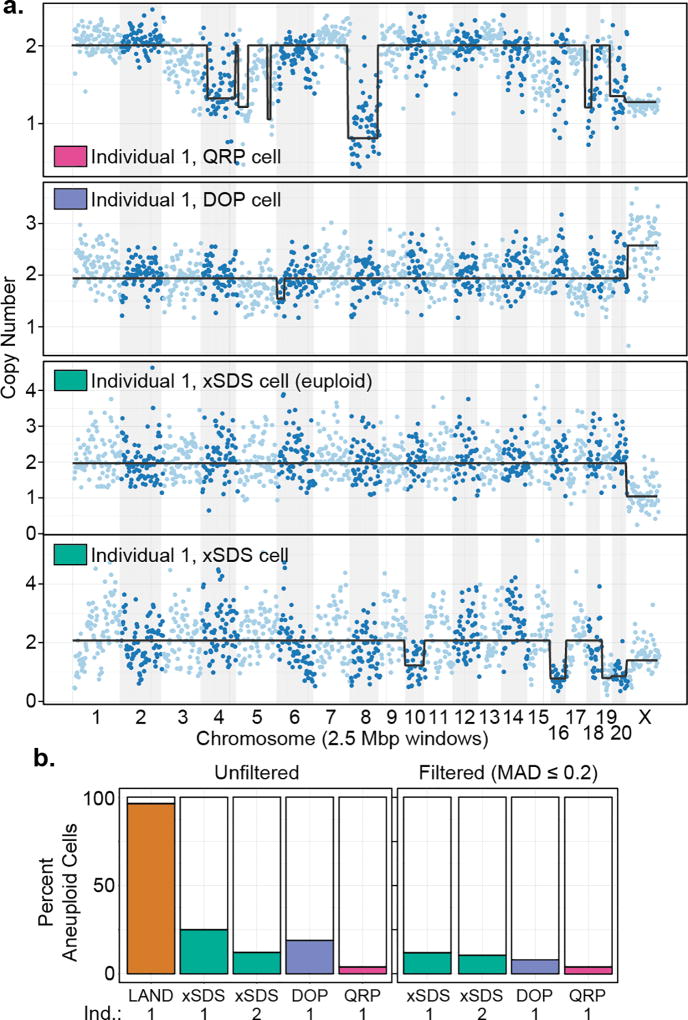
Across all methods of library construction we observed greater discrepancies between the three CNV calling approaches than in the human analyses (Supplementary Fig. 11–14), likely due to the lower quality of the Rhesus reference genome (284,705 contigs < 1 Mbp), emphasizing the need for “platinum” quality reference genomes27. We therefore focused on the HMM results for sub-chromosomal calls (Fig. 3a) and performed aneuploidy analysis using the intersection of CBS and HMM calls. Consistent with our cell line results, the LAND preparation produced a much higher aneuploidy rate (95.1%), suggestive of false positives stemming from coverage nonuniformity (Supplementary Fig. 15,16). The xSDS SCI-seq unfiltered aneuploidy rate (25.0%) was close to the DOP preparation (18.5%), with QRP producing a much lower rate (3.1%; Fig. 3b). After imposing a variance filter for cells with a MAD score of 0.2 or lower, the aneuploidy rates dropped to 12.0% for the xSDS preparation, 8.7% for the DOP, and stayed the same for the QRP preparation at 3.1%. These rates were similar to those produced by xSDS SCI-seq on a 200 mm3 section of frontal cortex from a second individual (381 single cells, median read count of 62,731, 213 cells ≥ 50,000 unique reads) which produced unfiltered and filtered aneuploidy rates of 12.1% and 10.3% respectively (Supplementary Fig. 17).
Figure 3. Somatic CNVs in the Rhesus brain.
(a) Three single cell examples showing copy number variants, and one representative euploid cell for the SCI-seq preparation (HMM). (b) Frequency of aneuploidy as determined by each of the methods with and without filtering.
SCI-seq on primary tumor samples reveals clonal populations
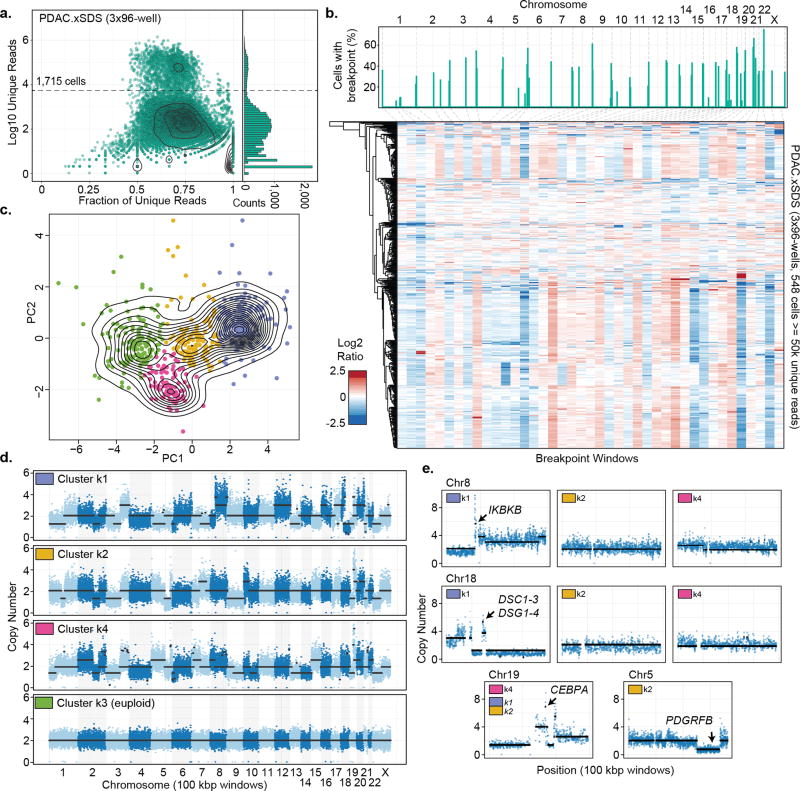
One of the primary applications of single cell genome sequencing is in the profiling of tumor heterogeneity and understanding clonal evolution in cancer as it relates to treatment resistance5–8. We carried out a single xSDS SCI-seq preparation on a freshly acquired stage III pancreatic ductal adenocarcinoma (PDAC) sample measuring approximately 250 mm3 which resulted in 1,715 single cell libraries sequenced to a median unique read count of 49,272 per cell (M50 of 71,378; 846 cells ≥ 50,000 unique reads at the depth the library was sequenced; Fig. 4a). We first performed CNV calling using our GM12878 library as a euploid baseline for comparison to identify a set of high-confidence euploid cells (298, 35.2%) which were then used as a new baseline specific to the individual and preparation (Supplementary Fig. 17–19). Assuming that subchromosomal copy number alterations (caused by genome instability) are more informative for identifying subclonal populations than whole chromosome aneuploidy (due to errors during cell division), we developed a strategy to identify putative copy number breakpoints at low resolution to be used as new window boundaries (Methods, Supplementary Fig. 20) followed by stratification via principle components analysis (PCA) and k-means clustering. We initially applied this method to our HeLa libraries (2,361 single cells in total), revealing no distinct heterogeneity and further supporting the stability of the HeLa cell line20 (Supplementary Fig. 21–24), and then on our primary PDAC sample, which revealed an optimum cluster count of 4 by silhouette analysis (Fig. 4b,c).
Figure 4. SCI-seq analysis of a stage III human Pancreatic Ductal Adenocarcinoma (PDAC).
(a) SCI-seq library complexity. Right panel, histogram showing distribution of read counts. Dashed line represents single cell read cutoff. (b) Breakpoint calls (top) and breakpoint window matrix of log2 sequence depth ratio. (c) Principle component analysis and k-means clustering on breakpoint matrix. (d) 100 kbp resolution CNV calling on aggregated cells from each cluster. (e) Cluster specific CNVs and CEBPA amplification present in all clusters (k4 shown).
The first of these clusters (k3) is a population of euploid cells that were not considered high confidence euploid in the initial analysis, and thus not removed. When including these, the euploid population rises to 389 for a final tumor cell purity of 46.0%, within the expected range for PDAC28. For the remaining clusters k1 (199 cells), k2 (115 cells) and k4 (91 cells), we aggregated all reads from cells proximal to each centroid (Methods) and carried out CNV calling using 100 kbp windows, a 25-fold greater resolution than the initial analysis, and then determined absolute copy number states20 (Fig. 4d).
Across the three tumor clusters, a substantial portion of copy number segments were shared (44.8%), suggesting that they arose from a common progenitor population. This includes a highly rearranged chromosome 19 which harbors a focal amplification of CEBPA, which encodes an enhancer binding protein, at copy number 7 which is frequently mutated in AML29, and has recently been shown to have altered epigenetic regulation in pancreatic tumors30 (Fig. 4e). An all-by-all pairwise comparison revealed clusters k2 and k4 as the most similar, sharing 65.9% of copy number segments, followed by k1 and k4 at 58.3%, and k1 and k2 at 55.0%. Several cluster-specific CNVs contain genes of potential functional relevance (Fig. 4e). These include a focal amplification to copy number 6 of IKBKB in cluster k1, which encodes a serine kinase important in the NF-κB signaling pathway31; another focal amplification to copy number 5 in cluster k1 containing genes DSC1,2,3 and DSG1,2,3,4 all of which encode proteins involved in cell-cell adhesion and cell positioning and are often mis-regulated in cancer32; and the deletion of a region containing PDGRFB specific to cluster k2, which encodes a tyrosine kinase cell surface receptor involved in cell proliferation signaling, and is frequently mutated in cancer33.
Lastly, we applied xSDS SCI-seq to a frozen stage II rectal adenocarcinoma measuring 500 mm3. During preparation we noticed a high abundance of nuclear debris and ruptured nuclei which likely attributed to the decreased yield of the preparation (16 PCR indexes) of 146 single cell libraries (median unique read count of 71,378; M50 of 352,168; 111 cells ≥ 50,000 unique reads). We carried out the same CNV calling approach as with the PDAC sample; however high frequency breakpoints were not observed and subclonal populations could not be identified (Supplementary Fig. 25). This may be a result of nuclear deterioration due to irradiation, a common treatment for rectal cancers, underscoring the challenge of producing high-quality single cell or nuclei suspensions shared by all single cell methods12.
Discussion
We developed SCI-seq, a method which utilizes nucleosome depletion in a combinatorial indexing workflow to produce thousands of single cell genome sequencing libraries. Using SCI-seq, we produced 16,698 single cell libraries (of which 5,395 were sequenced to a depth sufficient for CNV calling) from myriad samples, including primary tissue isolates representative of the two major areas of single cell genome research: somatic aneuploidy and cancer. In addition to the advantages of throughput, the platform does not require specialized microfluidics equipment or droplet emulsification techniques. Using our more uniform nucleosome depletion strategy, xSDS, we were able to achieve resolution on the order of 250 kbp, though we suspect further optimization, such as alternative crosslinking agents, may provide sufficient depth for improved resolution. We also demonstrate the ability to identify clonal populations that can be aggregated to facilitate high resolution CNV calling by applying this strategy to a pancreatic ductal adenocarcinoma which revealed subclone-specific CNVs that may impact proliferation, migration, or possibly drive other molecular subtypes34.
While the technology is currently limited to copy number variant detection, it may be possible to include in situ pre-amplification within the nuclear scaffold prior to SCI-seq or the incorporation of T4 in vitro transcription, such as in THS-seq35, an ATAC-seq variant, to boost the resulting coverage and facilitate single nucleotide variant detection. While optimization is possible, as with any new method, we believe that the throughput provided by SCI-seq will open the door to deep quantification of mammalian somatic genome stability as well as serve as a platform to assess other properties of single cells including DNA methylation and chromatin architecture.
Online Methods
Sample preparation and nuclei isolation
Tissue culture cell lines were trypsinized then pelleted if adherent (HeLa S3, ATCC CCL-2.2; NIH/3T3, ATCC CRL-1658) or pelleted if grown in suspension (GM12878, Coriell; karyotyped at the OHSU Research Cytogenetics Laboratory), followed by one wash with ice cold PBS. They were then carried through crosslinking (for the xSDS method) or directly into nuclei preparation using Nuclei Isolation Buffer (NIB, 10 mM TrisHCl pH7.4, 10 mM NaCl, 3 mM MgCl2, 0.1% igepal, 1× protease inhibitors (Roche, Cat. 11873580001)) with or without nucleosome depletion. Tissue samples (RhesusFcx1, RhesusFcx2, PDAC, CRC) were dounce homogenized in NIB then passed through a 35µm cell strainer prior to nucleosome depletion. The frozen Rhesus frontal cortex samples, RhesusFcx1 (4 yr. female) and RhesusFcx2 (9 yr. female), were obtained from the Oregon National Primate Research Center as a part of their aging nonhuman primate resource.
Standard Single Cell Library Construction
Single cell libraries constructed using quasi-random priming (QRP) and degenerate oligonucleotide primed PCR (DOP) were prepared from isolated nuclei without nucleosome depletion and brought up to 1 mL of NIB, stained with 5 µL of 5 mg/ml DAPI (Thermo Fisher, Cat. D1306) then FANS sorted on a Sony SH800 in single cell mode. One nucleus was deposited into each single well containing the respective sample buffers. QRP libraries were prepared using the PicoPlex DNA-seq Kit (Rubicon Genomics, Cat. R300381) according to the manufacturer’s protocol and using the indexed PCR primers provided in the kit. DOP libraries were prepared using the SeqPlex DNA Amplification Kit (Sigma, Cat. SEQXE-50RXN) according to the manufacturer’s protocol, but with the use of our own custom PCR indexing primers that contain 10 bp index sequences. To avoid over-amplification, all QRP and DOP libraries were amplified with the addition of 0.5 µL of 100× SYBR Green (FMC BioProducts, Cat. 50513) on a BioRad CFX thermocycler in order to monitor the amplification and pull reactions that have reached mid-exponential amplification.
Nucleosome Depletion
Lithium assisted nucleosome depletion (LAND): Prepared Nuclei were pelleted and resuspended in NIB supplemented with 200 µL of 12.5 mM lithium 3,5-diiodosalicylic acid (referred to as Lithium diiodosalycylate in main text, Sigma, Cat. D3635) for 5 minutes on ice prior to the addition of 800 µL NIB and then taken directly into flow sorting.
Crosslinking and SDS nucleosome depletion (xSDS): Crosslinking was achieved by incubating cells in 10 mL of media (cell culture) or nuclei in 10 mL of HEPES NIB (20 mM HEPES, 10 mM NaCl, 3mM MgCl2, 0.1% igepal, 1× protease inhibitors (Roche, Cat. 11873580001)) (tissue samples) containing 1.5% formaldehyde at room for 10 minutes. The crosslinking reaction was neutralized by bringing the reaction to 200 mM Glycine (Sigma, Cat. G8898-500G) and incubating on ice for 5 minutes. Cell culture samples were crosslinked and then washed once with 10 ml ice cold 1× PBS and had nuclei isolated by incubating in NIB buffer on ice for 20 minutes and pelleted once again. Nuclei were then resuspended in 800 uL 1× NEBuffer 2.1 (NEB, Cat. B7202S) with 0.3% SDS (Sigma, Cat. L3771) and incubated at 42°C with vigorous shaking for 30 minutes in a thermomixer (Eppendorf). SDS was then quenched by the addition of 200 µL of 10% Triton-X100 (Sigma, Cat. 9002-93-1) and incubated at 42°C with vigorous shaking for 30 minutes.
Combinatorial indexing via tagmentation and PCR
Nuclei were stained with 5 µL of 5mg/ml DAPI (Thermo Fisher, Cat. D1306) and passed through a 35 µm cell strainer. A 96 well plate was prepared with 10 µL of 1× Nextera® Tagment DNA (TD) buffer from the Nextera® DNA Sample Preparation Kit (Illumina, Cat. FC-121-1031) diluted with NIB in each well. A Sony SH800 flow sorter was used to sort 2,000 single nuclei into each well of the 96 well tagmentation plate in fast sort mode. Next, 1 µL of a uniquely indexed 2.5 µM transposase-adaptor complex (transposome) was added to each well. These complexes and associated sequences are described in Amini et. al. 2015, Ref. 14. Reactions were incubated at 55°C for 15 minutes. After cooling to room temperature, all wells were pooled and stained with DAPI as previously described. A second 96 well plate, or set of 96 well plates, were prepared with each well containing 8.5 µL of a 0.058% SDS, 8.9 nM BSA solution and 2.5 µL of 2 uniquely barcoded primers at 10 µM. 22 post-tagmentation nuclei from the pool of 96 reactions were then flow sorted on the same instrument but in single cell sort mode into each well of the second plate and then incubated in the SDS solution at 55°C for 5 minutes to disrupt the nuclear scaffold and disassociate the transposase enzyme. Crosslinks were reversed by incubating at 68°C for an hour (xSDS). SDS was then diluted by the addition of 7.5 µL of Nextera® PCR Master mix (Illumina, Cat. FC-121-1031) as well as 0.5 µL of 100× SYBR Green (FMC BioProducts, Cat. 50513) and 4 µL of water. Real time PCR was then performed on a BioRad CFX thermocycler by first incubating reactions at 72°C for 5 minutes, prior to 3 minutes at 98°C and 15–20 cycles of [20 sec. at 98°C, 15 sec. at 63°C, and 25 sec. at 72°C]. Reactions were monitored and stopped once exponential amplification was observed in a majority of wells. 5 µL of each well was then pooled and purified using a Qiaquick PCR Purification column (Qiagen, Cat. 28104) and eluted in 30 µL of EB.
Library quantification and sequencing
Libraries were quantified between the range of 200bp and 1 kbp on a High Sensitivity Bioanalyzer kit (Agilent, Cat. 5067-4626). Libraries were sequenced on an Illumina NextSeq® 500 loaded at 0.8 pM with a custom sequencing chemistry protocol (Read 1: 50 imaged cycles; Index Read 1: 8 imaged cycles, 27 dark cycles, 10 imaged cycles; Index Read 2: 8 imaged cycles, 21 dark cycles, 10 imaged cycles; Read 2: 50 imaged cycles) using custom sequencing primers described in Amini et. al. 2015, Ref.14. QRP and DOP libraries were sequenced using standard primers on the NextSeq® 500 using high-capacity 75 cycle kits with dual-indexing. For QRP there is an additional challenge that the first 15 bp of the read are highly enriched for “G” bases, which are non-fluorescent with the NextSeq® 2-color chemistry and therefore cluster identification on the instrument fails. We therefore sequenced the libraries using a custom sequencing protocol that skips this region (Read 1: 15 dark cycles, 50 imaged cycles; Index Read 1: 10 imaged cycles; Index Read 2: 10 imaged cycles).
Sequence Read Processing
Software for processing SCI-seq raw reads can be found in the accompanying Supplementary Software or downloaded from http://sci-seq.sourceforge.net. Sequence runs were processed using bcl2fastq (Illumina Inc., version 2.15.0) with the --create-fastq-for-index-reads and --with-failed-reads options to produce fastq files. Index reads were concatenated (36 bp total) and used as the read name with a unique read number appended to the end. These indexes were then matched to the corresponding index reference sets allowing for a hamming distance of two for each of the four index components (i7-Transposase (8 bp), i7-PCR (10 bp), i5-Transposase (8 bp), and i5-PCR (10 bp)), reads matching a quad-index combination were then renamed to the exact index (and retained the unique read number) which was subsequently used as the cell identifier. Reads were then adaptor trimmed, then paired and unpaired reads were aligned to reference genomes by Bowtie2 and merged. Human preparations were aligned to GRCh37, Rhesus preparations were aligned to RheMac8, and Human/Mouse mix preparations were aligned to a combined human (GRCh37) and mouse (mm10) reference. Aligned bam files were subjected to PCR duplicate removal using a custom script that removes reads with identical alignment coordinates on a per-barcode basis along with reads with an alignment score less than 10 as reported by Bowtie2.
Single Cell Discrimination
For each PCR plate, a total of 9,216 unique index combinations are possible (12 i7-Transposase indexes × 8 i5-Transposase indexes × 12 i7-PCR indexes × 8 i5-PCR indexes), for which only a minority should have a substantial read count, as the majority of index combinations should be absent – i.e. transposase index combinations of nuclei that were not sorted into a given PCR well. These “empty” indexes typically contain very few reads (1–3% of a run) with the majority of reads falling into bona fide single cell index combinations (97–99% of a run). The resulting histogram of log10 unique read counts for index combinations (Supplementary Fig. 3) produces a mix of two normal distributions: a noise component and a single cell component. We then used the R package “mixtools” to fit a mixed model (normalmixEM) to identify the proportion (λ) mean (μ) and standard deviation (σ) of each component. The read count threshold to qualify as a single cell library was taken to be the greater of either one standard deviation below the mean of the single cell component in log10 space, or 100 fold greater than the mean of the noise component (+2 in log10 space), and had to be a minimum of 1,000 unique reads.
Human-Mouse Mix Experiments
We took one of two approaches to mix human (GM12878 or HeLa S3) and mouse (3T3) cells: i) mixing at the cell stage (HumMus.LAND1 and HumMus.LAND2) or ii) mixing at the nuclei stage (HumMus.LAND3, HumMus.LAND4, and HumMus.xSDS). The reason we employed the latter was to control for nuclei crosslinking or agglomerating together that could result in doublets. Libraries were constructed as described above, for instances where two distinct DAPI-positive populations were observed during flow sorting, included both populations in the same gate so as not to skew proportions. Reads were processed as in other experiments, except reads were instead aligned to a reference comprised of GRCh37 (hg19) and mm10. The mapping quality 10 filter effectively removed reads that aligned to conserved regions in both genomes and then for each identified single cell, reads to each species were tallied and used to estimate collision frequency. For our early LAND preparations we sorted 25 indexed nuclei per PCR well and produced total collision rates (i.e. twice the human-mouse collision rate) of 28.1% and 10.4%. For the second two LAND preparations we sorted 22 nuclei per PCR well, which produced a total collision rate of 4.3% for one preparation and no detectable collisions in another. We also tested two FANS sorting conditions for our xSDS preparation, one was permissive and allowed a broader range of DAPI fluorescence, and the other more restrictive, and carried out both preparations on separate sides of the same PCR plate. For the permissive gating we observed a total collision rate of 23.6% with a substantial reduction for the more restrictive gating at 8.1%. Based on these results we decided to continue sorting 22 nuclei per PCR well using the more restrictive FANS
Library Depth Projections
To estimate the performance of a library pool if, or when, it was sequenced to a greater depth, we incrementally sampled random reads from each SCI-seq preparation across all index combinations including unaligned and low quality reads without replacement at every one percent of the total raw reads. For each point we identified the total number reads that are aligned with high quality (MQ ≥ 10) assigned to each single cell index and the fraction of those reads that are unique, non-PCR duplicates, as well as the corresponding fraction of total reads sampled that were assigned to that index. Using these points we fit both a nonlinear model and a Hanes-Woolfe transformed model to predict additional sequencing for each individual single cell library within the pool and projected out to a median unique read percentage across cells of 5%. To determine the accuracy of the models, we determined the number of downsampled raw reads of each library that would reach the point in which the median unique read percentage per cell was 90%, which is somewhat less than what was achieved for libraries that were sequenced at low coverage. We then subsampled the pre-determined number of reads for 30 iterations and built a new model for each cell at each iteration and then predicted the unique read counts for each cell out to the true sequencing depth that was achieved. The standard deviation of the true read count across all iterations for all cells was then calculated.
Genome Windowing
Genomic windows were determined on a per-library basis using custom tools. For each chromosome the size of the entire chromosome was divided by the target window size to produce the number of windows per chromosome. The total read count for the chromosome summarized over the pool of all single cells (GM12878 for all human samples where absolute copy number was determined, as well as for each pooled sample where amplifications or deletions relative to the mean copy number were determined) was then divided by the window count to determine the mean read count per window. The chromosome was then walked and aligned reads from the pool tallied and a window break was made once the target read count per window was reached. Windows at chromosome boundaries were only included if they contained more than 75% of the average reads per window limit for that chromosome. By using dynamic windows we accounted for biases, such as highly repetitive regions, centromeres and other complex regions that can lead to read dropout in the case of fixed size bins22.
GC Bias Correction
Reads were placed into the variable sized bins and GC corrected based on individual read GC content instead of the GC content of the dynamic windows. We posit that the large bin sizes needed for single cell analysis average out smaller scale GC content changes. Furthermore, SCI-seq does not involve pre-amplification where large regions of the genome are amplified, therefore GC bias originates solely from the PCR and is amplicon-specific. To calculate correction weights for the reads we compared the fraction of all reads with a given GC to the fraction of total simulated reads with the average insert size at the same GC fraction. This weight was then used in lieu of read counts and summed across all reads in a given window. All regions present in DAC blacklisted regions were excluded from analysis for the human sample analyses (http://genome.ucsc.edu/cgi-bin/hgFileUi?db=hg19&g=wgEncodeMapability)19. Following GC correction, all reads were normalized by the average number of reads per bin across the genome. Finally for each window we took the normalized read count of each cell and divided it by the pooled sample baseline to produce a ratio score.
Measures of data variation
To measure data quality, we calculated two different measures of coverage dispersion: the median absolute deviation (MAD), the median absolute pairwise difference (MAPD). For each score we calculated the median of the absolute values of all pairwise differences between neighboring bins that have been normalized by the mean bin count within the cell (log2 normalized ratios for the MAPD scores). These scores measure the dispersion of normalized binned reads due to technical noise, rather than due copy number state changes, which are less frequent2,22.
Copy Number Variant Calling
CNV calling was performed on the windowed, GC corrected and bulk sample normalized reads with two available R packages that employ two different segmentation strategies: a Hidden Markov Model approach (HMMcopy, version 3.3.0, Ref. 25) and Circular Binary Segmentation (DNAcopy, version 1.44.0, Ref. 24). Values were Log2 transformed for input (2*log2 for CBS) and copy number calls were made based on the optimized parameters from Knouse et al. 2016, Ref. 11. For optimal sensitivity and specificity to detect copy number calls with sizes ≥5Mb we set the probability of segment extension (E) to 0.995 for HMM and for CBS we chose the significance level to accept a copy number change (α) to be 0.0001. The Log2 cutoffs for calling losses or gains were 0.4 and −0.35 for HMM and 1.32 and 0.6 for CBS. As an additional tool for CNV calling we used Ginkgo22, which uses an alternative method for data normalization. We uploaded bed files for each cell and a bulk down sampled bed file, which we created with Picard Tools (we used a down sample probability of 0.1). For the analysis we chose to segment single cells with the down sampled bulk bed file and when ploidy was known for the samples we created FACS files to force Ginkgo to normalize to that ploidy. Calls for the three methods were intersected either on a per-window basis or were filtered to only include calls that span ≥ 80% of a chromosome arm and then intersected for aneuploidy analysis.
Tumor breakpoint analysis
Unlike the assessment of sporadic aneuploidy, tumor structural variation is much more complex with a large portion of breakpoints within chromosomes. Further, sporadic aneuploidy within any given subclone of a tumor is less pertinent than an accurate profile of the subpopulations that are present. We therefore used the HMM and CBS segmented ratio score matrixes to identify breakpoints by tallying up the boundaries of segmented regions across cells. We then used the resulting distribution of shared chromosomal breakpoints across the genome to identify local maxima to account for variability in which specific window the call was made, and then retained those that are present in at least 5% of cells. We then merged all windows within each breakpoint span and calculated the new log2 ratio of each aneuploid cell over the mean values of the euploid population. We then carried out principle components analysis prior to k-means clustering with a k value determined by Silhouette analysis. To minimize the effect of doublets which can account for ~10% of putative single cells and also to exclude low-performance cells, we retained only those in the close proximity to their respective centroids. We then merged sequence reads for all cells within each cluster and then carried out a higher resolution CNV analysis (target window size of 100 kbp) using an HMM strategy followed by absolute copy number state identification and the identification of focal amplifications and deletions using a sliding window outlier strategy20. Intra-tumoral clonal relationships are most accurately captured by shared breakpoints as opposed to the drift in copy number of a segment based on the assumption that structural changes involving breaks in the DNA as being more impactful on the cell. We therefore compared cells by assessing the proportion of segments between breakpoints that were identified using the high resolution (100 kbp) CNV analysis that overlapped by at least 90% (to account for noise in the exact window that was called as the copy number change) out of the total number of segments.
Editor’s Summary
Single-cell Combinatorial Indexed Sequencing (SCI-seq) resolves genomic heterogeneity by generating thousands of low-pass single-cell libraries at once for somatic copy number variant detection.
Supplementary Material
Acknowledgments
The genome sequence described/used in this research was derived from a HeLa cell line. Henrietta Lacks, and the HeLa cell line that was established from her tumor cells without her knowledge or consent in 1951, have made significant contributions to scientific progress and advances in human health. We are grateful to Henrietta Lacks, now deceased, and to her surviving family members for their contributions to biomedical research. The data generated from this research were submitted to the database of Genotypes and Phenotypes (dbGaP), as a substudy under accession number phs000640.
We thank the aging nonhuman primate resource at the Oregon National Primate Research Center for the banked Rhesus samples, the Brenden-Colson Center for Pancreatic Care for the pancreatic ductal adenocarcinoma sample, and the Knight Tissue Bank for the rectal adenocarcinoma sample. We thank Jay Shendure and Shendure lab members Darren Cusanovich, and Riza Daza for helpful advice and comments, and Martin Kircher for providing PCR-stage index sequences. We also thank Brian J. O’Roak and Ryan Mulqueen for helpful discussions and manuscript suggestions. A.A. is supported by an Oregon Medical Research Foundation New Investigator Award. J.L.R. is supported by the Collins Medical Trust Foundation and Glenn/AFAR Scholarship for Research in the Biology of Aging. L. Carbone is supported by the Office of the Director/Office of Research Infrastructure Programs (OD/ORIP) of the NIH (grant no. OD011092).
Footnotes
Accession Codes
NCBI BioProject ID: PRJNA326698
HeLa dbGaP Accession: phs000640
Data Availability
GM12878 and Rhesus sequence data are accessible through the NCBI Sequence Read Archive (SRA) under BioProject ID: PRJNA326698 for unrestricted access. HeLa sequence data are undergoing submission to the database of Genotypes and Phenotypes (dbGaP), as a substudy under accession number phs000640. Human tumor samples are undergoing submission to dbGaP and are awaiting study accession assignment. Software developed specifically for this project can be found at http://sci-seq.sourceforge.net or in Supplementary Software. All methods for making the transposase complexes are described in (Ref. 14); however, Illumina will provide transposase complexes in response to reasonable requests from the scientific community subject to a material transfer agreement.
Author Contributions
A.A. designed and supervised all aspects of the study. A.A., S.A.V., and K.A.T. wrote the manuscript. All authors contributed and edited the manuscript. S.A.V. carried out all SCI-seq and GM12878 DOP library preparations, designed experiments, and performed all sequencing. A.A. and K.A.T. processed all sequence data and analyzed data. K.A.T. performed all copy number calling. J.L.R. constructed QRP and DOP libraries on Rhesus samples. A.J.F. prepared all GM12878 QRP library construction and co-prepared all SCI-seq libraries using xSDS for nucleosome depletion. M.H.W. provided tumor samples and aided in the analyses of those samples. L. Carbone supervised and provided all samples for Rhesus work. F.J.S. contributed to experimental design and contributed to the manuscript, L. Christiansen produced all transposase complexes used in this study.
Competing Financial Interests
F.J.S. and L. Christiansen declare competing financial interests in the form of paid employment by Illumina, Inc. One or more embodiments of one or more patents and patent applications filed by Illumina may encompass the methods, reagents, and data disclosed in this manuscript. Some work in this study is related to technology described in patent applications WO2014142850, 2014/0194324, 2010/0120098, 2011/0287435, 2013/0196860, and 2012/0208705. A.A. and S.A.V. have a provisional patent filed for some of the methods pertaining to this study.
References
- 1.McConnell MJ, et al. Mosaic Copy Number Variation in Human Neurons. Science (80-.) 2013;342:632–637. doi: 10.1126/science.1243472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cai X, et al. Single-Cell, Genome-wide Sequencing Identifies Clonal Somatic Copy-Number Variation in the Human Brain. Cell Rep. 2014;8:1280–1289. doi: 10.1016/j.celrep.2014.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Knouse KA, Wu J, Whittaker CA, Amon A. Single cell sequencing reveals low levels of aneuploidy across mammalian tissues. Proc Natl Acad Sci U S A. 2014;111:13409–13414. doi: 10.1073/pnas.1415287111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rehen SK, et al. Chromosomal variation in neurons of the developing and adult mammalian nervous system. Proc. Natl. Acad. Sci. U. S. A. 2001;98:13361–6. doi: 10.1073/pnas.231487398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Navin N, et al. Tumour evolution inferred by single-cell sequencing. Nature. 2011;472:90–94. doi: 10.1038/nature09807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eirew P, et al. Dynamics of genomic clones in breast cancer patient xenografts at single-cell resolution. Nature. 2014;518:422–6. doi: 10.1038/nature13952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gawad C, Koh W, Quake SR. Dissecting the clonal origins of childhood acute lymphoblastic leukemia by single-cell genomics. Proc. Natl. Acad. Sci. U. S. A. 2014;111:17947–52. doi: 10.1073/pnas.1420822111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gao R, et al. Punctuated copy number evolution and clonal stasis in triple-negative breast cancer. Nat. Genet. 2016:1–15. doi: 10.1038/ng.3641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zong C, Lu S, Chapman AR, Xie XS. Genome-Wide Detection of Single Nucleotide and Copy Number Variations of a Single Human Cell. Science (80-.) 2012;338:1622–1626. doi: 10.1126/science.1229164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baslan T, et al. Optimizing sparse sequencing of single cells for highly multiplex copy number profiling. Genome Res. 2015;125:714–724. doi: 10.1101/gr.188060.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Knouse KA, Wu J, Amon A. Assessment of megabase-scale somatic copy number variation using single cell sequencing. Genome Res. 2016 doi: 10.1101/gr.198937.115. gr.198937.115- [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gawad C, Koh W, Quake SR. Single-cell genome sequencing: current state of the science. Nat. Rev. Genet. 2016;17:175–88. doi: 10.1038/nrg.2015.16. [DOI] [PubMed] [Google Scholar]
- 13.Adey A, et al. Rapid, low-input, low-bias construction of shotgun fragment libraries by high-density in vitro transposition. Genome Biol. 2010;11:R119. doi: 10.1186/gb-2010-11-12-r119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Amini S, et al. Haplotype-resolved whole-genome sequencing by contiguity-preserving transposition and combinatorial indexing. Nat. Genet. 2014;46:1343–9. doi: 10.1038/ng.3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Adey A, et al. In vitro, long-range sequence information for de novo genome assembly via transposase contiguity. Genome Res. 2014;24:2041–2049. doi: 10.1101/gr.178319.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods. 2013;10:1213–8. doi: 10.1038/nmeth.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cusanovich Da, et al. Epigenetics. Multiplex single-cell profiling of chromatin accessibility by combinatorial cellular indexing. Science. 2015;348:910–4. doi: 10.1126/science.aab1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stergachis AB, et al. Developmental fate and cellular maturity encoded in human regulatory DNA landscapes. Cell. 2013;154:888–903. doi: 10.1016/j.cell.2013.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.The ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Adey A, et al. The haplotype-resolved genome and epigenome of the aneuploid HeLa cancer cell line. Nature. 2013;500:207–211. doi: 10.1038/nature12064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Macosko EZ, et al. Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets. Cell. 2015;161:1202–1214. doi: 10.1016/j.cell.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Garvin T, et al. Interactive analysis and quality assessment of single-cell copy-number variations. bioRxiv. 2014:11346. doi: 10.1101/011346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goryshin IY, Miller JA, Kil YV, Lanzov VA, Reznikoff WS. Tn5/IS50 target recognition. Proc. Natl. Acad. Sci. U S A. 1998;95:10716–10721. doi: 10.1073/pnas.95.18.10716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Olshen AB, Venkatraman ES, Lucito R, Wigler M. Circular binary segmentation for the analysis of array-based DNA copy number data. Biostatistics. 2004;5:557–572. doi: 10.1093/biostatistics/kxh008. [DOI] [PubMed] [Google Scholar]
- 25.Ha G, et al. Integrative analysis of genome-wide loss of heterozygosity and monoallelic expression at nucleotide resolution reveals disrupted pathways in triple-negative breast cancer. Genome Res. 2012;22:1995–2007. doi: 10.1101/gr.137570.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rosenkrantz J, Carbone L. Investigating somatic aneuploidy in the brain: why we need a new model. Chromosoma. 2016 doi: 10.1007/s00412-016-0615-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Callaway E. ‘Platinum’ genome takes on disease. Nat. News. 2014;515:323. doi: 10.1038/515323a. [DOI] [PubMed] [Google Scholar]
- 28.Waddell N, et al. Whole genomes redefine the mutational landscape of pancreatic cancer. Nature. 2015;518:495–501. doi: 10.1038/nature14169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.De Kouchkovsky I, Abdul-Hay M. ‘Acute myeloid leukemia: a comprehensive review and 2016 update’. Blood Cancer J. 2016;6:e441. doi: 10.1038/bcj.2016.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kumagai T, et al. Epigenetic regulation and molecular characterization of C/EBPalpha in pancreatic cancer cells. Int J Cancer. 2009;124:827–833. doi: 10.1002/ijc.23994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Perkins ND. Integrating cell-signalling pathways with NF-kappaB and IKK function. Nat. Rev. Mol. Cell Biol. 2007;8:49–62. doi: 10.1038/nrm2083. [DOI] [PubMed] [Google Scholar]
- 32.Stahley SN, Kowalczyk AP. Desmosomes in acquired disease. Cell Tissue Res. 2015;360:439–56. doi: 10.1007/s00441-015-2155-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Forbes SA, et al. COSMIC: Exploring the world’s knowledge of somatic mutations in human cancer. Nucleic Acids Res. 2015;43:D805–D811. doi: 10.1093/nar/gku1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bailey P, et al. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature. 2016;531:47–52. doi: 10.1038/nature16965. [DOI] [PubMed] [Google Scholar]
- 35.Sos B, et al. Characterization of chromatin accessibility with a transposome hypersensitive sites sequencing (THS-seq) assay. Genome Biol. 2016;17:20. doi: 10.1186/s13059-016-0882-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.