Abstract
Mitochondrial neurogastrointestinal encephalomyopathy (MNGIE) is an autosomal recessive disorder caused by thymidine phosphorylase (TP) deficiency resulting in systemic accumulation of thymidine (d-Thd) and deoxyuridine (d-Urd) and characterized by early-onset neurological and gastrointestinal symptoms. Long-term effective and safe treatment is not available. Allogeneic bone marrow transplantation may improve clinical manifestations but carries disease and transplant-related risks. In this study, lentiviral vector-based hematopoietic stem cell gene therapy (HSCGT) was performed in Tymp−/−Upp1−/− mice with the human phosphoglycerate kinase (PGK) promoter driving TYMP. Supranormal blood TP activity reduced intestinal nucleoside levels significantly at low vector copy number (median, 1.3; range, 0.2–3.6). Furthermore, we covered two major issues not addressed before. First, we demonstrate aberrant morphology of brain astrocytes in areas of spongy degeneration, which was reversed by HSCGT. Second, long-term follow-up and vector integration site analysis were performed to assess safety of the therapeutic LV vectors in depth. This report confirms and supplements previous work on the efficacy of HSCGT in reducing the toxic metabolites in Tymp−/−Upp1−/− mice, using a clinically applicable gene transfer vector and a highly efficient gene transfer method, and importantly demonstrates phenotypic correction with a favorable risk profile, warranting further development toward clinical implementation.
Keywords: MNGIE, thymidine phosphorylase, hematopoietic stem cells, lentiviral vectors, gene therapy
Introduction
Mitochondrial neurogastrointestinal encephalomyopathy (MNGIE) is an autosomal recessive disease caused by mutations in the thymidine phosphorylase gene (TYMP),1 which result in partial or complete deficiency of the enzyme thymidine phosphorylase (TP).2 TP deficiency results in systemic accumulation of the substrates d-Thd and d-Urd in plasma and tissues of MNGIE patients3 and thereby to imbalances in intra-mitochondrial deoxyribonucleoside triphosphate pools (dNTPs), which has been considered to induce somatic mitochondrial DNA alterations in tissues of MNGIE patients.4, 5 The clinical spectrum of MNGIE ranges from mild symptoms in late-onset cases6 to early-teenage onset with fatal malabsorption and intestinal dysmotility.7 The typical symptoms include ptosis and ophthalmoplegia, gastrointestinal dysmotility, neuropathy with reduced sensory-motor conduction, as well as central nervous system anomalies.4, 8 The latter manifest on brain MRI as a unique pattern of progressive symmetrical cerebral (and occasionally cerebellar) hyper-intense signal changes on T2-weighted images.9 The available treatment options for MNGIE such as peritoneal dialysis, platelet infusion, and enzyme replacement therapy fail to achieve persistent clinical improvement.10, 11, 12 As a long-term solution, allogeneic hematopoietic stem cell transplantation (HSCT) has been proposed; however, this is associated with high mortality rates due to disease and transplant-related complications.13, 14 Recently, liver transplantation has emerged as a promising treatment option, preferably for patients with pre-existing liver failure; this, however, requires a matched organ donor and long-term immunosuppression.15 Autologous hematopoietic stem cell gene therapy (HSCGT) could be beneficial for MNGIE patients as a single intervention with low-dose conditioning and poses no risks of graft rejection or graft-versus-host disease. Third-generation self-inactivating lentiviral (LV) vectors are currently used successfully in clinical trials for other metabolic disorders.16, 17, 18 The present study supplements and confirms previous reports19, 20 by using (1) a highly efficient transduction method, (2) clinically applicable LV vectors to evaluate the therapeutic efficacy to reverse the biochemical phenotype and extends these reports by demonstrating (3) therapeutic efficacy of HSGCT in reversing pathological changes, particularly those of the brain, and assessing (4) potential adverse effects such as pheno- and genotoxicity.
Results
LV Vectors, Experimental Design, TP Expression, and Enzyme Activity in Target Cells
Similar to previous studies,19, 20, 21 Tymp−/−Upp1−/− mice (referred to here as KO [knockout]) were used. Unlike in humans, murine uridine phosphorylase degrades Thd and d-Urd, therefore Tymp−/−Upp1−/−, rather than Tymp−/− mice,22 reflect the biochemical phenotype of MNGIE patients. A LV vector containing both TP and GFP sequences (PGK-TP-GFP) was reported previously19 for biochemical correction in Tymp−/−Upp1−/− mice and is used in this study as a control. In the present study, third-generation self-inactivating LVs containing a similar backbone as currently used in clinical trials for metachromatic leukodystrophy (MLD)17 and Wiskott-Aldrich syndrome23 were constructed for expression of TP. The human cellular phosphoglycerate kinase (hPGK) promoter was used to drive expression of the native human TYMP sequence (TP) as well as the re-coded human TYMP sequence (TPco) in the therapeutic vectors, as well as to drive expression of the GFP transgene in the sham control vector (Figure 1A). The rationale for using TPco was to enhance protein production with a minimum number of transduced donor cells and vector copy number, as reported for IL2RG and RAG2.24, 25 The strong spleen focus forming (SF) virus promoter, which may contribute to leukemic events by transactivating proto-oncogenes,26 was used to evaluate potential phenotoxicity by excessive overexpression of TYMP and/or genotoxicity rather than an option for potential clinical application (Figure 1A).
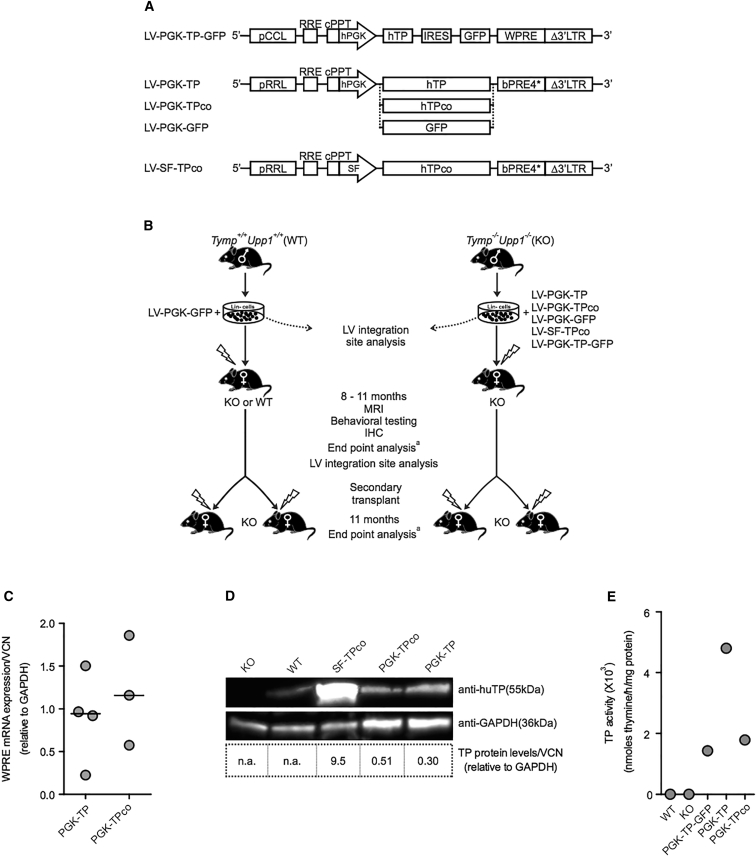
Figure 1.
Lentiviral Vectors and Design of Transplantation Experiments and TP Expression in the Target Cells
(A) Third-generation self-inactivating LV vectors used in the study and their elements are shown. pCCL and pRRL, LV vectors containing CMV-HIV or RSV-HIV 5′ long terminal repeats, respectively; RRE, Rev response element; cPPT, central polypurine tract; hPGK, human phosphoglycerate kinase promoter; hTP, native coding sequence of the human TYMP cDNA; hTPco, codon-optimized sequence; IRES, internal ribosome entry site; SF, spleen focus forming virus promoter; WPRE, Woodchuck hepatitis posttranscriptional regulatory element; bPRE4*, adapted form of WPRE.38 (B) Design of HSC transplantation experiments. The hematopoietic stem and progenitor cells (Lin−) from male donor mice were ex vivo transduced overnight by the LV vector constructs and transplanted into female recipient mice. To determine the integration site profiles, LAM-PCR and integration site analysis were performed on a fraction of the transduced cells in vitro and BM cells from transplanted primary mice in vivo; see also Figure S3A. To evaluate LV vector-related insertional transformation in vivo, Lin− cells from each primary recipient were transplanted into two secondary KO female recipient mice. All transplanted mice received 6Gy total body irradiation 24 hr prior to transplantation. All experiments included age-matched untreated KO and WT controls. aEnd point analysis: sample collection for biochemical and molecular analysis and flow cytometry analysis. MRI, magnetic resonance imaging; IHC, immunohistochemistry. (C–E) TP expression and activity in KO Lin− cells transduced (MOI 10) and cultured in vitro. (C) qPCR analysis of WPRE mRNA expression normalized to GAPDH levels and corrected for VCN/cell. The horizontal line represents the median, n = 3–4 replicates. (D) Western blot of total protein extracts from Lin− cells and their content corrected for VCN/cell, n = 1; n.a., not applicable. Included also are lysates of SF-TPco-treated cells confirming the higher TP protein with this promoter. (E) TP activity in the cultured Lin− cells.
Two virus concentrations (MOI) were used for transduction, i.e., MOIs of 3 or 10. By transplantation experiments (Figure 1B), the efficiency of the therapeutic LVs (PGK-TP, TPco) were evaluated for (1) biochemical correction (MOI 3 or 10) and (2) neurological correction as assessed by brain MRI, pathology, and behavioral tests (MOI 10). To assess the safety of HSCGT, primary recipients were subjected to long-term follow-up for potential transgene overexpression-related phenotoxicity (in particular in the SF-TPco treatment group) and LV-related genotoxicity, as well as an integration site analysis of gene-modified cells prior to and after engraftment in primary recipient mice aged 13 months, i.e., 11 months after HSCGT, to estimate the risk of insertional oncogenesis, and finally by secondary transplantation, a useful pre-clinical tool to detect vector induced hematological transformations.26, 27 Woodchuck posttranslational regulatory element (WPRE) mRNA and TP protein levels were detectable in PGK-TP and TPco-transduced Lin− cells (Figures 1C and 1D), and TP enzyme activity was enhanced in Lin− cells, granulocyte-monocyte progenitors, and 293T cells (Table S1). The two vectors were used interchangeably and referred to as PGK-TP(co) hereafter.
Biochemical Correction and Molecular Chimerism following Ex Vivo HSCGT
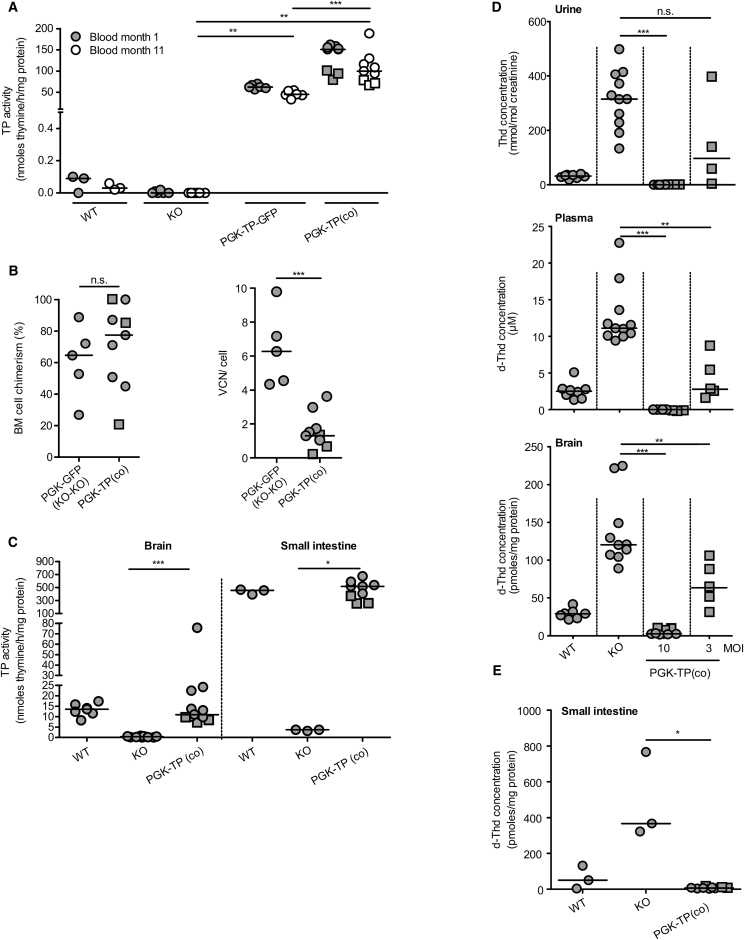
Stable blood TP activity was observed both in the PGK-TP-GFP group as previously reported20 and in recipients of the therapeutic LV-PGK-TP(co) (Figure 2A). Lin− cells successfully engrafted both PGK-TP(co) and PGK-GFP recipient KO mice, and integrated vector copies were detected (Figure 2B; Table S2). We next measured TP activity in diseased tissues of our mice. Transduction resulted in increased TP activity to normal levels in brain and small intestine in the PGK-TP(co)-treated mice (Figure 2C). As a consequence of the high-TP activity, urine and plasma nucleosides were undetectable in almost all recipients of PGK-TP(co) with extensive reduction in brain nucleosides compared to both KO and wild-type (WT) animals (Figures 2D and S1A). Overall, median nucleoside levels were reduced insufficiently at the lower MOI (Figures 2D and S1A; MOI 3, range 0.1–0.8 VCN/cell and percentage chimerism 47%–76%, n = 5; Table S2). Eleven months after transplantation, recipients of PGK-TP(co) displayed 98% and 57% reduction in median intestinal d-Thd and d-Urd levels, respectively (Figures 2E and S1B). Moreover, HSCGT provided high-TP enzyme activity and reduced nucleoside levels in skeletal muscle and liver (Figure S1C). All together, the transduction efficiency, integrated vector copies, and engraftment levels determine the outcome of biochemical correction (Table S2; Figures 2D and S1A). At an MOI of 3, significant reduction of nucleosides was observed, full correction of the biochemical phenotype only occurred at transplantation of 6 Gy irradiated recipients of 5 × 105 MOI 10 transduced cells, particularly in the small intestine, in contrast to the previous study.20
Figure 2.
Long-Term Biochemical Correction and Molecular Chimerism following HSCGT
Lin− cells were transduced with the therapeutic and control LV vectors at MOI 10, and 5 × 105 cells were transplanted into six Gy pre-conditioned 2-month-old KO mice. (A) TP enzyme activity was measured in blood at months 1 and 11 after transplantation, n = 3–9 mice/group. (B) BM cell chimerism and vector copy number of recipient mice, n = 5–9 mice/group. (C) Brain and intestine TP activity were measured 11 months after transplantation, n = 3–12 mice/group. (D) Quantification of Thd in urine, d-Thd in plasma, brain 8–11 months after transplantation (MOI 10 or 3), n = 4–11 mice/group, and (E) in intestines 11 months after transplantation, n = 3–9 mice/group. The horizontal line represents the median, *p < 0.05, **p < 0.01, and ***p < 0.001; n.s., not significant. Mice in the PGK-TPco treatment group are identified (square symbols).
MRI and Rescue of Brain White Matter Damage after HSCGT
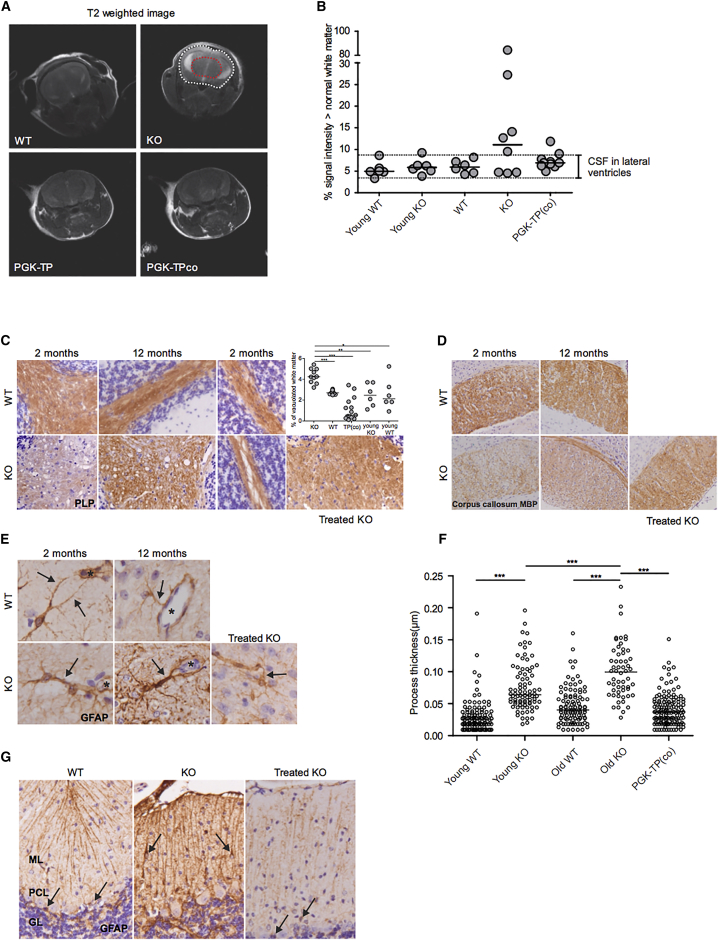
Neurological changes are consistently reported in MNGIE patients, and minor changes were reported in old Tymp−/−Upp1−/− mice aged ≥18 months.22 To characterize neurological abnormalities at earlier ages and to address the impact of HSCGT as early as at 2 months of age, brain MRI scans and immunohistochemistry (IHC) were performed. In contrast to WT mice, an increase in percentage of fluid content was observed as a hyper-intense signal in the brains of KO mice aged 6 to 12 months (five out of eight) (Figure 3A). Increased T2 signals are indicative for fluid accumulation in the white matter surrounding the lateral ventricles, suggesting edema. White matter edema was absent in most treated mice; 8 out of 10 treated mice fell in the same hyper-intense volume range of WT animals (Figure 3B). Brain sections from KO mice stained with mature myelin proteolipid protein (PLP) and myelin basic protein (MBP) revealed the presence of progressive vacuolization in the cerebellar and cerebral white matter structures absent in WT mice. White matter vacuolizations were lined and crossed by thin immunopositive tissue strands, consistent with intramyelinic edema (Figures 3C and 3D). PLP and MBP staining revealed disappearance of white matter vacuolization in the cerebellar white matter and corpus callosum of PGK-TP(co)-treated group (Figures 3C and 3D). IHC staining for the astrocyte-specific glial fibrillary acidic protein (GFAP) revealed increased thickness of astrocyte processes abutting the blood vessels in KO mice as young as 2 months, compared with age-matched WT mice, which was further increased in KO mice aged 12 months (Figures 3E and 3F). The thickness of astrocyte perivascular processes in cerebellum, corpus callosum, and hippocampus white matter of PGK-TP(co)-treated mice were reduced to normal levels (p < 0.0001) (Figures 3E and 3F). In addition, GFAP staining of the cerebellar cortex revealed that Bergmann glia were localized to the molecular layer in KO mice in contrast to the normal localization in the Purkinje cell layer in WT mice and after treatment (Figure 3G). All together, brain MRI and IHC revealed the classical morphological changes in KO mice at a younger age and further demonstrated an underlying edema linked to changes of the brain astrocytes, which are reversed upon HSCGT.
Figure 3.
Brain Magnetic Resonance Imaging and Characterization of Brain White Matter Damage and Rescue by HSCGT
Lin− cells were transduced with the therapeutic LV vectors at MOI 10, and 5 × 105 Lin− cells were transplanted into six Gy pre-conditioned 2-month-old KO mice. MRI scans were performed on 2-month-old (young controls) and 6- to 12-month-old controls and treated mice. (A) Representative T2 scans of WT, KO, and treated mice. Hyper-intense voxels within the brain (white dotted) were quantified by using a reference ROI (red dotted). (B) Results of the quantification of cerebrospinal fluid (CSF) and white matter edema. The data displayed is the percentage of voxels with high signal intensity of the whole brain, n = 6–10 mice/group. The horizontal line represents the median. Immunohistochemical staining was performed on brain tissue collected from young controls and 12-month-old controls, n = 2–3/group, and treated mice 9–10 months after transplantation, n = 4 mice. (C) Staining for the mature myelin protein proteolipid protein (PLP) of cerebellum of young (left and right at higher magnification) and old (middle) KO and WT female mice shows normal myelin amounts in the cerebellar white matter of a PGK-TP-treated mouse. Quantification shows significantly more vacuoles in the cerebellar white matter of KO animals. The horizontal line represents the median, *p < 0.05, **p < 0.01, and ***p < 0.001. (D) Staining for the mature myelin protein myelin basic protein (MBP) of the corpus callosum shows myelin pallor in KO compared to WT mice, which is rescued after gene therapy in treated female mice; a section from a recipient of PGK-TPco transduced HSCs is shown. (E) Staining for the astrocyte-specific intermediate filament protein glial fibrillary acidic protein (GFAP) shows thicker cellular processes abutting the blood vessels in the cerebellar white matter of KO mice as young as 2 months compared to WT mice. The asterisk indicates the blood vessel lumen. Labeling against GFAP shows thinner perivascular astrocytic processes in the brain of a recipient of PGK-TPco transduced HSCs. (F) Thickness of astrocyte processes surrounding blood vessels, n ≥ 56 processes/group. The horizontal line represents the median, ***p < 0.0001. (G) Stain against the astrocyte-specific intermediate filament protein glial fibrillary acidic protein (GFAP) shows that Bergmann glia (arrows) are located in Purkinje cell layer (PCL) of the cerebellar cortex of WT and a recipient mouse of PGK-TPco transduced HSCs (left and right, respectively), whereas in KO mice, they are mislocalized to the molecular layer (ML) (middle); GL, granular layer.
Safety Evaluation
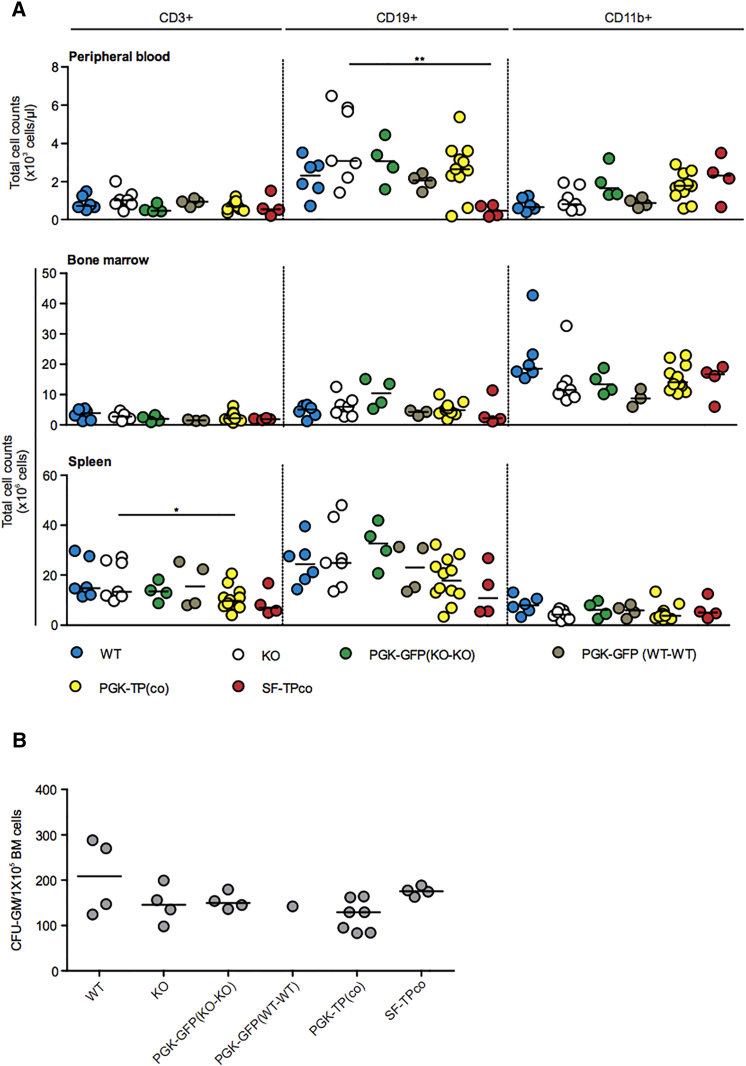
Excessive transgene expression-related phenotoxicity and/or genotoxicity caused by LV insertional mutagenesis are potential drawbacks for clinical application of HSCGT. These two issues were not addressed in previous studies19, 20 of HSCGT for MNGIE. A total of 106 controls and primary recipients of 5 × 105 Lin− cells transduced with therapeutic and GFP LV were monitored for 11 months. Survival was not significantly reduced in the group combining PGK-TP or TPco recipients (70%) compared with PGK-GFP (92%) or untreated KO controls (94.4%) (Figure S2A). Some recipients of PGK-TP or TPco were sacrificed and analyzed prematurely because of death or high discomfort scores. We have been unable to determine the actual cause of wasting, but importantly, vector-positive hematological clonal expansion was not observed (Table 1; Figure S2). At termination, hematopoietic reconstitution between groups was similar, except for a reduction in numbers of spleen CD3+ T cells in PGK-TP(co) and peripheral blood CD19+ B cells in SF-TPco-treated mice (Figure 4; Table S3). Excessive increase in TP activity was detected in Lin− cells, granulocyte-monocyte progenitors, and 293T cells transduced with LV-SF-TPco (Table S4A), in blood and brain (Table S4B). This increase was substantially higher than in the PGK-TP and TPco treatment groups (Table S1; Figures 2A and 2C). As expected, nucleosides were not detectable in urine and plasma and extremely reduced in the brains of SF-TPco mice (Table S4B). Although percentage donor chimerism in SF-TPco-treated mice was high (Table S4B), 12 out of 14 primary recipients were sacrificed prematurely mainly due to severe body weight loss (>15%) observed at death (n = 6 out of 12). This increased incidence of weight loss was not observed in PGK-TP or TPco treatment groups (Table 1; Figure S2). To evaluate genotoxic risk, we analyzed the therapeutic and GFP containing LV vector integration site profiles in mouse Lin− cells in vitro and in bone marrow (BM) cells from primary recipient mice (Figures 1B and S3). In total, 7,435 unique integration sites (IS) were retrieved by linear amplification mediated-PCR (LAM-PCR) and sequencing from 4 in vitro samples and 10 transplanted mice (Table 2). A polyclonal integration pattern was observed with 1,174 common IS (CIS).28 In total, 225 IS near oncogenes were identified, accounting for 3.03% of all genes (Table 2). We analyzed the IS patterns for the oncogene frequency between groups classified based on promoter, i.e., PGK versus SF, or pre-transplantation lineage negative cells versus bone marrow in vivo or transgene GFP versus TP (co), but no differences were observed (Table 3). To assess genotoxicity further, BM Lin− cells from primary recipients (n = 44) were transplanted into 88 secondary recipients. In both the primary and secondary recipients, we did not observe any vector-positive hematological clonal expansion in the therapeutic PGK-TP or PGK-TPco groups (Table S5). In nine recipients of cells LV transduced by PGK-GFP, TP, TPco, and SF-TPco, vectors altered leukocyte levels and counts revealed hematological aberrations. These cases had very low median vector copy number (VCN)/cell (0.03 with range 0.01–0.07) and donor chimerism (0.7% with range 0.2%–7.1%, n = 8; Table S5) and therefore were classified as non-vector-related. Two PGK-TP-GFP recipients of Lin− cells of a single primary mouse developed vector-positive diffuse intermediate to large B220+GFP+ B cell lymphoma (Table S5) at 9 and 11 months after transplantation. The VCN/cell and percentage chimerism were 0.86 and 67% in one mouse and 0.76 and 48% in the other mouse. An abundance of abnormally large B cell population of B220+GFP+ cells was observed in BM (Figure S4A) of both mice, i.e., >80% of total BM cells (Table S5), spleen, and peripheral blood of one mouse. Pathological analysis showed infiltration of malignant B cells in spleen and liver (Figure S4B). The B cell clone contained a single dominant integration site (IS) in the chromosome 11 gene Zfp207. No known oncogenes were found within 500 kb of this IS (Figure S4C). Of note, the primary donor mouse had two dominant IS located in the genes Aplp2 on chromosome 9 and Csmd3 on chromosome 15, but the Zfp207 IS was not detected by LAM-PCR. Primer-specific PCR amplification confirmed very low presence of this IS in bone marrow and spleen (data not shown). Besides the miscellaneous symptoms observed in therapeutic PGK-TP(co)-treated mice, which are not likely to be related to the gene therapy procedure, incidence of phenotoxicity was observed in the SF treatment group, i.e., severe weight loss in the majority of the treated cohort. Hematological abnormalities or increased risk of insertional mutagenesis were not observed in the group of mice receiving the therapeutic vectors.
Table 1.
Hematological Aberrations and Clonal Expansion of Transduced Cells Were Not Observed in Primary Recipient Mice
| LV Vector | Number of Vector-Positive Hematological Aberrationsa | Number of Non-Vector-Positive Hematological Aberrationsa | Number of Prematurely Dead Mice or Mice Sacrificed with High Discomfort Scoresb |
|---|---|---|---|
| WT CTR (n = 13) | 0 | 0 | 0 |
| KO CTR (n = 18) | 0 | 0 | 1 (macrocephaly) |
| PGK-GFPc (n = 25) | 0 | 0 | 1 (found dead) |
| 1 (rectal prolapse) | |||
| 1 (passive) | |||
| PGK-TP-GFP (n = 6) | 0 | 0 | 0 |
| PGK-TP (n = 19) | 0 | 0 | 1 (plagiocephaly) |
| 1 (wounds) | |||
| 1 (passive) | |||
| 1 (epileptic seizure during behavioral test) | |||
| 2 (found dead) | |||
| 1 (body weight loss)d | |||
| PGK-TPco (n = 11) | 0 | 0 | 1 (epileptic seizure during behavioral test) |
| 1 (found dead) | |||
| 2 (passive) | |||
| SF-TPco (n = 14) | 0 | 0 | 5 (body weight loss) |
| 1 (kyphosis, body weight loss) | |||
| 2 (passive) | |||
| 1 (edema) | |||
| 1 (found dead) | |||
| 2 (kyphosis) |
Mice were transplanted with LV transduced 5 × 105 BM Lin− cells (MOI 10) and followed for 11 months after transplantation. CTR, control untreated mice.
Hematological aberrations and clonal expansion of transduced cells were assessed based on altered FACS phenotypes and/or elevated WBCs, and/or enlarged spleen, thymus, or lymph nodes. BM was tested for male donor cells and vector copy number.
The right column includes mice that were found dead or mice sacrificed prematurely during the experiment due to high discomfort scores and for which hematological aberrations could not be identified (details are specified between parenthesis).
PGK-GFP group includes recipient mice of KO or WT Lin− cells transduced by PGK-GFP.
Body weight loss at the time of death >15% over a time span of 1 to 2 months.
Figure 4.
Hematopoietic Reconstitution after HSCGT
The following analyses were performed 9–11 months after transplantation of Lin− cells transduced at MOI 10. (A) FACS analysis of peripheral blood, bone marrow, and spleen cells. Cells were stained with antibodies against CD45.2 and CD3, CD19, and CD11b for detection of, respectively, T, B, and myeloid leukocytes (n = 3–12 mice/group). The horizontal line represents the median; *p < 0.05, **p < 0.01. (B) Colony-forming unit assay. BM cells were cultured and granulocyte-monocyte progenitors (CFU-GM) were counted, n = 1–6 mice /group. The horizontal line represents the median.
Table 2.
Summary of LV Vector IS Retrieved by LAM-PCR and Sequencing
| Cell Type (Donor-Recipient) | LV Vector | Number of Samples | Number of IS | Number of Oncogenes | % Oncogenes |
|---|---|---|---|---|---|
| In Vitro | |||||
| Lin− (KO) | PGK-TP | 2 | 3,147 | 102 | 3.24 |
| Lin− (KO) | SF-TPco | 2 | 2,489 | 74 | 2.97 |
| In Vivo | |||||
| BM (WT-WT) | PGK-GFP | 2 | 782 | 20 | 2.56 |
| BM (KO-KO) | PGK-TP | 5 | 488 | 3 | 0.61 |
| BM (KO-KO) | PGK-TP-IRES-GFP | 3 | 529 | 26 | 4.91 |
| total | 14 | 7,435 | 225 | 3.03 | |
Table 3.
Summary of LV IS Classified Based on Promoter, Cell Type, and Transgene
| Group | Number of Samples | Number of IS | Unique Genes | CIS | Unique Oncogenes | Number of Oncogenes | % Oncogenes |
|---|---|---|---|---|---|---|---|
| All | 14 | 7,435 | 4,104 | 1,174 | 110 | 226 | 3.03 |
| PGK only | 12 | 4,946 | 3,082 | 702 | 87 | 152 | 3.07 |
| SF | 2 | 2,489 | 1,869 | 241 | 52 | 74 | 2.97 |
| Lin− (in vitro) | 4 | 5,636 | 3,408 | 815 | 102 | 177 | 3.14 |
| BM (in vivo) | 10 | 1,799 | 1,379 | 178 | 35 | 49 | 2.72 |
| GFP only | 2 | 782 | 666 | 48 | 17 | 20 | 2.56 |
| TP (co) | 12 | 6,653 | 3,799 | 1,030 | 106 | 206 | 3.10 |
Discussion
The present study, using a clinically applicable self-inactivating transfer vector (SIN) LV vector with a modified WPRE and without GFP and a highly efficient, overnight gene transfer procedure, demonstrates not only full biochemical correction, but, importantly, also complete clearance of intestinal nucleosides and a remarkable phenotype correction in the brain of Tymp−/−Upp1−/− mice. Overall, the biochemical correction obtained in blood and tissues reported earlier by using PGK-TP-GFP20 was confirmed here in a larger cohort of mice by using the therapeutic vectors LV-PGK-TP(co). The additional complete clearance of intestinal nucleosides and remarkable phenotypic correction in the brain demonstrates the feasibility of a curative gene therapy approach for MNGIE patients.
In MNGIE patients, the intestines are severely affected, resulting in cachexia, severe infections, and death. The intestine facilitates uptake of nutrients possibly requiring high TP enzyme activities to reduce incoming thymidine levels locally. Of note, intestinal TP activity in WT mice is substantially higher than that in blood and brain tissue (Figures 2A and 2C), which emphasizes that high TP activity levels in intestine are likely required to normalize nucleosides. In our study, therapeutically relevant transduction efficiencies by PGK-TP(co) (median VCN/cell = 1.3, range, 0.2–3.6; Figure 2B; Table S2) resulted in complete clearance of intestinal nucleosides, in contrast to the earlier study,20 in which d-Thd was significantly reduced in plasma and brain, but not in the intestine. In addition to the high liver and muscle TP activity, this finding of an improved biochemical correction supports our low vector MOI (3 or 10 versus 10020) and pre-transplant conditioning protocol.
The current experiments, performed with high transduction efficiency and dose, resulted in supranormal levels of TP activity and in near-complete elimination of systemic nucleosides, i.e., below WT levels, indicating that lower enzyme levels should be sufficient to correct the biochemical imbalance. For gene-modified and vector-dose-titration experiments, relatively large cohorts of mice are required to demonstrate a dose-response relationship. However, whether or not lower cell and LV dose would be efficient is of doubt, since the small intestine requires a high cell dose and an MOI of 10 to normalize TP activity and nucleosides (Figures 2C, 2E, and S1B). The absence of a clear clinical phenotype is a limitation of the only murine disease model of MNGIE available, and very scanty pathological findings have been reported, mainly in Tymp−/−Upp1−/− mice aged ≥18 months.22 In the present study, we observed pathological white matter changes in KO animals as early as 2 months of age and MRI changes at 6 months of age, which recapitulate the findings in the brain of MNGIE patients, i.e., white matter edema without demyelination or gliosis and hyperintense T2-weighted signal changes.8, 29 In this, our mouse model is representative of the human MNGIE syndrome. Importantly, HSCGT resulted in elevated TP activity and restored brain white matter integrity and reduced edema as evident by both MRI and IHC. Another study on MNGIE patients demonstrated that loss of brain TP was associated with loss of blood-brain barrier integrity, which resulted in activation of astrocytes.30 We showed increased astrocytic process thickness in Tymp−/−Upp1−/− mice, which is limited to the cell processes abutting blood vessels. This argues against reactive astrogliosis and rather suggests swelling of the astrocyte perivascular endfeet. Such a phenotype might be responsible for the observed progressive intramyelinic edema in Tymp−/−Upp1−/− mice as reported in other white matter diseases31 and could be explained by an interference with the normal function of astrocytes in maintaining ion-water homeostasis.32 The latter finding prompts further research aimed at investigating the molecular mechanisms underlining brain pathophysiology in MNGIE, as this knowledge could be exploited for further therapeutic intervention.
We did not demonstrate TP-positive hematopoietic cells in the brain, however, previous murine gene therapy studies using AAV2/8 vectors21 transducing the liver showed reversal of nucleoside imbalance without the local presence of TP in the brain. However, in HSCGT, gene-modified monocyte progenitors may migrate to the brain and differentiate into microglia.33 Since TP is a cytoplasmic protein, we measure this TP activity in the brain (Figure 2C), indicating that transduced microglia might reside in the brain after long-term follow-up.
Despite the brain pathology in Tymp−/−Upp1−/− mice, we could not detect significant differences in several memory and motor function assessments, which involved cerebellum, cortex, and hippocampus, between the Tymp+/+Upp1+/+, Tymp−/−Upp1−/−, and gene therapy-treated mice (data not shown). This could be attributed to differences in physiology and lifespan between mice and man. Other leukodystrophy mouse models, including those for megalencephalic leukoencephalopathy with subcortical cysts similarly lack a clinical phenotype despite the presence of severe pathological abnormalities.31 Similar to the analysis of MNGIE human brains,30 we did not detect mtDNA depletion, deletions, or any changes in mtDNA topology or strand breaks. Although the variation between individual mice is large, Tymp−/−Upp1−/− and treated mice showed signs of replication stalling in brain tissue (data not shown), which requires further investigation.
Of major concern, as yet unaddressed, is the potential transgene or LV-related phenotoxicity or genotoxicity. The results indicate that overexpression has no direct effect on hematopoietic system. First, we did not observe loss of vector-transduced cells in the hematopoietic compartment or donor-derived cells (Figure 2B; Table S2). Second, blood TP activity was not extremely reduced 11 months after transplantation (Figure 2A), so there is no clear selection for low expressor cells. Finally, there was no difference in the polyclonality between GFP and therapeutic vectors in vitro and 11 months after transplantation and therefore no apparent reduction in the number of integrated vector (Table 2). The toxicity is not related to the LV vector, since LV-PGK-GFP did not show toxicity (Table 1; Figure S2). Additionally, the higher death rates with the clinical vector included only one mouse that had wasting. For the other deaths, the symptoms or potential causes of death were miscellaneous. Only when highly overexpressed by the SF vector was the main observation before death the wasting (Table 1; Figure S2). Therefore, we assume that the observed toxicity is related to the excessively expressed TP by the strong SF promoter and the increased mortality (Table 1; Figure S2A) likely due to complete elimination of d-Thd and/or d-Urd affecting downstream mitochondrial dNTPs. dNTP pool imbalance triggers cell cycle arrest and apoptosis in vitro in hematopoietic cell lines and yeast variants,34, 35 which, however, needs further investigation in Tymp−/−Upp1−/− mice. Using the therapeutic vectors, oncogenic adverse effects were not observed after a total follow-up in primary and secondary recipient mice of 22 months, demonstrating the safety profile of PGK-driven TP expression. A single, possibly insertional, oncogenic event was observed in the PGK-TYMP-IRES-GFP-treated mice. Two secondary recipients acquired the same B cell lymphoma clone with the PGK-TP-GFP vector integrated near the Zfp207 gene, which could be traced back to one primary recipient mouse. Zfp207 or its human homolog is not reported as a proto-oncogene but has a role in cell cycle and mitotic nuclear division: in particular, ZFP207 is required for proper chromosome alignment,36 and Zfp207 knockdown has been associated with lethal chromosome congression defects in transformed cells.37 The B cell lymphoma contained a LV integration with an early version Woodchuck hepatitis virus posttranslational regulatory element (PRE). Although reports have never shown unequivocally that the use of such a PRE carries additional insertional oncogenic risks, our therapeutic LV vectors contain a mutated element (bPRE4*) to reduce risks further.38 Since the lymphomas in the secondary transplanted mice were identical, the oncogenic event necessarily occurred in the primary transplanted mouse. The integration near the Zfp207 gene might have contributed to the development or expansion of this clone, but it is not excluded that the observed lymphoma is not related to vector integration at all, which would require further investigation of potentially oncogenic mutations.
In summary, the therapeutic PGK-TP(co) vectors provided sufficient expression for biochemical and phenotypic correction without detectable phenotoxicity and a favorable safety profile, similar to the LV vectors currently in clinical trial for other inherited disorders. Nonetheless, excessive TP expression in the LV-SF treatment group, not intended for clinical application, resulted in reduced survival (Table 1; Figure S2), a finding in need of further investigation and underlining the requirement of a moderately strong housekeeping gene promoter such as PGK for clinical application in MNGIE, possibly in combination with low-intensity or non-myeloablative conditioning.
It is concluded that the present study warrants further development including cell and vector dose-response experiments with a larger cohort of mice, biodistribution studies, and assessment in human hematopoietic cells to substantiate any claims that the therapeutic vector in any way may be toxic or to develop this further to clinical trials for MNGIE.
Materials and Methods
LV Vectors
The SIN pCCL-hPGK-TP-IRES-GFP-WPRE was described before and is used in our study as a control.19 The human TYMP sequence from this LV vector, a re-coded TYMP sequence (human TPco, GenScript, Piscataway, NJ, USA)24 with optimized open reading frame (Genscript algorithm) with a consensus Kozak sequence and an additional stop codon, and the green fluorescent protein GFP sequence were all cloned into the pRRL-SIN-bPRE4* backbone under the human PGK promoter.38 Human TPco was also cloned under the SF promoter (Figure 1A). Third-generation packaging plasmids pMDL-g/pRRE, pRSVREV and envelop plasmid pMD2-VSVg were used.39 LV vectors were generated by calcium-phosphate precipitation of HEK293T cells.40 Forty-eight hours after transfection, LV vectors were concentrated by ultracentrifugation at 20, 000 rpm for 2 hr at 4°C and the pellets resuspended in PBS. LV vector titration was performed on HeLa cells. After 72 hr, cells were analyzed on an LSR-II flow cytometer and FACS diva software to calculate the titers of a GFP containing batch. Titers of TP and TPco containing vectors were determined by qPCR.
HSC Transduction and Primary and Secondary Transplantation
The generation of Tymp−/−Upp1−/− (KO) and Tymp+/+Upp1+/+ WT mice on a C57BL/6J genetic background have been described before.22 The animals were bred in the Erasmus MC Experimental Animal Center (Rotterdam, the Netherlands). During the experiments, mice were kept in filter top cages, fed with autoclaved water, and irradiated chow ad libitum. The animal experiments were reviewed and approved by an ethical committee of Erasmus MC, Rotterdam in accordance with legislation in the Netherlands.
Bone marrow cells were harvested aseptically from femurs and tibiae of 4- to 19-week-old male mice by flushing with PBS, followed by lineage depletion (Lin−) according to manufacturer recommendations (BD Biosciences). Lin− cells were cultured overnight in the presence of the LV vector constructs at an MOI of 10 or 3 in serum-free modified Dulbecco’s medium with supplements41 and stimulated with murine stem cell factor (SCF; 100 ng/mL), human FMS-like tyrosine kinase 3 ligand (Flt3-L, 50 ng/mL), murine thrombopoietin (TPO, 20 ng/mL). Subsequently, Lin− transduced cells (5 × 105) were injected into the tail vein of 4 to 17 weeks of age recipient KO or WT female mice, subjected to six Gy irradiation 24 hr prior to transplantation. For the secondary transplantation, similarly each of two 5- to 14-week-old KO female recipients were transplanted with 2 × 105 enriched Lin− cells from viable cryopreserved bone marrow of a single primary recipient mouse (Figure 1B).
Sample Collection, Analysis, and Termination of Experiments
During the course of experiments, primary, and secondary recipient mice were monitored carefully for any signs of discomfort. Body weights and leukocyte phenotypes were measured monthly up to 6 months post-transplantation in some primary recipients. Complete blood cell counts (CBCs) and leukocyte phenotypes of secondary recipients were monitored 3 months after transplantation. When applicable, blood and urine samples for biochemical analysis were collected. Mice were removed from experiments if they appeared ill, had severe body weight loss of >15%, had changes in behavior, or for other reasons, for example malocclusion or fight wounds. The following analyses were performed at experimental end point or at death when possible. EDTA peripheral blood and urine samples were collected on ice and stored at −80°C for biochemical analysis. Mice were sacrificed by inhalation of a CO2/O2 mixture and were transcordially perfused with PBS. The following was performed: assessment of body weights, physical appearance, presence of abnormality such as pale feet, rectal prolapse, injuries, lumps (tumors, cysts), visual examination of major organs (heart, lung, brain, liver, kidney, gastrointestinal tract, spleen, thymus, lymph nodes), documentation of organomegaly (spleen, thymus, lymph nodes), and processing of samples from lesions and enlarged organs for H&E staining/IHC. Tissue samples were snap-frozen in liquid nitrogen and stored at −80°C for biochemical analysis. CBCs were measured using automated equipment (scil Vet ABC Hematology Analyzer, Gurnee, IL). Leukocytes, spleen cells, and fractions of total bone marrow were used for FACS analysis. In some experiments, bone marrow cells were cultured to assess progenitor cell content by colony assays. Aliquots of bone marrow cells were stored at −80°C for molecular analysis as described below, and the remainder of the bone marrow cells was cryopreserved for retransplantation. For testing the risk of insertional oncogenesis, hematological aberrations were based on abnormal FACS phenotype of at least one of the hematopoietic tissues, BM, spleen, or peripheral blood and/or white blood cell counts (WBC) (further analyzed for leukemia in case WBC > 25.0 × 103 cells/μL) and /or enlarged spleen, thymus, or lymph nodes. The hematological aberration was considered a potential oncogenic clone if both donor cells and VCN were prominent in the recipient’s BM sample.
Flow Cytometry
Erythrocytes were removed with lysis buffer (0.15 M NH4Cl, 0.1 M EDTA), and leukocytes, bone marrow, and spleen cells were washed with Hank’s balanced salt solution (Invitrogen) containing 0.5% (w/v) BSA and 0.05% (w/v) sodium azide (HBN) followed by 30 min incubation at 4°C in HBN containing 2% heat-inactivated normal mouse serum and antibodies against CD45.2, CD3, CD19, and CD11b, directly conjugated respectively to APC-Cy7, APC, PE, and PerCP-Cy5.5; or, against CD8, CD45R/B220, Gr-1, c-Kit or CD4, IgD, CD19, Sca-1 or CD3, IgM, CD11b, directly conjugated respectively to APC, PE or PerCP (Cy5.5) (BD Biosciences and Biolegend, London, UK). Cells were washed and measured on a Canto or LSR-II flow cytometer (BD Biosciences).
Progenitor Cell Assay
To detect colony-forming unit granulocyte monocyte (CFU-GM), 5 × 104 total bone marrow cells in 10% of the total volume were seeded in semisolid methylcellulose in 80% of the total volume (MethoCult M3231, Stem Cell Technology, France) with 10% of the total volume of Iscove’s modified Dulbecco’s medium (IMDM) supplemented with rm-SCF (100 ng/mL), rm-interleukin-3 (30 ng/mL) and rm-GM-CSF (30 ng/mL) (R&D Systems). Cultures were incubated at 37°C in a humidified atmosphere containing 5% CO2, and colonies were scored 7 days later under an inverted microscope.
Biochemical Analysis of TP Enzyme Activity and Nucleoside Levels
TP enzyme activity in blood cells, tissue samples, and plasma d-Thd and d-Urd levels were measured by liquid chromatography (UPLC-UV), while tissue d-Thd and d-Urd levels were measured by liquid chromatography-mass spectrometry (HPLC-MS) as described before.19, 21 Urinary Thd and d-Urd were measured by HPLC.42 Urine samples were thawed to room temperature by incubation at 37C° for 30 min prior to measurement of concentrations. One volume of sample plus 1 volume of internal standard solution was diluted with 1 volume of eluent A (30 mM ammonium acetate, 1% methanol [pH 4.80]). The samples were incubated at 60°C for 30 min, followed by centrifugation on filter units at room temperature at 13,000 × g for 20 min. Twenty microliters of filtrates were then injected into an HPLC system (Shimadzu, LC20 series with a binary pump and Photodiode array detector), equipped with an Alltima C18 5μ, 250 mm × 4.6 mm column and Alltima C18 5μ guard column. Chromatography was performed as described previously.42 Concentrations were calculated by comparison of the Thd and d-Urd peak areas in samples to the corresponding peak areas of standards using caffeine as an internal standard to correct for injection volume. Creatinine concentrations of the samples were measured in the same runs.
qPCR and Western Blotting
KO Lin− cells were transduced by PGK-TP, PGK-TPco, or SF-TPco (MOI 10) and cultured in vitro for 7 days. Total RNA was extracted, and 1 μg was reverse transcribed into cDNA (RNeasy MicroKit and QuantiTect Reverse Transcription Kit, QIAGEN, Hilden, Germany) following manufacturer’s instructions. Genomic DNA isolation from both cultured Lin− cells and bone marrow cells was performed by using NucleoSpin Tissue kits (Bioké, Leiden, the Netherlands) following manufacturer’s protocol. RNA and DNA concentration and purity were determined by using Nanodrop (Nano Drop 1000Spectrophotometer). Template cDNA and 100 ng template genomic DNA were subject to qPCR by using SYBR Green PCR master mix and the primers described before26, 43, 44 and summarized in Table S6 (Applied Biosystems, Foster City, CA; Eurogentec, Maastricht, the Netherlands). The PCR reactions were carried out in the ABI7900 Taqman machine, and results were analyzed with SDS2.2.2 software (Applied Biosystems). The average vector copy per cell were calculated by comparing the cycle threshold values obtained against a standard curve obtained from mouse 3T3 or HeLa cells, and to calculate Y chromosome chimerism a standard curve was obtained from male mouse bone marrow samples.
Western blotting was performed by using Novex midi gel system for electrophoresis and blotting (Invitrogen, Carsland, CA) following manual’s instructions. Cultured Lin− cells were lysed by sonication, protein concentrations were determined by using BCA protein assay kit (Thermo Fisher), and a total of 15 μg proteins were separated by 4%–12% Bis-Tris SDS-PAGE, transferred to nitrocellulose and PVDF membranes and blotted with primary and secondary antibodies (summarized in Table S7), and finally signals were detected with enhanced chemiluminescent substrates detection kit (Thermo Fisher). ImageJ software was used to quantify protein levels.
Vector IS
In order to identify vector IS, vector LTR-genome junctions were amplified by LAM-PCR as previously described.45, 46 In brief, after linear amplification with biotinylated LTR-specific primers, amplification products were purified using streptavidin magnetic beads. The following steps included complementary strand synthesis, parallel digestion with three different restriction enzymes (Tsp509 I, HpyCH4 IV, and Aci I), and ligation to a linker cassette. The fragments generated were then amplified by two additional exponential PCR steps. LAM-PCR products were separated by gel electrophoresis on Spreadex high-resolution gels (Elchrom Scientific), underwent a fusion PCR for the addition of barcoded adaptors, were pooled into libraries, and were Illumina sequenced (MiSeq) as previously described.47 The resulting sequences were processed through a customized informatics pipeline (Biasco et al.46; L. Leonardelli et al., 2016, ASGCT, abstract), which performs cleaning for quality and collisions, mapping, and annotating of IS on the mouse genome (December 2011, GRCm38/mm 10).
mtDNA Analysis
MtDNA copy number, conformation, and integrity were evaluated in total and mitochondrial DNA extracted from tissue samples.48, 49 In brief, mtDNA copy number was determined by real time-PCR using primers and Taqman probes for mitochondrial and NDUVF1 and the AccuStartII PCR supermix (QuantaBio). The respective primer sequences are summarized in Table S6. To quantify the levels of existing mtDNA strand breaks, mitochondrial DNA and free 3′ ends were labeled in vitro using radioactive incorporation of dCTP by terminal deoxynucleotide transferase. 0.5 μg of purified mtDNA was incubated with 5 μCi dCTP (3000 Ci/mmol) and 7.5 U terminal deoxynucleotidyl transferase (TdT) in 30 μL 1× TdT buffer at 37°C for 30 min and separated over a 1% Tris-borate-EDTA (TBE)/agarose gel containing 0.1 μg/μL ethidium bromide. Equal loading was confirmed by UV visualization, the gel was Southern blotted onto Hybond-XL membrane, and the intensity of each lane was quantified by phosphor imaging (Molecular Imager FX, Bio-Rad). The abundance of replication intermediates was visualized using Brewer/Fangman 2D neutral/neutral agarose electrophoresis. MtDNA deletions were detected by long-range PCR as previously described50 and visualized by Phosporimager.
MRI
Imaging was performed on a 7.0 T dedicated animal scanner (Discovery MR901, Agilent Technologies/GE Healthcare) using a 4-channel surface receiver coil (Rapid MR International, Ohio, USA) and a 72-mm transmit body coil. Single-shot fast-spin echo (SSFSE) images were acquired with settings repetition time/echo time (TR/TE) = 1,600/8 ms, field of view (FOV) = 5 cm, voxel size 0.2 × 0.2 × 1 mm. T2 images were additionally acquired using a fast pin echo sequence for anatomical reference, settings TR/TE = 2,500/25 ms, FOV = 5 cm, voxel size 0.2 × 0.2 × 1 mm.
Image analysis was as follows: The SSFSE is very sensitive for high liquid content and therefore, by using these images, the signal intensity (SI) of voxels with high liquid content is much higher than that of other tissue. This allowed us to determine the fraction of edematic tissue by selecting voxels through thresholding after segmentation of the whole brain. The threshold was determined by quantifying normal tissue values by determining a reference region of interest (ROI) within normal brain tissue. This ROI was manually drawn in a slice of brain without hyper-intense areas. Voxels with SI of 4 standard deviations above the reference ROI SI were considered to have high liquid content (4 SD = 99.9% confidence interval). The analysis was performed using in-house developed software based on MATLAB (MathWorks, Natick, MA, USA).
IHC
Mice were euthanized by inhalation of a CO2/O2 mixture and transcardially perfused with PBS to remove blood. Brains were extracted and immersion-fixed in 4% paraformaldehyde for 24 hr, paraffin-embedded, and cut longitudinally to obtain 6-μm-thick sections. Staining was performed as previously described.31 Antigen retrieval was performed by treatment with 0.01 M citrate buffer (pH 6.00) in a steamer and endogenous peroxidase blocked with 0.3% H2O2 in methanol. Sections were blocked in 5% normal goat serum then incubated overnight at 4°C with the primary and secondary antibodies summarized in Table S7. Immunoreactivity was detected with 3-3′-diaminobenzidine as chromogen. Sections were finally counterstained with hematoxylin and visualized under a Leica DM6000B microscope. Omitting the primary antibodies yielded no significant staining. Image series from brain sections stained with H&E or against protein PLP or the GFAP were obtained with a ×40 objective. Quantification of vacuole and astrocyte morphology was performed blind to the genotype using ImageJ. The percentage of cross-sectioned area occupied by vacuoles was quantified in the cerebellar and callosal white matter. The thickness of perivascular astrocytic cell processes at the maximal width was measured.51
Statistical Analysis
Significance of differences between groups was determined by two-tailed Mann-Whitney U test. Data was graphed and analyzed by Graph Pad-Prism5 software (version 5.03).
Author Contributions
Conceptualization, R.Y., N.P.v.T., M.S., I.F.M.d.C., G.W.; Investigation, R.Y., R.C.-P., J.T.-T., M.B., J.C.H., E.B., S.G., E.H.J., R.M.V.; Formal Analysis, R.Y., R.C.-P., J.T.-T., M.B., N.P.v.T., J.C.H., M.W.H., E.B., S.G., E.H.J., L.L.; Writing – Original Draft, R.Y.; Writing – Review & Editing, R.Y., N.P.v.T., I.F.M.d.C., G.W.; Funding Acquisition, I.F.M.d.C., G.W.; Resources, L.B., M.R.B., G.R., R.M.; Supervision, G.W., N.P.v.T., I.F.M.d.C.
Conflicts of Interest
The authors declare no conflicts of interest.
Acknowledgments
The authors acknowledge the financial support for this study by Join4energy, Ride4Kids, the Sophia Foundation (SSW0645), Stichting NeMo, and the Spanish Instituto de Salud Carlos III and FEDER funds (grant 15/000465 to R.M.), in the context of funding provided by the European Commission’s 5th, 6th, and 7th Framework Programs (contracts QLK3-CT-2001-00427-INHERINET, LSHB-CT-2004-005242-CONSERT, LSHB-CT-2006-19038-Magselectofection, and grant agreements 222878-PERSIST and 261387 CELL-PID), and by the Netherlands Health Research and Development Organization ZonMw (Translational Gene Therapy program projects 43100016 and 43400010). We thank Dr. Michio Hirano (Department of Neurology, Columbia University Medical Center, New York, USA) for providing the murine model, Louis Boon (Epirus Biopharmaceuticals, Utrecht, the Netherlands) for kindly providing anti-B220 antibody, Prof. Peter A.E. Sillevis Smitt (Department of Neurology, Erasmus MC, Rotterdam, the Netherlands), Pier.G. Mastroberardino and Chiara Milanese (Department of Molecular Genetics, Erasmus MC), Kees Schoonderwoerd (Department of Clinical Genetics, Erasmus MC), and Jeroen de Vrij (Department of Neurosurgery, Erasmus MC) for valuable discussions, Lidia Hussaarts (Department of Clinical Genetics, Erasmus MC) for technical support, King Lam (Department of Pathology, Erasmus MC) for pathology evaluation, and F. Dionisio and A. Aiuti from HSR-TIGET, Milan, for the support to the integration site analysis.
Footnotes
Supplemental Information includes four figures and seven tables and can be found with this article online at https://doi.org/10.1016/j.omtm.2018.01.001.
Supplemental Information
References
- 1.Hirano M., Nishigaki Y., Martí R. Mitochondrial neurogastrointestinal encephalomyopathy (MNGIE): a disease of two genomes. Neurologist. 2004;10:8–17. doi: 10.1097/01.nrl.0000106919.06469.04. [DOI] [PubMed] [Google Scholar]
- 2.Massa R., Tessa A., Margollicci M., Micheli V., Romigi A., Tozzi G., Terracciano C., Piemonte F., Bernardi G., Santorelli F.M. Late-onset MNGIE without peripheral neuropathy due to incomplete loss of thymidine phosphorylase activity. Neuromuscul. Disord. 2009;19:837–840. doi: 10.1016/j.nmd.2009.08.013. [DOI] [PubMed] [Google Scholar]
- 3.Valentino M.L., Martí R., Tadesse S., López L.C., Manes J.L., Lyzak J., Hahn A., Carelli V., Hirano M. Thymidine and deoxyuridine accumulate in tissues of patients with mitochondrial neurogastrointestinal encephalomyopathy (MNGIE) FEBS Lett. 2007;581:3410–3414. doi: 10.1016/j.febslet.2007.06.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hirano M., Silvestri G., Blake D.M., Lombes A., Minetti C., Bonilla E., Hays A.P., Lovelace R.E., Butler I., Bertorini T.E. Mitochondrial neurogastrointestinal encephalomyopathy (MNGIE): clinical, biochemical, and genetic features of an autosomal recessive mitochondrial disorder. Neurology. 1994;44:721–727. doi: 10.1212/wnl.44.4.721. [DOI] [PubMed] [Google Scholar]
- 5.González-Vioque E., Torres-Torronteras J., Andreu A.L., Martí R. Limited dCTP availability accounts for mitochondrial DNA depletion in mitochondrial neurogastrointestinal encephalomyopathy (MNGIE) PLoS Genet. 2011;7:e1002035. doi: 10.1371/journal.pgen.1002035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Martí R., Verschuuren J.J., Buchman A., Hirano I., Tadesse S., van Kuilenburg A.B., van Gennip A.H., Poorthuis B.J., Hirano M. Late-onset MNGIE due to partial loss of thymidine phosphorylase activity. Ann. Neurol. 2005;58:649–652. doi: 10.1002/ana.20615. [DOI] [PubMed] [Google Scholar]
- 7.Chapman T.P., Hadley G., Fratter C., Cullen S.N., Bax B.E., Bain M.D., Sapsford R.A., Poulton J., Travis S.P. Unexplained gastrointestinal symptoms: think mitochondrial disease. Dig. Liver Dis. 2014;46:1–8. doi: 10.1016/j.dld.2013.04.008. [DOI] [PubMed] [Google Scholar]
- 8.Simon L.T., Horoupian D.S., Dorfman L.J., Marks M., Herrick M.K., Wasserstein P., Smith M.E. Polyneuropathy, ophthalmoplegia, leukoencephalopathy, and intestinal pseudo-obstruction: POLIP syndrome. Ann. Neurol. 1990;28:349–360. doi: 10.1002/ana.410280308. [DOI] [PubMed] [Google Scholar]
- 9.Morató L., Bertini E., Verrigni D., Ardissone A., Ruiz M., Ferrer I., Uziel G., Pujol A. Mitochondrial dysfunction in central nervous system white matter disorders. Glia. 2014;62:1878–1894. doi: 10.1002/glia.22670. [DOI] [PubMed] [Google Scholar]
- 10.la Marca G., Malvagia S., Casetta B., Pasquini E., Pela I., Hirano M., Donati M.A., Zammarchi E. Pre- and post-dialysis quantitative dosage of thymidine in urine and plasma of a MNGIE patient by using HPLC-ESI-MS/MS. J. Mass Spectrom. 2006;41:586–592. doi: 10.1002/jms.1013. [DOI] [PubMed] [Google Scholar]
- 11.Lara M.C., Weiss B., Illa I., Madoz P., Massuet L., Andreu A.L., Valentino M.L., Anikster Y., Hirano M., Martí R. Infusion of platelets transiently reduces nucleoside overload in MNGIE. Neurology. 2006;67:1461–1463. doi: 10.1212/01.wnl.0000239824.95411.52. [DOI] [PubMed] [Google Scholar]
- 12.Bax B.E., Bain M.D., Scarpelli M., Filosto M., Tonin P., Moran N. Clinical and biochemical improvements in a patient with MNGIE following enzyme replacement. Neurology. 2013;81:1269–1271. doi: 10.1212/WNL.0b013e3182a6cb4b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Halter J.P., Michael W., Schüpbach M., Mandel H., Casali C., Orchard K., Collin M., Valcarcel D., Rovelli A., Filosto M. Allogeneic haematopoietic stem cell transplantation for mitochondrial neurogastrointestinal encephalomyopathy. Brain. 2015;138:2847–2858. doi: 10.1093/brain/awv226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yadak R., Sillevis Smitt P., van Gisbergen M.W., van Til N.P., de Coo I.F.M. Mitochondrial neurogastrointestinal encephalomyopathy caused by thymidine phosphorylase enzyme deficiency: from pathogenesis to emerging therapeutic options. Front. Cell. Neurosci. 2017;11:31. doi: 10.3389/fncel.2017.00031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.De Giorgio R., Pironi L., Rinaldi R., Boschetti E., Caporali L., Capristo M., Casali C., Cenacchi G., Contin M., D’Angelo R. Liver transplantation for mitochondrial neurogastrointestinal encephalomyopathy. Ann. Neurol. 2016;80:448–455. doi: 10.1002/ana.24724. [DOI] [PubMed] [Google Scholar]
- 16.Cartier N., Hacein-Bey-Abina S., Bartholomae C.C., Veres G., Schmidt M., Kutschera I., Vidaud M., Abel U., Dal-Cortivo L., Caccavelli L. Hematopoietic stem cell gene therapy with a lentiviral vector in X-linked adrenoleukodystrophy. Science. 2009;326:818–823. doi: 10.1126/science.1171242. [DOI] [PubMed] [Google Scholar]
- 17.Biffi A., Montini E., Lorioli L., Cesani M., Fumagalli F., Plati T., Baldoli C., Martino S., Calabria A., Canale S. Lentiviral hematopoietic stem cell gene therapy benefits metachromatic leukodystrophy. Science. 2013;341:1233158. doi: 10.1126/science.1233158. [DOI] [PubMed] [Google Scholar]
- 18.Wagemaker G. Lentiviral hematopoietic stem cell gene therapy in inherited metabolic disorders. Hum. Gene Ther. 2014;25:862–865. doi: 10.1089/hum.2014.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Torres-Torronteras J., Gómez A., Eixarch H., Palenzuela L., Pizzorno G., Hirano M., Andreu A.L., Barquinero J., Martí R. Hematopoietic gene therapy restores thymidine phosphorylase activity in a cell culture and a murine model of MNGIE. Gene Ther. 2011;18:795–806. doi: 10.1038/gt.2011.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Torres-Torronteras J., Cabrera-Pérez R., Barba I., Costa C., de Luna N., Andreu A.L., Barquinero J., Hirano M., Cámara Y., Martí R. Long-term restoration of thymidine phosphorylase function and nucleoside homeostasis using hematopoietic gene therapy in a murine model of mitochondrial neurogastrointestinal encephalomyopathy. Hum. Gene Ther. 2016;27:656–667. doi: 10.1089/hum.2015.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Torres-Torronteras J., Viscomi C., Cabrera-Pérez R., Cámara Y., Di Meo I., Barquinero J., Auricchio A., Pizzorno G., Hirano M., Zeviani M., Martí R. Gene therapy using a liver-targeted AAV vector restores nucleoside and nucleotide homeostasis in a murine model of MNGIE. Mol. Ther. 2014;22:901–907. doi: 10.1038/mt.2014.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.López L.C., Akman H.O., García-Cazorla A., Dorado B., Martí R., Nishino I., Tadesse S., Pizzorno G., Shungu D., Bonilla E. Unbalanced deoxynucleotide pools cause mitochondrial DNA instability in thymidine phosphorylase-deficient mice. Hum. Mol. Genet. 2009;18:714–722. doi: 10.1093/hmg/ddn401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aiuti A., Biasco L., Scaramuzza S., Ferrua F., Cicalese M.P., Baricordi C., Dionisio F., Calabria A., Giannelli S., Castiello M.C. Lentiviral hematopoietic stem cell gene therapy in patients with Wiskott-Aldrich syndrome. Science. 2013;341:1233151. doi: 10.1126/science.1233151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huston M.W., van Til N.P., Visser T.P., Arshad S., Brugman M.H., Cattoglio C., Nowrouzi A., Li Y., Schambach A., Schmidt M. Correction of murine SCID-X1 by lentiviral gene therapy using a codon-optimized IL2RG gene and minimal pretransplant conditioning. Mol. Ther. 2011;19:1867–1877. doi: 10.1038/mt.2011.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van Til N.P., de Boer H., Mashamba N., Wabik A., Huston M., Visser T.P., Fontana E., Poliani P.L., Cassani B., Zhang F. Correction of murine Rag2 severe combined immunodeficiency by lentiviral gene therapy using a codon-optimized RAG2 therapeutic transgene. Mol. Ther. 2012;20:1968–1980. doi: 10.1038/mt.2012.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Modlich U., Schambach A., Brugman M.H., Wicke D.C., Knoess S., Li Z., Maetzig T., Rudolph C., Schlegelberger B., Baum C. Leukemia induction after a single retroviral vector insertion in Evi1 or Prdm16. Leukemia. 2008;22:1519–1528. doi: 10.1038/leu.2008.118. [DOI] [PubMed] [Google Scholar]
- 27.Zhou S., Ma Z., Lu T., Janke L., Gray J.T., Sorrentino B.P. Mouse transplant models for evaluating the oncogenic risk of a self-inactivating XSCID lentiviral vector. PLoS ONE. 2013;8:e62333. doi: 10.1371/journal.pone.0062333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Deichmann A., Brugman M.H., Bartholomae C.C., Schwarzwaelder K., Verstegen M.M., Howe S.J., Arens A., Ott M.G., Hoelzer D., Seger R. Insertion sites in engrafted cells cluster within a limited repertoire of genomic areas after gammaretroviral vector gene therapy. Mol. Ther. 2011;19:2031–2039. doi: 10.1038/mt.2011.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bardosi A., Creutzfeldt W., DiMauro S., Felgenhauer K., Friede R.L., Goebel H.H., Kohlschütter A., Mayer G., Rahlf G., Servidei S. Myo-, neuro-, gastrointestinal encephalopathy (MNGIE syndrome) due to partial deficiency of cytochrome-c-oxidase. A new mitochondrial multisystem disorder. Acta Neuropathol. 1987;74:248–258. doi: 10.1007/BF00688189. [DOI] [PubMed] [Google Scholar]
- 30.Szigeti K., Sule N., Adesina A.M., Armstrong D.L., Saifi G.M., Bonilla E., Hirano M., Lupski J.R. Increased blood-brain barrier permeability with thymidine phosphorylase deficiency. Ann. Neurol. 2004;56:881–886. doi: 10.1002/ana.20302. [DOI] [PubMed] [Google Scholar]
- 31.Dubey M., Bugiani M., Ridder M.C., Postma N.L., Brouwers E., Polder E., Jacobs J.G., Baayen J.C., Klooster J., Kamermans M. Mice with megalencephalic leukoencephalopathy with cysts: a developmental angle. Ann. Neurol. 2015;77:114–131. doi: 10.1002/ana.24307. [DOI] [PubMed] [Google Scholar]
- 32.van der Knaap M.S., Boor I., Estévez R. Megalencephalic leukoencephalopathy with subcortical cysts: chronic white matter oedema due to a defect in brain ion and water homoeostasis. Lancet Neurol. 2012;11:973–985. doi: 10.1016/S1474-4422(12)70192-8. [DOI] [PubMed] [Google Scholar]
- 33.Hoogerbrugge P.M., Suzuki K., Suzuki K., Poorthuis B.J., Kobayashi T., Wagemaker G., van Bekkum D.W. Donor-derived cells in the central nervous system of twitcher mice after bone marrow transplantation. Science. 1988;239:1035–1038. doi: 10.1126/science.3278379. [DOI] [PubMed] [Google Scholar]
- 34.Oliver F.J., Collins M.K., López-Rivas A. dNTP pools imbalance as a signal to initiate apoptosis. Experientia. 1996;52:995–1000. doi: 10.1007/BF01920108. [DOI] [PubMed] [Google Scholar]
- 35.Kumar D., Viberg J., Nilsson A.K., Chabes A. Highly mutagenic and severely imbalanced dNTP pools can escape detection by the S-phase checkpoint. Nucleic Acids Res. 2010;38:3975–3983. doi: 10.1093/nar/gkq128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jiang H., He X., Wang S., Jia J., Wan Y., Wang Y., Zeng R., Yates J., 3rd, Zhu X., Zheng Y. A microtubule-associated zinc finger protein, BuGZ, regulates mitotic chromosome alignment by ensuring Bub3 stability and kinetochore targeting. Dev. Cell. 2014;28:268–281. doi: 10.1016/j.devcel.2013.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Toledo C.M., Herman J.A., Olsen J.B., Ding Y., Corrin P., Girard E.J., Olson J.M., Emili A., DeLuca J.G., Paddison P.J. BuGZ is required for Bub3 stability, Bub1 kinetochore function, and chromosome alignment. Dev. Cell. 2014;28:282–294. doi: 10.1016/j.devcel.2013.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schambach A., Bohne J., Baum C., Hermann F.G., Egerer L., von Laer D., Giroglou T. Woodchuck hepatitis virus post-transcriptional regulatory element deleted from X protein and promoter sequences enhances retroviral vector titer and expression. Gene Ther. 2006;13:641–645. doi: 10.1038/sj.gt.3302698. [DOI] [PubMed] [Google Scholar]
- 39.Dull T., Zufferey R., Kelly M., Mandel R.J., Nguyen M., Trono D., Naldini L. A third-generation lentivirus vector with a conditional packaging system. J. Virol. 1998;72:8463–8471. doi: 10.1128/jvi.72.11.8463-8471.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van Til N.P., Stok M., Aerts Kaya F.S., de Waard M.C., Farahbakhshian E., Visser T.P., Kroos M.A., Jacobs E.H., Willart M.A., van der Wegen P. Lentiviral gene therapy of murine hematopoietic stem cells ameliorates the Pompe disease phenotype. Blood. 2010;115:5329–5337. doi: 10.1182/blood-2009-11-252874. [DOI] [PubMed] [Google Scholar]
- 41.Wognum A.W., Visser T.P., Peters K., Bierhuizen M.F., Wagemaker G. Stimulation of mouse bone marrow cells with kit ligand, FLT3 ligand, and thrombopoietin leads to efficient retrovirus-mediated gene transfer to stem cells, whereas interleukin 3 and interleukin 11 reduce transduction of short- and long-term repopulating cells. Hum. Gene Ther. 2000;11:2129–2141. doi: 10.1089/104303400750001435. [DOI] [PubMed] [Google Scholar]
- 42.Van Acker K.J., Eyskens F.J., Verkerk R.M., Scharpé S.S. Urinary excretion of purine and pyrimidine metabolites in the neonate. Pediatr. Res. 1993;34:762–766. doi: 10.1203/00006450-199312000-00012. [DOI] [PubMed] [Google Scholar]
- 43.Huston M.W., Riegman A.R., Yadak R., van Helsdingen Y., de Boer H., van Til N.P., Wagemaker G. Pretransplant mobilization with granulocyte colony-stimulating factor improves B-cell reconstitution by lentiviral vector gene therapy in SCID-X1 mice. Hum. Gene Ther. 2014;25:905–914. doi: 10.1089/hum.2014.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Farahbakhshian E., Verstegen M.M., Visser T.P., Kheradmandkia S., Geerts D., Arshad S., Riaz N., Grosveld F., van Til N.P., Meijerink J.P. Angiopoietin-like protein 3 promotes preservation of stemness during ex vivo expansion of murine hematopoietic stem cells. PLoS ONE. 2014;9:e105642. doi: 10.1371/journal.pone.0105642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schmidt M., Schwarzwaelder K., Bartholomae C., Zaoui K., Ball C., Pilz I., Braun S., Glimm H., von Kalle C. High-resolution insertion-site analysis by linear amplification-mediated PCR (LAM-PCR) Nat. Methods. 2007;4:1051–1057. doi: 10.1038/nmeth1103. [DOI] [PubMed] [Google Scholar]
- 46.Biasco L., Pellin D., Scala S., Dionisio F., Basso-Ricci L., Leonardelli L., Scaramuzza S., Baricordi C., Ferrua F., Cicalese M.P. In vivo tracking of human hematopoiesis reveals patterns of clonal dynamics during early and steady-state reconstitution phases. Cell Stem Cell. 2016;19:107–119. doi: 10.1016/j.stem.2016.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Biasco L., Scala S., Basso Ricci L., Dionisio F., Baricordi C., Calabria A., Giannelli S., Cieri N., Barzaghi F., Pajno R. In vivo tracking of T cells in humans unveils decade-long survival and activity of genetically modified T memory stem cells. Sci. Transl. Med. 2015;7:273ra13. doi: 10.1126/scitranslmed.3010314. [DOI] [PubMed] [Google Scholar]
- 48.Goffart S., Cooper H.M., Tyynismaa H., Wanrooij S., Suomalainen A., Spelbrink J.N. Twinkle mutations associated with autosomal dominant progressive external ophthalmoplegia lead to impaired helicase function and in vivo mtDNA replication stalling. Hum. Mol. Genet. 2009;18:328–340. doi: 10.1093/hmg/ddn359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yasukawa T., Yang M.Y., Jacobs H.T., Holt I.J. A bidirectional origin of replication maps to the major noncoding region of human mitochondrial DNA. Mol. Cell. 2005;18:651–662. doi: 10.1016/j.molcel.2005.05.002. [DOI] [PubMed] [Google Scholar]
- 50.Ylikallio E., Tyynismaa H., Tsutsui H., Ide T., Suomalainen A. High mitochondrial DNA copy number has detrimental effects in mice. Hum. Mol. Genet. 2010;19:2695–2705. doi: 10.1093/hmg/ddq163. [DOI] [PubMed] [Google Scholar]
- 51.Bugiani M., Dubey M., Breur M., Postma N.L., Dekker M.P., Ter Braak T., Boschert U., Abbink T.E.M., Mansvelder H.D., Min R. Megalencephalic leukoencephalopathy with cysts: the Glialcam-null mouse model. Ann. Clin. Transl. Neurol. 2017;4:450–465. doi: 10.1002/acn3.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.