Abstract
Brassinosteroid (BR) mutants of Arabidopsis have pleiotropic phenotypes and provide evidence that BRs function throughout the life of the plant from seedling development to senescence. Screens for BR signaling mutants identified one locus, BRI1, which encodes a protein with homology to leucine-rich repeat receptor serine (Ser)/threonine (Thr) kinases. Twenty-seven alleles of this putative BR receptor have been isolated to date, and we present here the identification of the molecular lesions of 14 recessive alleles that represent five new mutations. BR-insensitive-1 (BRI1) is expressed at high levels in the meristem, root, shoot, and hypocotyl of seedlings and at lower levels later in development. Confocal microscopy analysis of full-length BRI1 fused to green fluorescent protein indicates that BRI1 is localized in the plasma membrane, and an in vitro kinase assay indicates that BRI1 is a functional Ser/Thr kinase. Among the bri1 mutants identified are mutants in the kinase domain, and we demonstrate that one of these mutations severely impairs BRI1 kinase activity. Therefore, we conclude that BRI1 is a ubiquitously expressed leucine-rich repeat receptor that plays a role in BR signaling through Ser/Thr phosphorylation.
Receptor protein kinases (RPKs) activate a complex array of intracellular signaling pathways in response to the extracellular environment (van der Geer et al., 1994; Padgett, 1999). RPKs are single-pass transmembrane proteins that contain an amino-terminal signal sequence, extracellular domains unique to each receptor, and a cytoplasmic kinase domain. In general, ligand binding induces homo- or heterodimerization of RPKs, and the resultant close proximity of the cytoplasmic domains results in kinase activation by transphosphorylation. Although plants have many proteins similar to RPKs, no ligand has been identified for these receptor-like kinases (RLKs). The majority of plant RLKs belong to the family of Ser/Thr kinases, and most have extracellular Leu-rich repeats (LRRs; Becraft, 1998). The LRRs form a solvent-exposed parallel β-sheet, which creates a surface that mediates protein-protein interactions in other systems (Kobe and Deisenhofer, 1995). The known interactors for mammalian LRR receptors are peptide hormones, such as nerve growth factor and gonadotropin (Braun et al., 1991; Kobe and Deisenhofer, 1995).
Plant LRR-RLKs are involved in multiple processes including regulation of development (ERECTA, HAESA, and CLV1), disease resistance (Xa21), and steroid hormone signaling (brassinosteroid [BR]-insensitive-1 [BRI1]) (Song et al., 1995; Torii et al., 1996; Clark et al., 1997; Li and Chory, 1997; Jinn et al., 2000). ERECTA is important for proper shaping of organs originating in the shoot apical meristem (Torii et al., 1996). CLV1 is involved in the control of cell division and differentiation in the shoot apical meristem with CLV3 being the putative peptide ligand (Fletcher et al., 1999). Missense mutations in either the LRRs or the kinase domains of CLV1 and ERECTA lead to loss of function, confirming the importance of these domains for function (Torii et al., 1996; Clark et al., 1997). BRI1 encodes a putative BR receptor (Li and Chory, 1997), and bri1 mutants display a BR-deficient phenotype but fail to be rescued by BR treatment (Clouse et al., 1996; Kauschmann et al., 1996; Li and Chory, 1997; Noguchi et al., 1999). BRs are a unique class of plant steroids found throughout the plant kingdom that exhibit multiple effects when applied exogenously, including cell expansion of young aerial tissues, especially the hypocotyl and leaf petioles (Mandova, 1988). Classical animal steroid hormone receptors belong to a subfamily of nuclear receptors that are ligand-dependent transcription factors that regulate gene expression (Beato et al., 1995). There is also evidence for action of steroid hormones outside the nucleus involving membrane receptors and protein phosphorylation (Wehling, 1997). In Xenopus oocytes, progesterone through an unidentified surface-associated receptor activates a Ser/Thr kinase, Eg2 (Andrésson and Ruderman, 1998). Additionally, progesterone stimulates Tyr phosphorylation in human sperm via a putative cell surface receptor (Tesarik et al., 1993; Mendoza et al., 1995), and in osteoblastic cells estrogen causes a rapid and transient MAP kinase activation (Endoh et al., 1997).
The BRI1 extracellular domain contains 21 tandem amino-terminal LRRs, a 70-amino acid island domain and four additional LRRs preceding the transmembrane domain. This organization is similar to that in tomato disease-resistance membrane-anchored LRR proteins, Cf-2, Cf-4, Cf-5, and Cf-9 (Jones and Jones, 1997; Dixon et al., 1998). These proteins contain “loop out” domains, similar to but smaller than the island domain, and these “loop out” domains interrupt tandem LRRs, creating four separate LRRs prior to the transmembrane domain. Three bri1 mutations are in glycines of the island domain, and one is a missense mutation in the first LRR following the 70-amino acid island domain (Li and Chory, 1997; Noguchi et al., 1999), supporting the necessity of these regions for function. Four missense mutations occur in the cytoplasmic Ser/Thr kinase domain of BRI1, implicating another essential domain for BRI1 function (Li and Chory, 1997; Noguchi et al., 1999).
In this paper we present the identity of additional recessive alleles of BRI1, which highlights the importance of certain domains for BRI1 function. Additionally, we used a green fluorescent protein fusion to show the localization of BRI1 to the plasma membrane and to determine its pattern of expression within the plant. Finally, the function of BRI1 as a Ser/Thr kinase is directly demonstrated by an in vitro kinase assay. Therefore, we conclude that BRI1, the putative brassinolide receptor, is a ubiquitously expressed, plasma membrane-localized, LRR Ser/Thr kinase.
RESULTS
Sequencing of Additional bri1 Alleles Implicates a New Domain in BRI1-Mediated Steroid Signaling
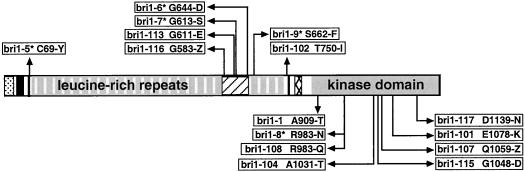
We previously conducted a screen for BR-insensitive mutants, and isolated 18 new bri1 alleles (Li and Chory, 1997). To identify regions of functional significance, we sequenced BRI1 from 14 of these mutants. The molecular lesions of bri1 alleles are reported in Table I and are schematically represented in Figure 1, including already published bri1 alleles (Li and Chory, 1997; Noguchi et al., 1999). We identified three new mutations in the kinase domain; bri1-1 and bri1-108 are missense alleles altering amino acids Ala-909 to Thr and Arg-983 to Gln, respectively. Ala-909 in subdomain II is present in all protein kinases, and Arg-983 in subdomain VIA is conserved among the putative plant LRR receptor kinases, including BRI1, CLV1, ERECTA, and Xa21 (Li and Chory, 1997). The bri1-117 allele has a mutation at codon 1,139 of a non-conserved Asp to Asn in a region of the protein that generally contains negatively charged residues in protein kinases. These new mutants re-emphasize the necessity of the BRI1 kinase domain. In the BRI1 extracellular domain, we identified two new mutations: a nonsense mutation in bri1-114 generates a stop codon early within the 70-amino acid island domain, and a missense mutation in bri1-102 results in the substitution of Thr-750 with an Ile. The latter mutation occurs after the 25th LRR and before the second Cys pair, indicating a new region in BRI1 whose integrity is necessary for function.
Table I.
bri1 alleles
| Allele | Lesion | Predicted Effect | Reference |
|---|---|---|---|
| bri1-1 | G → A | Ala-909 → Thr | This work; Clouse et al. (1996) |
| bri1-3 | 4-bp deletion | Premature stop | Noguchi et al. (1999) |
| bri1-4 | 10-bp deletion | Premature stop | Noguchi et al. (1999) |
| bri1-5 | G → A | Cys-69 → Tyr | Noguchi et al. (1999) |
| bri1-6, 119 | G → A | Gly-644 → Asp | This work; Noguchi et al. (1999) |
| bri1-7 | G → A | Gly-613 → Ser | Noguchi et al. (1999) |
| bri1-8 | G → A | Arg-983 → Asn | Noguchi et al. (1999) |
| bri1-9 | C → T | Ser-662 → Phe | Noguchi et al. (1999) |
| bri1-101 | G → A | Glu-1078 → Lys | Li and Chory (1997) |
| bri1-102 | C → T | Thr-750 → Ileu | This work |
| bri1-103, 104 | G → A | Ala-1031 → Thr | This work; Li and Chory (1997) |
| bri1-105-107 | C → T | Gln-1059 → Stop | This work; Li and Chory (1997) |
| bri1-108-112 | G → A | Arg-983 → Gln | This work |
| bri1-113 | G → A | Gly-611 → Glu | Li and Chory (1997) |
| bri1-114,116 | C → T | Gln-583 → Stop | This work |
| bri1-115 | G → A | Gly-1048 → Asp | Li and Chory (1997) |
| bri1-117,118 | G → A | Asp-1139 → Asn | This work |
Figure 1.
The majority of bri1 mutations cluster in the island and kinase domains. A schematic representation of BRI1 including all the known bri1 point mutations with their predicted effects. Symbols represent the following:  , Signal peptide;
, Signal peptide;  , putative Leu-zipper motif;
, putative Leu-zipper motif;  , Cys pair;
, Cys pair;  , LRRs;
, LRRs;  , 70-amino acid island;
, 70-amino acid island;  , transmembrane domain;
, transmembrane domain;  , kinase domain. Asterisk, These alleles were published by Noguchi et al. (1999).
, kinase domain. Asterisk, These alleles were published by Noguchi et al. (1999).
BRI1 Is a Ubiquitously Expressed Plasma Membrane-Localized Protein
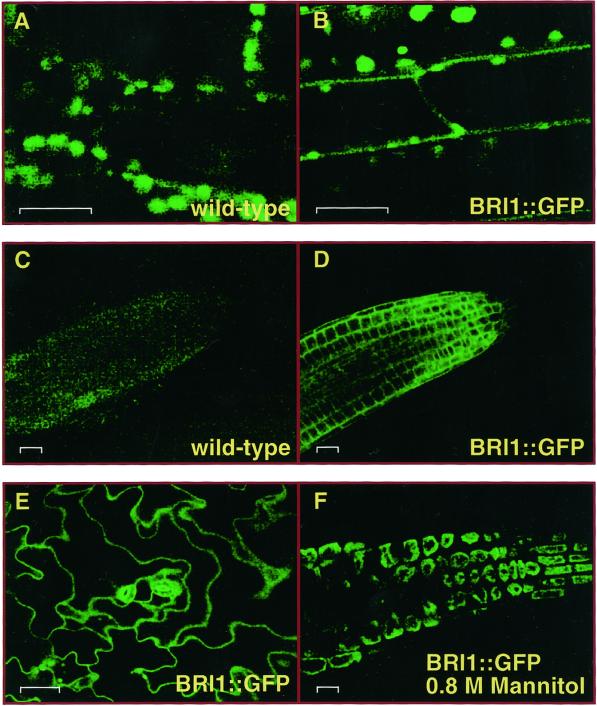
To determine the spatial pattern of expression and subcellular localization of BRI1, a green fluorescent protein (GFP) fusion with the C terminus of full-length BRI1 was made in the context of a BRI1 genomic clone containing, in addition to coding sequence, 1.7 kb of promoter DNA and upstream regulatory elements. This translational fusion (BRI1::GFP) was able to rescue bri1-104 (data not shown), indicating that the fusion protein was functional. The expression of BRI1::GFP is ubiquitous in young tissue, especially in the meristem. Figure 2 presents a confocal microscopy analysis of BRI1 expression and shows that the BRI1::GFP fluorescence is localized to the cell surface in the hypocotyl, root, and cotyledons of young light-grown seedlings. The hypocotyl cells of wild-type (vector alone) seedlings (Fig. 2A) exhibit only the background chlorophyll autofluorescence, whereas in the hypocotyls of BRI1::GFP transgenic plants, the surface of each cell is outlined by the intense GFP fluorescence. The cotyledons show a similar pattern with the BRI1::GFP fluorescence illuminating the surface of epidermal cells (Fig. 2E). Roots have very low background fluorescence due to the lack of chloroplasts, and in Figure 2C, the fluorescence of the image was enhanced to show that a root-tip was in the field. In stark contrast to this wild-type root is the transgenic root (Fig. 2D), in which the surface of each cell is apparent. In these young cells in the root tip, the cytoplasm is not pushed up against the plasma membrane because the vacuoles are small. Therefore, we conclude that the BRI1-directed GFP fluorescence is at the cell wall/plasma membrane and not in the cytoplasm of these cells. The BRI1 promoter drives expression of BRI1::GFP in all tissues in both light- and dark-grown seedlings (data not shown). Expression continues in younger tissue, but BRI1::GFP fluorescence decreases in non-growing older tissue (data not shown).
Figure 2.
BRI1-GFP is expressed ubiquitously during early seedling development and is localized to the plasma membrane. A BRI1::GFP fusion protein was expressed from the BRI1 promoter in stably transformed wild-type Arabidopsis. A, Background chlorophyll fluorescence in a wild-type hypocotyl; B, BRI1::GFP fluorescence along the cell surface of hypocotyl cells; C, background fluorescence of a wild-type Arabidopsis root; D, BRI1::GFP at the cell surface in young root; E, BRI1::GFP at the cell surface in cotyledon epidermal cells (wild type has only stomata and guard cell chloroplast autofluorescence, which is not shown); F, BRI1::GFP localizes with the cytoplasm if cells are collapsed in 0.8 m mannitol, indicating that BRI1::GFP is in the plasma membrane. Bar = 20 μm.
To determine if BRI1 is localized to the cell wall or the plasma membrane, a plasmolysis experiment was performed. Negative osmotic pressure results in the evacuation of fluid from inside the cell, and the plasma membrane is internalized with the cellular organelles, leaving the cell wall unaltered. When roots from a transgenic seedling were placed in 0.8 m mannitol, the BRI1::GFP fluorescence was internalized with the plasma membrane and the rest of the cell (Fig. 2F). Although many plant LRR-RLKs have putative signal sequences and transmembrane domains, this is the first evidence of visual localization of a LRR-RLK to the plasma membrane.
BRI1 Is a Ser/Thr Kinase
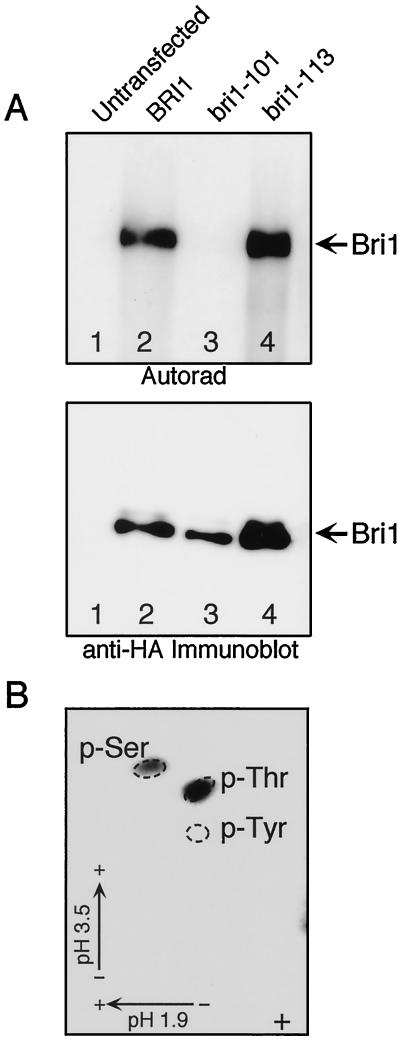
The BRI1 cytoplasmic domain is predicted to contain Ser/Thr kinase activity, and we performed an in vitro kinase assay to determine if BRI1 is an active kinase. Wild-type and mutant BRI1 cDNA constructs containing a C-terminal HA tag were expressed in human embryonic kidney 293T cells by transient transfection. The mutants tested were bri1-101, a kinase domain mutant altering Gly-1078 to Glu in subdomain IX, and bri1-113, which has a mutation in the extracellular island domain (Fig. 1). Transfected BRI1 is readily detectable as an approximately 150-kD protein by immunoblot of whole cell lysates using the 12CA5 monoclonal antibody and enhanced chemiluminescence (data not shown). The kinase activity of BRI1 proteins was determined by an in vitro kinase assay. In this assay, BRI1 is immunoprecipitated from transiently transfected 293T cells and is incubated with [γ-32P]ATP in a buffer containing Mg2+ and Mn2+. 32P-Labeled proteins are separated by SDS-PAGE and detected by autoradiography. Figure 3A shows that although the wild-type and bri1-113 proteins are competent for autophosphorylation, the kinase activity of bri1-101 is significantly reduced. Quantitation revealed that the kinase activity of bri1-101 was 45 times lower than that of the wild type in this assay, whereas bri1-101 expression was only one-half of the wild-type levels (Fig. 3A). The kinase- associated protein phosphatase (KAPP) binds in a phosphorylation-dependent manner to the kinase domains of the LRR-RLKs, HAESA (RLK5) and CLV1 (Stone et al., 1994, 1998; Williams et al., 1997). BRI1 was shown to phosphorylate KAPP as a substrate in vitro (J. Li, R. Williams, E. Meyerowitz, and J. Chory, unpublished data). The kinase domain sequence predicts that BRI1 belongs to the Ser/Thr kinase family (Li and Chory, 1997). Phosphoamino acid analysis performed with the in vitro autophosphorylated receptor showed that phospho-Ser and phospho-Thr were present, thus confirming this prediction (Fig. 3B).
Figure 3.
BRI1 is a Ser/Thr kinase. Immunoprecipitated HA-BRI1, wild type, bri1-101, and bri1-113 were used for an in vitro kinase assay. A, The top presents an autoradiogram of the kinase assay. The bottom presents an immunoblot using anti-HA antibody of the same gel. Lane 1, Untransfected 293T cells; lane 2, wild-type BRI1; lane 3, bri1-101 kinase mutant; lane 4, bri1-113 island domain mutant. B, Autophosphorylation of BRI1 occurs on Ser/Thr residues. Anti-HA-BRI1 autophosphorylation was subjected to phosphoamino acid analysis. The positions of the internal phospho-Ser, phospho-Thr, and phospho-Tyr standards (visualized by ninhydrin staining) are indicated. The origin (+) and the directions of electrophoresis with the pH are as indicated.
DISCUSSION
BRI1 Is a Functional Ser/Thr Kinase
Five plant LRR receptor Ser/Thr kinases have been shown to be active protein kinases. The kinase domain of HAESA (RLK5), for example, was expressed in Escherichia coli as a fusion protein and then was purified and used in in vitro kinase assays (Horn and Walker, 1994). The results from these assays showed that the fusion protein possessed kinetic parameters typical of protein kinases. We chose to express BRI1 in an animal cell system because similarities exist between animal and plant cells with regard to the expression and the peptide signal-mediated membrane targeting of large receptor proteins. We reasoned that proper membrane expression of the full-length protein in a cellular context would allow the study of hormonal regulation of receptor activation. BRI1 expressed in 293T cells by transient transfection was used for in vivo labeling experiments (data not shown) and was also immunoprecipitated and used for an in vitro kinase assay.
The in vitro kinase assay revealed that both the wild-type protein and the product of an allele that contains a mutation in the extracellular domain were active protein kinases, at least in vitro, under conditions of antibody-mediated dimerization. On the other hand, the product of the bri1-101 allele, which contains a mutation in the kinase domain, exhibited greatly reduced kinase activity. This assay confirms the predicted activity of the kinase domain of BRI1 and can now be used as a biochemical tool to identify direct substrates of the activated receptor.
The demonstration that BRI1 encodes an active protein kinase provides more substantial ground to explain the effects of the intracellular domain mutations in BRI1. Although mutations in the intracellular domain of a transmembrane receptor might affect several processes, such as receptor homo- and heterodimerization, interaction with other proteins, receptor stability, and enzymatic activity, most mutations found in the intracellular domain of BRI1 are consistent with the latter possibility: bri1-1 substitutes an Ala residue in the protein kinase subdomain II (Hanks et al., 1988) that is conserved in all protein kinases; bri1-8 and bri1-108 substitute an Arg in subdomain VIa that is conserved in LRR kinases; bri1-104 substitutes an Ala in subdomain VII that is frequently found at that position in other protein kinases; bri1-115 changes a Gly residue in subdomain VIII that is thought to be involved in substrate recognition and Ser/Thr specificity; bri1-107 creates a stop codon that truncates the kinase domain in subdomain VIII; bri1-117 replaces an Asp with Asn in a segment of subdomain XI in which negatively charged residues are frequently observed among protein kinases; and bri1-101, which revealed greatly reduced kinase activity in vitro, substitutes a Glu that is frequently found at that position in subdomain IX of protein kinases.
Phosphoamino acid analyses of in vitro phosphorylated BRI1 revealed the presence of phosphorylated Ser and Thr only, as originally predicted from the inspection of primary amino acid sequence of this kinase. Based on data from other protein kinases, we anticipate that some of the phosphorylated residues will lie in the activation loop of the kinase domain, which is in fact Ser/Thr–rich in BRI1. In addition to activation loop phosphorylation, which generally serves to increase kinase activity, sites elsewhere may be phosphorylated to recruit phosphopeptide-binding proteins into an activated receptor complex, as happens for receptor protein-Tyr kinases. Three modular protein domains are now known that selectively bind to peptide sequences containing phosphorylated Ser or Thr: FHA, WW, and 14-3-3 (Yaffe and Cantley, 1999). All of these domains are found in plants and, in fact, an FHA domain-containing protein phosphatase, KAPP, has been shown to selectively bind to activated LRR receptors (Stone et al., 1994; Williams et al., 1997; Braun et al., 1997; Li et al., 1999).
We have not detected either basal or steroid-induced phosphorylation of BRI1 in 293T cells labeled with [32P]orthophosphate, despite the fact that many variables have been explored and that phosphorylation of several other protein kinases and kinase substrates was readily detected under similar conditions (data not shown). Based on what has been learned from RPKs in other systems and on the observation that at least part of the autophosphorylation of HAESA (RLK5) is intermolecular (Horn and Walker, 1994), we speculate that BRI1 is found in the inactive monomeric state when expressed in 293T cells and that the formation of kinase-active BRI1 dimers is induced during the immunoprecipitation reaction that precedes the in vitro kinase assay. This model explains why, under the conditions of the latter assay, a ligand is not required to promote stimulation of receptor kinase activity. Brassinolide and other steroids may have failed to promote BRI1 dimerization and activation in intact cells for several reasons, such as a missing accessory steroid carrier protein or presenting molecule in the heterologous 293T cell system.
Role of the Extracellular Domain
There are several putative domains in the extracellular region of BRI1. Analysis of mutant alleles indicates domains of functional importance, including the amino-terminal Cys pair, the 70-amino acid island, the LRR domain, and the region between the LRR and the second Cys pair (Fig. 1).
The island domain contains three missense mutations, bri1-6, bri1-7, and bri1-113, which affect three separate Gly residues. Although the bri1-113 mutant fails to respond to the steroid, it was shown in this paper to be an active kinase in vitro. This mutant could be an active kinase in vitro due to antibody-mediated dimerization, but in vivo a mutation of Gly in the island domain Gly might inhibit positive regulation of the kinase domain. The requirement for this Gly could be due to the small size of this residue, which allows for a conformational change of the protein when the ligand is bound to activate the kinase domain. Alternatively, mutation of these glycines could interfere with protein/ligand binding to BRI1 or with extracellular dimerization. Although the importance of the Gly residues is unclear, the Cf-2, Cf-4, Cf-5, and Cf-9 defense genes contain “loop out” domains that are similar to the island domain of BRI1 but that are much smaller, 32, 27, and 39 amino acids, respectively. All of these proteins contain a Gly for the −6 amino acid relative to the most carboxy-terminal amino acid in each domain. This Gly is mutated in the bri1-6 allele, thereby supporting the hypothesis that the Glys represent a structural requirement for LRR transmembrane protein function.
The LRR domain is another region possibly involved in protein/ligand interactions. The predicted site for this interaction is in the solvent exposed parallel β-sheet (Kobe and Deisenhofer, 1995). The LxxLxLxx (x is any amino acid) domain within the LRR corresponds with the solvent face of the protein with the Leu resides facing away from the solvent face (Jones and Jones, 1997). The x or variable amino acids could lead to specificity of the protein binding to the LRR. The strong clv1 alleles, clv1-4 and clv1-8, are both missense mutations in this solvent face domain. In contrast to this, bri1-9 is a weak allele (Noguchi et al., 1999) altering a Ser in the conserved BRI1 solvent face, LxxLxLSx (Li and Chory, 1997). This mutation could alter protein/ligand interactions or dimerization. It is interesting that this mutation is in the first of the four LRR after the island domain. This could indicate that many of the important molecular interactions occur close to the transmembrane domain. This idea is supported by bri1-102, a mutation also in this region. Conversely, the clustering of mutations closer to the transmembrane domain could indicate that the protein interactions with the first 21 LRRs are strong enough that altering one amino acid does not significantly destabilize binding.
BRI1 Expression Is Not Spatially Regulated
BRI1 is expressed in all tissues in the seedling (root, hypocotyl, cotyledons, and leaves; Fig. 2 and data not shown) and in adult organs, including cauline leaves and inflorescent stems (data not shown). The expression pattern of BRI1-GFP fusion protein is consistent with previous mRNA expression data (Li and Chory, 1997), which revealed the presence of BRI1 transcripts in all tissues. Although there is no tissue-specific expression of BRI1, there is temporal regulation. Fully expanded leaves and elongated root or inflorescence cells express BRI1::GFP at low levels (data not shown). This expression of the transgene correlates with physiological data that showed that exogenously applied BRs promote growth only in younger tissues (Mandova, 1988). The lack of response of older tissues to BRs may be because BRI1, the putative BR receptor, is present at significantly diminished levels in fully expanded tissues.
Model of BRI1 Signaling
We propose two models for the function of BRI1 in brassinolide signaling. First, similar to animal RPKs, the binding of ligand to the LRR or the island domain may result in the dimerization of BRI1 with itself or another receptor kinase. This dimerization would result in transphosphorylation and activation of the kinase domain. The activated kinase would then send a phosphorylation signal to alter gene expression and induce cell expansion, among other effects. Conversely, BRI1 may not be the receptor itself but may be a protein in the brassinolide receptor complex whose extracellular domain is involved in interactions with other receptor complex proteins. The formation of an active complex results in activation of the kinase phosphorylation signal.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Arabidopsis Columbia was the wild-type ecotype. Seeds were surface sterilized by washing for 20 min in 70% (v/v) ethanol containing 0.05% (v/v) Triton X-100, followed by a wash with 95% (v/v) ethanol. Seeds were dried on filter papers under sterile conditions and sown on 0.5× Murashige-Skoog medium (Gibco-BRL, Cleveland) supplemented with 1% (v/v) Suc and 0.8% (v/v) phytoagar. The plates were wrapped in aluminum foil and left at 4°C overnight to induce germination. Seedlings were grown in growth chambers at 21°C under long-day conditions (16 h of light).
Sequence Analysis of bri1 Alleles
The alleles described were isolated from ethyl methane-sulfonate-mutagenized Arabidopsis Columbia seeds carrying the homozygous mutation glabrous1 (Lehle Seeds, Round Rock, TX), except for bri1-119, which was obtained from the Arabidopsis Biological Resource Center (Ohio State University, Columbus; stock no. CS399, ecotype Enkheim-2). Genomic DNA was isolated from various bri1 mutants using a plant DNA miniprep method (Li and Chory, 1997). Based on the wild-type BRI1 sequence, three pairs of gene-specific primers (forward 1, 5′-AGTTACCATTGCAGACGA-3′; reverse 1, 5′-AACCCAACCAACGACGTT-3′; forward 2, 5′-GAATTCAATCTCCGGTGCTA-3′; reverse 2, 5′-GAAGAGGATAACCACAGA-3′; forward 3, 5′-TGGTTCGATTCCTGATGA-3′; and reverse 3, 5′-GAAT-TAATAGGTCACGTGC-3′) were designed to amplify three overlapping PCR fragments covering approximately 2.0 kb of 5′-untranscribed/untranslated sequence, the complete protein-coding sequence (3, 588 bp), and 420 bp of 3′-untranslated region. PCR amplifications were conducted in 100-μL reaction volumes containing 50 mm Tris [Tris-(hydroxymethyl)-aminomethane]-HCl (pH 8.3), 2 mm MgCl2, 0.01% (w/v) gelatin, approximately 100 ng of Arabidopsis DNA, 200 μm dNTPs, 250 ng each of a specific pair of BRI1 primers, and 5 units of Taq DNA polymerase (Fisher Scientific, Pittsburgh).
The PCR reactions were performed in a thermocycler (Trio-Thermoblock, Biometra, Germany) by denaturing the template DNA for 5 min at 95°C followed by 40 cycles of 45 s at 94°C, 45 s at 55°C, 90 s at 72°C, and a 10-min extension at 72°C. The PCR products were size-fractionated by electrophoresis in 0.8% (w/v) agarose gel, purified using the QIAEX II gel extraction kit (Qiagen, Chatsworth, CA), and directly sequenced on an ABI PRISM 310 genetic analyzer using the dRhodamine terminator cycle sequencing kit (PE-Applied Biosystems, Foster City, CA). Putative mutations were identified by comparing the DNA sequences of mutant bri1 alleles with the wild-type BRI1 sequence using the Lasergene Sequence Analysis System (DNASTAR, Inc., Madison, WI), and they were confirmed by sequencing at least two independently amplified PCR fragments or by conducting cleaved-amplified polymorphic sequence/derived cleaved-amplified polymorphic sequence analysis (Konieczny and Ausubel, 1993; Neff et al., 1998). The wild-type BRI1 gene of the ecotype Enkheim-2 was sequenced and used to identify the bri1-119 mutation.
Construction of BRI1::GFP
The entire BRI1 coding region, including 1,690 bp of promoter relative to the translation start, was fused to GFP5.1 by replacing the BRI1 stop codon with a three-amino acid linker: Trp, Asp, Pro. GFP5.1 was created by inserting the NdeI-BstBI fragment from mGFP5 (GenBank accession no. U87974; Siemering et al., 1996) into a S65T mutated version of GFP. This fusion construct was cloned into pCHF4, a pPZP212-derived vector with a pea ribulose 1,5-bisphosphate carboxylase terminator SacI-EcoRI fragment. An ASE Agrobacterium tumefaciens strain containing the BRI1::GFP translational fusion construct was used to transform wild-type Arabidopsis plants by vacuum infiltration. Transformed seedlings were selected on 0.5× Murashige-Skoog medium, 1% (w/v) Suc, 0.8% (w/v) phytagar, and 50 μg/mL kanamycin and were propagated on soil.
Fluorescence Microscopy
Confocal and conventional fluorescence microscopy were performed on an IX70 inverted microscope (Olympus, Tokyo). Fluorescence was filtered with fluorescein isothiocyanate filter sets (Olympus).
In Vitro Kinase Assay
Hemagglutinin (HA) tag was added to the 3′ end of BRI1, bri1-101, and bri1-113 by cloning the PCR product (BRI1-N, 5′-CCCCGGGTACCTTGAGAAATGAAGACT-3′; BRI1-HAC, 5′-GGGCTAGCGTAATCTGGAACATCGTATG-GGTATAATTTTCCTTCAGGAACTTC-3′) from wild-type BRI1 DNA and bri1-101 and bri1-113 mutant DNA into the pCMX-PL2 vector. The products were sequenced to confirm that no mutations were introduced by the PCR. Human embryonic kidney 293T cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% (v/v) fetal calf serum. Five micrograms of HA-tagged BRI1 plasmid DNA and 5 μg of carrier DNA were transfected into 2 × 106 cells with the calcium phosphate method (Sambrook et al., 1989). Forty-eight hours after transfection, cells were lysed in buffer containing 25 mm Tris-Cl (pH 8.0), 1% (v/v) Nonidet-P40, 10 mm phenylmethylsulfonyl fluoride, 1 mm dithiothreitol, 10 units/mL aprotinin, 20 μg/mL leupeptin, 20 mm EDTA, 10 mm β-glycerophosphate, 1 mm sodium orthovanadate, and 10 mm sodium fluoride for 20 min on ice. Cell lysates were cleared by centrifugation after a 20-min incubation with fixed Staphylococcus aureus at 4°C. HA-tagged BRI1 proteins were immunoprecipitated with the 12CA5 anti-HA tag monoclonal antibody and protein A agarose beads (RepliGen, Needham, MA). The immune complexes were washed twice with lysis buffer, once with phosphate-buffered saline, and twice with kinase reaction buffer (20 mm HEPES [4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid], pH 7.4, 10 mm MgCl2, 10 mm MnCl2, and 1 mm dithiothreitol). After a 20-min incubation at room temperature in 20 μL of kinase reaction buffer containing 5 μm ATP and 5 μCi [γ-32P]ATP (1 Ci = 37 GBq), the kinase reaction was terminated by the addition of 1 mL of phosphate-buffered saline containing 20 mm EDTA. Immune complexes were collected by centrifugation, resuspended in sample buffer, separated on denaturing SDS/polyacrylamide gel, transferred to Immobilon-P membrane (Millipore, Bedford, MA), reacted with primary and secondary antibodies (12CA5 and horseradish per-oxidase-conjugated sheep anti-mouse, respectively), and visualized by enhanced chemiluminescence (Amersham, Buckinghamshire, UK). Kinase reaction products were detected in the same membrane by autoradiography.
Phosphoamino Acid Analysis
The experiment was performed as described by Boyle et al. (1991).
ACKNOWLEDGMENTS
The authors would like to thank Jeff Plautz and Steve Kay for assistance with the confocal microscope.
Footnotes
This work was supported by grants from the U.S. Department of Agriculture (to J.C.) and the National Institutes of Health (to T.H.). J.C. is an associate investigator of the Howard Hughes Medical Institute. T.H. is a Frank and Else Schilling American Cancer Society research professor. D.M.F. was partially supported by a National Institutes of Health Training Grant. C.A.P.J. and J.L. were American Cancer Society Postdoctoral Fellows.
LITERATURE CITED
- Andrésson T, Ruderman JV. The kinase Eg2 is a component of the Xenopus oocyte progesterone-activated signaling pathway. EMBO J. 1998;17:5627–5637. doi: 10.1093/emboj/17.19.5627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beato M, Herrlich P, Schutz G. Steroid hormone receptors: many actors in search of a plot. Cell. 1995;83:851–857. doi: 10.1016/0092-8674(95)90201-5. [DOI] [PubMed] [Google Scholar]
- Becraft PW. Receptor kinases in plant development. Trends Plant Sci. 1998;3:384–388. [Google Scholar]
- Boyle WJ, van der Geer P, Hunter T. Phosphopeptide mapping and phosphoamino acid analysis by two-dimensional separation on thin-layer cellulose plates. Methods Enzymol. 1991;201:110–149. doi: 10.1016/0076-6879(91)01013-r. [DOI] [PubMed] [Google Scholar]
- Braun DM, Stone JM, Walker JC. Interaction of the maize and Arabidopsis kinase interaction domains with a subset of receptor-like protein kinases: implications for transmembrane signaling in plants. Plant J. 1997;12:83–95. doi: 10.1046/j.1365-313x.1997.12010083.x. [DOI] [PubMed] [Google Scholar]
- Braun T, Schofield PR, Sprengel R. Amino-terminal leucine-rich repeats in gonadotropin receptors determine hormone selectivity. EMBO J. 1991;10:1885–1890. doi: 10.1002/j.1460-2075.1991.tb07714.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark SE, Williams RW, Meyerowitz EM. The CLAVATA1 gene encodes a putative receptor kinase that controls shoot and floral meristem size in Arabidopsis. Cell. 1997;89:575–585. doi: 10.1016/s0092-8674(00)80239-1. [DOI] [PubMed] [Google Scholar]
- Clouse SD, Langford M, McMorris TC. A brassinosteroid-insensitive mutant in Arabidopsis thaliana exhibits multiple defects in growth and development. Plant Physiol. 1996;111:671–678. doi: 10.1104/pp.111.3.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon MS, Hatzixanthis K, Jones DA, Harrison K, Jones JDG. The tomato Cf-5 disease resistance gene and six homologs show pronounced allelic variation in leucine-rich repeat copy number. Plant Cell. 1998;10:1915–1925. doi: 10.1105/tpc.10.11.1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endoh H, Sasaki H, Maruyama K, Takeyama K, Waga I, Shimizu T, Kato S, Kawashima H. Rapid activation of MAP kinase by estrogen in the bone cell line. Biochem Biophys Res Commun. 1997;235:99–102. doi: 10.1006/bbrc.1997.6746. [DOI] [PubMed] [Google Scholar]
- Fletcher JC, Brand U, Running MP, Simon R, Meyerowitz EM. Signaling of cell fate decisions by CLAVATA3 in Arabidopsis shoot meristems. Science. 1999;283:1911–1914. doi: 10.1126/science.283.5409.1911. [DOI] [PubMed] [Google Scholar]
- Hanks SK, Quinn AM, Hunter T. The protein kinase family: conserved features and deduced phylogeny of the catalytic domains. Science. 1988;241:42–52. doi: 10.1126/science.3291115. [DOI] [PubMed] [Google Scholar]
- Horn MA, Walker JC. Biochemical properties of the autophosphorylation of RLK5, a receptor-like protein kinase from Arabidopsis thaliana. Biochem Biophys Acta. 1994;1208:65–74. doi: 10.1016/0167-4838(94)90160-0. [DOI] [PubMed] [Google Scholar]
- Jinn T-L, Stone JM, Walker JC. HAESA, an Arabidopsis leucine-rich repeat receptor kinase, controls floral organ abscission. Genes Dev. 2000;14:108–117. [PMC free article] [PubMed] [Google Scholar]
- Jones DA, Jones JDG. The role of leucine-rich repeat proteins in plant defenses. Adv Bot Res. 1997;24:89–167. [Google Scholar]
- Kauschmann A, Jessop A, Koncz C, Szekeres M, Willmitzer L, Altmann T. Genetic evidence for an essential role of brassinosteroids in plant development. Plant J. 1996;9:701–713. [Google Scholar]
- Kobe B, Deisenhofer J. A structural basis of the interactions between leucine-rich repeats and protein ligands. Nature. 1995;374:183–186. doi: 10.1038/374183a0. [DOI] [PubMed] [Google Scholar]
- Konieczny A, Ausubel FM. A procedure for mapping Arabidopsis mutations using codominant ecotype-specific PCR-based markers. Plant J. 1993;4:403–410. doi: 10.1046/j.1365-313x.1993.04020403.x. [DOI] [PubMed] [Google Scholar]
- Li J, Chory J. A putative leucine-rich receptor kinase involved in brassinosteroid signal transduction. Cell. 1997;90:929–938. doi: 10.1016/s0092-8674(00)80357-8. [DOI] [PubMed] [Google Scholar]
- Li J, Smith GP, Walker JC. Kinase interaction domain of kinase-associated protein phosphatase, a phosphoprotein-binding domain. Proc Natl Acad Sci USA. 1999;96:7821–7826. doi: 10.1073/pnas.96.14.7821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandova NB. Plant growth-promoting brassinosteroids. Annu Rev Plant Physiol Plant Mol Biol. 1988;39:23–52. [Google Scholar]
- Mendoza C, Solar A, Tesarik J. Nongenomic steroid action: independent targeting of a plasma membrane calcium channel and a tyrosine kinase. Biochem Biophys Res Comm. 1995;210:518–523. doi: 10.1006/bbrc.1995.1690. [DOI] [PubMed] [Google Scholar]
- Neff MM, Neff JD, Chory J, Pepper AE. dCAPS, a simple technique for the genetic analysis of single nucleotide polymorphisms: experimental applications in Arabidopsis thaliana genetics. Plant J. 1998;14:387–392. doi: 10.1046/j.1365-313x.1998.00124.x. [DOI] [PubMed] [Google Scholar]
- Noguchi T, Fujioka S, Choe S, Takatsuto S, Yoshida S, Yuan H, Feldmann FA, Tax FE. Brassinosteroid-insensitive dwarf mutants of Arabidopsis accumulate brassinosteroids. Plant Physiol. 1999;121:743–752. doi: 10.1104/pp.121.3.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padgett RW. Intracellular signaling: fleshing out the TGFβ pathway. Curr Biol. 1999;9:R408–R411. doi: 10.1016/s0960-9822(99)80255-3. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Expression of cloned genes in cultured mammalian cells. In: Sambrook J, Fritsch EF, Maniatis T, editors. Molecular Cloning: A Laboratory Manual. Ed 2. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. pp. 16.30–16.40. [Google Scholar]
- Siemering KR, Golbik R, Sever R, Haseloff J. Mutations that suppress the thermosensitivity of green fluorescent protein. Curr Biol. 1996;6:1653–1663. doi: 10.1016/s0960-9822(02)70789-6. [DOI] [PubMed] [Google Scholar]
- Song W-Y, Wang G-L, Chen L-L, Kim H-S, Pi L-Y, Holsten T, Gardner J, Wang B, Zhao W-X, Zhu L-H, Fauquet C, Ronald P. A receptor kinase-like protein encoded by the rice disease resistance gene, Xa21. Science. 1995;270:1804–1806. doi: 10.1126/science.270.5243.1804. [DOI] [PubMed] [Google Scholar]
- Stone JM, Collinge MA, Smith RD, Horn MA, Walker JC. Interaction of a protein phosphatase with an Arabidopsis serine-threonine receptor kinase. Science. 1994;266:793–795. doi: 10.1126/science.7973632. [DOI] [PubMed] [Google Scholar]
- Stone JM, Trotochaud AE, Walker JC, Clark SE. Control of meristem development by CLAVATA1 receptor kinase and kinase-associated protein phosphatase interactions. Plant Physiol. 1998;117:1217–1235. doi: 10.1104/pp.117.4.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tesarik J, Moos J, Mendoza C. Stimulation of protein tyrosine phosphorylation by a progesterone receptor on the cell surface of human sperm. Endocrinology. 1993;133:328–335. doi: 10.1210/endo.133.1.7686481. [DOI] [PubMed] [Google Scholar]
- Torii KU, Mitsukawa N, Oosumi T, Matsuura Y, Yokoyama R, Whittier RF, Komeda Y. The Arabidopsis ERECTA gene encodes a putative receptor protein kinase with extracellular leucine-rich repeats. Plant Cell. 1996;8:735–746. doi: 10.1105/tpc.8.4.735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Geer P, Hunter T, Lindberg RA. Receptor protein-tyrosine kinases and their signal transduction pathways. Annu Rev Cell Biol. 1994;10:251–337. doi: 10.1146/annurev.cb.10.110194.001343. [DOI] [PubMed] [Google Scholar]
- Wehling M. Specific, nongenomic actions of steroid hormones. Annu Rev Physiol. 1997;59:365–393. doi: 10.1146/annurev.physiol.59.1.365. [DOI] [PubMed] [Google Scholar]
- Williams RW, Wilson JM, Meyerowitz EM. A possible role for kinase-associated protein phosphatase in the Arabidopsis CLAVATA1 signaling pathway. Proc Natl Acad Sci USA. 1997;94:10467–10472. doi: 10.1073/pnas.94.19.10467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaffe MB, Cantley LC. Signal transduction: grabbing phosphoproteins. Nature. 1999;402:30–31. doi: 10.1038/46925. [DOI] [PubMed] [Google Scholar]