Abstract
Oral biofilm and its molecular analysis provide a basis for investigating various dental research and clinical questions. Knowledge of biofilm composition leads to a better understanding of cariogenic and periopathogenic mechanisms. Microbial changes taking place in the oral cavity during childhood are of interest for several reasons. The evolution of the child oral microbiota and shifts in its composition need to be analyzed further to understand and possibly prevent the onset of disease. At the same time, advanced knowledge of the natural composition of oral biofilm is needed. Early stages of caries-free permanent dentition with healthy gums provide a widely unaffected subgingival habitat that can serve as an in situ baseline for studying features of oral health and disease. Analysis of children's oral biofilm during different stages in life is thus an important theme in the field. Modern molecular analysis methods can provide comprehensive information about the bacterial diversity of such biofilms. To enable microbiota data comparison, it is important to standardize each step in the procedure for molecular data generation. This procedure spans from clinical sampling, Next Generation Sequencing (NGS), bioinformatic data processing, to taxonomic interpretation. One of the most critical factors here is biofilm sampling. Sampling in children is even more challenging in particular due to limited space in subgingival areas. We thus focus on the use of paper points for subgingival sampling. This article provides a detailed protocol for oral biofilm sampling of the subgingival sulcus, the mucosa, and saliva in children.
Keywords: Medicine, Issue 130, Subgingival, mucosal, paper point, sampling, oral biofilm, saliva, microbiota, microbiome, child, orthodontics
Introduction
Human oral biofilm comprises a broad community consisting mostly of commensals and beneficial microorganisms1,2,3. Species found here colonize all niches that the oral cavity offers4,5,6. Biofilm composition in these niches varies as widely as the habitats. Saliva for example displays different bacterial profiles than plaque samples. In plaque samples of healthy adults, the relative abundance of Actinobacteria is over 20%, while less than 7% is found in saliva. Bacteroidetes and Firmicutes in contrast appear in much higher numbers in saliva7. Thus, it is important to sample at different locations in the mouth in order to get the whole picture. Additionally, various factors such as geographic and ethnic differences, age, sex, and many other factors make it difficult to identify general rules for biofilm development and disease onset8,9. For many years the investigation of periopathogenic and cariogenic biofilm has been a central concern8,10,11,12,13.
In recent years, research on healthy subjects has gained importance not only for a broader understanding of disease but also for the implementation of preventive measures14. New technologies and molecular analyses further elucidate oral biofilm formation and function15,16, and enable the complete profiling of microbial diversity17,18. This is expected to also lead to a new understanding of microbial changes during orthodontic therapy19. An impact on the development of orthodontic biomaterials is foreseeable. The enhanced perspective will shed new light on complications of orthodontic therapy associated with biofilm, such as enamel demineralization and periodontal diseases12,20,21. To permit worldwide data comparisons, it is crucial to standardize all steps in data generation. Minor changes in laboratory procedures can strongly influence the results. In addition, the use of varying computational platforms in data processing can lead to non-comparable datasets. Lastly, the choice of statistical tests and corrections has an influence on the results. However, sampling bias can already occur long before any lab work or bio-computing commences. Non-standardized sampling methods lead to an inherently biased study. In view of low to moderate levels of standardization in the relevant studies, little evidence exists on the relationship between orthodontics and microbiomes19. Therefore, the development of standardized methods would facilitate qualified comparison of data in the field.
This article presents standardized oral biofilm sampling procedures. The protocol is intended to contribute towards generating globally comparable collections of microbial sequence data. A step-by-step protocol for oral biofilm sampling in healthy children is presented. As the method of choice, paper points are inserted atraumatically into the sound subgingival sulcus. To facilitate habitat comparisons, cheek mucosa is sampled according to the same protocol. This article additionally demonstrates parallel and pooled sampling. Furthermore, saliva sampling is shown. A simple color-coded transport and storage system is also presented, facilitating specimen management for further processing. The choice of downstream operations regarding storage medium, metagenomic analyses, and bioinformatics depends on the clinical questions raised by different fields in oral research. In this manuscript, DNA sampling for NGS has been chosen as an example of a possible application for orthodontic research.
Protocol
Protocol and video shooting were approved by the institutional review board of the Medical University of Graz (Votum 27-126ex14/15). Written consent for this video publication was obtained from the child and her parent.
1. Instruments and Materials
Cut calibrated (ISO 015/02) and sterile paper points, usually used in endodontic therapy, at the first ring mark in order to standardize their length (Figure 1).
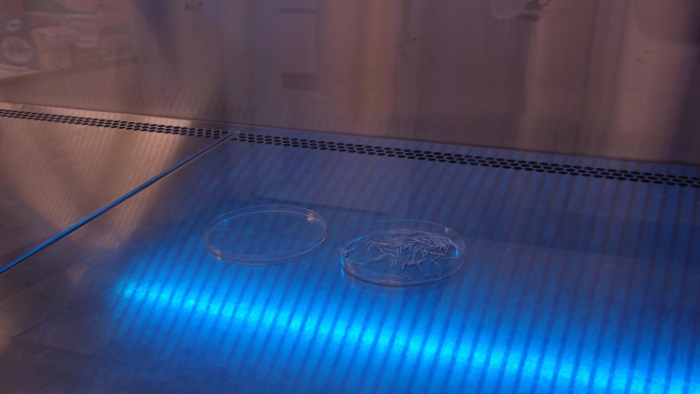
Apply UV light irradiation at 260 nm for 30 min in order to avoid DNA and RNA contamination of the paper points (Figure 2).
Autoclave the tubes for storage at 120 °C and 1.2 bar for 20 min and UV-irradiate them as well or employ ready-to-use gamma irradiated tubes.

2. Preparation of Subjects
Note: Written informed consent was obtained from the child and her parent prior to enrollment.
Apply cocoa butter to the lips.
Mount the cheek and tongue retractor for full access to both dental arches.
Stain the dental plaque with plaque disclosure.
Remove the dry field apparatus.
Rinse the mouth with water until the water is colorless.
Brush teeth thoroughly with an electric tooth brush at 45 ° angulation. Apply water only but no toothpaste, as this would alter the oral biofilm.
Mount the dry field apparatus again, including the tongue guard to keep the mouth open and dry.
Clean and dry the index teeth with sterile cotton swabs to avoid absorption of supragingival fluid during paper point sampling.
3. Biofilm Sampling
- Paper point sampling of the subgingival sulcus
- Grasp the paper points with sterile dental tweezers.
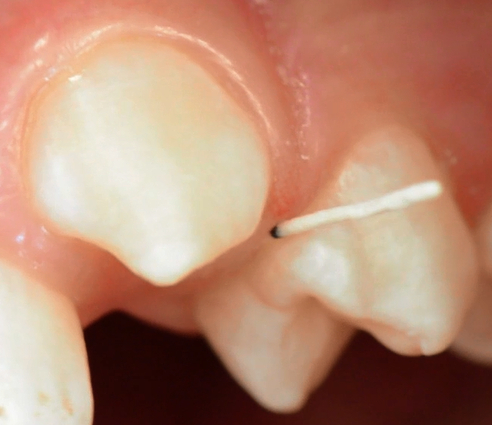
- Insert paper points tangentially up to a defined length of 4 mm. Take special care not to traumatize the junctional epithelium (Figure 3).
- Remove paper points after 20 s.
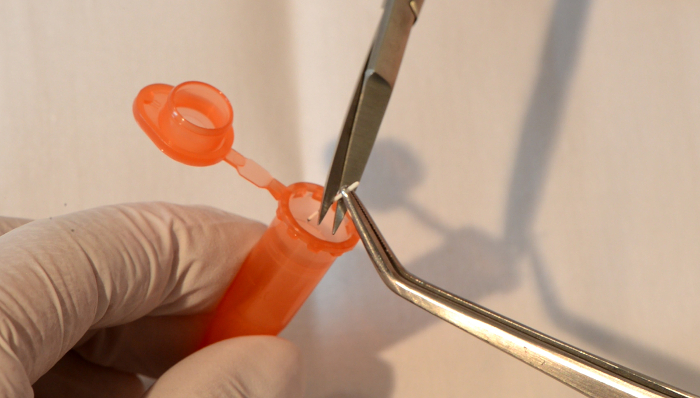
- Collect paper points directly into prepared tubes: place the paper point tip directly in a sterile and DNA-free vial, and cut it at the third mark to a standardized length of 4 mm (Figure 4).
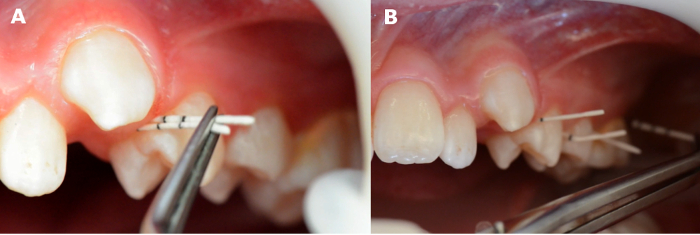
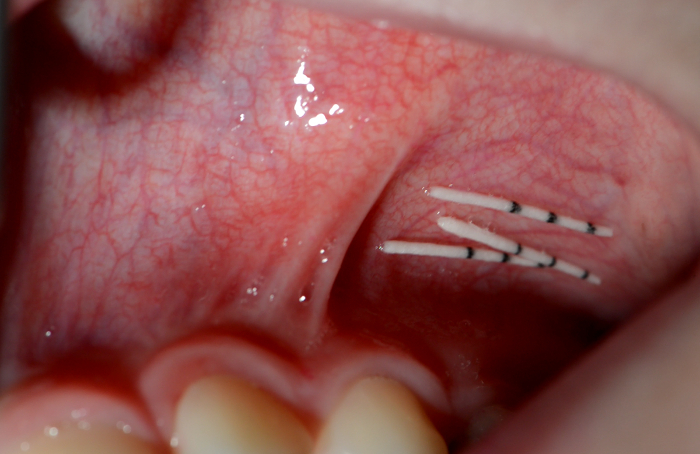
- If indicated by the study protocols, insert two paper points parallel and simultaneously at the same site (Figure 5A).
- Alternatively, collect paper points from several sites if the study protocol requires "pooled samples" (Figure 5B).
- Remove the dry field apparatus for mucosal and saliva sampling.
- Mucosal paper point sampling
- Apply paper points to the vestibular fold of the upper cheek (Figure 6).
- Close the cheek and massage it briefly.
- Open the cheek and remove the paper points after 20 s.
- Cut paper points at the third mark and place them in a sterile tube as indicated for subgingival sampling.
- Saliva sampling
- Let the child spit unstimulated saliva into a sterilized collection vessel (Figure 7).

4. Transfer and Storage
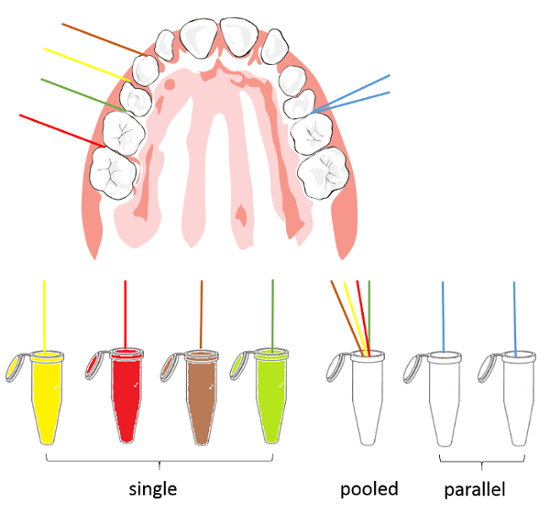
Collect paper points as single, pooled, or parallel samples depending on the study design (Figure 8).
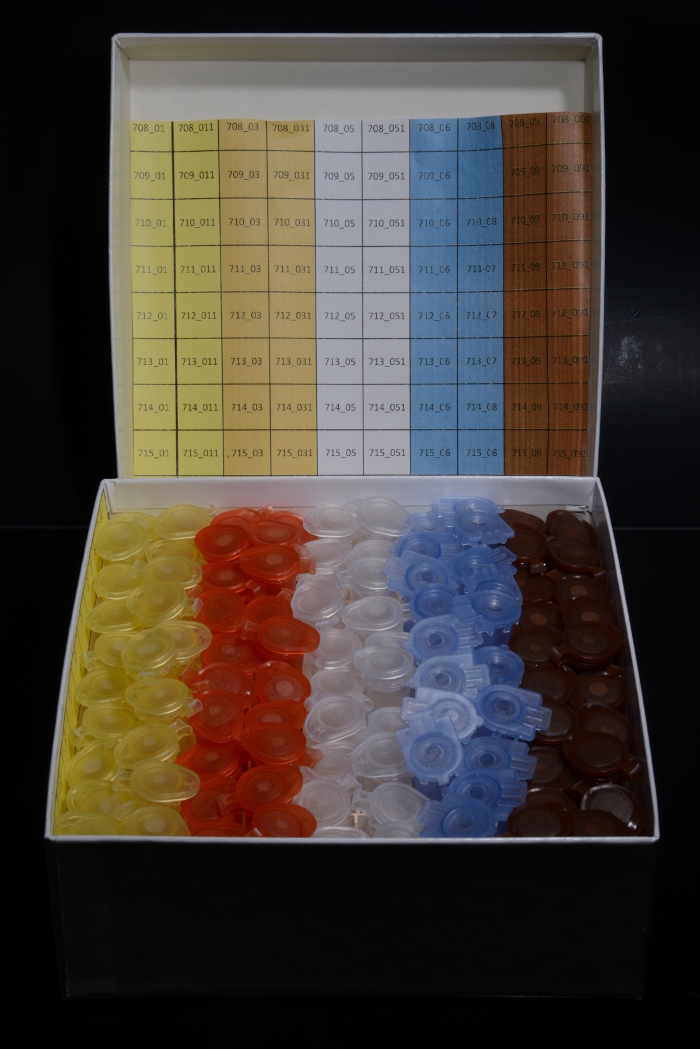
Apply a color-coded storage system to facilitate further sample management (Figure 9).
Finally, store the samples at −80 °C pending microbiome analysis.
Representative Results
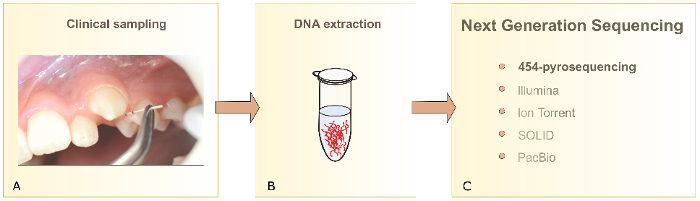
In the present publication, DNA sampling for NGS is demonstrated as an example of a possible application for orthodontic research. Bacterial DNA is extracted directly from the paper point samples. This DNA can then be used in studies of the microbial composition for clinical or research questions (as summarized in Figure 10).
Modern molecular analysis methods such as the NGS method 454-pyrosequencing give an overview of the complete microbiota of a sample. The DNA of thousands of bacteria is identified simultaneously at different phylogenetic levels over the 16s rRNA sequence differences. Bioinformatic platforms need to be generated as a means of structuring into clusters the information retrieved from the large data sets. Statistical analysis can then be applied to microbiota datasets.
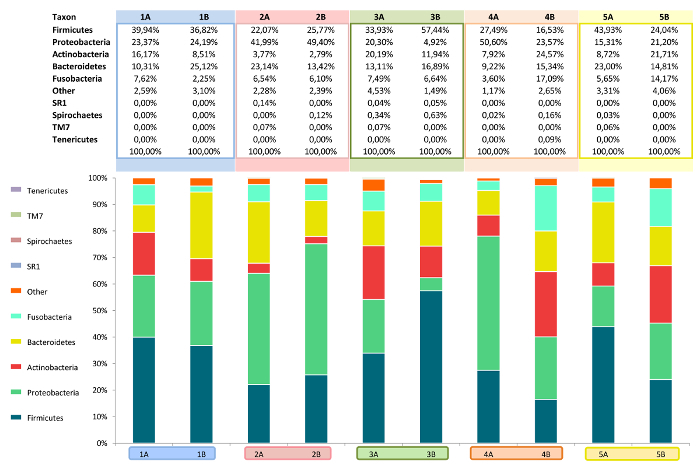
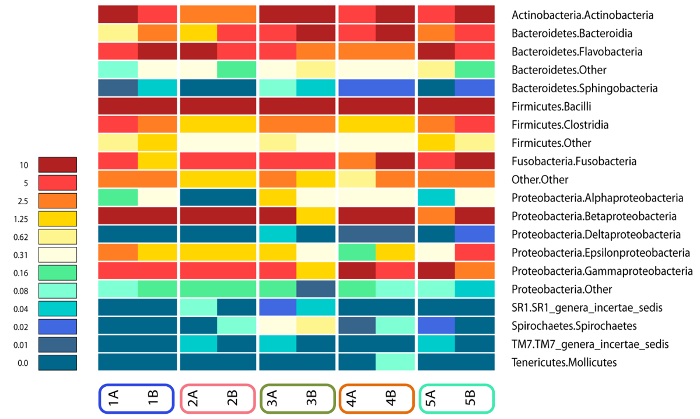
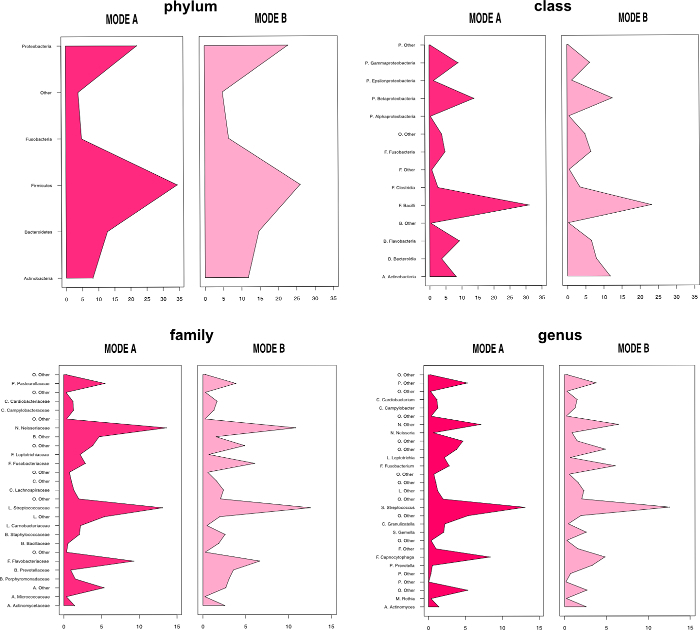
The overall relative abundance of certain bacterial species can be analyzed as can shifts in the microbiota due to changing habitat conditions. Bar charts (Figure 11) and heatmaps (Figure 12) display the subgingival bacterial composition at phylum level or at order level. Various individuals and treatments can be compared by descriptive and inferential statistics. Figure 13 presents a histogram comparison of two modes of subgingival paper point sampling at different taxonomic levels. Table 1 shows the shift in biofilm composition during an in vitro study comparing two time points (T1 and T3). Statistically significant changes were calculated with Wilcoxon signed-rank test and following Bonferroni correction from the 454-pyrosequencing data.
Finally, Principal Coordinate Analysis (PCoA) enables direct 3D comparison at Operational Taxonomic Unit (OTU) level as identified in different samples. The PCoA plots in Figure 14 display intra- and inter-individual differences of the subgingival microbiome in a case series of five children. Figure 15 shows the results of an orthodontic case-control study: pink to red dots are the cases, light to dark blue dots are the controls, all sampled repeatedly over a period of four months. A clear clustering of the cases after the orthodontic intervention (red dots) represents a shift in the microbiome. Blue dots, representing the control group, are evenly distributed.
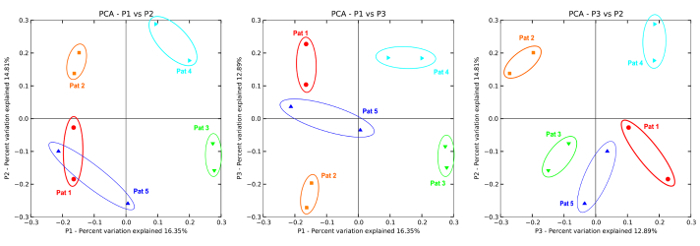
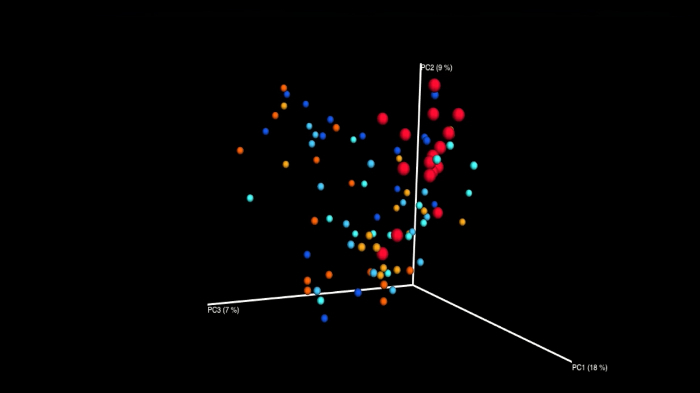
Figure 1: Prepare paper points of standardized length. Sterile calibrated paper points are cut at the first ring mark to standardize their length. Please click here to view a larger version of this figure.
Figure 2:Sterilize and free paper points from DNA. Standardized paper points are sterilized and UV irradiated in a clean bench. Please click here to view a larger version of this figure.
Figure 3: Subgingival paper point insertion. Sterile calibrated paper points are inserted tangentially into the subgingival sulcus until the 4 mm ring mark for 20 s.
Figure 4: Cutting paper point into tube. Directly after sampling, paper points are cut at the third ring mark (4 mm) into a sterile and DNA-free 1.5 mL vial. Please click here to view a larger version of this figure.
Figure 5: Pooled and parallel sampling. Two paper points are inserted simultaneously into the subgingival sulcus of one tooth (A) or several paper points into the subgingival sulci of several teeth (B). Please click here to view a larger version of this figure.
Figure 6: Mucosal sampling. Sterile calibrated paper points are laid into the mucosa of the vestibular fold. Please click here to view a larger version of this figure.
Figure 7: Saliva collection. Unstimulated saliva is spit into a sterile beaker. Please click here to view a larger version of this figure.
Figure 8: Sampling scheme. Samples can be collected as single (left side, one tooth each), pooled (several to all stripes in one vial), or parallel (blue stripes, two paper points at one tooth) samples. Please click here to view a larger version of this figure.
Figure 9:Color-coded storage. A referring color code for sheets and vials facilitates further sample handling. Please click here to view a larger version of this figure.
Figure 10: Subgingival biofilm analysis. Schematic presentation of clinical biofilm sampling (A), DNA extraction (B), and different NGS technologies (C). Please click here to view a larger version of this figure.
Figure 11: Bar chart. Showing relative abundance of bacteria at phylum level analyzed using 454-pyrosequencing. Image modified from Santigli et al.22 Please click here to view a larger version of this figure.
Figure 12: Heatmap. Showing relative abundances of bacterial orders analyzed using 454-pyrosequencing. Dark red represents a high relative abundance (> 10%). Dark blue represents a very low to no relative abundance of the respective bacterial order. Image modified from Santigli et al.22 Please click here to view a larger version of this figure.
Figure 13: Histogram. Comparison of two modes of paper point sampling (modes A and B) at different taxonomic levels. Data generated with 454-pyrosequencing. Image modified from Santigli et al.22 Please click here to view a larger version of this figure.
Figure 14: 2D PCoA plot. A case series (n=5) showing intra-individual and inter-individual differences as a result of two subgingival sampling methods (1 color is 1 subject).Data generated with 454-pyrosequencing. Image modified from Santigli et al.22 Please click here to view a larger version of this figure.
Figure 15: Snapshot of a 3D PCoA plot animation. Showing subgingival microbiome shifts in an orthodontic case control study (n = 16; each group, 3 points in time; cases: pink to red, controls: light to dark blue). Data generated with 454-pyrosequencing. Unpublished data from working group Santigli. Please click here to view a larger version of this figure.
| Taxon: Bacteriae | Time point 1 [%] | Time point 3 [%] | Wilcoxon signed-rank test | |||||
| Phylum/Genus | 25th | Median | 75th | 25th | Median | 75th | p-value | p-value adjusted# |
| Other | ||||||||
| Other | 0.70 | 1.07 | 1.28 | 1.32 | 1.63 | 1.93 | .000 | .005 |
| Actinobacteria | ||||||||
| Actinomyces | 0.00 | 0.04 | 0.13 | 0.06 | 0.30 | 0.97 | .001 | .021 |
| Rothia | 0.00 | 0.00 | 0.05 | 0.00 | 0.13 | 0.32 | .001 | .009 |
| Bacteroidetes | ||||||||
| Prevotella | 0.00 | 0.01 | 0.22 | 0.15 | 1.02 | 2.44 | .000 | .006 |
| Firmicutes | ||||||||
| Other (Bacilli) | 0.00 | 0.01 | 0.06 | 0.00 | 0.04 | 0.10 | .706 | 1.000 |
| Staphylococcus | 0.00 | 0.01 | 0.12 | 0.00 | 0.07 | 0.17 | .548 | 1.000 |
| (Gemellaceae) | 0.00 | 0.00 | 0.07 | 0.12 | 0.77 | 1.24 | .005 | .091 |
| (Lactobacillales) | 0.00 | 0.00 | 0.04 | 0.00 | 0.12 | 1.50 | .004 | .073 |
| Granulicatella | 0.09 | 0.23 | 0.37 | 0.64 | 1.59 | 2.28 | .000 | .003 |
| Lactobacillus | 0.00 | 0.00 | 0.18 | 0.00 | 0.00 | 0.04 | .700 | 1.000 |
| Other (Streptococcaceae) | 0.00 | 0.04 | 0.13 | 0.00 | 0.10 | 0.19 | .768 | 1.000 |
| (Streptococcaceae) | 0.04 | 0.10 | 0.23 | 0.12 | 0.37 | 0.69 | .005 | .083 |
| Streptococcus | 53.42 | 61.96 | 88.38 | 48.44 | 60.83 | 73.97 | .055 | .930 |
| Other (Lachnospiraceae) | 0.00 | 0.00 | 0.01 | 0.00 | 0.00 | 0.11 | .003 | .058 |
| Dialister | 0.00 | 0.00 | 0.04 | 0.00 | 0.00 | 0.04 | .240 | 1.000 |
| Veillonella | 3.43 | 27.15 | 42.78 | 6.69 | 13.38 | 27.05 | .157 | 1.000 |
| Proteobacteria | ||||||||
| Haemophilus | 0.00 | 0.00 | 0.03 | 0.31 | 0.84 | 2.38 | .000 | .008 |
| #p-value Wilcoxon signed-rank test adjusted according Bonferroni correction | ||||||||
| A p<0.0026 was considered significant |
Table 1: Statistical analysis. Differences in the oral microbiome of healthy children at two points in time. Data generated with 454-pyrosequencing. Image modified from Klug et al.23
Discussion
Bacteria are found at all sites within the oral cavity24,25,26. Many studies have focused on the use of saliva as a sampling medium to map oral colonization as it can easily be sampled through spitting27,28,29,30,31,32. However, salivary biofilm does not reflect subgingival biofilm composition. Thus, the use of other sampling methods is crucial for creating the whole picture, and will evoke new discussions in dental research.Several articles compare the use of curettes and paper points for subgingival sampling of periodontally diseased subjects8,33,34. Yet no research group is known to have focused on the application of standardized sampling devices for different age groups, especially for children19.
In this video, oral biofilm sampling of three oral habitats in healthy children is presented.
Modifications and Troubleshooting:
The aim of the manuscript focuses on the clinical protocol for subgingival biofilm sampling of healthy children. The method of choice refers to sound subgingival sulci where limited space makes sampling especially challenging. In this video the method was applied to early permanent dentition. It can also be applied to mixed or mature dentition, and to children and/or adults. Our sampling method allows for modifications such as single or pooled samples. The latter might become a crucial factor for molecular analyses due to the small amount of DNA collectable from the healthy subgingival sulcus. The choice of the index teeth may be modified on demand; in particular, another tooth may be sampled as an alternative when the epithelium is traumatized and bleeding, thus making the paper point unusable. Parallel sampling using two paper points simultaneously at one site renders sample replicates in one step, in addition to shortening the chair time for children.
With slight modifications diseased periodontal pockets can be sampled. Here paper points will need to be inserted to the full length of the pocket, which can reach up to 10 mm. The length and conus of paper points as well as the depth of insertion must be adapted to the intraoral habitat of interest, but should always be standardized by material and dimensions. Samples can also be taken from the mucosa using paper points. Identical protocols allow comparisons of different oral habitats and various dental materials. Patient preparation and sample storage can be performed as described in this video. For metatranscriptome research, RNA could be sampled with the same method. Thus, after sampling, the paper points should be inserted directly in RNA stabilization solution.
Limitations of the Technique:
Regardless of modifications, the clinical sampling method we describe is limited to the sound subgingival sulcus. Other sampling sites, as for example periodontally diseased pockets, were not part of our study design. As sampling to the full depth of such pockets is needed, paper points might not be large enough to collect samples sufficiently. If the experimental goal is to compare data sampled from sound subgingival sulci and periodontally diseased pockets, it is important to be aware of the inherent bias that is associated with different clinical sampling methods. It therefore is not advisable to compare such data.
A limitation of the method presented here is sampling without the subsequent use of propidium monoazide. Adding this agent directly after sampling allows for specific analysis of only the living cells in the biofilm analyzed. The sampling method described here reflects numbers of living and dead cells. For research of severe periodontitis, paper points will probably need to be replaced by curettes, as biofilm formation in deep pockets is strong. Paper point sampling might not reflect the entire microbial spectrum in these cases, as their surface might be saturated too quickly.
Significance with Respect to Existing Methods:
The proposed manuscript is the first of its kind to standardize subgingival sampling in the healthy sulcus with paper points. Other reports have referred to the use of metal curettes35,36 or paper points 37,38,39, but did not describe the processes that take place prior to sampling, such as plaque control, tooth cleaning, tooth isolation, and drying, as well as the processes that result from inadequate specifications on the sampling technique and time lines.
In two previous articles, we show that the use of paper points is a reproducible method for subgingival biofilm sampling in children22,40. Due to their design, curettes are too large for the shallow sulcus with limited space and too sharp for the tender junctional epithelium. It is crucial to access the subgingival sulcus without traumatizing the epithelium. In this way, bias arising from bleeding is avoided. Thus, the use of the slim and tender paper points is preferred for this area. Paper points are functional to monitor changes in biofilm composition during orthodontic treatment21,41. They are slim and flexible enough to fit between the elements of fixed orthodontic appliances. This is a major advantage to other sampling methods like curettes. Atraumatic sampling is simplified and sampling of small amounts of DNA is possible.
Future Applications:
In this video we demonstrated the method on non-diseased, early, permanent dentition. It also can be applied to mixed or mature dentition. With some adaptations it is possible to sample periodontally diseased sites like teeth or implants and other niches due to dental materials by following the same protocol. Caries research could benefit from this standardized protocol, in particular studies on root caries. The most important lecture from our sampling video is standardization of the protocols to make data comparable, no matter what and how samples are measured. Future video demonstrations could enhance the field of clinical sampling in general.
Oral biofilm obtained as shown in the video is used in clinics and for scientific investigations. This in vivo approach represents a good addition to in vitro molecular techniques such as fluorescence in situ hybridization and confocal laser scanning microscopy15. NGS methods, as the pyrosequencing shown here, applied to the collected biofilm, allow the analysis of the complete microbiota. Calculating the relative abundances of certain bacterial species can be used to support treatments or to compare different patients or different treatments. Together with a standardized sequencing protocol and sequence analysis workflow that we have previously described40, this sampling method allows sequence analysis down to the genus level. Further "meta"-studies such as metatranscriptomic or metabolomic studies are possible based on the sampling protocol presented in this video.
Critical Steps:
Avoiding contamination is a critical step: sterile instruments have to be applied in a non-sterile in vivo environment. The transfer from the mouth to the bench has to be performed quickly and securely by an experienced examiner to keep chair time for study participants as short as possible. The most critical step in the protocol is the atraumatic insertion of the paper point into the subgingival sulcus. Disruption of the junctional epithelium and bleeding absolutely have to be avoided. Further care has to be taken in order to not contaminate paper points and storage vials with exogenous DNA or RNA. The storage itself then also is a critical step. Samples need to be frozen directly, at best at −80 °C. This avoids changes in bacterial composition post sampling, which would falsify sequencing results.
Disclosures
The authors report no conflicts of interest related to this scientific video production.
Acknowledgments
The authors are grateful to Joachim Theussl (Center of Medical Research, Medical University of Graz) for the excellent technical assistance in the video production and to Monica Farrell (Research Management, Medical University of Graz) as voice talent.
References
- Paster B, et al. Bacterial Diversity in Human Subgingival Plaque. J Bacteriol. 2001;183(12):3770–3783. doi: 10.1128/JB.183.12.3770-3783.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aas J, Paster B, Stokes L, Olsen I, Dewhirst F. Defining the Normal Bacterial Flora of the Oral Cavity. J Clin Microbiol. 2005;43(11):5721–5732. doi: 10.1128/JCM.43.11.5721-5732.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ledder R, et al. Molecular Analysis of the Subgingival Microbiota in Health and Disease. Appl Environ Microb. 2007;73(2):516–523. doi: 10.1128/AEM.01419-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaura E, Keijser B, Huse S, Crielaard W. Defining the healthy 'core microbiome' of oral microbial communities. BMC Microbiol. 2009;9(259):1–12. doi: 10.1186/1471-2180-9-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcaraz LD, et al. Identifying a healthy oral microbiome through metagenomics. Clin Microbiol Infec. 2012;18(11):54–57. doi: 10.1111/j.1469-0691.2012.03857.x. [DOI] [PubMed] [Google Scholar]
- Dewhirst F, et al. The Human Oral Microbiome. J Bacteriol. 2010;192:5002–5017. doi: 10.1128/JB.00542-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keijser BJF, et al. Pyrosequencing analysis of the Oral Microflora of healthy adults. Journal of Dental Research. 2008;87:1016–1020. doi: 10.1177/154405910808701104. [DOI] [PubMed] [Google Scholar]
- Könönen E, Müller H. Microbiology of aggressive periodontitis. Periodontol 2000. 2014;65(1):46–78. doi: 10.1111/prd.12016. [DOI] [PubMed] [Google Scholar]
- Nasidze I, Li J, Quinque D, Tang K, Stoneking M. Global diversity in the human salivary microbiome. Biotechfor. 2009;19(4):636–643. doi: 10.1101/gr.084616.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jervøe-Storm PM, Koltzscher M, Falk W, Dörfler A, Jepsen S. Comparison of culture and real-time PCR for detection and quantification of five putative periopathogenic bacteria in subgingival plaque samples. J Clin Periodontol. 2005;32(7):778–783. doi: 10.1111/j.1600-051X.2005.00740.x. [DOI] [PubMed] [Google Scholar]
- Ouhara K, et al. Susceptibilities of periopathogenic and cariogenic bacteria to antibacterial peptides, β-defensins and LL37, produced by human epithelial cells. J Antimicrob Chemoth. 2005;55:888–896. doi: 10.1093/jac/dki103. [DOI] [PubMed] [Google Scholar]
- Belda-Ferre P, et al. The oral metagenome in health and disease. ISME J. 2011;6(1):46–56. doi: 10.1038/ismej.2011.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt S, Ebersole J. Porphyromonas gingivalis, Treponema denticola, and Tannerella forsythia: the 'red complex', a prototype polybacterial pathogenic consortium in periodontitis. Periodontol 2000. 2005;38:72–122. doi: 10.1111/j.1600-0757.2005.00113.x. [DOI] [PubMed] [Google Scholar]
- Gomez A, Nelson K. The Oral Microbiome of Children: Development, Disease, and Implications Beyond Oral Health. Microb Ecol. 2017;73(2):492–503. doi: 10.1007/s00248-016-0854-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klug B, et al. Oral Biofilm Analysis of Palatal Expanders by Fluorescence In-Situ Hybridization and Confocal Laser Scanning Microscopy. J Vis Exp. 2011. p. e2967. [DOI] [PMC free article] [PubMed]
- Schlafer S, Dige I. Ratiometric Imaging of Extracellular pH in Dental Biofilms. J Vis Exp. 2016. [DOI] [PMC free article] [PubMed]
- Kilian M, et al. The oral microbiome - an update for oral healthcare professionals. British Dental Journal. 2016;221(10):657–666. doi: 10.1038/sj.bdj.2016.865. [DOI] [PubMed] [Google Scholar]
- Razzouk S, Termechi O. Host genome, epigenome, and oral microbiome interactions: toward personalized periodontal therapy. J Periodontol. 2012;84(9):1266–1271. doi: 10.1902/jop.2012.120531. [DOI] [PubMed] [Google Scholar]
- De Freitas A, Marquezan M, da Nojima M, Alviano D, Maia L. The influence of orthodontic fixed appliances on the oral microbiota: A systematic review. Dent Press J Orthod. 2014;19(2):46–55. doi: 10.1590/2176-9451.19.2.046-055.oar. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akin M, Tezcan M, Ileri Z, Ayhan F. Incidence of white spot lesions among patients treated with self- and conventional ligation systems. Clin Oral Invest. 2015;19(6):1501–1506. doi: 10.1007/s00784-014-1382-3. [DOI] [PubMed] [Google Scholar]
- Ren Y, Jongsma M, Mei L, van der Mei H, Busscher H. Orthodontic treatment with fixed appliances and biofilm formation-a potential public health threat? Clin Oral Invest. 2014;18(7):1711–1718. doi: 10.1007/s00784-014-1240-3. [DOI] [PubMed] [Google Scholar]
- Santigli E, Trajanoski S, Eberhard K, Klug B. Sampling Modification Effects in the Subgingival Microbiome Profile of Healthy Children. Frontiers Microbiol. 2017;7:2142. doi: 10.3389/fmicb.2016.02142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klug B, et al. From Mouth to Model: Combining in vivo and in vitro Oral Biofilm Growth. Frontiers Microbiol. 2016;7:1448. doi: 10.3389/fmicb.2016.01448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ximénez-Fyvie L, Haffajee A, Socransky S. Microbial composition of supra- and subgingival plaque in subjects with adult periodontitis. J Clin Periodontol. 2000;27(10):722–732. doi: 10.1034/j.1600-051x.2000.027010722.x. [DOI] [PubMed] [Google Scholar]
- Zijnge V, et al. Oral Biofilm Architecture on Natural Teeth. PLoS ONE. 2010;5(2):e9321. doi: 10.1371/journal.pone.0009321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz PI, et al. Using high throughput sequencing to explore the biodiversity in oral bacterial communities. Mol Oral Microbiol. 2012;27(3):182–201. doi: 10.1111/j.2041-1014.2012.00642.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goode M, Cheong S, Li N, Ray W, Bartlett C. Collection and extraction of saliva DNA for next generation sequencing. J Vis Exp. 2014. [DOI] [PMC free article] [PubMed]
- Hodgson N, Granger D. Collecting saliva and measuring salivary cortisol and alpha-amylase in frail community residing older adults via family caregivers. J Vis Exp. 2013. p. e50815. [DOI] [PMC free article] [PubMed]
- Luo AH, Yang DQ, Xin BC, Paster BJ, Qin J. Microbial profiles in saliva from children with and without caries in mixed dentition. Oral Dis. 2012;18(6):595–601. doi: 10.1111/j.1601-0825.2012.01915.x. [DOI] [PubMed] [Google Scholar]
- Nasidze I, et al. Comparative analysis of human saliva microbiome diversity by barcoded pyrosequencing and cloning approaches. Anal Biochem. 2009;391(1):64–68. doi: 10.1016/j.ab.2009.04.034. [DOI] [PubMed] [Google Scholar]
- Pfaffe T, Cooper-White J, Beyerlein P, Kostner K, Punyadeera C. Diagnostic Potential of Saliva: Current State and Future Applications. Clin Chem. 2011;57(5):675–687. doi: 10.1373/clinchem.2010.153767. [DOI] [PubMed] [Google Scholar]
- Zhu W, Gallo R, Huang CM. Sampling human indigenous saliva peptidome using a lollipop-like ultrafiltration probe: simplify and enhance peptide detection for clinical mass spectrometry. J Vis Exp. 2012. p. e4108. [DOI] [PMC free article] [PubMed]
- Belibasakis G, Schmidlin P, Sahrmann P. Molecular microbiological evaluation of subgingival biofilm sampling by paper point and curette. Apmis. 2014;122(4):347–352. doi: 10.1111/apm.12151. [DOI] [PubMed] [Google Scholar]
- Hayashi F, Okada M, Soda Y, Miura K, Kozai K. Subgingival distribution of Campylobacter rectus and Tannerella forsythensis in healthy children with primary dentition. Arch Oral Biol. 2006;51(1):10–14. doi: 10.1016/j.archoralbio.2005.05.004. [DOI] [PubMed] [Google Scholar]
- Papaioannou W, et al. The microbiota on different oral surfaces in healthy children. Oral Microbiol Immunol. 2009;24(3):183–189. doi: 10.1111/j.1399-302X.2008.00493.x. [DOI] [PubMed] [Google Scholar]
- Abusleme L, et al. The subgingival microbiome in health and periodontitis and its relationship with community biomass and inflammation. ISME J. 2013;7(5):1016–1025. doi: 10.1038/ismej.2012.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffen A, et al. Distinct and complex bacterial profiles in human periodontitis and health revealed by 16S pyrosequencing. ISME J. 2011;6(6):1176–1185. doi: 10.1038/ismej.2011.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jünemann S, et al. Bacterial Community Shift in Treated Periodontitis Patients Revealed by Ion Torrent 16S rRNA Gene Amplicon Sequencing. PLoS ONE. 2012;7(8):e41606. doi: 10.1371/journal.pone.0041606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortelli J, et al. Detection of periodontal pathogens in newborns and children with mixed dentition. European J Clin Microbiol Infect Dis. 2012;31:1041–1050. doi: 10.1007/s10096-011-1405-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trajanoski S, et al. Next-generation sequencing in microbiome analysis: factors affecting reproducibility of repeated biofilm sampling of the gingival sulcus of children. JDOCE. 2013;1(3):34–46. [Google Scholar]
- Jongsma M, et al. Biofilm formation on stainless steel and gold wires for bonded retainers in vitro and in vivo and their susceptibility to oral antimicrobials. Clin Oral Invest. 2013;17(4):1209–1218. doi: 10.1007/s00784-012-0807-0. [DOI] [PubMed] [Google Scholar]


