Supplemental Digital Content is available in the text.
Abstract
Background:
Craniofacial malformations are among the most common congenital anomalies. Cranial neural crest cells (CNCCs) form craniofacial structures involving multiple cellular processes, perturbations of which contribute to craniofacial malformations. Adhesion of cells to the extracellular matrix mediates bidirectional interactions of the cells with their extracellular environment that plays an important role in craniofacial morphogenesis. Talin (tln) is crucial in cell-matrix adhesion between cells, but its role in craniofacial morphogenesis is poorly understood.
Methods:
Talin gene expression was determined by whole mount in situ hybridization. Craniofacial cartilage and muscles were analyzed by Alcian blue in Tg(mylz2:mCherry) and by transmission electron microscopy. Pulse-chase photoconversion, 5-ethynyl-2’-deoxyuridine proliferation, migration, and apoptosis assays were performed for functional analysis.
Results:
Expression of tln1 was observed in the craniofacial cartilage structures, including the palate. The Meckel’s cartilage was hypoplastic, the palate was shortened, and the craniofacial muscles were malformed in tln1 mutants. Pulse-chase and EdU assays during palate morphogenesis revealed defects in CNCC proliferation in mutants. No defects were observed in CNCC migration and apoptosis.
Conclusions:
The work shows that tln1 is critical for craniofacial morphogenesis in zebrafish. Loss of tln1 leads to a shortened palate and Meckel’s cartilage along with disorganized skeletal muscles. Investigations into the cellular processes show that tln1 is required for CNCC proliferation during palate morphogenesis. The work will lead to a better understanding of the involvement of cytoskeletal proteins in craniofacial morphogenesis.
BACKGROUND
More than three-fourths of structural congenital anomalies involve malformation of the craniofacial region.1 The craniofacial region is assembled during early development by complex cellular processes involving cranial neural crest cells (CNCCs). The CNCCs comprise a group of pluripotent cells that delaminate from the neural tube and migrate through defined pathways to contribute to craniofacial cartilage.2,3 While undergoing migration, the CNCCs also proliferate and undergo controlled apoptosis to form distinct craniofacial structures such as the palate and the Meckel’s cartilage.1 Perturbations in the cellular mechanisms of CNCCs contribute to most craniofacial malformations. Hence, the molecular factors involved in CNCC formation, migration, and maintenance are subjects of much interest.
The cellular processes are influenced by mechanical stimuli via extensive interactions between the cell surface structures and extracellular matrix (ECM). The ECM consists of multiple matrix molecules such as glycoproteins, fibronectin, proteoglycans, and collagens.4 Cells within tissues and organs adhere to the ECM scaffold that in turn induces discrete cell surface structures. The cell-matrix adhesions mediate bidirectional interactions of the cell with its extracellular environment and play central roles in embryonic development. Many genes of the cytoskeletal network and the cell-matrix adhesion complex have been implicated in perturbing cellular processes in CNCCs, and thereby result in craniofacial anomalies. For example, caldesmon is an actin modulator, which when mutated, causes severe cranial cartilage defects with aberrant CNCC migration in Xenopus.5 In zebrafish, knockdown of caldesmon causes defective cardiovascular development.6 Myosin-X is required for CNCC migration in Xenopus and for cell–cell adhesion in in vitro cell cultures.7 We and others showed that SPECC1L, 1 of the first genes implicated in Tessier oblique facial cleft, encodes for a cytoskeletal protein known to interact with actin filaments.8,9 Mutations in FILAMIN A (FLNA) are known to be associated with craniosynostosis.10 Loss-of-function (LOF) of capzb, an actin-capping protein causes cleft palate and micrognathia.11 Stickler syndrome, which includes cleft lip and palate as 1 of its major presentations, is caused by mutations in COL2A1, a gene that encodes collagen, a component of the ECM.12
Integrins are the principle cell surface adhesion receptors that regulate interactions between cells and the ECM. Integrin proteins have a large extracellular domain that interacts with ECM proteins, and a small intracellular domain that interacts with various proteins involved in cell signaling, and adapter proteins that provide connections to the cytoskeleton.13 Talin (tln) is 1 of the key cytoskeletal proteins involved in integrin-mediated cell signaling.14 Biochemical and cellular studies indicate that Tln activates integrins by binding to the cytoplasmic domain of the integrin β-subunit (Fig. 1, above).15–17 In addition to its role in signaling cascades inside the cell, Tln links integrins to the actin cytoskeletal network (Fig. 1, above), which is crucial to transmit force from the actin cytoskeleton to the ECM.18
Fig. 1.
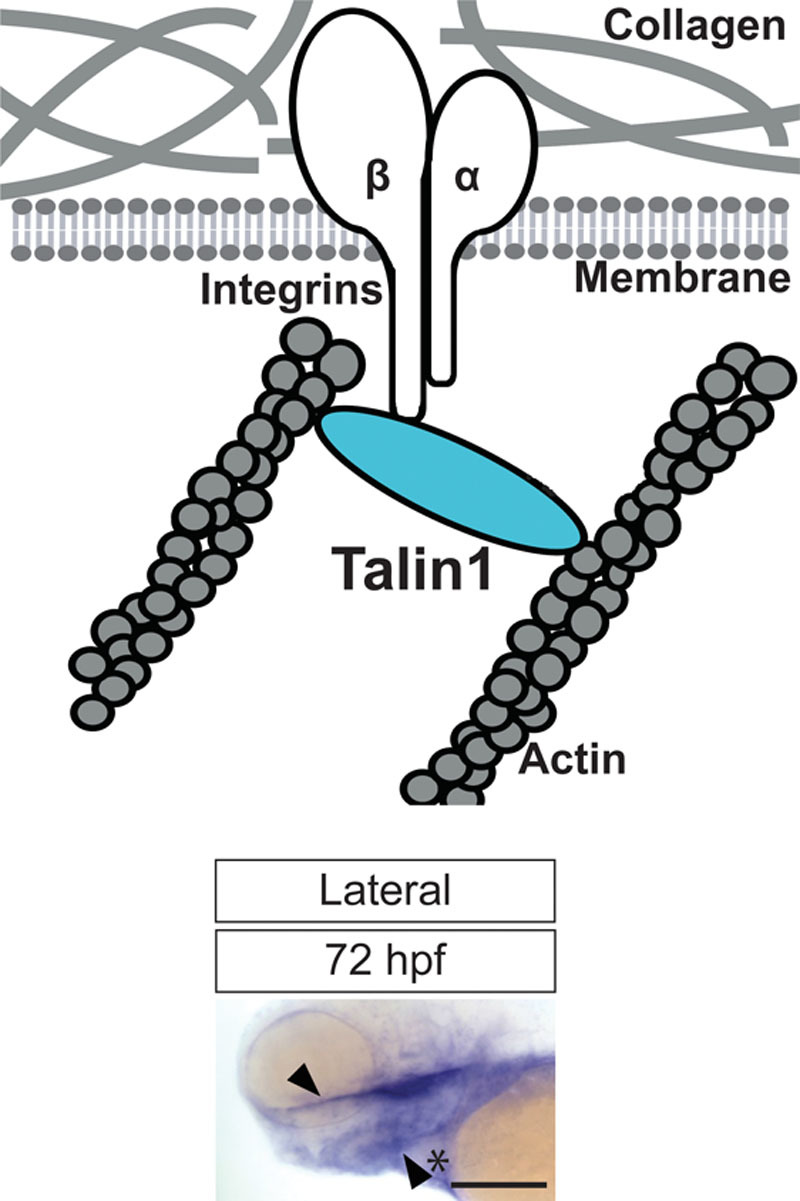
Diagram depicting Talin1 (Tln1) protein with respect to the ECM (depicted by collagen), and the cell-surface (above). Tln1 links the actin cytoskeleton inside the cells to the ECM via the cytoplasmic domain of the β-subunit of integrins. Image of whole-mount embryo at 72 hpf shows the gene expression of tln1 in the craniofacial region during embryogenesis (below). Specific expression is observed in the palate (depicted by black arrowhead) and Meckel’s cartilage (depicted by black arrowhead and asterix). Scale bar: 500 μm.
There are 2 Tln isoforms in most vertebrates.19 In zebrafish, there are 3 tln isoforms: tln1, tln2, and tln2a.19,20 Global LOS of tln1 results in embryonic lethality in mice due to defects in cell migration during gastrulation, whereas Tln2-null mice are viable and fertile with only mild dystrophy.21,22 These studies suggest that Tln1 plays a critical role during embryonic development. Elucidating the role of Tln1 will provide an important vantage point for understanding craniofacial morphogenesis.
Zebrafish (Danio rerio) is a powerful vertebrate model to study the molecular factors involved in craniofacial morphogenesis. We and others have shown that the zebrafish ethmoid plate (hereafter palate) is analogous to the human primary palate.9 In addition, many genes involved in human craniofacial morphogenesis are conserved in the zebrafish.9,11,23–26 Here, we explore the role of tln1 in the development of the craniofacial cartilage and muscle in zebrafish. We also elucidate the role of tln1 in the cellular processes involved in palate development to investigate how forces generated by the cytoskeletal scaffold affect CNCCs. We find that tln1 is required for the formation of the craniofacial complex by regulating CNCC proliferation.
MATERIALS AND METHODS
Zebrafish Lines and Maintenance
The retroviral insertion mutant line Tln1hi3093Tg/+ (hereafter referred to as Tln1+/-) was obtained from the Zebrafish International Resource Center. Wildtype (WT), Tg(sox10:kaede),27 Tg(sox10:mcherry),28 and Tg(mylz2:mcherry)29 were used for various experiments. Embryos were obtained by natural spawning and staged. All embryos and fish were raised and cared for by using established protocols in accordance with the Subcommittee on Research Animal Care, Massachusetts General Hospital.
Total RNA Extraction and cDNA Synthesis
Total RNA extraction and cDNA synthesis was performed as described previously.11 Forty-eight hours postfertilization (hpf) embryos were used for total RNA synthesis for generation of tln1 riboprobe. Forty-eight hpf, 72 hpf, and 5 days postfertilization (dpf) embryos were used for quantitative real time polymerase chain reaction (qRT-PCR).
Whole-mount RNA In Situ Hybridization
The cDNA template for probe synthesis was generated by RT-PCR (see Fig. 2 for primer sequences). The (digoxin) DIG-labeled riboprobe was synthesized using the mMESSAGE mMACHINE T3 Transcription Kit (Ambion, Foster City, Calif.) according to the manufacturer’s instructions. Whole-mount RNA in situ hybridization (WISH) was performed with staged Tü embryos as previously described.30
Fig. 2.
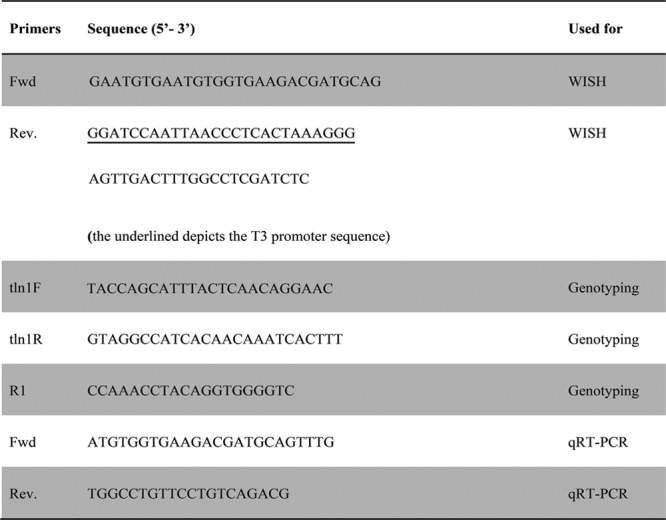
Primers used for tln1 WISH, genotyping and qRT-PCR.
Genotyping of tln1hi3093Tg/+ Retroviral Insertion Line
Genomic DNA was prepared from adult zebrafish tail-clips and embryos as described previously.11 Briefly, 5 μl genomic DNA was used for separate amplification reaction with primer pairs tln1F-tln1R and tln1F-R1 (see Fig. 2 for primer sequences), respectively. The following conditions were used: 94°C for 3 minutes; 34 cycles of 94°C for 30 seconds, 58°C for 40 seconds, 72°C for 40 seconds; 72°C for 5 minutes; and hold at 8°C. Five hundred eight base pairs in tln1F-tln1R PCR reaction confirms wild-type (WT), and 256 base pairs in tln1F-R1 PCR reaction confirms presence of the hi3093Tg insertion.
qRT-PCR
tln1 Transcripts were quantified in whole tln1- /- and WT embryos via qRT-PCR (see Fig. 2 for primers sequences) and normalized with respect to ef1α expression as described previously.11
Cartilage Staining
Alcian blue staining was performed at 4 dpf to analyze the craniofacial cartilage as described previously.31
mRNA Rescue Injections
Full-length tln1 (NM_001009560.1) cDNA was synthesized using GeneArt (Thermo Fisher Scientific, Waltham, MA). Eight synonymous substitutions were made to inactivate BamHI sites present in tln1 cDNA (see Table, Supplemental Digital Content 1, http://links.lww.com/PRSGO/A661). The cDNA was cloned into the pCS2+ vector at the BamHI sites in the multiple cloning site by Thermo Fisher Scientific. The pCS2+tln1 plasmid was linearized with NotI (New England Biolabs, Ipswich, MA), and 1 μg of the linearized template was used to synthesize mRNA using SP6 mMESSAGE mMACHINE RNA synthesis kit (Thermo Fisher Scientific) following the manufacture’s instructions. Approximately 200 pg of RNA was injected into 1-cell stage tln1+/- intercross embryos. Injected embryos were grown to 4.5 days postinjection at 28°C and fixed in 4% PFA. Tails of all embryos were clipped for individual genotyping, and heads were used to perform Alcian blue staining.
Visualization of Craniofacial Skeletal Muscles
WT; mylz2:mCherry and Tln1-/-; mylz2:mCherry embryos at 4 dpf were imaged at 10× on the confocal microscope (Nikon A1R Si Confocal Eclipse Ti series).
Transmission Electron Microscopy
Transmission electron microscopy (TEM) was performed on WT and tln1-/- embryos at 4.5 dpf, at the Microscopy Core at the Program of Membrane Biology at MGH (Mass.) as described previously.11
Cell Migration and Apoptosis Assays
These were performed with WT and mutants in Tg(sox10:kaede) and Tg(sox10:mCherry) transgenic as described previously.11
Cell Proliferation Assays
The pulse-chase photoconversion assay was performed in WT;sox10:kaede and tln1-/-;sox10:kaede embryos at 60 hpf (pulse) and 72 hpf (chase) as described previously.11 5-ethynyl-2’-deoxyuridine (EdU) assay was performed in WT;sox10:mCherry and tln1-/-;sox10:mCherry embryos at 72 dpf with the Click-iT EdU Alexa Fluor 488 Imaging Kit (Thermo Fisher Scientific) according to manufacturer’s instructions. The number of CNCCs proliferating at the given time point in WT and tln1-/- was quantified by counting the EdU-positive CNCCs (sox10+-red and EdU+-green, resulting in yellow CNCCs) in the proliferative, distal most part of the palate (region bound by the white dotted line, Fig. 3, right).
Fig. 3.
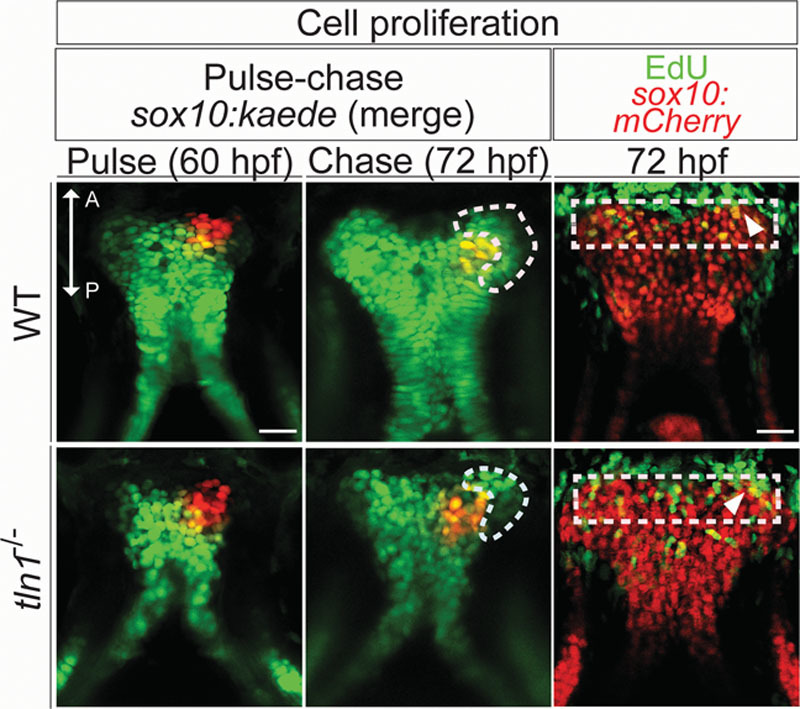
Pulse-chase assay in Tg(sox10:kaede) transgenic to investigate palatal CNCC proliferation (above). Anterior (A) is up in all images. The distal tip of the palate is photoconverted from green to red at 60 hpf (pulse), in WT (left and above) and the mutant (left and below), with the contralateral side as control. Confocal images are taken after chase, at 72 hpf in both WT and tln1 (center). The palate is imaged in the green and the red channels in all instances. Green cells added onto the distal part of the palate during the chase depict the proliferative front, marked with white dotted line (center). Reduced cell proliferation is observed in tln1 mutants, (center and below) when compared with WT (center and above). EdU staining (green) reveals a proliferative front in both the WT and tln1 palate (red), marked by a white dotted rectangle (right). The proliferative cells are in yellow (overlap of green EdU+ and mCherry sox10+ CNCCs), depicted by white arrowheads. Scale bar: 100 μm. A, anterior; P, posterior.
Imaging, Measurements, and Data Processing
Embryos were mounted in 95% glycerol in 1× PBS with Tween 20 and imaged at 10× on Nikon 80i compound microscope (Nikon Instruments, Melville, NY) for WISH and cartilage staining experiments. The palate and mandible were dissected and flat mounted before imaging them at 40×. Embryos were mounted in 3% methylcellulose and imaged at 20× on the confocal microscope (Nikon A1R Si Confocal Eclipse Ti series) for cell proliferation, migration, and apoptosis assays. All images were processed with NIS-Elements advanced image acquisition and analysis system (Nikon Instruments). Figures were composed with Adobe Photoshop CS6 and Adobe Illustrator CS6 (Adobe, San Jose, CA). All measurements were performed using ImageJ (NIH).
Statistical Analysis
All statistical analyses were performed by two-tailed Student’s t test using the GraphPad software (https://graphpad.com/quickcalcs/). A threshold of P ≤ 0.001 was considered to be statistically significant in all cases. All results are represented as mean ± SD.
RESULTS
Gene Expression of tln1 in Craniofacial Region of Zebrafish
Examination of gene expression using WISH revealed that tln1 transcripts were detected at the 2-cell stage and strongly expressed in the craniofacial region at 48 hpf (see Figure, Supplemental Digital Content 2, http://links.lww.com/PRSGO/A662). From 60 to 72 hpf, tln1 gene expression was restricted to the palate and the Meckel’s cartilage (Fig. 1, below; see Figure, Supplemental Digital Content 2, http://links.lww.com/PRSGO/A662).
Characterization of tln1 Retroviral Insertional Mutant Line
Analysis of tln1hi3093Tg/+ intercross revealed morphological anomalies, where genotyping confirmed that the anomalies were exhibited by tln1 homozygotes observing Mendelian ratios (see Table, Supplemental Digital Content 3, http://links.lww.com/PRSGO/A663). The mutant phenotype began to be apparent at 48 hpf when tln1-/- embryos displayed cardiac edema that became more severe by 60 hpf (data not shown). The homozygotes did not survive past 5.5 dpf. qRT-PCR demonstrated a 60% reduction in tln1 mRNA in mutants by 2 dpf, a 70% reduction by 3 dpf, and nearly 75% reduction by 5 dpf, when compared with WT embryos (see Figure, Supplemental Digital Content 4, http://links.lww.com/PRSGO/A664).
Craniofacial Cartilage Defects in tln1 Mutants
Examination of the craniofacial cartilages showed that the tln1-/- mutants exhibited malformed craniofacial structures (Fig. 4, above and center). The Meckel’s cartilage was highly hypoplastic (Fig. 4, left, above, and center). The length/width (L/W) ratio for the Meckel’s cartilage of the tln1-/- mutants was 0.30 ± 0.045, which was significantly smaller than in WT (0.61 ± 0.0733; see Figure, Supplemental Digital Content 5, http://links.lww.com/PRSGO/A665). Similarly, the palate was foreshortened with a dysmorphic leading edge (Fig. 4, right, above, and center). The L/W ratio for the mutant palate was 0.59 ± 0.04, which was significantly lower than that in WT (1.10 ± 0.08; see Figure, Supplemental Digital Content 5, http://links.lww.com/PRSGO/A665).
Fig. 4.
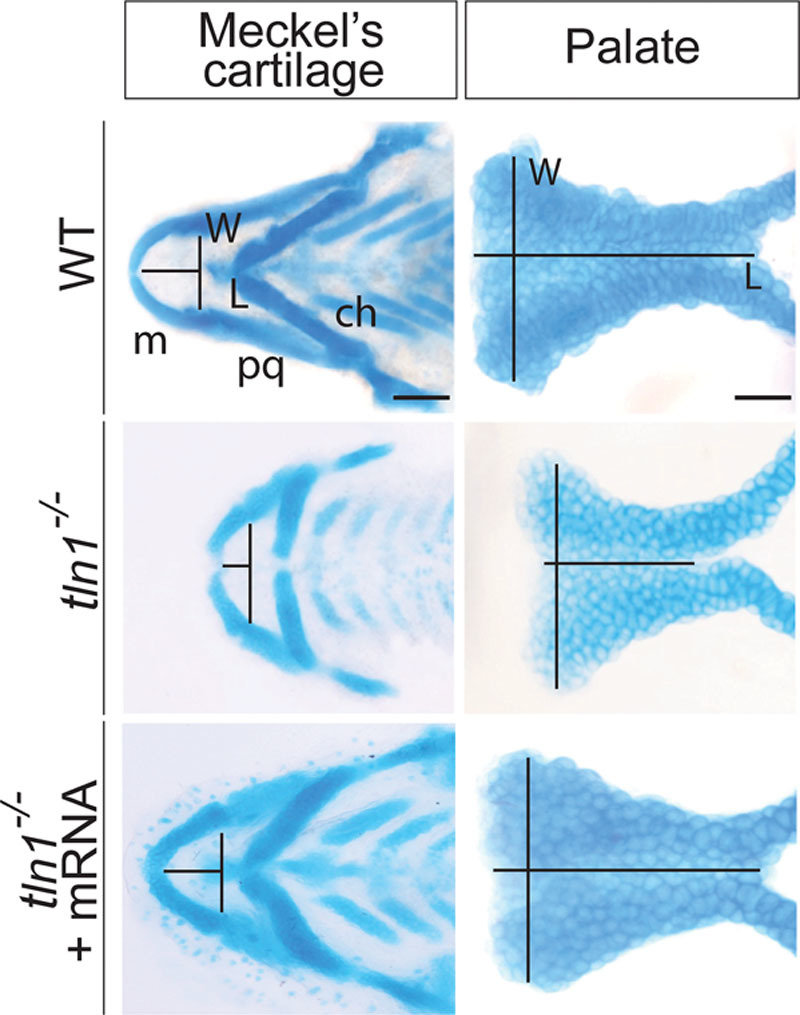
Alcian blue images of flat mounted Meckel’s cartilage and the palate of the WT (above), tln1 (center), and rescued mutants (below) at 4 dpf are shown. The length (L) of the Meckel’s cartilage is measured from the midline of the Meckel’s cartilage to the midline of an imaginary line drawn joining the joints between the Meckel’s cartilage and the palatoquadrate. Likewise the width (W) is across the joints between the Meckel’s cartilage and the palatoquadrate. The L of the palate is measured from the anterior to the posterior of the palate through its mid point, whereas W is measured as the maximum distance between the 2 lateral edges at the anterior most region. Tln1 mutants have a shorter Meckel’s cartilage (center and left) and palate (center and right). Injection of full-length tln1 mRNA partially rescues the phenotypes (below). Scale bars: 500 μm (Meckel’s cartilage) and 100 μm (palate). ch, ceratohyal; L, length; m, Meckel’s cartilage; pq, palatoquadrate; W, width.
Defects in Craniofacial Skeletal Muscles in tln1 Mutants
Severe defects were observed in the craniofacial skeletal muscles of the tln1 mutants (Fig. 5, above and center). Although all the craniofacial muscle elements were present, the overall muscle architecture was malformed. The angles between the 2 interhyoideus (ih) and that of between the 2 hyohyoideus (hh) muscles were significantly increased in the mutants (Fig. 5, left, above, and center). The angle between the 2 ih muscles in the mutants was 131.29 ± 6.43 degrees, which was higher than that in WT (63.74 ± 4.43 degrees; see Figure, Supplemental Digital Content 6, http://links.lww.com/PRSGO/A666). Likewise, the angle between the 2 hh muscles in the mutants was 167.76 ± 3.02 degrees, whereas in the WT, it was 112.86 ± 3.27 degrees (see Figure, Supplemental Digital Content 6, http://links.lww.com/PRSGO/A666). Ultrastructure analysis of ih muscles using TEM revealed significant disruption of the sarcomere architecture, with loss of Z-disk alignment and absence of A and I-bands in the tln1-/- mutants (Fig. 5, right, above, and center).
Fig. 5.
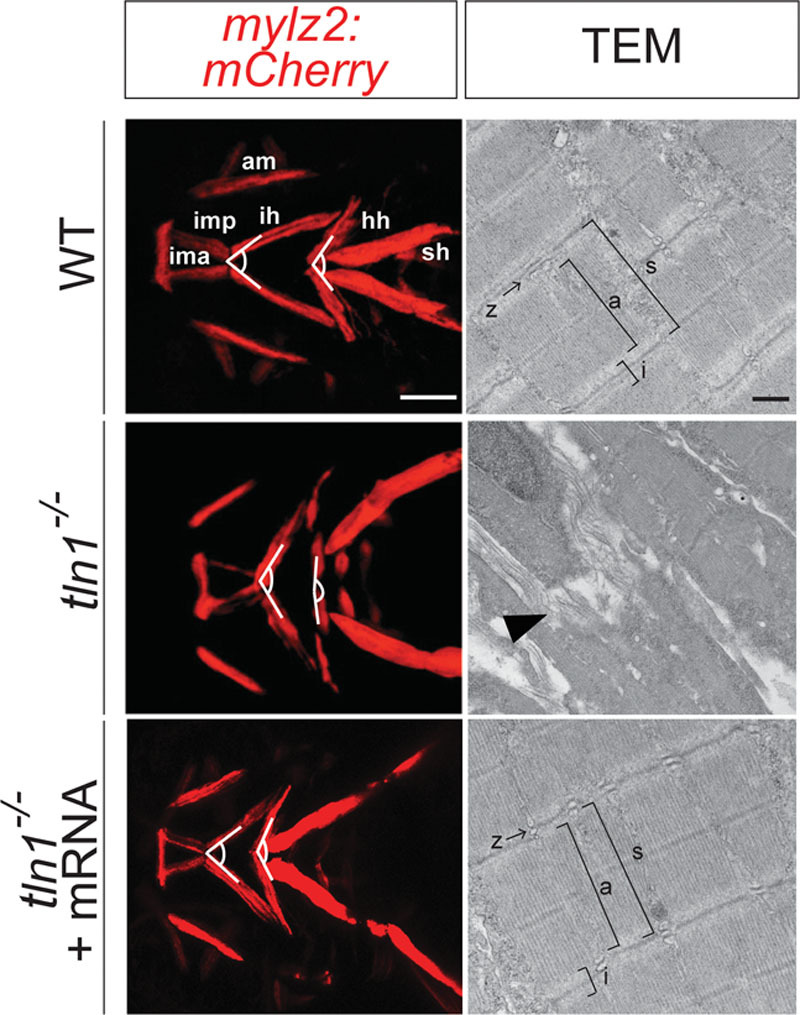
Defects in craniofacial skeletal muscles in tln1 mutants. Confocal images of craniofacial skeletal muscles in Tg(mylz2:mCherry) transgenic reveal increased intermuscular tension in the craniofacial muscles in the mutants (left and center) when compared with WT (left and above). TEM images of the transverse section of the ih (depicted by white dotted line, above and center) show complete disruption of the sarcomere structure in the mutants, depicted by black arrowhead (right and center) when compared with WT (right and above). The Z-line and the A-and I-bands, observed in the WT (right and above) are completely absent in the mutants (right and center). Injection of full-length tln1 mRNA partially rescues the phenotypes as observed by decreased intermuscular tension in the craniofacial muscles in the rescued mutants (left and below). TEM images of the transverse section of the ih in the rescued mutants show reorganized sarcomere structure, similar to the WT (right and below). Scale bar: 500μm (Tg(mylz2: mCherry)) and 500nm (TEM). a, A-band; am, adductor mandibulae; i, I-band; ima, intermandibularis anterior; imp, intermandibularis posterior; s, sarcomere unit; sh, sternohyoideus.
Injection of Full-length tln1 mRNA Partially Rescues Craniofacial Defects in Mutants
Injection of the full-length tln1 mRNA partially rescued the craniofacial cartilage and muscle phenotypes of tln1-/- mutants (Fig. 4, below; Fig. 5, below). The L/W ratio of both the Meckel’s cartilage and the palate were measured in the rescued tln1-/- embryos. The L/W ratio for the Meckel’s cartilage of the rescued mutants was 0.59 ± 0.0322, which was significantly higher than that of the tln1-/- mutants and closer to WT ratio (see Figure, Supplemental Digital Content 5, http://links.lww.com/PRSGO/A665). The L/W ratio for the palate of the rescued tln1-/- was 1.09 ± 0.05, which was significantly more than that of the mutants and varied nonsignificantly with the WT (see Figure, Supplemental Digital Content 5, http://links.lww.com/PRSGO/A665). The angle between the 2 ih and hh muscles in the rescued embryos were 79.72 ± 4.26 degrees and 135.90 ± 5.63 degrees, respectively (see Figure, Supplemental Digital Content 6, http://links.lww.com/PRSGO/A666), which were both significantly less than the mutants and closer to the WT measurements. The ultrastructure images of the ih muscle in the rescued mutant revealed improved organization of the sarcomere architecture, with clearly defined A and I-bands and the Z-disk (Fig. 5, right and below).
tln1 Mutants Exhibit Reduced Proliferation of CNCCs during Palate Formation
CNCCs migration and apoptosis assays did not show any defects in the tln1 mutants (see Figure, Supplemental Digital Content 7, http://links.lww.com/PRSGO/A667). Proliferation of CNCCs during palatal morphogenesis was examined using 2 methods: pulse-chase experiment using the Tg(sox10:kaede) reporter (Fig. 3, left and center) and by EdU assay (Fig. 3, right). In the pulse-chase assay, green fluorescent cells added onto the distal part of the maxillary process during the chase period, also known as the proliferative front (marked with white dotted line, Fig. 3, center), which was notably deficient in the tln1-/- mutants (Fig. 3, below and center). Similarly, in the EdU experiment, the number of EdU+ (proliferative) CNCCs at the leading edge of the palate was reduced in the tln1-/- mutants (6.2 ± 1.48) when compared with the WT (11.8 ± 1.3; Fig. 3, right; see Figure, Supplemental Digital Content 8, http://links.lww.com/PRSGO/A668).
DISCUSSION
tln1 Is Critical during Zebrafish Embryogenesis
Knockout of Tln1 in mice results in developmental arrest at gastrulation proving that tln1 has an important role in early mouse embryonic development.21 We show that zebrafish tln1 is expressed in very early stages of embryogenesis. At 48 hpf, tln1 is expressed strongly in the craniofacial region, localizing to the maxillary and mandibular structures by 72 hpf, suggesting that tln1 is required for development of craniofacial structures.
Zebrafish Model of tln1 LOF
Reduction of tln1 transcripts in the homozygous mutants observed by qRT-PCR suggests that the tln1hi3093Tg is a null allele. Homozygous tln1 embryos were initially indistinguishable from WT or heterozygous siblings until ~48 hpf, at which point the homozygotes exhibited cardiac edema. Indeed, previous studies in zebrafish showed that tln1 is required for cardiac development.20 Earlier work showed that maternal gene products contribute to early embryonic development in zebrafish.32,33 The maternally contributed tln1 protein masked the zygotic requirement of tln1 at earlier stages of embryonic development till 48 hpf. However, as maternal tln1 becomes slowly depleted over time, the consequence of tln1 deficiency manifested in multiple organ systems progressively, as the embryo developed cardiac edema, craniofacial cartilage hypoplasia, and muscle disorganization, leading to eventual embryonic lethality by 5.5 dpf.
Loss of tln1 Leads to Defects in Craniofacial Cartilage and Skeletal Muscles
Cartilage staining of tln1 mutants showed that the loss of tln1 resulted in significantly underdeveloped Meckel’s cartilage and a shortened palate. Although the craniofacial muscles were all present, the tension between the different muscle structures was increased in the mutants. This is evident by the increased angles between the ih and hh muscle pair in the mutants when compared with the WT. Of note, the angle between the pair of sternohyoideus was not measured as tln1 mutants showed significant cardiac edema that can in turn affect the sternohyoideus muscles located dorsal to the zebrafish heart. The craniofacial skeletal muscles in the mutants were disorganized with disruption of the sarcomere structure. The Z-disk, the A and I- bands were completely absent in the mutants.
CNCC Proliferation is Affected during Palate Morphogenesis in tln1 Mutants
The CNCCs primarily undergo migration, proliferation, and controlled cell death to form the craniofacial cartilage, specifically the palate. In the absence of tln1, it was observed that the palate is shortened. The critical cellular processes were studied in the palate to elucidate the role of tln1 during palate morphogenesis. The processes of cell migration and apoptosis were found to be not defective in the tln1 mutants. Pulse-chase experiments suggested that CNCC proliferation is defective in tln1 mutants. Edu assay also confirmed that the palate of tln1 mutants have ~50% less proliferative cells than that of the WT suggesting that defective CNCC proliferation leads to shortened palate in the mutants.
tln1 has been shown to play a key role in integrin-mediated cellular events during the development of kidney, heart, and vascular system.20,34,35 However, the role of tln1 in craniofacial morphogenesis has not been studied. This study reveals that tln1 is essential for the formation of craniofacial cartilage and maintenance of the skeletal muscles, and that CNCC proliferation during palate morphogenesis requires function of tln1. This work provides novel insights into the role of tln1 in craniofacial development.
Perspective in the Clinical Field
Identification of the genetic requirement of tln1 in craniofacial development is immediately clinically helpful, where detection of human gene variants in TLN1 can now be associated with orofacial cleft anomalies. As next generation sequencing technology continues to move the needle of diagnosis to earlier period in gestation, we will soon be able to better impute clinical significance of human gene sequencing data, where the function of genes important to craniofacial development are assembled.36 Further, the major gap in human genetics is to determine the function of human gene variants, which we are able to assay rapidly in zebrafish rescue assays.37 This tln1 zebrafish mutant can be used as a functional assay to determine the function of human TLN1 gene variants in a injection rescue assay. Lastly, the zebrafish model can be used as a platform for small molecule screens, to discover drugs that one day may mitigate cleft anomalies.3
ACKNOWLEDGMENTS
The authors appreciate support by the Shriners Hospital for Children (to E.C.L.) for the work. K.M. was supported by a Research Fellowship Award (#84309) from Shriners Hospitals for Children. The authors thank Jenna Galloway, Liao, and Galloway lab colleagues for discussions and for providing valuable feedback in the review of this article. The authors also thank Renee Daigle for excellent care of the aquatics facility. Electron microscopy was performed in the Microscopy Core of the Center for Systems Biology/Program in Membrane Biology.
Supplementary Material
Footnotes
Presented at the 57th Annual Meeting of New England Society of Plastic and Reconstructive Surgeons, June 11, 2016, Bretton Woods, N.H.; 39th Annual Meeting of Society of Craniofacial Genetics and Developmental Biology, August 4, 2016, Boston, Mass.; 62nd Annual Meeting of Plastic Surgeon Research Council, May 5, 2017, Durham, N.C.
Drs. Ishii and Mukherjee contributed equally to this work.
Disclosure: The authors have no financial interest to declare in relation to the content of this article. The Article Processing Charge was paid for by the authors.
Supplemental digital content is available for this article. Clickable URL citations appear in the text.
REFERENCES
- 1.Cordero DR, Brugmann S, Chu Y, et al. Cranial neural crest cells on the move: their roles in craniofacial development. Am J Med Genet A. 2011;155A:270–279.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lumsden A, Sprawson N, Graham A. Segmental origin and migration of neural crest cells in the hindbrain region of the chick embryo. Development. 1991;113:1281–1291.. [DOI] [PubMed] [Google Scholar]
- 3.Kong Y, Grimaldi M, Curtin E, et al. Neural crest development and craniofacial morphogenesis is coordinated by nitric oxide and histone acetylation. Chem Biol. 2014;21:488–501.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berrier AL, Yamada KM. Cell-matrix adhesion. J Cell Physiol. 2007;213:565–573.. [DOI] [PubMed] [Google Scholar]
- 5.Nie S, Kee Y, Bronner-Fraser M. Caldesmon regulates actin dynamics to influence cranial neural crest migration in Xenopus. Mol Biol Cell. 2011;22:3355–3365.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zheng PP, Severijnen LA, Willemsen R, et al. Caldesmon is essential for cardiac morphogenesis and function: in vivo study using a zebrafish model. Biochem Biophys Res Commun. 2009;378:37–40.. [DOI] [PubMed] [Google Scholar]
- 7.Nie S, Kee Y, Bronner-Fraser M. Myosin-X is critical for migratory ability of Xenopus cranial neural crest cells. Dev Biol. 2009;335:132–142.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Saadi I, Alkuraya FS, Gisselbrecht SS, et al. Deficiency of the cytoskeletal protein SPECC1L leads to oblique facial clefting. Am J Hum Genet. 2011;89:44–55.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gfrerer L, Shubinets V, Hoyos T, et al. Functional analysis of SPECC1L in craniofacial development and oblique facial cleft pathogenesis. Plast Reconstr Surg. 2014;134:748–759.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fennell N, Foulds N, Johnson DS, et al. Association of mutations in FLNA with craniosynostosis. Eur J Hum Genet. 2015;23:1684–1688.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mukherjee K, Ishii K, Pillalamarri V, et al. Actin capping protein CAPZB regulates cell morphology, differentiation, and neural crest migration in craniofacial morphogenesis. Hum Mol Genet. 2016;25:1255–1270.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wilkin DJ, Liberfarb R, Davis J, et al. Rapid determination of COL2A1 mutations in individuals with Stickler syndrome: analysis of potential premature termination codons. Am J Med Genet. 2000;94:141–148.. [DOI] [PubMed] [Google Scholar]
- 13.Lo SH. Focal adhesions: what’s new inside. Dev Biol. 2006;294:280–291.. [DOI] [PubMed] [Google Scholar]
- 14.Campbell ID, Ginsberg MH. The talin-tail interaction places integrin activation on FERM ground. Trends Biochem Sci. 2004;29:429–435.. [DOI] [PubMed] [Google Scholar]
- 15.Calderwood DA, Fujioka Y, de Pereda JM, et al. Integrin beta cytoplasmic domain interactions with phosphotyrosine-binding domains: a structural prototype for diversity in integrin signaling. Proc Natl Acad Sci U S A. 2003;100:2272–2277.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tadokoro S, Shattil SJ, Eto K, et al. Talin binding to integrin beta tails: a final common step in integrin activation. Science. 2003;302:103–106.. [DOI] [PubMed] [Google Scholar]
- 17.García-Alvarez B, de Pereda JM, Calderwood DA, et al. Structural determinants of integrin recognition by talin. Mol Cell. 2003;11:49–58.. [DOI] [PubMed] [Google Scholar]
- 18.Critchley DR. Biochemical and structural properties of the integrin-associated cytoskeletal protein talin. Annu Rev Biophys. 2009;38:235–254.. [DOI] [PubMed] [Google Scholar]
- 19.Senetar MA, McCann RO. Gene duplication and functional divergence during evolution of the cytoskeletal linker protein talin. Gene. 2005;362:141–152.. [DOI] [PubMed] [Google Scholar]
- 20.Wu Q, Zhang J, Koh W, et al. Talin1 is required for cardiac Z-disk stabilization and endothelial integrity in zebrafish. FASEB J. 2015;29:4989–5005.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Monkley SJ, Zhou XH, Kinston SJ, et al. Disruption of the talin gene arrests mouse development at the gastrulation stage. Dev Dyn. 2000;219:560–574.. [DOI] [PubMed] [Google Scholar]
- 22.Debrand E, Conti FJ, Bate N, et al. Mice carrying a complete deletion of the talin2 coding sequence are viable and fertile. Biochem Biophys Res Commun. 2012;426:190–195.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dougherty M, Kamel G, Grimaldi M, et al. Distinct requirements for wnt9a and irf6 in extension and integration mechanisms during zebrafish palate morphogenesis. Development. 2013;140:76–81.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Britanova O, Depew MJ, Schwark M, et al. Satb2 haploinsufficiency phenocopies 2q32-q33 deletions, whereas loss suggests a fundamental role in the coordination of jaw development. Am J Hum Genet. 2006;79:668–678.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lindgren AM, Hoyos T, Talkowski ME, et al. Haploinsufficiency of KDM6A is associated with severe psychomotor retardation, global growth restriction, seizures and cleft palate. Hum Genet. 2013;132:537–552.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miyake N, Mizuno S, Okamoto N, et al. KDM6A point mutations cause Kabuki syndrome. Hum Mutat. 2013;34:108–110.. [DOI] [PubMed] [Google Scholar]
- 27.Dougherty M, Kamel G, Shubinets V, et al. Embryonic fate map of first pharyngeal arch structures in the sox10: kaede zebrafish transgenic model. J Craniofac Surg. 2012;23:1333–1337.. [DOI] [PubMed] [Google Scholar]
- 28.Kamel G, Hoyos T, Rochard L, et al. Requirement for frzb and fzd7a in cranial neural crest convergence and extension mechanisms during zebrafish palate and jaw morphogenesis. Dev Biol. 2013;381:423–433.. [DOI] [PubMed] [Google Scholar]
- 29.Storer NY, White RM, Uong A, et al. Zebrafish rhabdomyosarcoma reflects the developmental stage of oncogene expression during myogenesis. Development. 2013;140:3040–3050.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thisse C, Thisse B. High-resolution in situ hybridization to whole-mount zebrafish embryos. Nat Protoc. 2008;3:59–69.. [DOI] [PubMed] [Google Scholar]
- 31.Walker MB, Kimmel CB. A two-color acid-free cartilage and bone stain for zebrafish larvae. Biotech Histochem. 2007;82:23–28.. [DOI] [PubMed] [Google Scholar]
- 32.Kelly C, Chin AJ, Leatherman JL, et al. Maternally controlled (beta)-catenin-mediated signaling is required for organizer formation in the zebrafish. Development. 2000;127:3899–3911.. [DOI] [PubMed] [Google Scholar]
- 33.Pelegri F, Knaut H, Maischein HM, et al. A mutation in the zebrafish maternal-effect gene nebel affects furrow formation and vasa RNA localization. Curr Biol. 1999;9:1431–1440.. [DOI] [PubMed] [Google Scholar]
- 34.Monkley SJ, Kostourou V, Spence L, et al. Endothelial cell talin1 is essential for embryonic angiogenesis. Dev Biol. 2011;349:494–502.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tian X, Kim JJ, Monkley SM, et al. Podocyte-associated talin1 is critical for glomerular filtration barrier maintenance. J Clin Invest. 2014;124:1098–1113.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Talkowski ME, Ordulu Z, Pillalamarri V, et al. Clinical diagnosis by whole-genome sequencing of a prenatal sample. N Engl J Med. 2012;367:2226–2232.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li EB, Truong D, Hallett SA, et al. Rapid functional analysis of computationally complex rare human IRF6 gene variants using a novel zebrafish model. PLoS Genet. 2017;13:e1007009. [DOI] [PMC free article] [PubMed] [Google Scholar]


