Abstract
The dilution effect, where an increase in biodiversity results in a reduction in the prevalence of an infectious disease, has been the subject of speculation and controversy. Conversely, an amplification effect occurs when increased biodiversity is related to an increase in prevalence. We explore the conditions under which these effects arise, using multi species compartmental models that integrate ecological and epidemiological interactions. We introduce three potential metrics for quantifying dilution and amplification, one based on infection prevalence in a focal host species, one based on the size of the infected subpopulation of that species and one based on the basic reproduction number. We introduce our approach in the simplest epidemiological setting with two species, and show that the existence and strength of a dilution effect is influenced strongly by the choices made to describe the system and the metric used to gauge the effect. We show that our method can be generalized to any number of species and to more complicated ecological and epidemiological dynamics. Our method allows a rigorous analysis of ecological systems where dilution effects have been postulated, and contributes to future progress in understanding the phenomenon of dilution in the context of infectious disease dynamics and infection risk.
Keywords: dilution effect, ecological epidemiology, infectious disease dynamics, infection risk, biodiversity, compartmental models
1. Introduction
There is increasing attention on the way in which ecological interactions in ecosystems can influence the transmission of infectious disease agents, and the impact that pathogens and parasites have on ecology. In particular, the connection between biodiversity and the transmission of infection has been a topic of considerable scientific research and debate in recent years, focusing on the vector-borne transmission of Lyme disease [1], as well as in its more general sense [2–7]. That debate is essentially about the question of how, and to what extent, loss of biodiversity in an ecosystem can lead to changes in infection transmission, and potentially to changes in human zoonotic infection risk. The so-called dilution effect (in its ‘inclusive’ sense [8]) is used to describe the idea that infection increases in a specified host species when diversity decreases in the community of which that species is part. The name actually refers to the opposite idea: greater diversity leading to a decrease in infection in the host species by ‘diluting’ the spread of infection. It remains unclear, however, under what circumstances a more biodiverse community lowers infection transmission to a species, and under what circumstances more biodiversity leads to increased transmission (referred to as an ‘amplification effect’ in [8]), and which mechanisms underly such effects.
Discussion of the dilution effect is clouded by the multiple interpretations of several of the relevant terms. For example, how should one measure ‘biodiversity’? What is meant by ‘increased infection’, ‘increased transmission’ or ‘increased risk’? Do we mean, for example, increased prevalence of a specific infectious agent in a specific host species, or increased size of the infectious subpopulation of that species, or an increase in the basic reproduction number 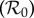 for the system as a whole? Should infection transmission increase in all species that are host to a given infectious agent, or only in a limited number of species, and if limited, what are the criteria (main host, reservoir host, host with highest zoonotic infection risk)? Is transmission between individuals of a host species dependent more on the density of infectious individuals, or more on the frequency of infectious individuals? Is the infectious agent directly transmitted between different host species, indirectly via environmental contamination, or via an insect vector? What is meant by the ‘ecosystem’ in which these effects should be measured and understood? The interpretation of all these aspects may well influence whether or not a dilution or amplification effect is said to occur in such circumstances, to what extent and generated by which underlying mechanisms. It is not surprising therefore that there is a wide range of evidence and opinion from experiments, field studies and mathematical models. Some recent reviews and meta-analyses of previously published studies highlight what is known, what is not known and what is unclear [1,9–11].
for the system as a whole? Should infection transmission increase in all species that are host to a given infectious agent, or only in a limited number of species, and if limited, what are the criteria (main host, reservoir host, host with highest zoonotic infection risk)? Is transmission between individuals of a host species dependent more on the density of infectious individuals, or more on the frequency of infectious individuals? Is the infectious agent directly transmitted between different host species, indirectly via environmental contamination, or via an insect vector? What is meant by the ‘ecosystem’ in which these effects should be measured and understood? The interpretation of all these aspects may well influence whether or not a dilution or amplification effect is said to occur in such circumstances, to what extent and generated by which underlying mechanisms. It is not surprising therefore that there is a wide range of evidence and opinion from experiments, field studies and mathematical models. Some recent reviews and meta-analyses of previously published studies highlight what is known, what is not known and what is unclear [1,9–11].
Discussion in this area is also hindered by methodological issues preventing an effective use of mathematical models. Without a way to objectively, uniformly and robustly quantify dilution and amplification, it remains difficult to study the problem and compare models, mechanisms and assumptions [8]. That this is important is shown in the examples in the present paper, where it becomes clear that one has to be very precise about what one assumes about the system and how one interprets the above terms. Different interpretations give different results, thus adding to confusion and controversy when comparing results from different studies. Here, we present a structured approach to studying the dilution effect for infectious disease agents in ecosystems, and three objective metrics to quantify the effect. We present this in the simplest possible setting, to allow us to introduce rigorous definitions of dilution and amplification in an intuitive way, and to allow analytic expressions. We also show how the definitions generalize to systems with any number of species.
In contrast with most studies using models to investigate the dilution effect, we explicitly integrate ecological and epidemiological interactions between species, while recognizing that any community of interacting species with an infectious agent will consist of species that are host to that agent and species that are not. The species that are not host to a particular infectious agent may, nevertheless, influence transmission dynamics among the host species in the same community through ecological interaction, directly through feeding relations or competition for resources, as well as indirectly via their interaction in the ecosystem or the (local) food web. The setting we use has previously been presented as an approach to integrate ecological and epidemiological interactions in one model, allowing for feeding relations and competition between multiple species, where only a subset of the species is host to the infectious agent under consideration [12]. There we focused on characterizing 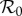 , showing the importance of both ecological and epidemiological stability in determining when a community with the infectious agent present can exist. We now adapt that model, restricting it initially to two competing species, both hosts to a given (micro-parasitic) infectious agent. This setting allows us to introduce all relevant aspects while retaining simplicity and clarity. We allow for frequency- and density-dependent transmission of an infectious agent within and between host species, and for density-dependent transmission via a common pool of infection (see [13]). This latter mechanism can also be used to approximate transmission via a vector [14]. For ease of exposition we model the pathogen using an susceptible–infectious (SI) model without recovery, but the method can be easily extended to more complicated compartmental descriptions of infection dynamics (SIR, SEIR, etc.). We initially derive results where the two species compete for resources, and the pathogen does not increase host mortality. We then show how these results may be modified if the species interact as predator and prey, and if infection increases mortality. While a two-species model may be regarded as representing the focal species and all other species in the ecosystem lumped together, most real-world applications would require the analysis of a larger number of interacting species. Hence we conclude by indicating how the analysis may be generalized to an arbitrary number of species.
, showing the importance of both ecological and epidemiological stability in determining when a community with the infectious agent present can exist. We now adapt that model, restricting it initially to two competing species, both hosts to a given (micro-parasitic) infectious agent. This setting allows us to introduce all relevant aspects while retaining simplicity and clarity. We allow for frequency- and density-dependent transmission of an infectious agent within and between host species, and for density-dependent transmission via a common pool of infection (see [13]). This latter mechanism can also be used to approximate transmission via a vector [14]. For ease of exposition we model the pathogen using an susceptible–infectious (SI) model without recovery, but the method can be easily extended to more complicated compartmental descriptions of infection dynamics (SIR, SEIR, etc.). We initially derive results where the two species compete for resources, and the pathogen does not increase host mortality. We then show how these results may be modified if the species interact as predator and prey, and if infection increases mortality. While a two-species model may be regarded as representing the focal species and all other species in the ecosystem lumped together, most real-world applications would require the analysis of a larger number of interacting species. Hence we conclude by indicating how the analysis may be generalized to an arbitrary number of species.
2. The two-host model
We introduce an approach to investigate hypotheses and mechanisms of dilution and amplification in ecosystems, as well as metrics to quantify the effects when they occur. We do this in a general framework, based on an earlier approach that allowed for the study of invasion of infectious agents in food webs and ecosystems, and of the way ecology and epidemiology influence each other to determine invasion success [12]. For ease of exposition, and to derive explicit expressions, we introduce our approach in the simplest setting of two interacting species. We first present the framework and the method of calculating appropriate metrics in a general setting with two-host species and SI infection dynamics. In §§2.1–2.3, we focus on different choices for modelling transmission within this setting.
Consider an ecosystem where just two species (numbered 1 and 2) interact, and each species is a potential host of the same infectious agent. Even here there are many choices to be made that can influence ecological and epidemiological dynamics and the occurrence and extent of dilution or amplification effects. Can one or both species sustain the infectious agent by itself? Do the species only compete for resources, or is one a predator on the other, or are there other mechanisms of ecological interaction? Is there between-species transmission and how does this relate to within-species transmission? What is the mechanism of transmission? What is the nature of contacts between the individuals related to transmission (both within the same species and between species)? What is the life history of the two species and how does density dependence act on various parts of that life history? How does infection influence behaviour, survival, reproduction, competition and hence life history? For many of the relevant factors mentioned here, the level or strength may matter. Think, for example, of the level of susceptibility and competence as a host for the infectious agent, competitive ability or level of infectivity. It is clear, for example, that the strength of predation of one species on another, together with the size of 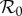 for an infectious agent in the predator, determines whether the three species (predator, prey, infectious agent) can coexist (see [12] and other papers cited therein).
for an infectious agent in the predator, determines whether the three species (predator, prey, infectious agent) can coexist (see [12] and other papers cited therein).
There is in principle no choice that is prohibited in our approach, but for exposition we start with some elementary choices. Assume that the two host species compete for resources, with the dynamics of their population densities described by
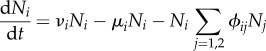 |
for i = 1, 2, with all parameters positive. Species i has maximum birth rate νi and minimum death rate μi. The population growth rate of species i is logistic with carrying capacity (νi − μi)/ϕii in the absence of species j≠i. The population growth rate of species i is reduced by ϕijNj due to competition for resources with species j.
When both species are present, steady-state solutions satisfy the linear system
| 2.1 |
As we are interested in studying the consequences of a reduction in biodiversity, we need to define a measure for quantifying changes in community composition. We designate species one to be the focal host species of interest. We then impose an increased mortality on species two, caused by some unspecified factor or mechanism (for example, an environmental or human-induced change in conditions, or a predator specific for species two), resulting in a reduced population density for that species. The interest is in gauging how infection in species one responds to that change. Define the (steady state) solutions of equation (2.1) to be Ni = N*i for i = 1, 2. Now suppose that the mortality of species two is increased to μ2 + ω, resulting in new values for the steady-state population densities of each species. We can characterize the change for the focal host species by defining
This is the elasticity of the population density of species one to changes in the population density of species two. We note that due to the linearity of our model (equation (2.1)), 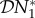 is independent of the value of ω. The derivative in the definition of elasticity implies a small increase in the mortality of species two, which would not be regarded as a change in biodiversity. However, this gives rise to the same calculated elasticity in species one that would be observed following an increase in mortality sufficient to eradicate species two. This will not necessarily be the case in more complicated settings.
is independent of the value of ω. The derivative in the definition of elasticity implies a small increase in the mortality of species two, which would not be regarded as a change in biodiversity. However, this gives rise to the same calculated elasticity in species one that would be observed following an increase in mortality sufficient to eradicate species two. This will not necessarily be the case in more complicated settings.
We now describe the epidemiological part of the system. We take into account both frequency-dependent and density-dependent transmission of the infectious agent, within and between species, but possibly (even probably) at (very) different rates. A choice for frequency-dependent transmission assumes that infectious contacts between individuals do not scale with population density. A choice for density-dependent transmission is typical for situations where transmission is the result of very brief encounters of susceptible individuals with infection. The assumption is that the encounters are so brief that their number will increase if the population density of the infected host species increases. This would not be the case if contacts, or transmission during contact, take a more substantial period of time, leading to saturation by time constraint or for other reasons. In many situations, ‘reality’ is a combination of the two extremes, with density dependence at low population densities and frequency dependence at higher densities.
We assume that the dynamics of the infectious populations are described by
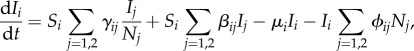 |
where Si = Ni − Ii for i = 1, 2. We assume, for ease of exposition, that competition and host mortality are unaffected by the infection status of the host species' individuals. In other words, we assume that epidemiology does not directly influence ecology at the individual level, with feeding rates and death rates being the same for susceptible and infected individuals. We relax these assumptions in §§3.2 and 3.3.
Steady-state prevalences of infection solve the equations
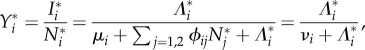 |
where the steady-state forces of infection are
The next-generation matrix K ([15]) has components
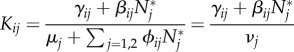 |
and the basic reproduction number 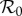 is the largest eigenvalue of K,
is the largest eigenvalue of K,
where trace K = K11 + K22 and 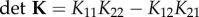 .
.
We now derive expressions for the magnitude of possible dilution or amplification effects. For this, we need to specify what we mean by an effect on infection in the focal host species and how we quantify such an effect. We focus on three different epidemiological interpretations of ‘effect on infection’, these are prevalence of infection in the focal species, Y*1; abundance of infected individuals of the focal species, I*1; and basic reproduction number of the system, 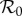 . We quantify the magnitude of changes in each of these, resulting from changes in the population density of species two, as an elasticity. We denote the sensitivity and elasticity of a measure X to changes in N*2 by X(2) and
. We quantify the magnitude of changes in each of these, resulting from changes in the population density of species two, as an elasticity. We denote the sensitivity and elasticity of a measure X to changes in N*2 by X(2) and 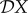 , respectively, where
, respectively, where
We can interpret these elasticities as follows. If 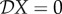 , then reducing species two has no effect on the measure X. If
, then reducing species two has no effect on the measure X. If 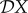 < 0, then a reduction in the density of species two (i.e. less diversity) leads to an increase in the epidemiological measure X: hence there is a dilution effect. As explained in the introduction, the name ‘dilution effect’ actually refers to the opposite: an increase in the density of species two leads to a decrease in X. If
< 0, then a reduction in the density of species two (i.e. less diversity) leads to an increase in the epidemiological measure X: hence there is a dilution effect. As explained in the introduction, the name ‘dilution effect’ actually refers to the opposite: an increase in the density of species two leads to a decrease in X. If 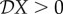 , then a reduction in the density of species two leads to a decrease in X, and an increase in the density of species two leads to an increase in X: hence there is an ‘amplification effect’. In addition to determining the direction of a potential effect, the elasticities also quantify the strength of such an effect relative to the initial values of X and N*2.
, then a reduction in the density of species two leads to a decrease in X, and an increase in the density of species two leads to an increase in X: hence there is an ‘amplification effect’. In addition to determining the direction of a potential effect, the elasticities also quantify the strength of such an effect relative to the initial values of X and N*2.
In this spirit, we define the elasticity of the prevalence of infection in species one with respect to changes in the population density of species two by
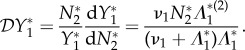 |
2.2 |
The elasticity of the abundance of infection is
| 2.3 |
With a judicious choice of the units of biomass and without loss of generality, the steady-state population densities can be set to N*1 = N*2 = 1. This rescaling simplifies the expression for the elasticity of the population density of species one to 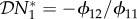 . The elasticity of the basic reproduction number may be defined as a function of the entries in K and their derivatives with respect to N*2. In general,
. The elasticity of the basic reproduction number may be defined as a function of the entries in K and their derivatives with respect to N*2. In general,
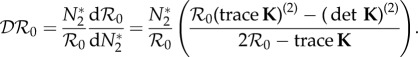 |
2.4 |
We now present three examples of the calculation of the elasticities of prevalence and abundance of infection, and of the elasticity of 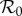 , for the two-host model. In §§2.1 and 2.2, we restrict the epidemiology of the pathogen to frequency- and density-dependent transmission, respectively. Then in §2.3, we show how infection transmission via the environment may be approximated by density-dependent transmission with separable mixing, and derive analytic expressions for the elasticities that arise in this case.
, for the two-host model. In §§2.1 and 2.2, we restrict the epidemiology of the pathogen to frequency- and density-dependent transmission, respectively. Then in §2.3, we show how infection transmission via the environment may be approximated by density-dependent transmission with separable mixing, and derive analytic expressions for the elasticities that arise in this case.
2.1. Example: frequency-dependent transmission only
Assume that βij = 0 for all i and j. In general,
We cannot derive explicit expressions for Y*1 and Y*2, but their values depend only on the parameters νi and γij. Hence, 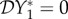 and there is no dilution or amplification effect on prevalence of infection. For this example, the elasticity of the abundance of infection is
and there is no dilution or amplification effect on prevalence of infection. For this example, the elasticity of the abundance of infection is  . As the parameters ϕij > 0, we have
. As the parameters ϕij > 0, we have 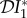 < 0, and hence (by this metric) there is a dilution effect of species two on species one. The next-generation matrix has entries Kij = γij/νj and so
< 0, and hence (by this metric) there is a dilution effect of species two on species one. The next-generation matrix has entries Kij = γij/νj and so 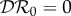 .
.
Note that these results can be understood as a direct consequence of our assumption that ecological interactions between species one and two are not affected by the epidemiological status of the individuals. Infected individuals are assumed not to differ in their competitive strength, either in relation to uninfected members of the same species, or with members of the other species, and irrespective of those members' infection status. So, the effect of competition on members of either species is independent of whether they belong to the Ii or Si compartment, leaving the ratio Y*i = I*i/N*i, i.e. the prevalence, unaffected. The apparent dilution effect on the abundance of infection, 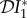 < 0, is due to the ecological interaction only. Similarly, as the expected number of new within-species and between-species cases generated by an infected individual during their infectious period is independent of population density of either host species, the elasticity of
< 0, is due to the ecological interaction only. Similarly, as the expected number of new within-species and between-species cases generated by an infected individual during their infectious period is independent of population density of either host species, the elasticity of 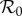 is zero.
is zero.
2.2. Example: density-dependent transmission only
Assume γij = 0 for all i, j. In general, we obtain
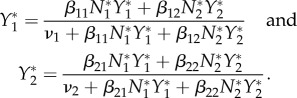 |
To analyse this example, we begin with the special case in which there is no cross-species transmission. We then have β12 = β21 = 0 and Y*1 = 1 − ν1/(β11N*1). Recalling that N*1 = N*2 = 1 and 0 ≤ Y*1 ≤ 1, we require β11 > ν1 for a non-trivial steady state Y*1 to exist. The elasticities of infection are
| 2.5 |
Therefore,  < 0 and
< 0 and 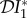 < 0 whenever a non-trivial steady state exists, and there is a dilution effect both in terms of prevalence and abundance of infected individuals of species one. The strength of the effect depends on ecological as well as epidemiological parameters. As explained before, in this section we assume no interaction between the effect of competition and the epidemiological status of individuals. If N2 is decreased, there is less competition felt by species one, resulting in a reduction in the loss rate of species one, leading to an increase in N1 compared to the unperturbed situation.
< 0 whenever a non-trivial steady state exists, and there is a dilution effect both in terms of prevalence and abundance of infected individuals of species one. The strength of the effect depends on ecological as well as epidemiological parameters. As explained before, in this section we assume no interaction between the effect of competition and the epidemiological status of individuals. If N2 is decreased, there is less competition felt by species one, resulting in a reduction in the loss rate of species one, leading to an increase in N1 compared to the unperturbed situation.
The next-generation matrix is diagonal with Kii = βiiN*i/νi and 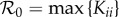 . We have
. We have 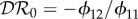 if K11 > K22 and
if K11 > K22 and 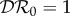 if K11 < K22. For this special case,
if K11 < K22. For this special case, 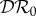 is piecewise constant, and discontinuous at K11 = K22.
is piecewise constant, and discontinuous at K11 = K22.
To move beyond the special case, we regard a gliding scale of increasing between-species transmission. For this, we introduce a quantity ε by setting  . We study the elasticities, obtained using equations (2.2)–(2.4), numerically by letting the value of ε increase from ε = 0 (no between-species transmission) to ε = 1 (between-species transmission rates equal to the geometric mean of the within-species transmission rates). Note that the case ε = 1, together with our assumption that β12 = β21, means that mixing is separable. Theoretically, one could also consider the case where ε > 1. We ignore this possibility, as between-species transmission is likely to be lower than within-species transmission, given that the contact rate for individuals of the same species is probably higher than that between individuals of different species.
. We study the elasticities, obtained using equations (2.2)–(2.4), numerically by letting the value of ε increase from ε = 0 (no between-species transmission) to ε = 1 (between-species transmission rates equal to the geometric mean of the within-species transmission rates). Note that the case ε = 1, together with our assumption that β12 = β21, means that mixing is separable. Theoretically, one could also consider the case where ε > 1. We ignore this possibility, as between-species transmission is likely to be lower than within-species transmission, given that the contact rate for individuals of the same species is probably higher than that between individuals of different species.
Numerical results obtained for 0 < ε < 1 are presented in figures 1–3, where we show how 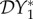 ,
, 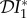 and
and 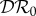 vary with ε. The special case ε = 0 has been explicitly calculated above; see equation (2.5). These results are indicated on the left-hand vertical axis in figures 1–3. When ε = 1, we have separable mixing. Results for this case may also be derived independently of the numerical calculations, and are indicated on the right-hand vertical axis. Their calculation is presented in §2.3 below.
vary with ε. The special case ε = 0 has been explicitly calculated above; see equation (2.5). These results are indicated on the left-hand vertical axis in figures 1–3. When ε = 1, we have separable mixing. Results for this case may also be derived independently of the numerical calculations, and are indicated on the right-hand vertical axis. Their calculation is presented in §2.3 below.
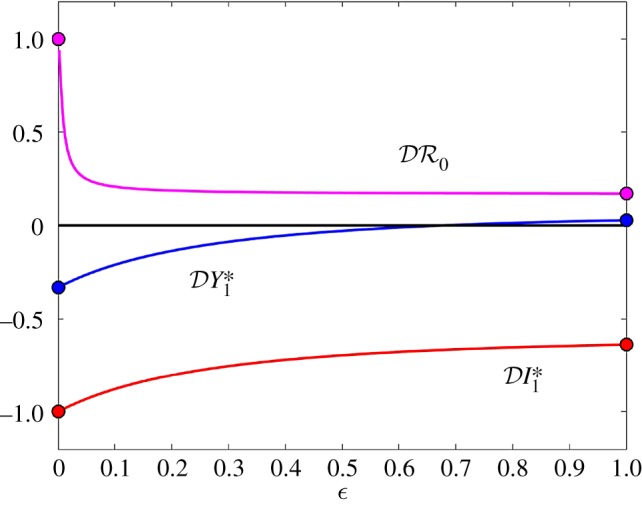
Figure 1.
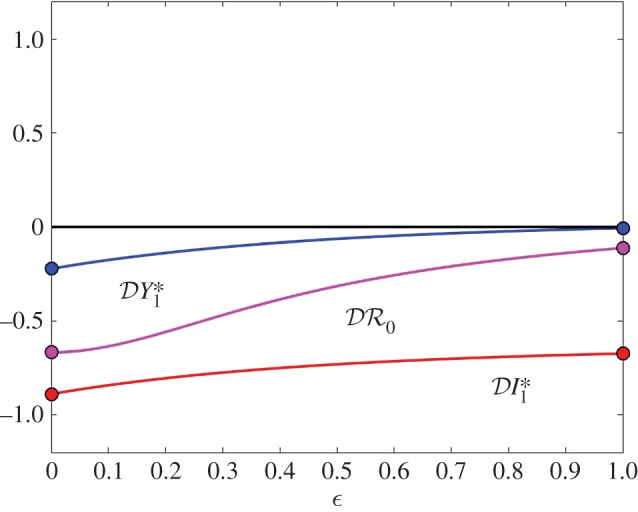
The elasticity of infection prevalence 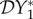 (blue), infection abundance
(blue), infection abundance 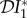 (red) and basic reproduction number
(red) and basic reproduction number 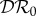 (magenta) as functions of ε. Transmission of infection is density-dependent with β11 = 5.0, β22 = 2.05 and
(magenta) as functions of ε. Transmission of infection is density-dependent with β11 = 5.0, β22 = 2.05 and 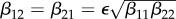 . Other parameter values are ϕ11 = ϕ22 = 0.15, ϕ12 = 0.1, ϕ21 = 0.075, μ1 = 1.0, μ2 = 0.8, ν1 = 1.25 and ν2 = 1.025. All parameters except ε have units time−1; ε is dimensionless. Circles at ε = 0 and ε = 1 are calculated from formulae in §§2.2 and 2.3, respectively. (Online version in colour.)
. Other parameter values are ϕ11 = ϕ22 = 0.15, ϕ12 = 0.1, ϕ21 = 0.075, μ1 = 1.0, μ2 = 0.8, ν1 = 1.25 and ν2 = 1.025. All parameters except ε have units time−1; ε is dimensionless. Circles at ε = 0 and ε = 1 are calculated from formulae in §§2.2 and 2.3, respectively. (Online version in colour.)
Figure 3.
The elasticity of infection prevalence 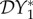 (blue), infection abundance
(blue), infection abundance 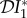 (red) and basic reproduction number
(red) and basic reproduction number 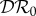 (magenta) as functions of ε. Transmission of infection is density-dependent with β11 = 3.750, β22 = 3.106 and
(magenta) as functions of ε. Transmission of infection is density-dependent with β11 = 3.750, β22 = 3.106 and 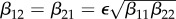 . Other parameter values as in figure 1. Circles at ε = 0 and ε = 1 are calculated from formulae in §§2.2 and 2.3, respectively. (Online version in colour.)
. Other parameter values as in figure 1. Circles at ε = 0 and ε = 1 are calculated from formulae in §§2.2 and 2.3, respectively. (Online version in colour.)
In evaluating the results presented in figures 1–3, recall that a negative value of 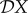 signifies a dilution effect on quantity X, and a positive value signifies an amplification effect. The parameter values used to generate the figures, apart from the values of β11 and β22 were chosen arbitrarily; see the caption to figure 1. The values of the βii were chosen so that transmission within species one was higher than transmission within species two (figure 1), lower than within species two (figure 2) or approximately equal (figure 3). In figure 1, where β11/ν1 = 2β22/ν2,
signifies a dilution effect on quantity X, and a positive value signifies an amplification effect. The parameter values used to generate the figures, apart from the values of β11 and β22 were chosen arbitrarily; see the caption to figure 1. The values of the βii were chosen so that transmission within species one was higher than transmission within species two (figure 1), lower than within species two (figure 2) or approximately equal (figure 3). In figure 1, where β11/ν1 = 2β22/ν2, 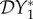 ,
, 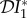 and
and 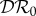 are negative for all 0 ≤ ε < 1, and increase with ε. Hence there is a dilution effect that is greater when there is less inter-species transmission of infection. In figure 2, where 2β11/ν1 = β22/ν2,
are negative for all 0 ≤ ε < 1, and increase with ε. Hence there is a dilution effect that is greater when there is less inter-species transmission of infection. In figure 2, where 2β11/ν1 = β22/ν2, 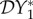 and
and 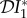 increase with ε, but
increase with ε, but 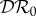 decreases. For these parameter values there is a dilution effect on the abundance of infection for all ε
decreases. For these parameter values there is a dilution effect on the abundance of infection for all ε
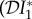 < 0), dilution of prevalence of infection for small ε changing to amplification for larger values, and an amplification effect on the basic reproduction number for all ε
< 0), dilution of prevalence of infection for small ε changing to amplification for larger values, and an amplification effect on the basic reproduction number for all ε
 . The change from dilution to amplification in the metric based on prevalence can be understood as follows. For small values of ε, i.e. small strength of cross-species transmission, the situation will probably remain the same as for the case ε = 0, where the analytical expression (equation (2.5)) showed that
. The change from dilution to amplification in the metric based on prevalence can be understood as follows. For small values of ε, i.e. small strength of cross-species transmission, the situation will probably remain the same as for the case ε = 0, where the analytical expression (equation (2.5)) showed that 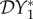 < 0. For ε = 0, decreasing N*2 will lead to an increase in N*1. Increasing cross-species transmission will cause I*1 to also increase, but for small values of ε possibly not as much as N*1 increases because of the reduction in abundance of species two. This can maintain
< 0. For ε = 0, decreasing N*2 will lead to an increase in N*1. Increasing cross-species transmission will cause I*1 to also increase, but for small values of ε possibly not as much as N*1 increases because of the reduction in abundance of species two. This can maintain 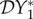 < 0. This continues until a value is reached where cross-species transmission becomes large enough to have a bigger influence in boosting I*1 than competition reduction has in boosting N*1, hence causing
< 0. This continues until a value is reached where cross-species transmission becomes large enough to have a bigger influence in boosting I*1 than competition reduction has in boosting N*1, hence causing 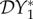 to become positive, and dilution to turn into amplification.
to become positive, and dilution to turn into amplification.
Figure 2.
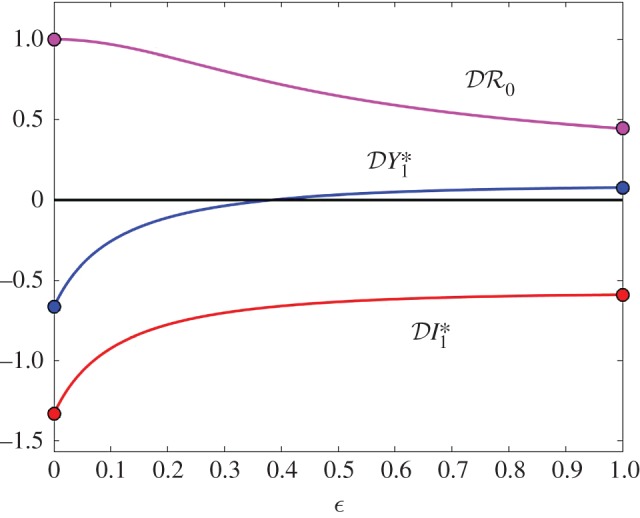
The elasticity of infection prevalence 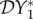 (blue), infection abundance
(blue), infection abundance 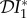 (red) and basic reproduction number
(red) and basic reproduction number 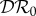 (magenta) as functions of ε. Transmission of infection is density-dependent, with β11 = 2.5, β22 = 4.1 and
(magenta) as functions of ε. Transmission of infection is density-dependent, with β11 = 2.5, β22 = 4.1 and 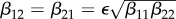 . Other parameter values as in figure 1. Circles at ε = 0 and ε = 1 are calculated from formulae in §§2.2 and 2.3, respectively. (Online version in colour.)
. Other parameter values as in figure 1. Circles at ε = 0 and ε = 1 are calculated from formulae in §§2.2 and 2.3, respectively. (Online version in colour.)
The results shown in figure 3, where β22/ν2 = 1.01β11/ν1, are similar to those shown in figure 2, except that 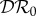 is approximately constant for 0.1 < ε < 1.0. This example was included to illustrate the result when β11/ν1 and β22/ν2 are approximately equal, but avoid the anomalous result previously noted for
is approximately constant for 0.1 < ε < 1.0. This example was included to illustrate the result when β11/ν1 and β22/ν2 are approximately equal, but avoid the anomalous result previously noted for 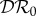 when ε = 0, which results in a discontinuity when β22/ν2 = β11/ν1.
when ε = 0, which results in a discontinuity when β22/ν2 = β11/ν1.
2.3. Example: density-dependent transmission with separable mixing
Density-dependent transmission with separable mixing is a special case of density dependence, equivalent to the example discussed in §2.2 with ε = 1. It is appropriate for modelling infection transmission via the environment or a vector. We assume that infected hosts of species i contribute to an environmental pool of pathogen, W, at rate σi, and are infected from the pool at rate κi. Infection events deplete W by a negligible amount, but the pathogen is lost from the environment at rate ρ. A simple equation for the dynamics of W would be
If W reaches equilibrium on a much faster timescale than the host–pathogen dynamics, then we can approximate the environmental contamination by the quasi-steady-state value W* = r1I*1 + r2I*2 where ri = σi/ρ. We then have βij = κirj and Y*i = κiW*/(νi + κiW*) for i = 1, 2. Hence W* solves
| 2.6 |
For this example, a potential fourth metric for dilution could be the elasticity of environmental contamination
By implicit differentiation of equation (2.6) we obtain
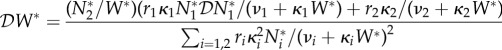 |
Using this metric, the criterion for dilution is 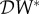 , which holds when
, which holds when
The elasticity of prevalence is 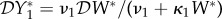 , so we have a dilution effect on prevalence
, so we have a dilution effect on prevalence 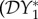 < 0 if
< 0 if 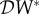 < 0.
< 0.
The next-generation matrix has components Kij = κirjN*j/νj and rank one. Hence
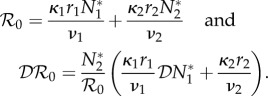 |
Results derived from these expressions coincide with those for the special case ε = 1, and are indicated on the right-hand vertical axes of figures 1–3.
3. Generalizations of the model
In §2, we analysed a model for the dynamics of a pathogen with two host species. We assumed that the host species competed for resources, and that a host's infection status did not change its population dynamics. In this section, we modify these assumptions. First, we comment on how the results would change if one host species were a predator on the other. We then present a model where infection with a pathogen increases a host individual's mortality, and present results showing how this increased mortality modifies the proposed metrics for dilution. Finally, we present a more general model with a variety of interactions between population dynamics and epidemiology as a basis for future studies of the dilution effect and related phenomena.
3.1. Predator–prey dynamics
In §2, we assumed that interaction between the two host species was due to competition for resources, hence an increase in host species one would cause a decrease in species two, and vice versa. We now examine the changes to our results when one species is a predator on the other, so that an increase in the population density of the prey species would result in an increase in the population density of the predator species. Consider equation (2.1). In §2, all the ϕij were positive, but if host species number one is a predator on host species two, then ϕ12 < 0 and 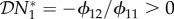 . Now consider our metrics for dilution in our different epidemiological scenarios. If transmission of infection is frequency-dependent only, then
. Now consider our metrics for dilution in our different epidemiological scenarios. If transmission of infection is frequency-dependent only, then 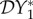 and
and 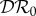 are unchanged at zero, but now
are unchanged at zero, but now 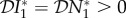 . Hence, there is an amplification effect on the abundance of infection, whereas when the species compete for resources there is a dilution effect. However, as in §2 this effect is entirely due to the ecological dynamics of the system, an increase in the prey species population results in an increase in the predator species population, and hence an increase in the population density of infected predators. If transmission of infection is density-dependent and within species only, then
. Hence, there is an amplification effect on the abundance of infection, whereas when the species compete for resources there is a dilution effect. However, as in §2 this effect is entirely due to the ecological dynamics of the system, an increase in the prey species population results in an increase in the predator species population, and hence an increase in the population density of infected predators. If transmission of infection is density-dependent and within species only, then 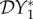 and
and 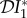 are positive (see equations (2.2) and (2.3)), so there is an amplification effect on prevalence and abundance of infection. If host species two is a predator on host species one, then ϕ21 < 0 and ϕ12 > 0. We then have the situation where an increase in the population density of species two results in a decrease in the population density of species one, a relationship superficially similar to that obtained when the species compete for resources. Hence, the results are qualitatively unchanged from those presented in §2.
are positive (see equations (2.2) and (2.3)), so there is an amplification effect on prevalence and abundance of infection. If host species two is a predator on host species one, then ϕ21 < 0 and ϕ12 > 0. We then have the situation where an increase in the population density of species two results in a decrease in the population density of species one, a relationship superficially similar to that obtained when the species compete for resources. Hence, the results are qualitatively unchanged from those presented in §2.
3.2. Infection-induced host mortality
In §2, we assumed that the pathogen had no effect on host population dynamics. We now relax one aspect of this assumption, by assuming that infected hosts have increased mortality. We assume that if a host of species i is infected, its death rate is increased from μi to μi + αi. The equations for the dynamics of the host population densities become
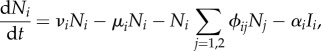 |
for i = 1, 2. Steady-state solutions now satisfy
| 3.1 |
The equations for population density and infection abundance no longer decouple, and the values of the Ni at the infection-free and infected steady-states are no longer equal. We denote the infection-free steady state by 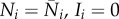 , and the infected steady state by Ni = N*i, Ii = I*i≠0. The equations for the dynamics of the infected populations are
, and the infected steady state by Ni = N*i, Ii = I*i≠0. The equations for the dynamics of the infected populations are
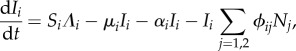 |
with 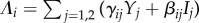 as before. Steady-state prevalences of infection now solve
as before. Steady-state prevalences of infection now solve
| 3.2 |
The next-generation matrix K has components
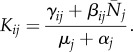 |
We now investigate how the increased host mortality may influence the dilution effect. The expressions for the elasticity of the prevalence and abundance of infection in species one with respect to changes in the population density of species two are unchanged,
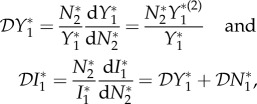 |
but now from equation (3.1)
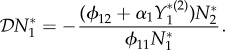 |
Substituting 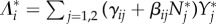 for i = 1, 2 in equation (3.2) and differentiating, we obtain
for i = 1, 2 in equation (3.2) and differentiating, we obtain
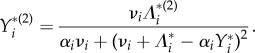 |
Given the steady-state values (N*1, N*2, Y*1, Y*2), the pair of equations
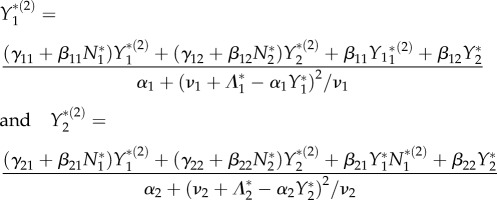 |
together with N*(2)1 = − (ϕ12 + α1Y*(2)1)/ϕ11 form a linear system for the three variables (N*(2)1, Y*(2)1, Y*(2)2). The elasticities 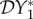 and
and 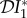 now follow, and the expression for the elasticity of
now follow, and the expression for the elasticity of 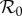 is given by equation (2.4).
is given by equation (2.4).
The elasticities 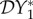 ,
, 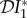 and
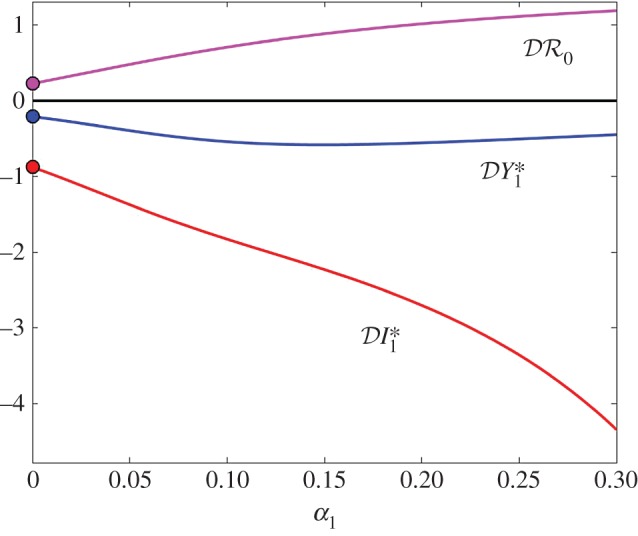
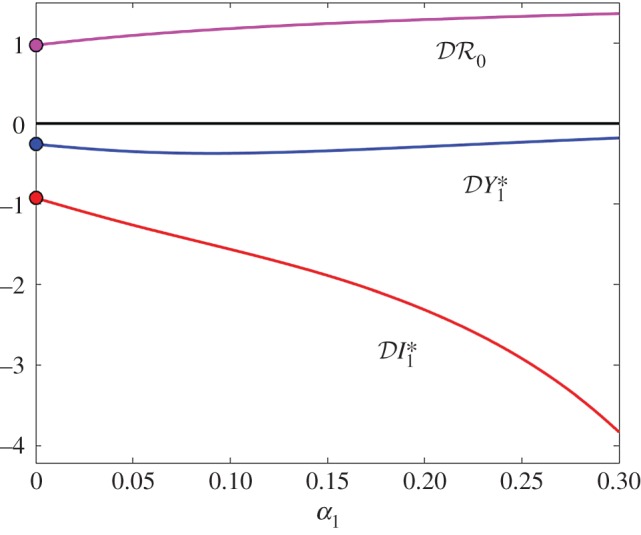
and 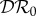 are shown as functions of the increased mortality α1 in figures 4–6. Parameter values are as in figures 1–3, respectively, except that α1≠0 and ε = 0.1 is fixed. In all three examples, the dilution effect on infection prevalence is enhanced for small values of α1, but then becomes relatively constant as α1 increases. By contrast, the effect on infection abundance is markedly increased (
are shown as functions of the increased mortality α1 in figures 4–6. Parameter values are as in figures 1–3, respectively, except that α1≠0 and ε = 0.1 is fixed. In all three examples, the dilution effect on infection prevalence is enhanced for small values of α1, but then becomes relatively constant as α1 increases. By contrast, the effect on infection abundance is markedly increased (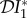 becomes more negative). In figures 4 and 5, there is little change in
becomes more negative). In figures 4 and 5, there is little change in 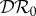 with α1, but for the example parameters in figure 6 the amplification of
with α1, but for the example parameters in figure 6 the amplification of 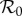 is enhanced.
is enhanced.
Figure 4.
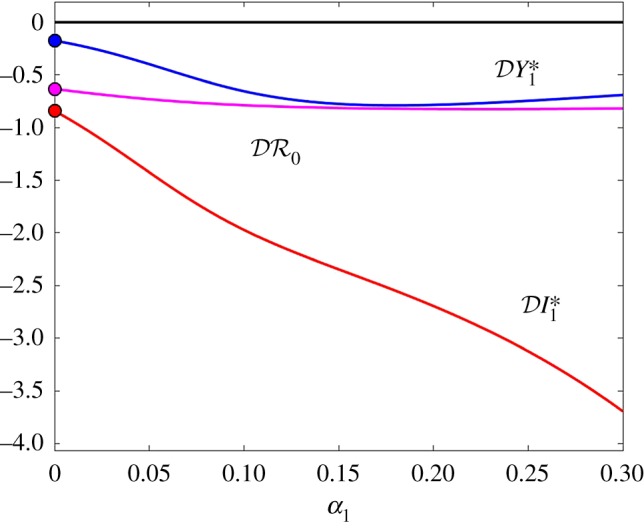
The elasticity of infection prevalence 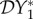 (blue), infection abundance
(blue), infection abundance 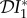 (red), and basic reproduction number
(red), and basic reproduction number 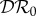 (magenta) as functions of α1. Transmission of infection is density-dependent with β11 = 5.0, β22 = 2.05 and
(magenta) as functions of α1. Transmission of infection is density-dependent with β11 = 5.0, β22 = 2.05 and 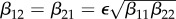 , ε = 0.1 and α2 = 0. Other parameter values as in figure 1. Circles at α1 = 0 are calculated from formulae in §2.2. (Online version in colour.)
, ε = 0.1 and α2 = 0. Other parameter values as in figure 1. Circles at α1 = 0 are calculated from formulae in §2.2. (Online version in colour.)
Figure 6.
The elasticity of infection prevalence 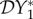 (blue), infection abundance
(blue), infection abundance 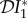 (red), and basic reproduction number
(red), and basic reproduction number 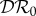 (magenta) as functions of α1. Transmission of infection is density-dependent with β11 = 3.750, β22 = 3.106 and
(magenta) as functions of α1. Transmission of infection is density-dependent with β11 = 3.750, β22 = 3.106 and 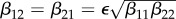 , ε = 0.1 and α2 = 0. Other parameter values as in figure 1. Circles at α1 = 0 are calculated from formulae in §2.2. (Online version in colour.)
, ε = 0.1 and α2 = 0. Other parameter values as in figure 1. Circles at α1 = 0 are calculated from formulae in §2.2. (Online version in colour.)
Figure 5.
The elasticity of infection prevalence 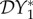 (blue), infection abundance
(blue), infection abundance 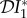 (red), and basic reproduction number
(red), and basic reproduction number 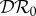 (magenta) as functions of α1. Transmission of infection is density-dependent with β11 = 2.5, β22 = 4.1 and
(magenta) as functions of α1. Transmission of infection is density-dependent with β11 = 2.5, β22 = 4.1 and 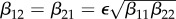 , ε = 0.1 and α2 = 0. Other parameter values as in figure 1. Circles at α1 = 0 are calculated from formulae in §2.2. (Online version in colour.)
, ε = 0.1 and α2 = 0. Other parameter values as in figure 1. Circles at α1 = 0 are calculated from formulae in §2.2. (Online version in colour.)
3.3. Multiple host species
When there are more than two host species it is in general more difficult, if not impossible, to derive analytic expressions for the quantities 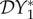 ,
, 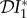 and
and 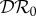 used to measure dilution and amplification. In this section we show how these quantities can be calculated, in order to provide a general method that can be used to analyse larger and more complicated ecosystems.
used to measure dilution and amplification. In this section we show how these quantities can be calculated, in order to provide a general method that can be used to analyse larger and more complicated ecosystems.
Consider n species interacting in an ecosystem. Let the population size of species i be Ni, in an appropriate unit (population density, number of animals, biomass, etc.). Assume that species i has maximum birth rate and minimum death rate, νi and μi, respectively, and that in the absence of other species its population growth rate would be νi − μi − ϕiiNi. In order that species i can sustain itself independently of the other species, we require νi > μi. Let those species that compete for resources with species i have indices contained in the set 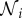 , where if
, where if 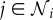 , then the growth rate of species i is reduced by an amount ϕijNj. Let those species that are consumers of species i have indices contained in the set
, then the growth rate of species i is reduced by an amount ϕijNj. Let those species that are consumers of species i have indices contained in the set 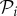 , and those species that are consumed by species i have indices contained in the set
, and those species that are consumed by species i have indices contained in the set 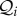 . Suppose that species i is consumed by species k, at a rate ϕikNk, and species k consequently increases its birth rate by ekiϕkiNi = − ϕikNi (note the order of subscripts). Hence eki is a measure of the efficiency of conversion of biomass of species i into biomass of species k. We do not allow cannibalism, so
. Suppose that species i is consumed by species k, at a rate ϕikNk, and species k consequently increases its birth rate by ekiϕkiNi = − ϕikNi (note the order of subscripts). Hence eki is a measure of the efficiency of conversion of biomass of species i into biomass of species k. We do not allow cannibalism, so 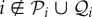 . We can, if necessary, adjust the ϕij so that
. We can, if necessary, adjust the ϕij so that 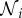 ,
, 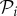 and
and 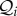 are disjoint subsets of Ω, the set of all species in the ecosystem.
are disjoint subsets of Ω, the set of all species in the ecosystem.
The population dynamics of the ecosystem species are described by the equations
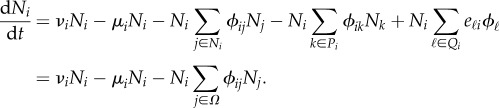 |
There are usually multiple steady states of these equations. A steady state 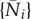 solves
solves
The stability of each steady state may be deduced from the eigenvalues of the Jacobian matrix,  , which has elements
, which has elements 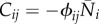 .
.
Let the ecosystem be infected by a pathogen, with prevalence Yi in species i. We now generalize the equations for the population dynamics of the ecosystem to include the possibility that the infection status of the host may change its ability to compete, predate or escape predation. Suppose that if species i competes for resources with species j
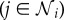 , then the reduction in the population growth rate of species i is pijϕij when hosts of species i are infected, and qijϕij when hosts of species j are infected. Suppose that if species i is consumed by species k
, then the reduction in the population growth rate of species i is pijϕij when hosts of species i are infected, and qijϕij when hosts of species j are infected. Suppose that if species i is consumed by species k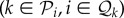 , then the rate of consumption is pikϕik when the predator only is infected, qikϕik when the prey only is infected and pikqikϕik when both are infected. For symmetry we require pki = qik and qki = pik. Suppose also that infected hosts of species i have their mortality increased by αi. The equations for population dynamics of the ecosystem species become
, then the rate of consumption is pikϕik when the predator only is infected, qikϕik when the prey only is infected and pikqikϕik when both are infected. For symmetry we require pki = qik and qki = pik. Suppose also that infected hosts of species i have their mortality increased by αi. The equations for population dynamics of the ecosystem species become
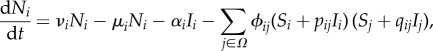 |
3.3 |
where Si = Ni − Ii = Ni(1 − Yi). Non-zero steady-state solutions for the population densities now satisfy
The equations for the infected species population densities are
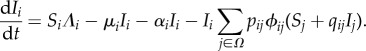 |
3.4 |
Steady-state prevalences of infection solve
where
We now find expressions for the elasticities of infection in one species with respect to changes in the population density of another. Define
Let n and i be vectors with components Ni and Ii, respectively, and write equations (3.3) and (3.4) as
Steady-state values satisfy F(n*, i*) = G(n*, i*) = 0. If the mortality of species ℓ is then increased by a small amount ω, then the new steady states n = n* + ωu and i = i* + ωv solve
Expanding and neglecting high-order terms in ω,
where J(n*, i*) is the Jacobian matrix evaluated at the steady state with ω = 0. The Jacobian matrix has the structure
and its components are
We can then compute the elasticities of infection by
Let 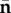 be the vector whose components are values of the Ni at an infection-free steady state,
be the vector whose components are values of the Ni at an infection-free steady state, 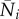 . Hence
. Hence 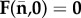 . The next-generation matrix K has components
. The next-generation matrix K has components
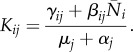 |
If we increase the mortality of species ℓ to μℓ + ω, then the infection-free steady state solution nω has components Nωi, and solves F(nω, 0) = ωNωℓeℓ. The NGM becomes Kω, with components
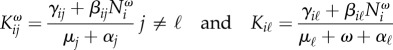 |
and spectral radius 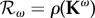 . Hence the elasticity of the basic reproduction number is
. Hence the elasticity of the basic reproduction number is
4. Discussion
We have introduced a flexible approach to the study of the dilution/amplification effect in compartmental eco-epidemiological models. We have focused on models describing the dynamics of a single pathogen species in a population consisting of host and non-host species for that pathogen. We have shown that metrics to quantify dilution and amplification can be clearly and objectively defined, based on the elasticity of a quantity related to infection, and in response to a decrease in the population density of a particular species. The infection quantities we discussed were the prevalence and incidence of infection in a focal species, and the basic reproduction number of the pathogen in the ecosystem.
We have illustrated the use of these metrics in simple settings involving just two species and assuming SI infection dynamics, for example contrasting the influence of different assumptions on the link between population density and transmission. We have done so in the realistic situation where the two species are allowed to interact both ecologically (competition, consumer–resource relationship) and epidemiologically (within-species and between-species transmission, infection-induced mortality). In this relatively simple setting, it is already clear that one needs to be specific about the system or model one is studying before making statements such as ‘this system/model shows a dilution effect’. Our simple examples show that, under the same circumstances, there can be a dilution effect as measured (by our definition) in terms of incidence, but not in terms of prevalence or 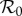 . In fact, there can be an amplification effect in one metric and a dilution effect in another. In addition, the assumptions regarding the way transmission scales with contact rate within species, the strength of interaction between species the relative efficiency of within-species transmission for the different species, and the strength of infection-induced host mortality all influence the outcome regarding dilution/amplification and the strength of such effects. For example, in the case of density-dependent transmission within host species, when within-species transmission is higher in species two, there is a dilution effect in species one in terms of the prevalence of infection if between-species transmission is weak. For increased between-species transmission, however, the strength of the dilution effect decreases and can, for relatively strong between-species transmission, change to an amplification effect. Several of these observations have been made before in pioneering modelling studies into dilution where special cases were treated [16–23], but the formalization in our general setting will allow future exploration directly contrasting a range of different assumptions, and greater flexibility to explore the many systems for which observational data and empirical work are now available [1,9–11,24,25].
. In fact, there can be an amplification effect in one metric and a dilution effect in another. In addition, the assumptions regarding the way transmission scales with contact rate within species, the strength of interaction between species the relative efficiency of within-species transmission for the different species, and the strength of infection-induced host mortality all influence the outcome regarding dilution/amplification and the strength of such effects. For example, in the case of density-dependent transmission within host species, when within-species transmission is higher in species two, there is a dilution effect in species one in terms of the prevalence of infection if between-species transmission is weak. For increased between-species transmission, however, the strength of the dilution effect decreases and can, for relatively strong between-species transmission, change to an amplification effect. Several of these observations have been made before in pioneering modelling studies into dilution where special cases were treated [16–23], but the formalization in our general setting will allow future exploration directly contrasting a range of different assumptions, and greater flexibility to explore the many systems for which observational data and empirical work are now available [1,9–11,24,25].
We have defined three different metrics for quantifying the dilution effect, and briefly discussed a fourth. Obviously, the choice to be made depends on the ecology and epidemiology of the system under study, and crucially on the question being asked. Where transmission of infection was frequency-dependent only (§2.1), the only non-zero elasticity was that of abundance of infection (population density of infected hosts), and that was entirely due to ecological changes. Although density-dependent transmission gave rise to dilution or amplification effects as measured by each of the proposed metrics, we have shown that choice of metric may determine the outcome. If one is concerned about infection in a particular species, then the quantities 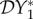 or
or 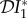 may be appropriate. If one is concerned about the connection between biodiversity and persistence of a pathogen, or invasion of an absent pathogen, then
may be appropriate. If one is concerned about the connection between biodiversity and persistence of a pathogen, or invasion of an absent pathogen, then 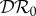 would be more appropriate. In §2.3, we introduced a potential fourth metric,
would be more appropriate. In §2.3, we introduced a potential fourth metric, 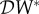 , which could be appropriate where the concern is risk of transmission to another species, maybe humans.
, which could be appropriate where the concern is risk of transmission to another species, maybe humans.
In §3.3, we show how the framework and metrics we have introduced generalize to n-species communities. One can imagine that the intricate relationship between biodiversity and infection that is seen in the two-species system will become even more complicated and subtle in larger communities, where the network of interaction between several species at different trophic levels adds many dimensions of interaction and complexity. On the other hand, one has to be careful with over-interpretation of two-species results because these represent only a small and special link in food webs and ecosystems. Larger interacting communities need to be studied to see whether these effects are annihilated in larger systems (through positive and negative feedback), changed, weakened or amplified. Our results provide an initial contribution and a way to gently and transparently introduce the framework and metrics, as well as an entry point for studying larger ecosystems.
We have chosen to interpret ‘reduction in biodiversity’ in our system as a reduction in the density of one species. This is a clear choice when the model consists of only two species. In the arbitrary n-species ecosystem, however, there is a wider range of options. One can imagine a reduction in population density or the complete elimination of a single species, but also of different sets of species. A relevant extension of our metrics is therefore to accommodate sets of species being reduced. Also, in our current framework, we chose to restrict quantifying a possible dilution/amplification effect for a single focal host species. It may be interesting to rotate the species that is the focal host and produce a matrix of effect sizes for an entire community. It remains a difficult issue that even with the generality we provide, the ‘real’ situation is that we not only have many interacting species for the study of possible dilution effects for a given infectious agent, but we have many different infectious agents acting at the same time and influencing species interactions (and vice versa).
Our aim was not to explore specific examples in great detail. As is already clear from the introduction, there are many factors involved if we want to obtain a deeper understanding of dilution/amplification. These factors involve both ecological and epidemiological aspects, as well as their interaction. The choices and combinations of mechanisms and processes that could be made are extensive, even without specifying ranges for parameter values in the descriptions used for such choices. Because of this, it would not be insightful to present a detailed analysis of the example systems we have used to introduce our approach. The value of the examples lies in illustrating the general approach, and in showing that being precise about many of the available options is important before drawing conclusions about dilution/amplification. We envisage that the framework and metrics presented here can now be used to study particular eco-epidemiological systems, where many of the choices that need to be made are dictated by the actual biology of those systems.
The study of infectious agents in ecosystems has a much broader relevance than understanding dilution/amplification effects. It is well known that ecosystems are changing. Habitat depletion and fragmentation, and other human-related activities, are threatening the viability of many species, and changing the population dynamics of just one species can have consequences for a number of other species. For examples of the direct and indirect effects of changing the populations of the largest carnivores and herbivores; see [26,27]. Many other examples of ecosystem dynamics and the interplay between species may be found in the literature. Studies that include pathogens as part of the ecosystem are less common, but this situation is rapidly changing; see [28] for a relatively recent review. Factors such as climate change may alter the dynamics of the ecosystem in a way that increases the potential exposure of humans to infection; see, for example, a study of monkeypox virus in the Congo Basin [29]. There is a broader need to study how infectious agents interact with the ecosystems of which they are integral parts, particularly if we want to understand how changes in ecosystems affect the distributions of infectious agents, the risk of host species jumps, and the risks and impact of future outbreaks [30].
Data accessibility
This article has no additional data.
Authors' contributions
The authors conceived this research, developed the method and prepared the manuscript. Both the authors contributed equally.
Competing interests
We declare we have no competing interests.
Funding
No funding has been received for this article.
References
- 1.Wood CL, Lafferty KD. 2013. Biodiversity and disease: a synthesis of ecological perspectives on Lyme disease transmission. Trends Ecol. Evol. 28, 239–247. ( 10.1016/j.tree.2012.10.011) [DOI] [PubMed] [Google Scholar]
- 2.Buhnerkempe M, Roberts MG, Dobson AP, Heesterbeek JAP, Hudson P, Lloyd-Smith JO. 2015. Eight challenges in modelling disease ecology in multi-host, multi-agent systems. Epidemics 10, 26–30. ( 10.1016/j.epidem.2014.10.001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Keesing F. et al. 2010. Impacts of biodiversity on the emergence and transmission of infectious diseases. Nature 468, 647–652. ( 10.1038/nature09575) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lafferty KD, Wood CL. 2013. It's a myth that protection against disease is a strong and general service of biodiversity conservation: response to Ostfeld and Keesing. Trends Ecol. Evol. 28, 503–504. ( 10.1016/j.tree.2013.06.012) [DOI] [PubMed] [Google Scholar]
- 5.Ostfeld RS, Keesing F. 2013. Straw men don't get Lyme disease: response to Wood and Lafferty. Trends Ecol. Evol. 28, 502–503. ( 10.1016/j.tree.2013.05.009) [DOI] [PubMed] [Google Scholar]
- 6.Randolph SE, Dobson ADM. 2012. Pangloss revisited: a critique of the dilution effect and the biodiversity-buffers-disease paradigm. Parasitology 139, 847–863. ( 10.1017/S0031182012000200) [DOI] [PubMed] [Google Scholar]
- 7.Roche B, Dobson AP, Guégan J-F, Rohani P. 2012. Linking community and disease ecology: the impact of biodiversity on pathogen transmission. Phil. Trans. R. Soc. B 367, 2807–2813. ( 10.1098/rstb.2011.0364) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Keesing F, Holt RD, Ostfeld RS. 2006. Effects of species diversity on disease risk. Ecol. Lett. 9, 485–498. ( 10.1111/j.1461-0248.2006.00885.x) [DOI] [PubMed] [Google Scholar]
- 9.Civitello DJ. et al. 2015. Biodiversity inhibits parasites: broad evidence for the dilution effect. Proc. Natl Acad. Sci. USA 112, 8667–8671. ( 10.1073/pnas.1506279112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Salkeld DJ, Padgett KA, Jones JH. 2013. A meta-analysis suggesting that the relationship between biodiversity and risk of zoonotic pathogen transmission is idiosyncratic. Ecol. Lett. 16, 679–686. ( 10.1111/ele.12101) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Strauss AT, Civitello DJ, Cáceras CE, Hall SR. 2015. Success, failure and ambiguity of the dilution effect among competitors. Ecol. Lett. 18, 916–926. ( 10.1111/ele.12468) [DOI] [PubMed] [Google Scholar]
- 12.Roberts MG, Heesterbeek JAP. 2013. Characterizing the next-generation matrix and basic reproduction number in ecological epidemiology. J. Math. Biol. 66, 1045–1064. ( 10.1007/s00285-012-0602-1) [DOI] [PubMed] [Google Scholar]
- 13.Fenton A, Streicker DG, Petchey OL, Pedersen AB. 2015. Are all hosts created equal? Partitioning host species contributions to parasite persistence in multihost communities. Am. Nat. 186, 610–622. ( 10.1086/683173) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Funk S, Nishiura H, Heesterbeek JAP, Edmunds WJ, Checchi F. 2013. Identifying transmission cycles at the human–animal interface: the role of animal reservoirs in maintaining gambiense human African trypanosomiasis. PLoS Comp. Biol. 9, e1002855 ( 10.1371/journal.pcbi.1002855) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Diekmann O, Heesterbeek JAP, Roberts MG. 2010. The construction of next-generation matrices for compartmental epidemic models. J. R. Soc. Interface 7, 873–885. ( 10.1098/rsif.2009.0386) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Begon M. 2008. Effects of host diversity on disease dynamics. In Infectious disease ecology: effects of ecosystems on disease and of disease on ecosystems (eds Ostfeld RS, Keesing F, Eviner VT), pp. 12–29. Princeton, NJ: Princeton University Press. [Google Scholar]
- 17.Begon M, Bowers RG. 1994. Host–host–pathogen models and microbial pest control: the effect of host self regulation. J. Theor. Biol. 169, 275–287. ( 10.1006/jtbi.1994.1148) [DOI] [PubMed] [Google Scholar]
- 18.Begon M, Bowers RG. 1995. Beyond host–pathogen dynamics. In Ecology of infectious diseases in natural populations (eds Grenfell BT, Dobson AP), pp. 478–509. Cambridge, UK: Cambridge University Press. [Google Scholar]
- 19.Bowers RG, Begon M. 1991. A host–host–pathogen model with free-living infective stages, applicable to microbial pest control. J. Theor. Biol. 148, 305–329. ( 10.1371/journal.pone.0066071) [DOI] [PubMed] [Google Scholar]
- 20.Dobson AP. 2004. Population dynamics of pathogens with multiple host species. Am. Nat. 164, S64–S78. ( 10.1086/424681) [DOI] [PubMed] [Google Scholar]
- 21.Holt RD, Dobson AP, Begon M, Bowers RG, Schauber EM. 2003. Parasite establishment in host communities. Ecol. Lett. 6, 837–842. ( 10.1046/j.1461-0248.2003.00501.x) [DOI] [Google Scholar]
- 22.Rudolf VHW, Antonovics A. 2005. Species coexistence and pathogens with frequency-dependent transmission. Am. Nat. 166, 112–118. ( 10.1086/430674) [DOI] [PubMed] [Google Scholar]
- 23.Ogden NH, Tsao JI. 2009. Biodiversity and Lyme disease: dilution or amplification? Epidemics 1, 196–206. ( 10.1016/j.epidem.2009.06.002) [DOI] [PubMed] [Google Scholar]
- 24.Johnson PTJ, Ostfeld RS, Keesing F. 2015. Frontiers in research on biodiversity and disease. Ecol. Lett. 18, 1119–1133. ( 10.1111/ele.12479) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Khalil H, Ecke F, Evander M, Magnusson M, H"e;ornfeldt B. 2016. Declining ecosystem health and the dilution effect. Sci. Rep. 6, 31314 ( 10.1038/srep31314) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ripple RJ. et al. 2014. Status and ecological effects of the world's largest carnivores. Science 343, 1241484 ( 10.1126/science.1241484) [DOI] [PubMed] [Google Scholar]
- 27.Ripple RJ. et al. 2015. Collapse of the world's largest herbivores. Sci. Adv. 1, e1400103 ( 10.1126/sciadv.1400103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Selaković S, de Ruiter PC, Heesterbeek JAP. 2014. Infectious disease agents mediate interaction in food webs and ecosystems. Proc. R. Soc. B 281, 20132709 ( 10.1098/rspb.2013.2709) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thomassen HA. et al. 2013. Pathogen–host associations and predicted range shifts of human monkeypox in response to climate change in central Africa. PLoS ONE 8, e66071 ( 10.1371/journal.pone.0066071) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cunningham AA, Daszak P, Wood JLN. 2017. One Health, emerging infectious diseases and wildlife: two decades of progress? Phil. Trans. R. Soc. B 372, 20160167 ( 10.1098/rstb.2016.0167) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This article has no additional data.


