Abstract
A new experimental platform for probing nanoscale molecular changes in living bacteria using atomic force microscopy–infrared (AFM–IR) spectroscopy is demonstrated. This near-field technique is eminently suited to the study of single bacterial cells. Here, we report its application to monitor dynamical changes occurring in the cell wall during cell division in Staphylococcus aureus using AFM to demonstrate the division of the cell and AFM–IR to record spectra showing the thickening of the septum. This work was followed by an investigation into single cells, with particular emphasis on cell-wall signatures, in several bacterial species. Specifically, mainly cell wall components from S. aureus and Escherichia coli containing complex carbohydrate and phosphodiester groups, including peptidoglycans and teichoic acid, could be identified and mapped at nanometre spatial resolution. Principal component analysis of AFM–IR spectra of six living bacterial species enabled the discrimination of Gram-positive from Gram-negative bacteria based on spectral bands originating mainly from the cell wall components. The ability to monitor in vivo molecular changes during cellular processes in bacteria at the nanoscale opens a new platform to study environmental influences and other factors that affect bacterial chemistry.
Keywords: atomic force microscopy–infrared, in vivo, cell wall, Gram-positive bacteria, Gram-negative bacteria
1. Introduction
In recent years, there has been an emerging focus on advancing research techniques to enable an ability to image samples with higher spatial resolution. This is of particular importance in the field of bacterial studies, as their small size significantly limits the use of conventional microscopic techniques. Transmission electron microscopy (TEM) and scanning electron microscopy (SEM) are commonly applied in research involving bacteria [1]. Super resolution microscopies, such as scanning near-field optical microscopy (SNOM), are also becoming popular for the study of a single bacterium [2]. Although these techniques enable one to achieve nanoscale resolution, they are limited to probing the morphology of the cell, without providing any information about the chemical composition of the sample. In addition, both techniques require dehydration of the sample prior to analysis. The chemical composition of a single bacterium can be probed using confocal fluorescence microscopy and other techniques such as fluorescent resonant energy transfer (FRET) and fluorescence recovery after photobleaching (FRAP) [3–5]. However, these approaches are often technically demanding (in particular, FRET and FRAP), and require the use of fluorescence stains, thus restricting their use to probing only a limited number of markers, usually associated with the cellular surface.
Atomic force microscopy–infrared (AFM–IR) absorption spectroscopy is a novel technique based on the detection of the thermal expansion caused by infrared absorption [6–8]. It combines AFM—designed to measure the local physical properties of a sample, such as height or probe deflection—with IR spectroscopy enabling spatially resolved chemical characterization. This approach enables the acquisition of IR spectra while overcoming the wavelength diffraction spatial resolution limit, resulting in a lateral spatial resolution approaching 20 nm. The fundamental process occurring during the measurement is the absorption of IR radiation [6–8], resulting in a local temperature increase. Although the lateral resolution of AFM–IR is in the order of tens of nanometres, the penetration of the IR beam into the sample can still be of the order of micrometres. The local increase in the temperature, caused by IR absorption, can be measured either directly as the change in temperature [9] or through measurement of the probe motion [6,7,8]. The latter occurs due to a force impulse created by the IR absorption and causes oscillation of the AFM cantilever probe [6–8]. The oscillation amplitude is proportional to the absorption coefficient [6,10]. During the measurement, the thermal expansion increases until the light pulse ends and subsequently decreases exponentially. The rate of this decrease depends on the sample properties; therefore, making the phenomenon of thermal expansion dependent directly on the sample composition. However, the strength of the signal depends also on the material properties, such as the thermal expansion coefficient, density, heat capacity, among other factors [6]. The AFM–IR technique enables the collection of single spectra and the generation of maps of the distribution of intensity of a signal at a chosen wavenumber, allowing one to study samples on a nanoscale.
The multiple benefits of the combination of AFM and IR spectroscopy have resulted in an increase in the application of the technique for a variety of samples [3,6,11–28]. In particular, AFM–IR spectroscopy has a broad application in the study of polymers [6,11–19] because the performance and behaviour of polymer materials is largely dependent on their quality at the nanoscale, which cannot be properly studied with conventional IR microspectroscopy. In addition to these applications, AFM–IR is finding a role in the nano-characterization of biological samples [3,20–27]. It has been applied to the study of plants [20], as well as to various mammalian tissues [21–23] and cells [24,25]. In the case of tissues, the majority of AFM–IR applications have focused on skin [21] and hair [22]; however, other types of tissue have also been studied, including the distribution of minerals in bone [23]. There is an increasing interest in applying AFM–IR to study bacteria, below the spatial resolution achievable by imaging with conventional IR spectroscopy [3,7,26,27]. Dazzi et al. [7] demonstrated the first successful application of AFM–IR mapping of Escherichia coli, showing a homogeneous distribution of proteins (via the amide I and amide III absorptions) and a good correspondence between AFM–IR spectra and FT–IR spectra. Since then, there have been several bacterial studies aimed at exploring the production of biofuel precursors by microorganisms (Streptomyces) [26] and the production of polyhydroxybutyrates, degradable polyesters with mechanical properties similar to thermoplastic synthetic polymers [3]. Furthermore, the high lateral spatial resolution of AFM–IR enables observation not only of bacteria as whole organisms, but also of minor nano-localized changes, such as the presence of a virus infecting a bacterium [27]. Bacterial cell walls are natural polymeric structures and thus are a logical extension of the techniques developed for in situ chemical analysis of synthetic polymers. A further step, undertaken in this study, is application of the technique in vivo and its use to monitor dynamical changes in the cell wall of a living bacterium.
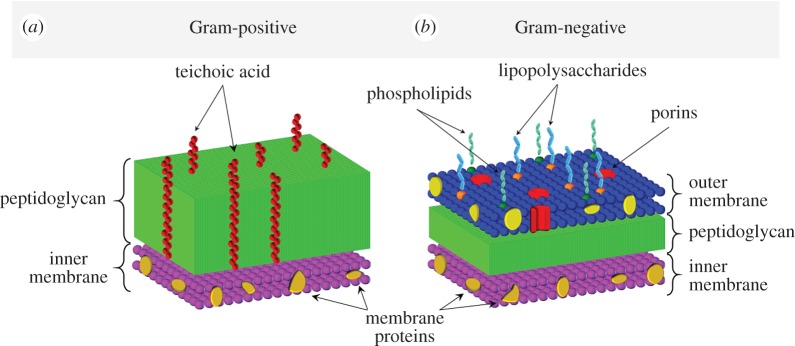
The composition of the cell wall is the basis for classification of bacterial species into two groups. Gram-positive bacteria have a thick cell wall consisting of peptidoglycans and teichoic acids (TAs) that cover the cytoplasmic membrane, while Gram-negative bacteria have an outer- and inner-membrane with a thin layer of peptidoglycans in between (figure 1) [28]. The outer membrane of Gram-negative bacteria contains proteins, phospholipids and lipopolysaccharides. It serves as a barrier to the external environment, at the same time enabling selective diffusion through porins, allowing nutrition uptake and waste removal [29]. The peptidoglycan component of Gram-negative bacteria consists of one to two layers [29]. In the case of E. coli, approximately 80% of the peptidoglycan is present in the form of a monolayer [30]. Owing to this, the overall thickness of the whole cell wall does not exceed a few nanometres for Gram-negative bacteria (e.g. for E. coli—4 nm) [31]. By contrast, Gram-positive bacteria are devoid of an outer membrane, but possess a thick peptidoglycan component containing 10–20 layers (figure 1) [28]. Peptidoglycan is a polymer composed of glycan strands, formed from disaccharide residues cross-linked with amino acids [28,31]. Each disaccharide contains a covalently bonded peptide that can bind to a peptide of another strand, thus enabling the formation of an elastic network. The average length of strands differs between bacteria (e.g. in E. coli—21 disaccharide units, in Staphylococcus aureus aureus—six disaccharide units) [32] and is strongly dependent on conditions, growth phase or even strain [31]. In addition, the Gram-positive bacterial cell walls contain TAs, absent in the cell wall of Gram-negative bacteria (figure 1). TAs are anionic glycerol phosphate-containing polymers attached to either the peptidoglycan layer or to membrane lipids (and present in the form of lipoteichoic acid, LTA). The bacterial cell wall, particularly its peptidoglycan layer, is a dynamical structure with many functions that ensures cell viability and is involved in maintaining cellular shape [31].
Figure 1.
Schematic illustration showing the composition of the cell wall from a Gram-positive (a) and Gram-negative (b) bacterium.
The aim of this work was to monitor dynamical changes in the bacterium in vivo, using AFM–IR. The high spatial resolution of AFM–IR enables the examination of a single live bacterium based mainly on the chemistry of their cell walls. However, prior to this application, aimed at real-time observation of biological processes, we focused on obtaining a detailed spectroscopic profile of single bacterial cells. The purpose of this was to identify the IR bands related to the cell wall and assess their contribution to the AFM–IR spectrum of a single, live bacterium in representative Gram-positive (S. aureus, Bacillus subtilis, Enterococcus faecalis) and Gram-negative (E. coli, Pseudomonas aeruginosa, Acinetobacter baumannii) bacteria.
2. Methods
2.1. Sample preparation
The bacterial strains used in this study were S. aureus A8819 [33], B. subtilis BS34A [34], En. faecalis ATCC 29212, En. faecalis ATCC19433, E. coli DC10B [35], P. aeruginosa PAO1 [36] and A. baumannii ATCC 17979. Prior to each measurement, the bacterial cells were plated on horse blood agar (HBA) plates (ThermoFisher Scientific) and incubated at 37°C for 24 h. Bacterial colonies were collected from HBA plates, washed with saline solution and the cells were pelleted using centrifugation at 2000 r.p.m. for 5 min. The resultant pellets were then washed with 500 µl of ultrapure water twice to remove saline and the remaining media. Subsequently, the pellets were re-suspended in 100 µl of ultrapure water, mixed for 1 min, placed on a CaF2 window and air-dried. The sample was then mounted on a flat magnetic stainless-steel substrate. For each bacterial strain, at least three technical replicates were prepared using cells grown on independent HBA plates. Peptidoglycan standard from S. aureus was purchased from Sigma Aldrich.
2.2. Attenuated total reflectance–Fourier transform infrared measurements
The ATR–FTIR measurements were performed using bacterial colonies transferred directly from the HBA plates onto the ATR crystal. For each bacterial strain, three replicates were measured. Spectra were recorded using a Bruker Alpha FTIR (Ettlingen, Germany) spectrometer with an attenuated total reflection (ATR) sampling device containing a single bounce diamond internal reflection element. The system is equipped with a globar source, KBr beam splitter and a deuterated triglycine sulfate detector. Spectra were collected over the spectral range of 1800–900 cm−1 with a spectral resolution of 6 cm−1 and 128 interferograms for both, the background and sample. A spectrum was recorded of the peptidoglycan standard by pressing the powder directly onto the ATR crystal and using the instrumental parameters stated above.
2.3. Atomic force microscopy–infrared measurements
The measurements were performed using a NanoIR2 system (Anasys Instruments Inc., Santa Barbara, CA, USA). The IR source was an optical parametric oscillator laser, generating a 10 ns pulse at a 1 kHz repetition rate. AFM–IR spectra were collected in contact mode, in the spectral range of 1800–900 cm−1 with a spectral resolution of 8 cm−1. For data collection contact mode, NIR2 probes were used (model: PR-EX-nIR2, Anasys Instruments, Inc.). For each bacterial strain, at least 30 single spectra were collected. The system was purged with N2 to control the humidity. For S. aureus and E. coli, IR maps at fixed wavenumber values were acquired in order to investigate the distribution of selected components. AFM height and deflection images as well as IR maps were acquired simultaneously.
2.4. Data analysis
The ATR–FTIR spectra were smoothed using the Savitzky–Golay (SG) algorithm with nine smoothing points. An ATR correction was performed using the Bruker Opus 7.5 Software. The AFM–IR spectra were normalized using the standard normal variate (SNV) method and smoothed using the SG algorithm with 15 smoothing points. The second derivatives of AFM–IR were calculated using the SG algorithm with 17 smoothing points. The dataset was mean centred prior to analysis. Principal component analysis (PCA) was computed on the set of all acquired spectra (n = 327) of different bacteria (n = 6) in the range 1400–950 cm−1. Matlab (Mathworks, Natik, USA), PLS_toolbox (Eigenvector research, Manson, USA), Bruker Opus 7.5, Anasys Studio and Origin Pro 9.1 were used for data pre-processing, analysis and presentation.
2.5. Bacterial viability after the atomic force microscopy–infrared measurements
To assess the viability of bacterial cells after the AFM–IR measurements, 100 µl of ultrapure water was placed on the CaF2 slide and gently mixed with a pipette tip to re-suspend the bacterial cells. The solution was then transferred into an Eppendorf tube, vortexed for 1 min and plated on HBA (3 × 20 µl) plates. These were then incubated at 37°C for 24 h and photographed.
3. Results and discussion
First, we characterized bacteria on a population level using ATR–FTIR spectroscopy (electronic supplementary material, figure S1). This technique has been consistently applied as an ideal tool for discriminating between different Gram-positive and Gram-negative bacterial species since the 1980s [37–41]. To confirm previous findings, live cells were transferred directly from colonies grown on HBA plates onto an ATR accessory of an FTIR spectrometer. The major differences between spectra of Gram-positive bacteria and Gram-negative bacteria were the intensities of bands around 1057, 1085 and 1117 cm−1. The spectral region 1200–900 cm−1 contains signals from several components, including DNA and phospholipids, as well as strong absorbance bands from complex sugar modes. Significant, intense bands within this region have been reported for different bacteria and assigned to the cell-wall components [37–40]. FTIR spectra of isolated cell walls from Gram-positive bacteria have previously been reported and show a substantially increased intensity of the aforementioned bands compared to intact organisms [41]. Some of these bands (in particular, approx. 1060 and approx. 1105 cm−1) are attributed to complex sugar modes of peptidoglycans based on isolation of peptidoglycan films from various bacteria (including E. coli, S. aureus and B. subtilis) [40,42,43]. It has been shown that the IR spectra of peptidoglycans isolated from Gram-positive and Gram-negative bacteria are very similar [40] and remain uniform for a wide range of temperatures, independent of the sample preparation [40]. The ATR–FTIR spectrum of the peptidoglycan standard from S. aureus (electronic supplementary material, figure S2) also exhibits bands at 1064 cm−1 and 1109 cm−1, among others. The consistent intensity increase of bands around 1057, 1085, 1117 cm−1 in the spectra of Gram-positive bacteria in comparison with the spectra of Gram-negative bacteria additionally indicates that the bands originate from the cell wall [42,43] and reflect directly its larger thickness in Gram-positive bacteria.
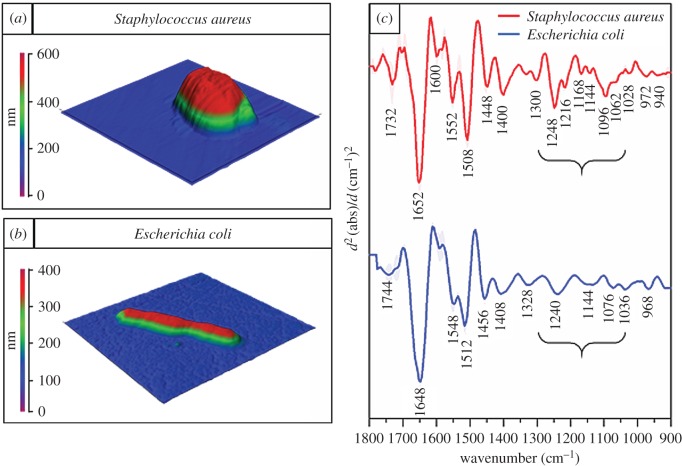
The ATR–FTIR spectra provided basic discrimination between the bacteria, mostly due to differences in their cell walls. However, in order to ensure a high signal to noise (S/N) ratio, complete coverage of the crystal surface with a layer of bacteria of minimal thickness equal to depth of penetration of the evanescent wave, is preferred. The depth of penetration of the evanescent wave is in the range of a few micrometres [44], whereas the thickness of a single bacterium falls in the range of few hundred nanometres. Therefore, each ATR–FTIR spectrum is a result of the average chemical composition of thousands of organisms. Thus, the spectra are dominated by the contributions of cytoplasm and nucleolus components and the spectral contribution from the cell wall of an individual organism is only a fraction of each spectrum. Although in some cases (e.g. Gram-positive versus Gram-negative bacteria) ATR–FTIR enables observations of differences based on the cell-wall contribution to the spectrum, it requires a significant change in the cell-wall composition (or thickness) and therefore is not particularly suitable for studies focused on bacterial cell walls and aiming at observing small changes in this structure. Furthermore, spectra recorded using ATR–FTIR provide information at the population level, without the ability to probe a single bacterium. For this purpose, AFM–IR was applied to specifically target individual bacterial cells and characterize their chemistry. Topographic AFM images of live S. aureus and E. coli together with the second derivatives of AFM–IR spectra of both organisms acquired in the centre of the cells are presented in figure 2. The live states of the bacteria were confirmed by the regrowth of bacterial colonies from plated samples taken after the measurements, which indicate a very high amount of living bacteria even after air-drying. Furthermore, the ability to observe bacteria dividing indicates that the sample preparation protocol provided in vivo conditions. Bacteria by nature show very high resistance to changes in external factors such as pH, temperature and osmotic pressure. It should be noted that bacteria can survive dehydration and in fact it has been shown using ATR–FTIR spectroscopy that DNA in living bacteria can undergo a conformational change from B-DNA (hydrated) to A-DNA (dehydrated). That study also showed that upon rehydration DNA reverts to the B-DNA form and reproduction occurs [45]. In the methodology applied here, the bacteria were not totally dehydrated and hence they could still undergo division.
Figure 2.
AFM–IR results obtained from representative Gram-positive and Gram-negative bacteria. AFM images recorded of live (a) S. aureus and (b) E. coli, showing the size of bacteria. Size of the measured areas: 2.3 × 2.4 µm (S. aureus) and 5.5 × 5.5 µm (E. coli). Colour scales are presented next to each AFM height image. (c) The second derivatives of average AFM–IR spectra of live S. aureus (in red) (n = 54) and E. coli (in blue) (n = 52) together with their standard deviations and with the most prominent bands labelled. Each single spectrum was obtained from the centre of the cell. Parentheses indicate the spectral range where prominent differences related to vibrations of phosphodiester groups and complex carbonyl modes are observed.
Consistent with known morphologies, S. aureus (figure 2a) were observed as round shaped cells of approximately 600 nm diameter, whereas E. coli cells (figure 2b) were rod-shaped with a length of approximately 4 µm and thickness of 400 nm. The second derivatives of AFM–IR spectra of the organisms (figure 2c) showed significant differences, particularly in the low wavenumber region (1300–900 cm−1). Clearly distinguishing features were intense bands located at 1062 and 1096 cm−1, apparent in the spectrum from S. aureus but significantly less intense (or entirely absent) in the spectrum of E. coli. Furthermore, the band located at 1096 cm−1 is significantly broadened towards the higher wavenumber values. The location of these bands correlates well with the ATR–FTIR results. These bands were assigned to complex sugar modes from compounds present in the cell wall (e.g. peptidoglycan layer), as well as contributions from symmetric stretching of phosphodiester groups, originating from phosphate-containing cell wall compounds (e.g. phospholipids and TA) [40–43]. Phosphodiester groups (present in various chemical components, such as DNA, phospholipids and TA/LTA) give rise to bands located at approximately 1080 cm−1 and approximately 1220 cm−1 corresponding to the symmetric and asymmetric PO2− stretching vibrations, respectively. TAs/LTAs are present in the Gram-positive cell wall providing an additional phosphate-containing compound compared to Gram-negative cell walls. This may contribute significantly to the increased intensity of the band at 1096 cm−1 (νsPO2−) along with increased intensities in bands at 1216 and 1248 cm−1 (νasPO2−). These bands may also contain contributions from phospholipids, present in the inner (cytosol) membrane of Gram-positive bacteria (figure 1); however, these contributions to the overall spectral information are expected to be small given this membrane is composed of only a bi-molecular lipid layer and associated proteins. Furthermore, the cytosol membrane (and related phospholipids) are present in all bacteria. In fact, the Gram-negative bacteria contain an additional outer membrane, rich in phospholipids (figure 1). The overall content of phospholipids in the cell wall is, therefore, higher in Gram-negative bacteria compared with Gram-positive bacteria, whereas the phosphodiester-related bands (1096, 1216, 1248 cm−1) are significantly more intense in the spectra of Gram-positive bacteria. This suggests that the phosphate-related bands in the AFM–IR spectrum of S. aureus contain significant contributions from vibrations of phosphodiester groups not related to phospholipids. In the case of E. coli, the intensity of the band from complex sugar modes is significantly smaller, nevertheless still notable (i.e. 1036 cm−1). This is consistent with the reduced thickness of the cell wall (and peptidoglycan layer) in E. coli (approx. 4 nm) compared with S. aureus (approx. 50 nm). However, the spectrum of a Gram-negative bacterium can be characterized not only by the absence (or decreased intensity) of bands originating from carbohydrate and phosphodiester groups of cell-wall components but also by the presence of specific chemical moieties. The band located at 1076 cm−1 indicates the presence of phosphate groups, even though the cell wall of E. coli does not contain TAs/LTAs. The prominent vibrational modes of the phosphate groups (1076 cm−1 and 1240 cm−1) in the spectrum of E. coli can originate from DNA, as well as contain a contribution from cell wall phospholipids. The significant contribution of DNA to the spectrum of E. coli is also manifested through the band from P-O-C stretching vibrations at 968 cm−1 [46]. The thinner cell wall in E. coli results in a larger contribution of signals from components in the cytoplasm, such as DNA. This is also apparent in the high intensity of the amide I band (1648 cm−1) compared with the rest of the bands, suggesting a large protein contribution to the spectrum. The remaining bands in both spectra are assigned to different vibrational modes of other components (mainly proteins) of the cell wall and cytoplasm. Interestingly, the AFM–IR spectrum of S. aureus shows an intense band at 1732 cm−1, more prominent than in case of E. coli. This band originates from a carbonyl stretch and is most commonly attributed to lipids. Even though the Gram-positive cell wall includes the cytosol membrane (containing various lipids), the overall lipid content is higher in the cell wall of Gram-negative bacteria due to the presence of the outer cell wall membrane, which is rich in lipopolysaccharides and phospholipids. The carbonyl band at 1744 cm−1 in the AFM–IR spectrum of E. coli is typical of vibrations of lipid ester groups and is assigned to lipoproteins and phospholipids. The carbonyl mode is more intense in S. aureus than in E. coli, despite the lower content of lipoproteins and phospholipids in Gram-positive cell walls (lack of outer membrane). However, this mode in S. aureus is also shifted to 1732 cm−1, indicating the presence of C=O groups in a different molecular environment to that in E. coli. Therefore, the intense carbonyl mode present in spectra of S. aureus originates from carbonyl groups mainly not related to lipoproteins and phospholipids (as in the case of E. coli). Carbonyl groups can be found in both peptidoglycans and TA (especially in its lipoteichoic acidic form) and their vibrational modes could contribute to the band at 1732 cm−1.
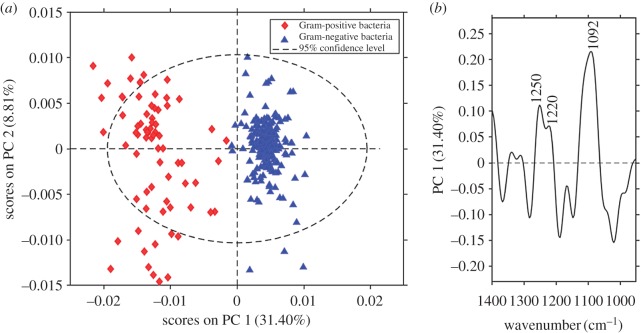
To confirm that the differences observed between AFM–IR spectra of S. aureus and E. coli are related mainly to the cell wall, we investigated four other representative species of Gram-positive (B. subtilis and En. faecalis) and Gram-negative bacteria (P. aeruginosa and A. baumannii). PCA was performed in the spectral range of 1400–950 cm−1 to examine the differences related to the cell wall (complex sugar modes and phosphodiester vibrations) and to avoid the influence of the band at 1640 cm−1 associated with variable water content (present in cytoplasm and related to the different cell wall thicknesses of the studied species) on the analysis. PCA scores and loading plots from the second derivatives of AFM–IR spectra of all studied bacteria are presented in figure 3. The separation of clusters of Gram-positive and Gram-negative bacteria occurs along PC1, which captured 32% of the spectral variance. The loadings plot depicted in figure 3b provides information about the bands involved in separation of the clusters. Several positive bands responsible for the separation are correlated to the positive values of scores for the Gram-positive bacteria; around 1092 cm−1 (carbohydrate modes), 1220 cm−1 (phosphodiester groups) and 1250 cm−1 (phosphodiester groups and amide III), consistent with the differences observed between the average spectra.
Figure 3.
PCA results for the dataset of the second derivatives of AFM–IR spectra. (a) Scores plot (PC1 versus PC2) together with (b) the loadings plot corresponding to PC1. Blue and red markers label AFM–IR spectra of Gram-negative and Gram-positive bacteria, respectively. PCA was performed on the set of AFM–IR spectra (n = 327) from six different bacteria in the spectral range 1400–950 cm−1. Loadings plot corresponding to PC2 is presented in electronic supplementary material, figure S3.
As shown above, the AFM–IR spectrum of S. aureus has a much higher contribution from cell wall signals compared with spectra from the Gram-negative bacterial species used in this study. The ability to observe the cell wall with nanoscale spatial resolution in living bacteria together with knowledge and understanding of AFM–IR spectral characteristics of this structure, provides a platform to observe dynamical changes in living cells. We were interested to see if the nanoscale changes in the cell wall during cell division could be observed and monitored over time with AFM–IR.
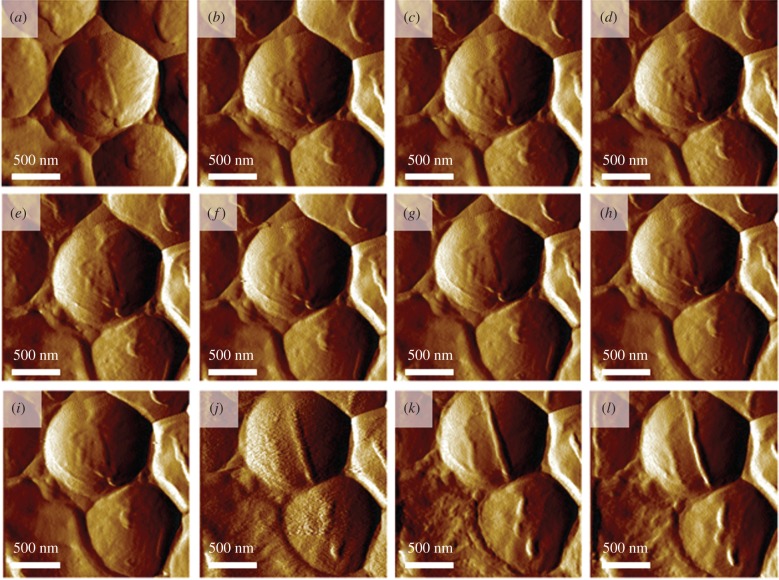
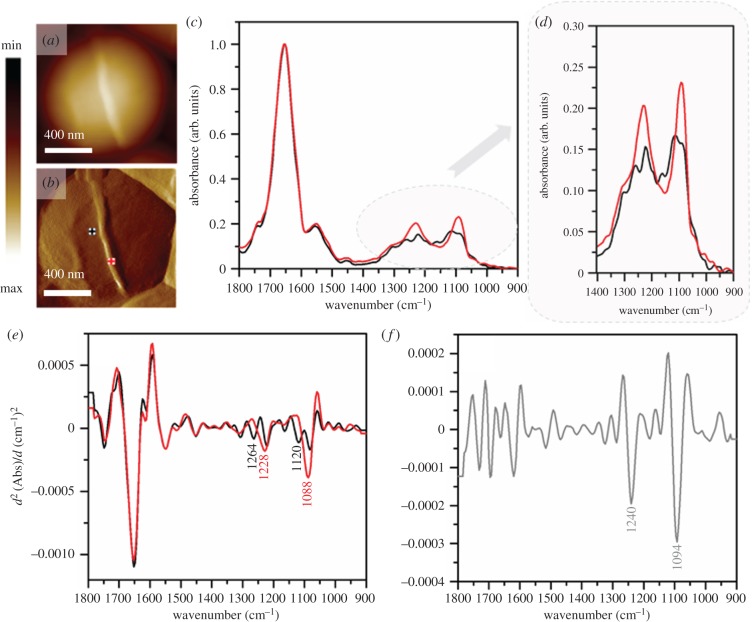
Accordingly, a set of AFM images of S. aureus were recorded within the time frame of 4 h over a group of S. aureus bacteria. These images clearly showed the formation of the cell septum in one of the cells prior to cell division (figure 4a–l). The diameter of the newly forming cell wall was approximately 45 nm. The AFM–IR spectrum recorded from the forming structure (figure 5c–e, red spectrum) clearly exhibits broadening and increased intensity of some bands (centred at 1088 cm−1 and 1228 cm−1 and attributed to carbohydrate and phosphodiester groups of cell-wall components), when compared with a spectrum from an area outside the septum (figure 5c–e, black spectrum). The areas from which spectra were obtained are marked in the AFM images collected directly prior to AFM–IR measurements and presented in figure 5a–b. The second derivative of a differential spectrum, obtained by subtraction of a non-septum spectrum from the septum spectrum, clearly exhibits bands centred at 1092 and 1240 cm−1 (carbohydrate and phosphodiester modes). It is worth noting that both spectra (from septum and non-septum) exhibit an unusual amide I/ amide II ratio. This could possibly result from high intracellular water content (contributing to the intensity of amide I band), although it is more likely artefactual (often observed in AFM–IR spectra). The chemical characterization of the septum structure at this spatial dimension in live bacteria during the division is possible due to the high lateral resolution of AFM–IR, relative to that achievable by conventional IR microspectroscopy, as well as increased density of septum and clearly demonstrates the ability of the new technique to gain molecular insights into biological processes in vivo at the nanoscale.
Figure 4.
S. aureus during division. (a–l) A series of AFM images of S. aureus recorded during division, demonstrating the formation of the septum. Images were recorded every 20 min. Size of the imaged area: 2 × 2 µm. The height of the newly forming septum is 45 nm higher than the rest of the cell. The height profile is shown in electronic supplementary material, figure S4.
Figure 5.
(a) AFM height and (b) AFM deflection image of the dividing cell collected prior to recording AFM–IR spectra at the marked points, from which spectra were recorded (non-septum in black and forming septum in red). Size of the area imaged by AFM: 1.17 × 1.15 µm. (c,d) Comparison of AFM–IR spectra recorded from septum (red) and non-septum (black) of the S. aureus cell during division in the spectral range (c) 1800–900 cm−1 and (d) 1400–900 cm−1. Spectra were normalized to amide I band. (e) The second derivatives of spectra presented in (c), with marked prominent differentiating bands. (f) The second derivative of a differential spectrum obtained by subtraction of non-septum spectrum (c, black) from the septum spectrum (c, red), with prominent bands marked.
Although the plating results do not unambiguously prove that every single bacterium analysed in this study is living, they indicate that the majority remained alive (data not shown). Figure 4 provides unequivocal proof that the bacterium survived AFM imaging and the corresponding AFM–IR spectra are consistent with a thickening septum providing the first example of AFM–IR applied to a living cell.
4. Conclusion
In conclusion, our work demonstrates that AFM–IR is a powerful technique enabling the chemical characterization of a sample with nanometre spatial resolution and is, therefore, particularly suitable for the study of bacteria. We were able to apply AFM–IR to study the dynamical nature of the bacterial cell wall in a live organism at a nanoscale. First, we investigated, in detail, various bacterial species to obtain their spectroscopic characteristics and identified signals from the cell-wall components in single live cells (approx. 1062, 1096, 1216, 1248 cm−1) due to complex carbohydrate modes (e.g. peptidoglycan) and phosphodiester-containing compounds (e.g. LTAs/TAs). We characterized spectroscopically several Gram-positive and Gram-negative representatives and demonstrated their similarity within groups, based primarily on the bands associated with cell wall components. We showed that the contribution of cell wall signals to the acquired spectra is dependent on the cell wall thickness, specifically on the ratio of the cell wall thickness to the overall thickness of bacteria. Therefore, the spectral differences between Gram-positive and Gram-negative bacteria were more distinct in the AFM–IR spectra and remained constant between species. Finally, we were able to observe the dynamical formation of a septum in living S. aureus, occurring prior to its division. This constitutes the first AFM–IR spectrum acquired from a newly forming cell wall of live bacteria during division and demonstrates the ability of the technique to monitor dynamical changes occurring in the bacterial cell wall, using the improved lateral resolution of this technique.
Supplementary Material
Acknowledgements
We acknowledge Mr Finlay Shanks for instrumental support.
Data accessibility
Electronic supplementary material is available online.
Authors' contributions
K.K. prepared samples for spectroscopic measurements, collected and analysed ATR–FTIR and AFM–IR data, performed the bacterial viability assessment and wrote the manuscript. D.P.-G. assisted in data analysis and interpretation. J.P. assisted in collection of AFM–IR data. J.-H.J. prepared the bacterial strains. J.-H.J. and A.Y.P. assisted in data interpretation (microbiology). P.H., D.Mc.N. and B.R.W. supervised and scoped the project. All authors read and corrected the manuscript.
Competing interests
The authors declare no competing interest.
Funding
This work is supported by an Australian Research Council (ARC) Future Fellowship grant FT120100926 and an ARC Discovery Project grant DP180103484.
References
- 1.Golding CG, Lamboo LL, Beniac DR, Booth TF. 2016. The scanning electron microscope in microbiology and diagnosis of infectious disease. Sci. Rep. 6, 26516 ( 10.1038/srep26516) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cefali E, Patane S, Arena A, Saitta G, Guglielmino S, Cappello S, Nicolo M, Allegrini M. 2002. Morphologic variations in bacteria under stress conditions: near-field optical studies. Scanning 24, 274–283. ( 10.1002/sca.4950240601) [DOI] [PubMed] [Google Scholar]
- 3.Mayet C, Dazzi A, Prazeres R, Ortega JM, Jaillard D. 2010. In situ identification and imaging of bacterial polymer nanogranules by infrared nanospectroscopy. Analyst 135, 2540–2545. ( 10.1039/c0an00290a) [DOI] [PubMed] [Google Scholar]
- 4.Mullineaux CW. 2007. Localization and mobility of bacterial proteins by confocal microscopy and fluorescence recovery after photobleaching. Methods Mol. Biol. 390, 3–15. ( 10.1007/978-1-59745-466-7_1) [DOI] [PubMed] [Google Scholar]
- 5.Mullineaux CW. 2007. Localization and mobility of bacterial proteins by confocal microscopy and fluorescence recovery after photobleaching. In Protein targeting protocols (ed. van der Giezen M.), pp. 3–16. Totowa, NJ: Humana Press. [DOI] [PubMed] [Google Scholar]
- 6.Dazzi A, Prater CB. 2016. AFM-IR: technology and applications in nanoscale infrared spectroscopy and chemical imaging. Chem. Rev. 117, 5146–5173. ( 10.1021/acs.chemrev.6b00448) [DOI] [PubMed] [Google Scholar]
- 7.Dazzi A, Prater CB, Hu Q, Chase DB, Rabolt JF, Marcott C. 2012. AFM-IR: combining atomic force microscopy and infrared spectroscopy for nanoscale chemical characterization. Appl. Spectrosc. 66, 1365–1384. ( 10.1366/12-06804) [DOI] [PubMed] [Google Scholar]
- 8.Marcott C, Awatani T, Ye J, Gerrard D, Lod M, Kjollerd K. 2014 Review of nanoscale infrared spectroscopy applications to energy related materials. Spectroscopy Europe26, 19–23.
- 9.Katzenmeyer AM, Holland G, Chae J, Band A, Kjoller K, Centrone A. 2015. Mid-infrared spectroscopy beyond the diffraction limit via direct measurement of the photothermal effect. Nanoscale 7, 17 637–17 641. ( 10.1039/c5nr04854k) [DOI] [PubMed] [Google Scholar]
- 10.Dazzi A, Glotin F, Carminati R. 2010. Theory of infrared nanospectroscopy by photothermal induced resonance. J. Appl. Phys. 107, 124519 ( 10.1063/1.3429214) [DOI] [Google Scholar]
- 11.Ye J, Midorikawa H, Awatani T, Marcott C, Lo M, Kjoller K, Shetty R. 2012. Nanoscale infrared spectroscopy and AFM imaging of a polycarbonate/acrylonitrile-styrene/butadiene blend. Microsc. Anal. 26, 24–27. [Google Scholar]
- 12.Marcott C, Lo M, Kjoller K, Prater C, Noda I. 2011. Spatial differentiation of sub-micrometer domains in a poly (hydroxyalkanoate) copolymer using instrumentation that combines atomic force microscopy (AFM) and infrared (IR) spectroscopy. Appl. Spectrosc. 65, 1145–1150. ( 10.1366/11-06341) [DOI] [PubMed] [Google Scholar]
- 13.Tang F, Bao P, Su Z. 2016. Analysis of nanodomain composition in high-impact polypropylene by atomic force microscopy-infrared. Anal. Chem. 88, 4926–4930. ( 10.1021/acs.analchem.6b00798) [DOI] [PubMed] [Google Scholar]
- 14.Eby T, Gundusharma U, Lo M, Sahagian K, Marcott C, Kjoller K. 2012. Reverse engineering of polymeric multilayers using AFM-based nanoscale IR spectroscopy and thermal analysis. Spectrosc. Eur. 24, 18. [Google Scholar]
- 15.Gong L, Chase DB, Noda I, Liu J, Martin DC, Ni C, Rabolt JF. 2015. Discovery of β-form crystal structure in electrospun poly [(R)-3-hydroxybutyrate-co-(R)-3-hydroxyhexanoate](PHBHx) nanofibers: from fiber mats to single fibers. Macromolecules 48, 6197–6205. ( 10.1021/acs.macromol.5b00638) [DOI] [Google Scholar]
- 16.Akyildiz HI, Lo M, Dillon E, Roberts AT, Everitt HO, Jur JS. 2014. Formation of novel photoluminescent hybrid materials by sequential vapor infiltration into polyethylene terephthalate fibers. J. Mater. Res. 29, 2817–2826. ( 10.1557/jmr.2014.333) [DOI] [Google Scholar]
- 17.Ghosh S, Ramos L, Remita S, Dazzi A, Deniset-Besseau A, Beaunier P, Goubard F, Aubert P-H, Remita H. 2015. Conducting polymer nanofibers with controlled diameters synthesized in hexagonal mesophases. New J. Chem. 39, 8311–8320. ( 10.1039/C5NJ00826C) [DOI] [Google Scholar]
- 18.Bandekar J, Sawyer A. 1996. FT-IR spectroscopic studies of polyurethanes: IV. Studies of the effect of the presence of processing aids on the hemocompatibility of polyurethanes. J. Biomater. Sci. Polym. Ed. 7, 485–501. ( 10.1163/156856295X00553) [DOI] [PubMed] [Google Scholar]
- 19.Sawyer A, Bandekar J, Li H. 1994. Examination of wax on surface of extruded Pellethane by scanning electron microscopy attenuated total reflection-infrared and x-ray photoelectron spectroscopy and its importance in blood compatibility. J. Vac. Sci. Technol. A Vac. Surf. Films 12, 2966–2970. ( 10.1116/1.578923) [DOI] [Google Scholar]
- 20.Janik E, et al. 2013. Molecular architecture of plant thylakoids under physiological and light stress conditions: a study of lipid–light-harvesting complex II model membranes. Plant Cell Online 25, 2155–2170. ( 10.1105/tpc.113.113076) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marcott C, Lo M, Kjoller K, Domanov Y, Balooch G, Luengo GS. 2013. Nanoscale infrared (IR) spectroscopy and imaging of structural lipids in human stratum corneum using an atomic force microscope to directly detect absorbed light from a tunable IR laser source. Exp. Dermatol. 22, 419–421. ( 10.1111/exd.12144) [DOI] [PubMed] [Google Scholar]
- 22.Marcott C, Lo M, Kjoller K, Fiat F, Baghdadli N, Balooch G, Luengo GS. 2014. Localization of human hair structural lipids using nanoscale infrared spectroscopy and imaging. Appl. Spectrosc. 68, 564–569. ( 10.1366/13-07328) [DOI] [PubMed] [Google Scholar]
- 23.Gourion-Arsiquaud S, Marcott C, Hu Q, Boskey AL. 2014. Studying variations in bone composition at nano-scale resolution: a preliminary report. Calcif. Tissue Int. 95, 413–418. ( 10.1007/s00223-014-9909-9) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Clede S, Policar C, Sandt C. 2014. Fourier transform infrared (FT-IR) spectromicroscopy to identify cell organelles: correlation with fluorescence staining in MCF-7 breast cancer cells. Appl. Spectrosc. 68, 113–117. ( 10.1366/13-07139) [DOI] [PubMed] [Google Scholar]
- 25.Baldassarre L, Giliberti V, Rosa A, Ortolani M, Bonamore A, Baiocco P, Kjoller K, Calvani P, Nucara A. 2016. Mapping the amide I absorption in single bacteria and mammalian cells with resonant infrared nanospectroscopy. Nanotechnology 27, 075101 ( 10.1088/0957-4484/27/7/075101) [DOI] [PubMed] [Google Scholar]
- 26.Vitry P, Rebois R, Bourillot E, Deniset-Besseau A, Virolle M-J, Lesniewska E, Dazzi A. 2016. Combining infrared and mode synthesizing atomic force microscopy: application to the study of lipid vesicles inside Streptomyces bacteria. Nano Res. 9, 1674–1681. ( 10.1007/s12274-016-1061-6) [DOI] [Google Scholar]
- 27.Dazzi A, Prazeres R, Glotin F, Ortega JM, Al-Sawaftah M, de Frutos M. 2008. Chemical mapping of the distribution of viruses into infected bacteria with a photothermal method. Ultramicroscopy 108, 635–641. ( 10.1016/j.ultramic.2007.10.008) [DOI] [PubMed] [Google Scholar]
- 28.Sadaie Y, Matsumoto K. 2012. Escherichia coli: and Bacillus subtilis: the frontiers of molecular microbiology revisited, pp.115–148. Thiruvananthapuram, Kerala: Research Signpost. [Google Scholar]
- 29.Beveridge TJ. 1999. Structures of gram-negative cell walls and their derived membrane vesicles. J. Bacteriol. 181, 4725–4733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gan L, Chen S, Jensen GJ. 2008. Molecular organization of Gram-negative peptidoglycan. Proc. Natl Acad. Sci. USA 105, 18 953–18 957. ( 10.1073/pnas.0808035105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang KC, Mukhopadhyay R, Wen B, Gitai Z, Wingreen NS. 2008. Cell shape and cell-wall organization in Gram-negative bacteria. Proc. Natl Acad. Sci. USA 105, 19282–19287. ( 10.1073/pnas.0805309105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hayhurst EJ, Kailas L, Hobbs JK, Foster SJ. 2008. Cell wall peptidoglycan architecture in Bacillus subtilis. Proc. Natl Acad. Sci. USA 105, 14 603–14 608. ( 10.1073/pnas.0804138105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peleg AY, Miyakis S, Ward DV, Earl AM, Rubio A, Cameron DR, Pillai S, Moellering RC Jr, Eliopoulos GM. 2012. Whole genome characterization of the mechanisms of daptomycin resistance in clinical and laboratory derived isolates of Staphylococcus aureus. PLoS ONE 7, e28316 ( 10.1371/journal.pone.0028316) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roberts AP, Hennequin C, Elmore M, Collignon A, Karjalainen T, Minton N, Mullany P. 2003. Development of an integrative vector for the expression of antisense RNA in Clostridium difficile. J. Microbiol. Methods 55, 617–624. ( 10.1016/S0167-7012(03)00200-8) [DOI] [PubMed] [Google Scholar]
- 35.Monk IR, Shah IM, Xu M, Tan MW, Foster TJ. 2012. Transforming the untransformable: application of direct transformation to manipulate genetically Staphylococcus aureus and Staphylococcus epidermidis. MBio 3 e00277-11 ( 10.1128/mBio.00277-11) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Holloway BW. 1955. Genetic recombination in Pseudomonas aeruginosa. J. Gen. Microbiol. 13, 572–581. ( 10.1099/00221287-13-3-572) [DOI] [PubMed] [Google Scholar]
- 37.Helm D, Naumann D. 1995. Identification of some bacterial cell components by FT-IR spectroscopy. FEMS Microbiol. Lett. 126, 75–79. ( 10.1016/0378-1097(94)00529-Z) [DOI] [Google Scholar]
- 38.Helm D, Labischinski H, Naumann D. 1991. Elaboration of a procedure for identification of bacteria using Fourier-transform IR spectral libraries: a stepwise correlation approach. J. Microbiol. Methods 14, 127–142. ( 10.1016/0167-7012(91)90042-O) [DOI] [Google Scholar]
- 39.Naumann D, Fijala V, Labischinski H, Giesbrecht P. 1988. The rapid differentiation and identification of pathogenic bacteria using Fourier transform infrared spectroscopic and multivariate statistical analysis. J. Mol. Struct. 174, 165–170. ( 10.1016/0022-2860(88)80152-2) [DOI] [Google Scholar]
- 40.Naumann D, Barnickel G, Bradaczek H, Labischinski H, Giesbrecht P. 1982. Infrared spectroscopy, a tool for probing bacterial peptidoglycan. Potentialities of infrared spectroscopy for cell wall analytical studies and rejection of models based on crystalline chitin. Eur. J. Biochem. 125, 505–515. ( 10.1111/j.1432-1033.1982.tb06711.x) [DOI] [PubMed] [Google Scholar]
- 41.Naumann D. 1984. Some ultrastructural information on intact, living bacterial cells and related cell-wall fragments as given by FTIR. Infrared Phys. 24, 233–238. ( 10.1016/0020-0891(84)90075-7) [DOI] [Google Scholar]
- 42.Kamnev AA. 2008. FTIR spectroscopic studies of bacterial cellular responses to environmental factors, plant–bacterial interactions and signalling. Spectroscopy 22, 83–95. ( 10.3233/spe-2008-0329) [DOI] [Google Scholar]
- 43.Jiang W, Saxena A, Song B, Ward BB, Beveridge TJ, Myneni SC. 2004. Elucidation of functional groups on Gram-positive and Gram-negative bacterial surfaces using infrared spectroscopy. Langmuir 20, 11 433–11 442. ( 10.1021/la049043%2B) [DOI] [PubMed] [Google Scholar]
- 44.Glassford SE, Byrne B, Kazarian SG. 2013. Recent applications of ATR FTIR spectroscopy and imaging to proteins. Biochim. Biophys. Acta 1834, 2849–2858. ( 10.1016/j.bbapap.2013.07.015) [DOI] [PubMed] [Google Scholar]
- 45.Whelan DR, Hiscox TJ, Rood JI, Bambery KR, McNaughton D, Wood BR. 2014. Detection of an en masse and reversible B- to A-DNA conformational transition in prokaryotes in response to desiccation. J. R. Soc. Interface 11, 20140454 ( 10.1098/rsif.2014.0454) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kochan K, Heraud P, Kiupel M, Yuzbasiyan-Gurkan V, McNaughton D, Baranska M, Wood BR. 2015. Comparison of FTIR transmission and transfection substrates for canine liver cancer detection. Analyst 140, 2402–2411. ( 10.1039/c4an01901f) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Electronic supplementary material is available online.