Supplemental Digital Content is available in the text
Keywords: glycoprotein, HCVcc, lectin microarray, MALDI-TOF-MS, N-linked glycans
Abstract
We used lectin microarray and mass spectrometric analysis to identify the N-linked glycosylation patterns of hepatitis C virus (HCV) particles. HCV J6/JFH-1 chimeric cell culture (HCVcc) in the culture supernatant was concentrated and purified by ultrafiltration and sucrose gradient ultracentrifugation. Twelve fractions were collected from the top and analyzed for viral infectivity and HCV RNA content after sucrose gradient separation. HCV RNA and proteins were separated by ultracentrifugation in a continuous 10% to 60% sucrose gradient to purify viral particles based on their sedimentation velocities. HCVcc particles were found mainly in fractions 6 to 8, as determined by quantitative polymerase chain reaction (qPCR) analysis for HCV RNA and ELISA of the HCV core protein. The N-glycans on HCV proteins were analyzed by lectin microarray and mass spectrometry. We identified that 32 of 37 lectins displayed the positive binding signals and 16 types of N-glycoforms of which the major HCV glycoforms were high mannose-type N-linked oligosaccharides, hybrid N-glycans, and fucosylated N-glycans. Our study provided new detailed information regarding the majority of the glycan–protein profile, complementing to previous findings of glycan–HCV protein interactions.
1. Introduction
Hepatitis C virus (HCV) is a major cause of chronic hepatitis, cirrhosis, and hepatocellular carcinoma and a prophylactic vaccine is not available.[1] New direct-acting antivirals drugs such as sofosbuvir and ledipasvir therapies increase the sustained virological response of up to 95% in patients infected with HCV genotype 1.[2] However, this treatment remains fairly expensive that limiting its application in many developing countries. While a vaccine would be the most cost-effective means of reducing the spread of HCV infection, its development has been complicated by the ability of the virus to escape the immune system.[3,4] A challenge for HCV vaccine development is to identify the conserved epitopes to elicit protective antibodies against the highly diverse virus. Glycan shielding has been found as a possible mechanism by which HCV masks broadly neutralizing epitopes on its viral glycoproteins.[5] The HCV genome encodes at least 10 different proteins including core, envelope proteins E1 and E2, p7, and nonstructural proteins.[6] The envelope proteins E1 and E2 play an important role in host–cell interactions, including receptor binding and internalization for vital biological functions.[7] Both HCV E1 and E2 are highly glycosylated, with up to 5 and 11 N-linked glycosylation sites in E1 and E2, respectively. Glycans play an important role in protein E1 and E2 interacting with cellular receptors and modulating sensitivity to neutralizing antibodies.[8] Some of these glycans are critical for protein folding and/or HCV infectivity. Extent and type of glycans are important factors for virulence and escape from the immune system. Glycoprotein analysis of HCV is problematic due to the lack of a robust cell culture system to amplify HCV in the past years.[9] Relevant studies have mainly used retroviral pseudotyped particles (HCVpp) or HCV envelope glycoprotein construct expression to study these glycans.[10] However, differences between HCVpp and HCV cell culture (HCVcc) have been reported due to differences in the assembly process.[11] HCVcc glycans have not yet been characterized in detail to date. Therefore, it is important to define the extent and type of glycan modification of HCVcc. In the present study, we focused on mapping the precise glycopatterns of HCV envelope glycoproteins by lectin microarray and MALDI-TOF/TOF-MS (matrix-assisted laser desorption/ionization (MALDI) time-of-flight (TOF) mass spectrometry [MS]). The high-throughput glycomic technology was performed to investigate the glycopatterns on the surface of whole virus particles. The N-linked glycosylation results were analyzed in detail. Our findings provided insights into differences in glycan status to develop of glycan-related prophylactic treating strategies for HCV infection.
2. Methods
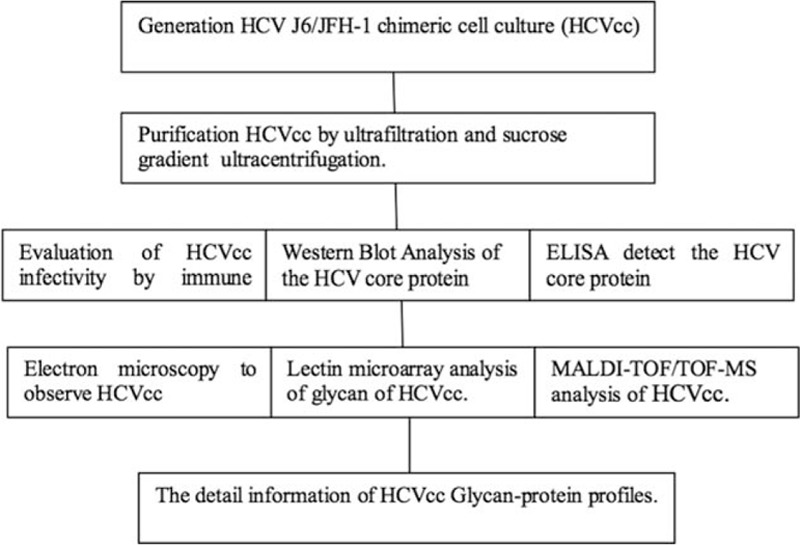
2.1. Generation and purification of cell culture-produced HCV particles
Huh7.5 cells and JFH-1/J6 plasmid were a generous gift from Dr. C. Rice (Laboratory of Virology and Infectious Disease, Center for the Study of Hepatitis C, The Rockefeller University, New York, NY). Huh7.5 cells were cultured in 5% CO2 at 37°C in Dulbecco Modified Eagle Medium (Hyclone, Logan City, UT) containing 10% fetal bovine serum (Gibco, Invitrogen, Carlsbad, CA). The J6/JFH-1 chimeric HCVcc cells were cultured as described in the Supplementary Materials and Methods sections, and as previously reported.[12] Supernatants containing HCVcc were harvested after 72 h, filtered through 0.45-μm-pore membranes and stored at −80°C before being used to infect Huh7.5 cells. J6/JFH-1 chimeric HCVcc was purified by ultrafiltration and ultracentrifugation as previously reported described in the Supplementary Materials and Methods sections.[4] There is no the ethics committee approval because the research only focus on HCVcc in vitro experiments and no involving in any human or animal.
2.2. Evaluation of HCVcc infectivity
HCVcc infectivity was assayed by immunofluorescence in cultured cells. Naive Huh7.5 cells were seeded in 6-well culture plates at 5 × 105 cells/well and infected 5 days later with J6/JFH-1 chimeric HCVcc (multiplicity of infection, 0.2) to determine HCV core protein expression. Naive Huh7.5 cells were served as a negative control. The cells were fixed in 4% paraformaldehyde for 20 min and permeabilized with 0.2% Triton X-100 for 10 min at room temperature (RT). After 3 times washes with PBST, the cells were incubated with phosphate buffer solution (PBS) containing 3% bovine serum albumin (BSA) (Sigma–Aldrich, St Louis, MO). Anti-HCV core protein (Abcam, Hong Kong, China) diluted 200-fold in PBS was added to the cells and incubated overnight at 4°C. Then, Alexa 488-conjugated goat antimouse antibody (Invitrogen) diluted 1000-fold was added to the samples and incubated for 30 min. Antibody binding was detected and visualized by fluorescence microscopy (Olympus, Tokyo, Japan).
2.3. SDS–PAGE and western blot analysis of the HCV core protein
Purified HCVcc was analyzed by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS–PAGE) and western blot. For SDS–PAGE, the sample was mixed with loading buffer containing SDS and boiled for 5 min at 100°C to denature the proteins. After electrophoresis, the gel was stained with silver nitrate (Jiancheng, Nanjing, China). For western blot analysis, the samples were transferred to a nitrocellulose membrane (Millipore, Billerica, MA). The membrane was blocked with 5% (w/v) skim milk in TBST (10 mM Tris–HCl, 150 mM NaCl, 0.2% v/v Tween-20, pH 7.6) at 4°C overnight. The membranes were incubated with mouse monoclonal anti-HCV core protein (Abcam, ab2740) diluted 1000-fold with gentle shaking for 1 h at RT. The membrane was washed 3 times with TBST and then incubated with horseradish peroxidase-labeled goat antimouse IgG (Abcam, ab136815) diluted 5000-fold for 1 h. The signals were developed with ECL Western blot detection reagents (Huamei Biotech, Xi’an, China). The purified HCVcc was observed by electron microscopy after negative staining as described elsewhere.
2.4. ELISA of the HCV core protein
The HCV core proteins were detected according to the protocol supplied with the HCV Core Antigen ELISA Kit (Cell Biolabs, San Diego, CA). One hundred microliters of inactivated sample or HCV-Ag standard was added to a coated plate and incubated at 37°C for 2 h. The microwell strips were washed 5 times with 250 μL of wash buffer per well between each step. Diluted antibody and HRP-conjugated antibody (100 μL) was incubated for 1 h at RT. Substrate solution (100 μL) was incubated for 15 min. The enzyme reaction was stopped by the addition of 100 μL of stop solution to each well, and the OD 450 of each well was determined.
Lectin microarray analysis of glycan of HCVcc. A lectin microarray was utilized to investigate the N-glycans of HCVcc. The purified HCV particles were inactivated by ultraviolet irradiation. Thirty-seven specific sugar-binding lectins were spotted on epoxysilane-coated slides according to our previous publications.[13–15] Each lectin was printed in triplicate per block on one slide. After incubation, the lectin microarrays were scanned using a confocal laser microarray scanner (AXON Instruments, Inc.), and the raw data were acquired with Genepix 6.0 (AXON Instruments, Inc.). Based on the screening criteria, lectin spots with a signal-to-noise ratio (SNR, calculated with Genepix 6.0) ≥2 were retained for further analysis. Detailed information could be found in the Supplementary Materials and Methods sections.
2.5. Identification of N-linked glycans from HCVcc by MALDI-TOF/TOF-MS
The purified HCVcc was lysed with 1% Triton X-100 for 2 h at 37°C, and the detergent-solubilized viral proteins were then transferred to a centrifuge filter (Amicon Ultra-0.5 3 KD device, Millipore) to centrifuge at 12,000g for 15 min. The HCVcc in solution was denatured with 8 M urea, 10 mM DTT, and 10 mM IAM (Sigma–Aldrich) and centrifuged. The ultrafiltration retentate was digested with sequencing-grade trypsin (Promega, Madison, WI) overnight at 37°C. The resulting polypeptides were collected by centrifugation and further digested with PNGase F (New England BioLabs, Ipswich, MA) overnight at 37°C. The envelope protein of purified HCVcc was digested with glycosidase as described in the Supplementary Materials and Methods sections. The released N-linked glycans were collected and desalted using Sepharose CL-4B (Sigma–Aldrich) and MS analysis of the N-linked glycans of HCVcc was conducted by MALDI-TOF/TOF-MS (UltrafleXtreme, Bruker Daltonics, Bremen, Germany) according to a previously described protocol.[16] Measurements were obtained in positive ion mode, and mass-to-charge ratio (m/z) data were annotated using GlycoWorkbench software. The relative intensity was analyzed and generated using FlexAnalysis software (Bruker Daltonics) based on the MALDI-TOF-MS intensity.
2.6. Statistical analysis
Statistical analyses were performed using GraphPad Prism 6 (GraphPad Software, San Diego, CA). The data are expressed as the mean ± standard error of the mean. Comparisons between groups were analyzed using an unpaired t test, and 1-way analysis of variance followed by Tukey test was used for comparisons within groups. The P value < .05 was considered significant.
3. Results
3.1. HCVcc particle purification
The purified HCVcc particles were concentrated by sucrose gradient ultracentrifugation. We found that most of the HCVcc particles were concentrated in 6 to 8 fractions after sucrose gradient centrifugation, as determined by quantitative polymerase chain reaction (qPCR) to detect HCV RNA copies and ELISA to detect HCV core protein (Fig. 1A–C). All fractions collected from the supernatant of uninfected Huh7.5 cells were negative for HCV core protein and RNA (Fig. 1A–C). Sucrose gradient purification removed most of the proteins from the solution of virus particles (Fig. 1D). Core protein from HCVcc was detected by western blot analysis (Fig. 1E). The HCV virions were determined by negative staining and observed by transmission electron microscopy (Fig. 1F).
Figure 1.
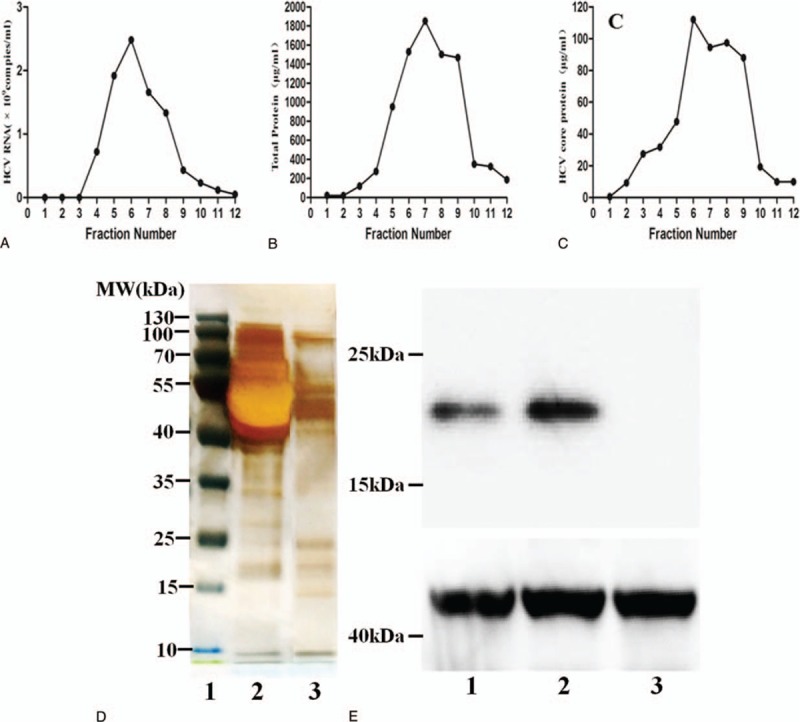
Purification and identification of infectious HCV particles. (A) Concentrated supernatants isolated from infected cells were subjected to ultracentrifugation using a sucrose step gradient. The collected particles were purified in a continuous 10% to 60% sucrose gradient. HCV RNA levels were determined by quantitative polymerase chain reaction, and the results are expressed as the number of HCV RNA copies per mL. (B) Total protein was determined by BCA, and the results are expressed in μg/mL. (C) HCV core protein levels were assessed by ELISA, and the results were expressed in μg/mL. (D) Representative plot of the gradient fractions showing sodium dodecyl sulfate polyacrylamide gel electrophoresis and silver staining (1: Marker, 2: HCVcc, 3: Purify HCVcc). Sucrose gradient purification removed most of the proteins from the solution of virus particles. (E) Western blot analysis of the HCV core protein with a molecular weight of 20 kDa (1: HCVcc, 2: Purify HCVcc, 3: Control). The HCVcc and purify HCVcc were detected with anti-HCV core protein by western blot, which revealed positive band; control: no band was detected in control. (F) The HCV virions were determined by negative staining and observed by transmission electron microscopy. Representative electron micrographs showing specific purified, negatively stained HCV particles. Scale bar, 200 nm. HCV = hepatitis C virus, HCVcc = HCV cell culture.
3.2. Infectivity of HCVcc
To further assess the infectivity of HCVcc, the cells were stained with anti-HCV core protein. We found that positive immunofluorescent staining in Huh7.5 cytoplasm infected with HCVcc after 5 d (Fig. 2). By contrast, no positive staining was detected in uninfected Huh7.5 cells. These findings indicated that Huh7.5 cells were infected with the infectious virus.
Figure 2.
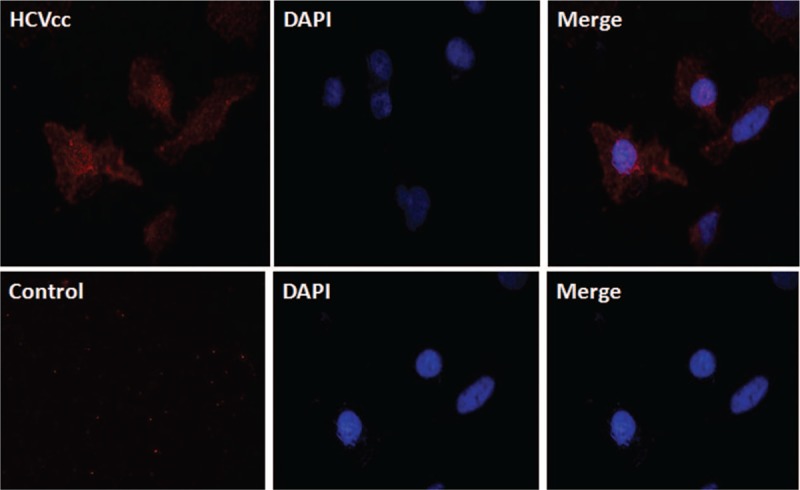
Immunofluorescence of Huh7.5 cells infected with HCVcc and control. Immunofluorescence indicating the HCV core protein (×400). HCVcc: The cells were stained with anti-HCV core protein, which revealed positive immunofluorescent staining in Huh7.5 cytoplasm infected with HCVcc. Control: no staining was detected in naive Huh7.5 cells. The nuclear was stained by DAPI. HCV = hepatitis C virus, HCVcc = HCV cell culture.
3.3. Lectin microarray analysis of N-glycans profile of HCVcc
To further investigate the glycan profile on the surface of HCVcc, we employed a sensitive lectin microarray technique to detect the purified HCV particles as described previously.[17] We found that 32 of 37 lectins displayed the positive binding signals (Fig. 3A). Among these lectins, the fluorescence intensities (FIs) of 4 lectins (GNA, MAL-II, PHA-E, and AAL) exceeded 1000 (Fig. 3A). GNA displayed the strongest signal, indicating that high mannose-type N-glycan was the predominant glycopattern in HCVcc. The other 3 lectins (MAL-II, PHA-E, and AAL) also showed strong binding signals. These results demonstrated that the glycopatterns of Siaα2-3Gal, bisecting GlcNAc, bi-antennary complex-type N-glycans, and α-fucose were highly abundant in HCVcc. Furthermore, 5 lectins (SJA, STL, DSA, VVA, and BPL) displayed the negative binding signals. Based on the specificity of lectins that exhibited positive binding signals, we obtained the following glycan profiles of HCVcc (Fig. 3B). High mannose-type N-glycan was a predominant glycopattern of HCVcc and associated with signals resulting from high mannose binders of GNA, HHL, and NPA. Oligosaccharides modified with sialic acid were also detected on HCVcc, based on the results for MAL-II, SNA, and WGA. In addition, the FIs of MAL-II were greater than those of SNA (approximately 1.8-fold), suggesting a greater abundance of Siaα2-3Gal than Siaα2-6Gal on HCVcc. The fucose content (core fucose, α1, 2/3-fucose) in glycans of HCVcc was evident on AAL, LTL, PSA, and LCA. LEL and PWM indicated the presence of (GlcNAc)2–4 glycopatterns on HCVcc. Among the 32 lectins, 10 bound to Gal/GalNAc, suggesting the presence of galactosylated N-glycans (digalactosylated or monogalactosylated) and O-linked glycans on HCVcc.
Figure 3.
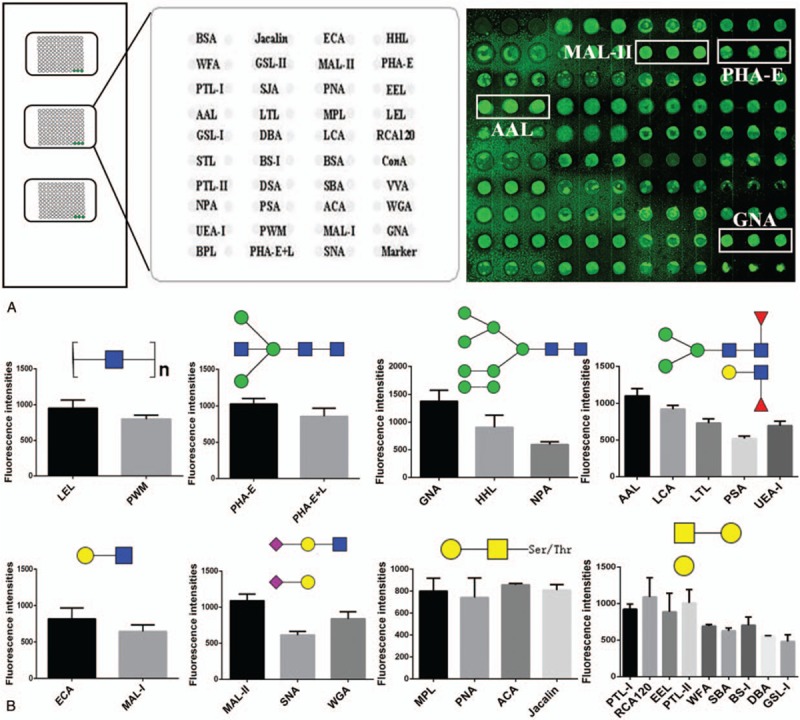
Glycan profiling of HCVcc by lectin microarrays. (A) Binding profile of HCVcc proteins to the lectin microarray. The lectin spots that showed significant binding to Cy-3-labeled HCVcc proteins were framed in white (Left: The layout of 37 lectins microarrays. Cy3 labeled BSA was spotted as location marker and BSA as negative control. Right: HCVcc detected by the lectin microarray). The average fluorescence intensity had significant differences in HCVcc group and control group (P < .05). (B) The FIs of the lectins that exhibited positive signals in the lectin microarrays. The data represent the average FI ± standard error of the mean of 3 biological replicates. BSA = bovine serum albumin, FIs = fluorescence intensities, HCVcc = hepatitis C virus cell culture.
3.4. Analysis of glycans released from HCVcc by PNGase F
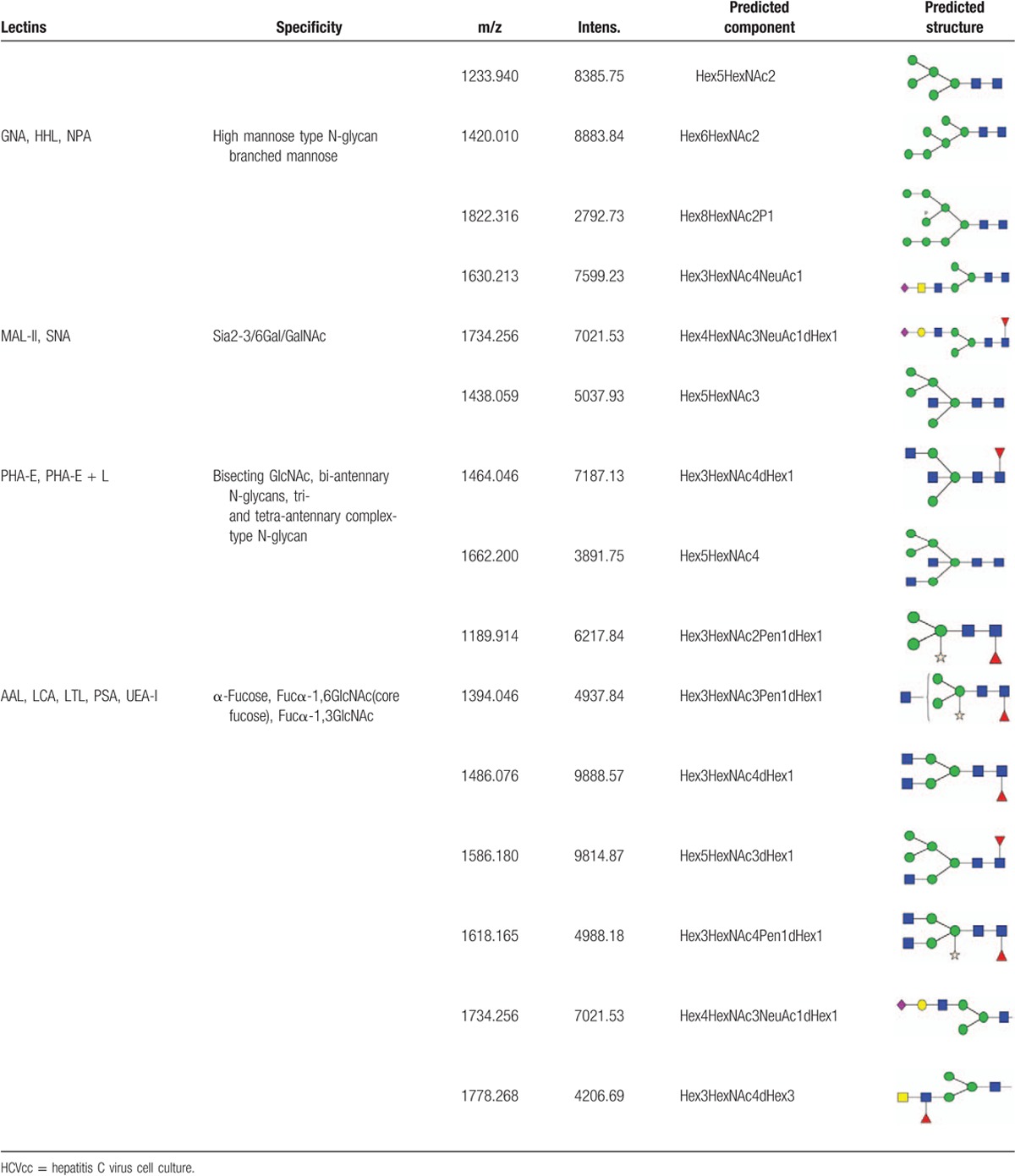
To precisely investigate N-glycan structure and composition, MALDI-TOF-MS spectra were employed to characterize N-linked oligosaccharides on HCVcc. N-linked glycans were released from the HCV proteins by digesting with the glycosidase PNGase F. After purification, the glycans were analyzed by MALDI-TOF-MS. The spectra of the N-linked oligosaccharides on HCVcc with SNR > 3 were annotated using GlycoWorkbench software. The total of 16 distinct m/z N-glycans were detected (Fig. 4). We found that high mannose-type N-glycan was an important class of glycans isolated from HCVcc, represented by the signals at m/z 1233.940((Man)2 + (Man)3(GlcNAc)2), 1420.010((Man)3 + (Man)3(GlcNAc)2), and 1822.316((Man)5 + (Man)3(GlcNAc)2). In addition, the signals at m/z 1438.059 ((Man)2(GlcNAc)1 + (Man)3(GlcNAc)2) and 1586.180((Man)2(GlcNAc)1(Fuc)1 + (Man)3(GlcNAc)2), 1662.200((Man)2(GlcNAc)2 + (Man)3(GlcNAc)2) in the MS spectrum corresponded to a hybrid-type N-glycan structure on HCVcc. N-glycans modified by bisecting GlcNAc were also detected at m/z 1438.059 ((Man)2(GlcNAc)1 + (Man)3(GlcNAc)2), 1464.046 ((GlcNAc)2(Fuc)1 + (Man)3(GlcNAc)2, and 1662.200((Man)2(GlcNAc)2 + (Man)3(GlcNAc)2). Furthermore, N-glycans modified by fucose, particularly core-fucose, were identified by the ion fragments at m/z 1189.914((Man)3(Xyl)1(Fuc)1(GlcNAc)2), 1394.046 ((GlcNAc)1 (Xyl)1(Fuc)1 + (Man)3(GlcNAc)2), 1486.076 ((GlcNAc)2(Fuc)1 + (Man)3(GlcNAc)2), 1586.180 ((Man)2(GlcNAc)1(Fuc)1 + (Man)3(GlcNAc)2), 1618.165 ((GlcNAc)2 (Xyl)1 (Fuc)1 + (Man)3(GlcNAc)2), 1734.256 ((NeuAc)1(Gal)1 (GlcNAc)1 (Fuc)1 + (Man)3(GlcNAc)2), and 1778.268 (Gal)1(GlcNAc)2(Xyl)1(Fuc)1 + (Man)3(GlcNAc)2). Moreover, the ion fragments at 1630.213 and 1734.265 suggested the presence of N-linked glycan modified by Neu5AC, while the presence of a monogalactosylated N-glycan was deduced from the ion at m/z 1778.268.
Figure 4.
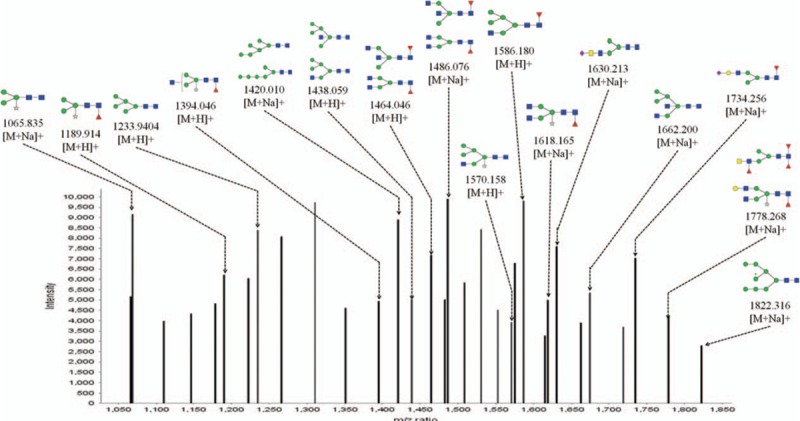
The HCVcc glycan profile was analyzed by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. The spectra of the N-linked oligosaccharides on HCVcc with signal to noise > 3 were annotated. A total of 16 distinct m/z N-glycans were detected. HCVcc = hepatitis C virus cell culture.
In the present study, a lectin microarray and MALDI-TOF-MS were employed to investigate both the composition and linkage of glycans on HCV. The integration of these results enabled a precise characterization of these glycans. A comparative analysis of these results is summarized in Table 1. Both the results of the lectin microarray and MALDI-TOF-MS indicated high mannose-type N-glycan and glycans with branched mannose on HCVcc. The lectin array revealed glycan modified by fucose, while 7-ion fragment peaks demonstrated the presence of fucose-modified N-glycan, particularly core fucosylation of N-linked glycans. Sialylated glycans were deduced by the sialic acid binders MALL-II and SNA, and the ion fragments at m/z 1630.213 and 1734.265 also confirmed this finding. Notably, 13 of the 32 lectins that displayed positive binding signals were Gal/GalNAc binders based on the results of the lectin array. However, only the ion fragment at m/z 1778.268 suggested the presence of monogalactosylated N-glycan. Based on the results of the lectin microarray, the T antigen (core 1 structure of O-linked glycan) binders MPL, PNA, ACA, and Jacalin exhibited positive binding signals, suggesting the presence of O-glycan on HCVcc. The O-linked glycans of HCV should be further characterized in future studies.
Table 1.
Comparative analysis of the HCVcc glycan profile on HCV by lectin microarray and mass spectrometry.
4. Discussion and conclusions
HCP is a positive-strand RNA virus that encodes a polyprotein of approximately 3000 amino acids. This polyprotein is cleaved into 10 viral proteins including 2 transmembrane envelope glycoproteins, E1 and E2, which are heavily N-glycosylated in the N-terminal ectodomains. Previous studies of HCV glycans, HCV pseudo-particles with wild-type glycoproteins and glycosylation mutants focused on HCVpp produced with E1 and E2 heterodimer or the expression of HCV envelope glycoproteins.[18–20] It indicates the importance of N-glycosylation for proper protein folding, receptor binding (e.g., CD81-LEL), HCV cell entry and evasion from the humoral immune response.[11,21–22] Although significant advances have been achieved in the study of HCV glycoproteins,[23–25] there are still no comprehensive studies to characterize the N-linked sugar structures at each of the potential glycosylation sites of HCV to date. Developments in the study of glycoproteins provide some new insights. The HCVcc system mimics the native HCV particle and facilitates the elucidation of the HCV life cycle and the characterization of unmodified glycoproteins.[26,27] This report provides evidence that the glycoproteins on the surface of HCVcc may more closely resemble HCV infection and HCV replicons. Detailed characterization of the N-linked glycan profiles of the HCVcc is helpful for the understanding of elucidating the molecular mechanism of viral entry as well as for the development of entry inhibitors as a novel therapeutic option.
HCV particles were purified from a cell culture of J6/JFH-1 HCV at first for detecting glycan profile on the surface of HCVcc. Briefly, concentrated supernatants were pelleted to ultracentrifugation in a continuous 10% to 60% sucrose gradient to purify viral particles based on their sedimentation velocities. The virus-like particles were observed only in the samples from HCVcc infected cells by HCV core protein immunostaining and HCV RNA qPCR. The majority of HCV RNA was migrated in 6 to 8 fractions (Fig. 1). All fractions collected from the supernatant of uninfected Huh7.5 cells were negative for HCV core protein and RNA.
HCV core protein and viral RNA were detected in the supernatant from HCV-infected naïve Huh7.5 cells (Fig. 1). As reported previously, the virus particles were mainly present in Fraction of 6 to 8 after sucrose gradient centrifugation. HCVcc immunofluorescence using anti-HCV core protein was used to compare HCVcc-transfected cells to the control. The infectivity of purified HCVcc makes the base for subsequent to acquire precise structural information regarding the glycan profile of HCVcc. The integration of the results of these analyses provided precise structural information about the glycans on the surface of HCVcc. High mannose-type N-linked oligosaccharides and N-glycans modified by fucose were the major structures detected. The results revealed that high mannose-type N-linked oligosaccharides and galactosylation were the major glycan structures on the surface of whole HCVcc particle. N-glycans share a common pentasaccharide core region and high mannose-type, complex-type, and hybrid-type. The bisected GlcNAc, bi- and tri-antennary complex-type N-glycans, and fucosylated, sialylated, and monogalactosylated N-glycans were also indicated.
The lectin micorarray and MALDI-TOF-MS were employed for the purified HCV particles. The lectin microarray provided detailed information about the partial structure of the glycans, whereas MS is a powerful analytical tool for the analysis of oligosaccharide composition.[28] We summarized the analyses results including precise structural information of the glycans on the surface of HCV in Fig. 4. Our findings were consistent with a previous report that a high heterogeneity associated with the N-glycans on HCV and that more than a dozen different types of glycans were identified. Based on our lectin arrays, the mannose high-mannose-type binders GNA, HHL, NPA, and ConA existed in HCV. As a result of MS/MS, a total of 16 distinct m/z N-glycans were detected (Fig. 4). The signals at m/z 1233.940, 1420.010, and 1822.316 suggested high mannose-type N-glycan was an important class of glycans isolated from HCVcc. In addition, the signals at m/z 1438.059, 1586.180, and 1662.200 in the MS spectrum corresponded to a hybrid-type N-glycan structure on HCVcc. N-glycans modified by bisecting GlcNAc were also detected at m/z 1438.059, 1464.046, and 1662.200. Furthermore, N-glycans modified by core-fucose were identified by the ion fragments at m/z 1189.914, 1394.046, 1486.076, 1586.180, 1618.165, 1734.256, and 1778.268. Moreover, the presence of N-linked glycan modified by Neu5AC was indicated by the ion fragments at 1630.213 and 1734.265. The lectins PHA-E and PHA-E + L were detected via moderate binding signals, which revealed that bisecting GlcNAc, biantennary or tetraantennary complex-type N-glycans existed in the glycome of HCVcc. Furthermore, α-fucose, especially α1,6-fucose, was present on HCVcc according to the glycans recognized by the lectins AAL, PSA, and LCA. Sialylation of HCV glycans was also identified according to the binding signals of the lectins MAL-II (Siaα2,3-Gal binder) and SNA (Siaα2,6-Gal binder). Moreover, MAL-II was greater than those of SNA (approximately 1.8-fold), suggesting a greater abundance of Siaα2-3Gal than Siaα2-6Gal on HCVcc.
Based on the annotation in Fig. 4, Glycoworkbench apparently assigned many structures deemed improbable for HCVcc glycosylation including the coreA xylosylated pauci mannose structure, core xylosylated complex type biantennary structures, fucosylated lacdiNAc, etc. Signals for several O-glycan binders (MPL, PNA, and ACA, among others) were observed, suggesting that O-linked glycans are present on HCVcc. The O-glycan of HCV should be characterized in future studies (Fig. 5).
Figure 5.
Flow diagram.
Certain lectins exhibit antiviral activity against HIV,[29,30] whereas the lectin-induced immune-mediated selective pressure of the HIV glycan shield has resulted in the loss of glycosylation sites. The HCV glycan shield does not appear to be evolving, and the glycosylation sites on E1 and E2 are highly conserved.[31] The plant-derived lectin GNA efficiently neutralizes human HCV infection and prevents viral entry into host cells.[32] Molecular docking of GNA and HCV glycoproteins (E1 and E2) has indicated that GNA inhibits HCV entry into cells by binding N-linked glycans.[33] Taken together, these data indicate that some glycans of HCV envelope glycoproteins play a major role in the entry of HCV in cells. Thus, lectins are an attractive target for therapeutic intervention.
In summary, the method described herein is effective for the analysis of N-glycan structures on the surface of whole HCV particles and can be applied to other viruses. We provide new detailed information regarding the majority of the glycan–protein profiles, complementing previous glycan–protein studies of HCV proteins. In future studies, we will screen lectins that exhibit a tendency to inhibit the attachment of HCV virions into target cells.
Our results support the development of lectin antivirals that target virion attachment. Several limitations of this study are worth noting. Since the viral glycosylation is dependent on host, an important consideration is how well the Huh7.5 cells transfected with HCV RNA represents the true host. The J6/J F H-1 chimeric HCVcc glycosylation which be purified will reflect that of Huh7.5 cell, which may or may not be the same as that of true infectious HCV infecting natural host. Future trials of lectin with large patient sample sizes are warranted to confirm our findings. Taken together, these results may provide insights into the development of a new therapeutic strategy for combating HCV infection.
Author contributions
Conceptualization: Yonghong Guo, Zhansheng Jia.
Data curation: Yonghong Guo, Hanjie Yu, Yaogang Zhong, Yuan Qin, zheng Li, Zhansheng Jia.
Formal analysis: Yonghong Guo, Hanjie Yu, Yaogang Zhong, Yu He, Yun Zhou, Ying Zhang, zheng Li.
Funding acquisition: Yonghong Guo, Zhansheng Jia.
Investigation: Ying Zhang, zheng Li.
Methodology: Yonghong Guo, Yaogang Zhong, Yu He, Xinmin Qin, Yuan Qin, Peixin Zhang, zheng Li, Zhansheng Jia.
Project administration: Yonghong Guo, Hanjie Yu, Ying Zhang, Zhansheng Jia.
Resources: Yu He, Xinmin Qin, Yuan Qin, Yun Zhou, Peixin Zhang.
Software: Hanjie Yu, Yaogang Zhong.
Supervision: Yonghong Guo.
Validation: Yonghong Guo.
Visualization: Yonghong Guo.
Writing – original draft: Yonghong Guo, Hanjie Yu, zheng Li, Zhansheng Jia.
Writing – review & editing: Yonghong Guo, zheng Li, Zhansheng Jia.
Supplementary Material
Footnotes
Abbreviations: HCVcc = hepatitis C virus cell culture, m/z = mass-to-charge ratio, MALDI-MS = matrix-assisted laser desorption/ionization-mass spectrometry, MS = mass spectrometry, qPCR = quantitative polymerase chain reaction, RT = room temperature, SDS–PAGE = sodium dodecyl sulfate polyacrylamide gel electrophoresis, TOF = time of flight.
Yonghong Guo and Hanjie Yu contributed equally to this work.
The work was supported by the National Natural Science Foundation of China grant (81670577 grant to YG) and Natural Science Foundation of Shaaxi province (2017JM8116 grant to YG).
The authors have no conflicts of interest to disclose.
Supplemental Digital Content is available for this article.
Supplemental digital content is available for this article. Direct URL citations appear in the printed text and are provided in the HTML and PDF versions of this article on the journal's Website (www.md-journal.com).
References
- [1].Thomas DL. Global control of hepatitis C: where challenge meets opportunity. Nat Med 2013;19:850–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Lawitz E, Poordad FF, Pang PS, et al. Sofosbuvir and ledipasvir fixed-dose combination with and without ribavirin in treatment-naive and previously treated patients with genotype 1 hepatitis C virus infection (LONESTAR): an open-label, randomised, phase 2 trial. Lancet 2014;383:515–23. [DOI] [PubMed] [Google Scholar]
- [3].Wong JA, Bhat R, Hockman D, et al. A recombinant HCV envelope glycoprotein vaccine elicits antibodies targeting multiple epitopes on the envelope glycoproteins associated with broad cross-neutralization. J Virol 2014;88:14278–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Akazawa D, Moriyama M, Yokokawa H, et al. Neutralizing antibodies induced by cell culture-derived hepatitis C virus protect against infection in mice. Gastroenterology 2013;145:447–55. [DOI] [PubMed] [Google Scholar]
- [5].Pantua H, Diao J, Ultsch M, et al. Glycan shifting on hepatitis C virus (HCV) E2 glycoprotein is a mechanism for escape from broadly neutralizing antibodies. J Mol Biol 2013;425:1899–914. [DOI] [PubMed] [Google Scholar]
- [6].Reed KE, Rice CM. Overview of hepatitis C virus genome structure, polyprotein processing, and protein properties. Curr Top Microbiol Immunol 2000;242:55–84. [DOI] [PubMed] [Google Scholar]
- [7].Edwards VC, Tarr AW, Urbanowicz R, et al. The role of neutralizing antibodies in hepatitis C virus infection. J Gen Virol 2012;93:1–9. [DOI] [PubMed] [Google Scholar]
- [8].Li Y, Pierce BG, Wang Q, et al. Structural basis for penetration of the glycan shield of hepatitis C virus E2 glycoprotein by a broadly neutralizing human antibody. J Biol Chem 2015;290:10117–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Krey T, d’Alayer J, Kikuti CM, et al. The disulfide bonds in glycoprotein E2 of hepatitis C virus reveal the tertiary organization of the molecule. PLoS Pathog 2010;6:e1000762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Basu A, Meyer K, Ray R. Influence of N-linked glycans on intracellular transport of hepatitis C virus E1 chimeric glycoprotein and its role in pseudotype virus infectivity. Virology 2004;324:273–85. [DOI] [PubMed] [Google Scholar]
- [11].Helle F, Vieyres G, Elkrief L, et al. Role of N-linked glycans in the functions of HCV envelope proteins incorporated into infectious virions. J Virol 2010;84:11905–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Lindenbach BD, Evans MJ, Syder AJ, et al. Complete replication of hepatitis C virus in cell culture. Science 2005;22:623–6. [DOI] [PubMed] [Google Scholar]
- [13].Hu S, Wong DT. Lectin microarray. Proteomics Clin Appl 2009;3:148–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Qin Y, Zhong Y, Dang L, et al. Alteration of protein glycosylation in human hepatic stellate cells activated with transforming growth factor-beta 1. J Proteomics 2012;75:4114–23. [DOI] [PubMed] [Google Scholar]
- [15].Zhong Y, Qin Y, Yu H, et al. Avian influenza virus infection risk in humans with chronic diseases. Sci Rep 2015;5:8971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Reiding KR, Blank D, Kuijper DM, et al. High-throughput profiling of protein N-glycosylation by MALDI-TOF-MS employing linkage-specific sialic acid esterification. Anal Chem 2014;86:5784–93. [DOI] [PubMed] [Google Scholar]
- [17].Zhong Y, Guo Y, Liu X, et al. Serum glycopatterns as novel potential biomarkers for diagnosis of acute-on-chronic hepatitis B liver failure. Sci Rep 2017;7:45957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Whidby J, Mateu G, Scarborough H, et al. Blocking hepatitis C virus infection with recombinant form of envelope protein 2 ectodomain. J Virol 2009;83:11078–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Iacob RE, Perdivara I, Przybylski M, et al. Mass spectrometric characterization of glycosylation of hepatitis C virus E2 envelope glycoprotein reveals extended microheterogeneity of N-glycans. J Am Soc Mass Spectrom 2008;19:428–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Goffard A, Callens N, Bartosch B, et al. Role of N-linked glycans in the functions of hepatitis C virus envelope glycoproteins. J Virol 2005;79:8400–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Boisvert M, Zhang W, Elrod EJ, et al. Novel E2 glycoprotein tetramer detects hepatitis C virus-specific memory B cells. J Immunol 2016;197:4848–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Helle F, Goffard A, Morel V, et al. The neutralizing activity of anti-hepatitis C virus antibodies is modulated by specific glycans on the E2 envelope protein. J Virol 2007;81:8101–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Chen PC, Chuang PK, Chen CH, et al. Role of N-linked glycans in the interactions of recombinant HCV envelope glycoproteins with cellular receptors. ACS Chem Biol 2014;9:1437–43. [DOI] [PubMed] [Google Scholar]
- [24].Bräutigam J, Scheidig AJ, Egge-Jacobsen W. Mass spectrometric analysis of hepatitis C viral envelope protein E2 reveals extended microheterogeneity of mucin-type O-linked glycosylation. Glycobiology 2013;23:453–74. [DOI] [PubMed] [Google Scholar]
- [25].Albecka A, Monserret R, Krey T, et al. Identification of new functional regions in hepatitis C virus envelope glycoproteins E2. J Virol 2011;85:1777–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Zhang P, Zhong L, Struble EB, et al. Depletion of interfering antibodies in chronic hepatitis C patients and vaccinated chimpanzees reveals broad cross-genotype neutralizing activity. Proc Natl Acad Sci USA 2009;106:7537–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Helle F, Duverlie G, Dubuisson J. The hepatitis C virus glycan shield and evasion of the humoral immune response. Viruses 2011;3:1909–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Kobayashi Y, Masuda K, Banno K, et al. Glycan profiling of gestational choriocarcinoma using a lectin microarray. Oncol Rep 2014;31:1121–6. [DOI] [PubMed] [Google Scholar]
- [29].Shahzad-Ul-Hussan S, Gustchina E, Ghirlando R, et al. Solution structure of the monovalent lectin microvirin in complex with Man((1-2)Man provides a basis for anti-HIV activity with low toxicity. J Biol Chem 2011;286:20788–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Hoorelbeke B, Van Damme EJ, Rouge P, et al. Differences in the mannose oligomer specificities of the closely related lectins from Galanthus nivalis and Zea mays strongly determine their eventual anti-HIV activity. Retrovirology 2011;8:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Bertaux C, Daelemans D, Meertens L, et al. Entry of hepatitis C virus and human immunodeficiency virus is selectively inhibited by carbohydrate-binding agents but not by polyanions. Virology 2007;366:40–50. [DOI] [PubMed] [Google Scholar]
- [32].Kachko A, Loesgen S, Shahzad-Ul-Hussan S. Inhibition of hepatitis C virus by the cyanobacterial protein Microcystis viridis lectin: mechanistic differences between the high-mannose specific lectins MVL, CV-N, and GNA. Mol Pharm 2013;10:4590–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Ashfaq UA, Masoud MS, Khaliq S. Inhibition of hepatitis C virus 3a genotype entry through Glanthus nivalis agglutinin. Virol J 2011;8:248. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


