Summary
The PRC2 and PRC1 complexes are aberrantly expressed in human cancers that have been linked with decreases in patient survival. MUC1-C is an oncoprotein that is overexpressed in diverse human cancers and is associated with a poor prognosis. Recent studies have supported a previously unreported function for MUC1-C in activating PRC2 and PRC1 in cancer cells. In the regulation of PRC2, MUC1-C (i) drives transcription of the EZH2 gene, (ii) binds directly to EZH2, and (iii) enhances occupancy of EZH2 on target gene promoters with an increase in H3K27 trimethylation. Regarding PRC1, which is recruited to PRC2 sites in the hierarchical model, MUC1-C induces BMI1 transcription, forms a complex with BMI1, and promotes H2A ubiquitylation. MUC1-C thereby contributes to the integration of PRC2- and PRC1-mediated repression of tumor suppressor genes, such as CDH1, CDKN2A, PTEN and BRCA1. Like PRC2 and PRC1, MUC1-C is associated with the epithelial-mesenchymal transition (EMT) program, the cancer stem cell (CSC) state, and the acquisition of anticancer drug resistance. In concert with these observations, targeting MUC1-C downregulates EZH2 and BMI1, inhibits EMT and the CSC state, and reverses drug resistance. These findings emphasize the significance of MUC1-C as a therapeutic target for inhibiting aberrant PRC function and reprogramming the epigenome in human cancers.
Keywords: MUC1-C, PRC2, PRC1, epigenome, EMT, CSC, DNA repair
Polycomb complexes and repression of gene expression
The Polycomb Group (PcG) proteins constitute Polycomb Repressive Complexes (PRCs; PRC1, PRC2) that function as epigenetic suppressors of gene expression in cell fate, development and cancer 1–3. The PRCs are recruited throughout the genome by transcription factors and non-coding RNAs, and at sites with CpG islands 4, 5. Components of PRC2 are EZH2, SUZ12 and EED, among others 6. EZH2 is a histone methyltransferase, which in association with SUZ12 and EED, catalyzes the mono-, di- and tri-methylation of histone H3 on K27 (H3K27me1, H3K27me2 and H3K27me3) and thereby the repression of target genes 7. In the canonical hierarchical model, H3K27me3 marks function as sites for the recruitment of PRC1, such that PRC2 and PRC1 co-localize with the resulting maintenance of chromatin in a transcriptionally suppressed state 4. BMI1 is a component of PRC1, which in concert with RING1 binds to the catalytic RING 2 subunit to form a ubiquitin E3 ligase 4, 8. In this way, PRC1 catalyzes the ubiquitylation of histone H2A and confers silencing of homeobox (HOX) genes and the CDNK2A locus, which encodes the p16INK4a and p14ARF tumor suppressors 8–10. In addition to the recruitment of PRC1 to sites of H3K27 trimethylation, PRC2 interacts with DNA methyltransferases (DNMTs) and directly controls DNA methylation with the downregulation of tumor suppressor genes (TSGs), including CDH1 11–13. PRC2 thus integrates H3K27 trimethylation with recruitment of PRC1 and DNMTs in a hierarchical program of epigenetic gene silencing. Of note, the canonical model is likely an oversimplification given the complexities of PRC interactions that are cell context dependent 3, 4.
Aberrant expression of PcG proteins in cancer
EZH2 is overexpressed in human tumors and promotes the proliferation of transformed human cells 14. Increases in EZH2 have been associated with aggressive breast cancers with a poor prognosis 15–19. Overexpression of EZH2 has also been linked to poor outcomes for patients with non-small cell lung cancer (NSCLC), prostate cancer and other types of carcinomas 20–26. Like EZH2, BMI1 is overexpressed in breast, lung and other carcinomas, and is associated with poor survival outcomes 8, 27–29. In this context, BMI1 induces a gene signature that is associated with highly invasive tumors which are resistant to treatment 30. Based on these findings, EZH2 and BMI1 have emerged as attractive targets for cancer treatment. Tazemetostat, CPI-1205 and GSK2816126 are SAM-competitive inhibitors of EZH2, which are presently in early stages of evaluation for the treatment of different cancer types 26, 31. PTC-209 is an inhibitor of BMI1 expression that is active preclinically against colorectal and lung adenocarcinomas 32, 33, but has yet to undergo clinical evaluation. Another potential approach for inhibiting EZH2, BMI1 or other essential PRC components is to target upstream effectors that contribute to their aberrant expression in cancer. Along these lines, E2F and MYC have been identified as activators of EZH2 and BMI1 transcription, respectively 14, 34. Moreover, recent studies have shown that the oncogenic MUC1-C protein drives EZH2 35 and BMI1 36 expression and thereby contributes to their regulation of the epigenome in human cancer cells.
MUC1-C transduces stress signals from the cell membrane to the nucleus
Mucin 1 (MUC1) emerged in mammals to protect the integrity of polarized epithelia from stress at the interface with the external environment 37, 38. Implicit to our understanding of MUC1 is that it encodes two subunits in epithelial cells; an extracellular N-terminal subunit that contributes to a physical mucous barrier at apical borders, and a C-terminal transmembrane subunit (MUC1-C) that activates stress responses for repair, proliferation and survival of the critical epithelial cell layer 37, 39. MUC1 is overexpressed in diverse human carcinomas 37, 39. Moreover, the initial finding that the MUC1-C subunit induces transformation 40 provided the basis for defining how MUC1-C functions as an oncoprotein 39, 41. MUC1-C interacts with receptor tyrosine kinases (RTKs), such as EGFR 42, 43, FGFR3 44, PDGFR 45, MET 46 and HER2 47 and, in certain settings, activates the downstream AKT and ERK signaling pathways 39, 41. As one example, MUC1-C→AKT signaling increases glucose uptake, lactate production and pyruvate kinase M2 activity 48, findings in concert with the stimulation of glycolysis. In addition, MUC1-C→AKT signaling upregulates the TIGAR protein, providing further evidence that MUC1-C controls the glycolytic and pentose phosphate pathways 49–51. MUC1-C also interacts with the cell membrane xCT light chain of the cysteine/glutamate transporter and thereby contributes to the dependency of cancer cells on glutamine metabolism 52, 53.
In addition to its impact on cell membrane signaling, MUC1-C is imported to the nucleus 54, where it associates with multiple transacting factors, including β-catenin/TCF4 55, 56, p53 57, NF-κB p65 58, STAT1/3 59, 60 and HIF-1α 61, 62. In this respect, MUC1-C activates gene signatures associated with tumorigenesis, which are significantly predictive of clinical outcomes 63, 64. MUC1-C induces gene signatures linked to metabolic reprogramming 53, 61, 65. The interaction between MUC1-C and NF-κB is of interest in that MUC1-C activates the proinflammatory TAK1→IKK→NF-κB p65 pathway, binds directly to these effectors and promotes the induction of NF-κB target genes 58, 66, 67. For example, MUC1-C associates with NF-κB p65 on the ZEB1 promoter with the induction of ZEB1 transcription 68. MUC1-C also binds directly to ZEB1 and contributes to the function of ZEB1 as an EMT-inducing transcription factor by suppressing miR-200c expression 68. These direct interactions with transcription factors and the initial observations that MUC1-C recruits the p300 histone acetyltransferase to target gene promoters 56, 69 supported the notion that this oncoprotein plays a role in reprogramming the epigenomes of cancer cells.
MUC1-C activates EZH2 expression and PRC2 function
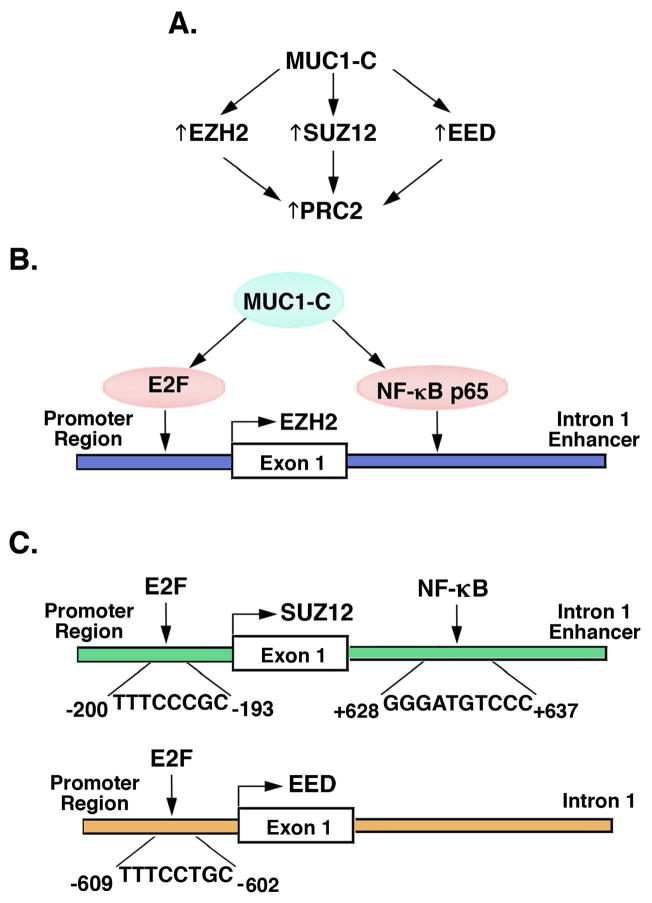
Aberrant EZH2 expression has been linked to breast and other types of carcinomas 7. Downregulating MUC1-C with genetic and pharmacologic approaches in triple-negative breast cancer (TNBC) cells results in the suppression of EZH2 expression and, interestingly, that of SUZ12 and EED, demonstrating that MUC1-C induces multiple components of the PRC2 complex (Fig. 1A) 35. MUC1-C is also necessary for EZH2 expression in NSCLC and prostate cancer cells, indicating that this MUC1-C function is broadly applicable to different types of carcinomas. MUC1-C has been associated with the activation of CDK4 and phosphorylation of pRB 69. In this way, MUC1-C induces EZH2 transcription by a pRB→E2F-mediated mechanism (Fig. 1B) 35. MUC1-C also enhances EZH2 transcription by associating with NF-κB p65 on NF-κB consensus sites in the EZH2 intron 1 enhancer region (Fig. 1B). Interestingly, MUC1-C-induced EZH2 expression is mediated by both E2F and NF-κB p65 35. The EZH2 promoter and enhancer regions include CpG islands with CTCF binding motifs that may contribute to a promoter-enhancer loop structure in an insulated neighborhood (Fig. 1B). MUC1-C also activates (i) SUZ12 by a mechanism involving E2F and NF-κB, and (ii) EED by E2F, but not NF-κB (Fig. 1C). Of further interest, the MUC1-C cytoplasmic domain interacts directly with EZH2 through binding to the EZH2 CXC region adjacent to the catalytic SET domain (Figs. 2A and 2B). The functional significance of MUC1-C binding to EZH2 is supported by the demonstration that MUC1-C increases CDH1 promoter H3K27 trimethylation in concert with repression of E-cadherin expression (Fig. 2C), a hallmark for passage through the EMT program. Breast cancer cells that overexpress EZH2 tend to have associated EMT gene signatures and phenotypic characteristics of cell invasion and metastases 17, 70. Aberrant EZH2 expression has also been linked to BRCA1 downregulation and DNA repair defects 71, 72. Importantly, the MUC1-C→EZH2 pathway is upstream to repression of the BRCA1 and RAD51 genes, which encode essential components of the homologous recombination (HR) DNA repair pathway 35. These findings and the suppression of other DNA damage response pathways, including non-homologous end-joining (NHEJ), have provided new insights into potential a potential role for MUC1-C in promoting genomic instability of cancer cells 35.
Figure 1. MUC1-C induces expression of PRC2 components, EZH2, SUZ12 and EED.
A. Targeting MUC1-C genetically or pharmacologically with the GO-203 inhibitor downregulates EZH2, SUZ12 and EED expression in TNBC, NSCLC and prostate cancer cells, demonstrating that MUC1-C induces multiple PRC2 components. B. MUC1-C drives EZH2 transcription by two mechanisms; (i) E2F-mediated activation of the EZH2 promoter, and (ii) binding of NF-κB p65 complexes to consensus sites in the EZH2 intron 1 enhancer region 35. The EZH2 promoter and enhancer regions include CpG islands (−1046 to −56; +161 to +914) 119 and CTCF binding sites (−603 to −598, −468 to −463; +292 to +297, +702 to +707) for forming a potential loop structure by the CTCF-cohesin complex. C. MUC1-C also activates SUZ12 and EED transcription by E2F, in concert with previous work 14, 120, and by NF-κB p65 (unpublished data). MUC1-C thereby integrates EZH2, SUZ12 and EED expression by E2F signaling and by the inflammatory NF-κB pathway.
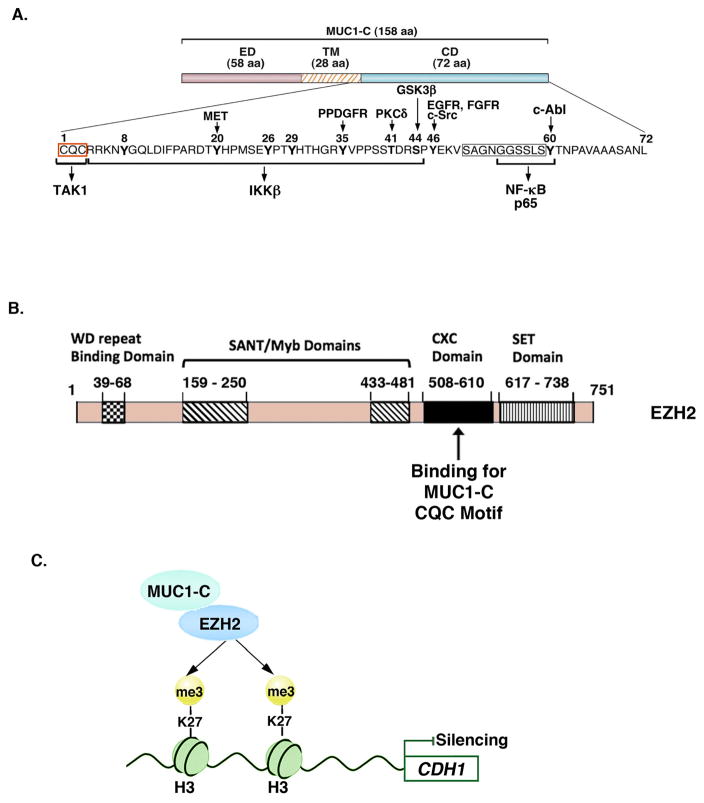
Figure 2. MUC1-C binds directly to EZH2 and promotes EZH2-induced H3K27 trimethylation.
A. Structure of the MUC1-C subunit, which includes a 58-aa extracellular domain (ED) and a 28-aa transmembrane domain (TM). The MUC1-C 72-aa cytoplasmic domain (CD) includes a CQC motif located immediately following the TM region that is necessary for MUC1-C homodimerization in the response to oxidative stress and for nuclear import 39, 54, 108. The CQC motif is the target for the GO-203 inhibitor. The remainder of the MUC1-C cytoplasmic domain is an intrinsically disordered region (IDR). IDRs have been identified in other oncoproteins, such as p53, that function as nodes for the integration of signaling cascades and are also prevalent in transcription factors and the transcriptional coactivators, p300 and CBP 109, 121. The MUC1-C cytoplasmic domain IDR is modified by diverse kinases and, as highlighted, interacts directly with multiple effectors of the inflammatory NF-κB p65 pathway 39, 108. The MUC1-C cytoplasmic domain CQC and the SAGNGGSSLS (boxed) motifs also bind directly to TCF4 and β-catenin, respectively, with activation of the WNT pathway. B. Schema of the EZH2 protein and the indicated domains. The MUC1-C CQC motif binds directly to the EZH2 CXC domain 35. C. MUC1-C forms a complex with EZH2, enhances EZH2 occupancy on the CDH1 promoter, and thereby increases H3K27 trimethylation with repression of E-cadherin expression 35.
MUC1-C integrates PRC2 with PRC1 and DNA methylation
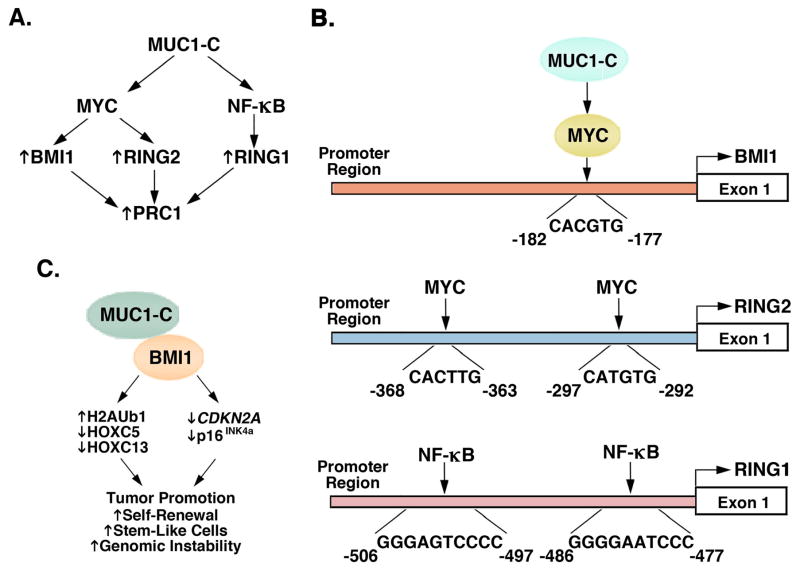
In the hierarchical model described above, a CBX-containing protein in PRC1 binds to the PRC2-mediated H3K27me3 mark with recruitment of PRC1 at PRC2-specified target sites 3, 7. As found for PRC2, MUC1-C also induces expression of the PRC1 components, BMI1, RING1 and RING2 36. MUC1-C interacts with the β-catenin/TCF4 pathway 55, 56, 73 and drives the WNT target gene, MYC 69, 74. In turn, MYC activates BMI1 and RING2 (Figs. 3A and 3B)36, 75. In contrast, MUC1-C→NF-κB p65 signaling induces RING1 expression (Figs. 3A and 3B) 36, 75. Interestingly, and as reported for EZH2, the MUC1-C cytoplasmic domain interacts directly with BMI1 36. In addition, MUC1-C promotes H2A ubiquitylation, downregulation of HOX genes, and BMI1 occupancy on the CDKN2A promoter, supporting a direct role in repressing BMI1 target genes (Fig. 3C). Consistent with this paradigm, targeting MUC1-C induces the p16INK4a tumor suppressor 36. These findings collectively support the premise that MUC1-C represses TSGs by activating and integrating PRC2 and PRC1 functions.
Figure 3. MUC1-C drives expression of PRC1 components, BMI1, RING1 and RING2.
A. MUC1-C activates (i) BMI1 and RING2 by MYC-mediated mechanisms, and (ii) RING1 through the NF-κB p65 pathway 36. Thus, targeting MUC1-C is associated with downregulation of BMI1, RING2 and RING1 expression in TNBC and NSCLC cells 36. B. Schemas of the BMI1, RING2 and RING1 promoters with highlighting of the MYC and NF-κB binding sites. C. MUC1-C binds directly to BMI1 by an interaction dependent of the MUC1-C CQC motif 36. Complexes of MUC1-C and BMI1 have been detected on the CDKN2A promoter 36. In support of the depiction, targeting MUC1-C genetically or with the GO-203 inhibitor (i) decreases H2A ubiquitylation, (ii) increases HOXC5 and HOXC13 expression, and (iii) activates CDKN2A and expression of p16INK4a 36. The findings thus support a role for MUC1-C in contributing to BMI1-driven tumor promotion, self-renewal capacity, the CSC state, and genomic instability.
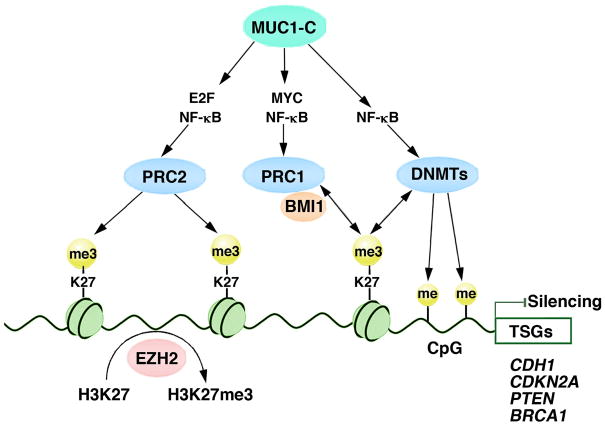
PcG-mediated repression of gene expression has been linked to the DNA methylation process 11, 76. Specifically, EZH2 interacts with DNMTs and is necessary for methylation of EZH2-target gene promoters 11. The H3K27me3 mark recruits DNMTs to CpG islands, leading to DNA methylation 12. Of potential relevance here is that MUC1-C→NF-κB p65 signaling activates the DNMT1 and DNMT3b genes in human cancer cells 77, 78. Targeting MUC1-C also induces changes in DNA methylation patterns in concert with depression of the CDH1, PTEN and BRCA1 TSGs 77, 78. These findings, when taken with the demonstration that MUC1-C activates EZH2, hold potentially important implications for MUC1-C involvement in integrating the PRC and DNA methylation systems in repression of TSG expression (Fig. 4).
Figure 4. Schema of the proposed model in which MUC1-C integrates functions of PRC2, PRC1 and DNA methylation in the repression of tumor suppressor genes.
In this model, MUC1-C induces expression of the PRC2 components, EZH2, SUZ12 and EED, and thereby drives H3K27 trimethylation on target gene promoters 35. In addition, MUC1-C induces expression of the PRC1 components, BMI1, RING2 and RING1, and in turn the potential recruitment of PRC1 to H3K27me3 sites 36. In addition, MUC1-C activates DNMT1 and DNMT3b expression and changes in DNA methylation patterns 77. EZH2 and the H3K27me3 mark recruit DNMTs, leading to DNA methylation 11, 12. In concert with this model of gene repression, targeting MUC1-C with the downregulation of EZH2 35, BMI1 36, 75, and DNMT1/3b 77 is associated with induction of the target CDH1, CDKN2A, PTEN and BRCA1 TSGs.
MUC1-C activates PRCs in concert with induction of the EMT program
A considerable body of evidence has supported the premise that epigenetic regulatory mechanisms control epithelial-mesenchymal plasticity in cancer 79. For instance, aberrant EZH2 expression promotes EMT, invasion and metastasis in diverse carcinomas 31. As a result, targeting EZH2 with an inhibitor downregulates EMT signaling 80. MUC1-C drives EZH2 expression, and increases EZH2 occupancy and H3K27 trimethylation on the CDH1 promoter 35. These findings and the role of MUC1-C in suppressing E-cadherin expression have given traction for the concept that MUC1-C could integrate PRC2 function with induction of the EMT program 35. Along these lines, MUC1-C induces EMT by activating the inflammatory TAK1→IKK→NF-κB pathway, which in turn drives ZEB1 and repression of the ZEB1-target gene miR-200c 68. The MUC1-C/ZEB1 interaction has also been associated with repression of the CDH1 gene, and downregulation of genes encoding cell polarity factors, such as CRB3, necessary for apical-basal polarity 68, 81. Moreover and in concert with induction of EZH2 and EMT, MUC1-C has been widely linked to cancers with more aggressive, invasive and metastatic phenotypes 37, 39.
Of additional interest, EZH2 and the EMT program are closely associated with the cancer stem cell (CSC) state by presently unclear mechanisms 82–84. Indeed, the role of MUC1-C in activating PRC2 and PRC1 may provide new insights into this association. EZH2 is linked to EMT, invasion and metastases 31. Additionally, aberrant BMI1 expression promotes self-renewal and tumorigenic potential of CSCs 8, 32, 85, 86. BMI1 has also been linked to EMT induction 8, 87, 88, and EZH2 to the CSC state 84, further strengthening the associations among PRCs, EMT and CSCs. Along these lines, MUC1-C signaling could contribute to the integration of PRCs, EMT and the CSC state. As such, in a simplified model, MUC1-C-induced PRC2 activation contributes to EMT induction of EMT and recruitment of PRC1 to PRC2 target sites. In turn, MUC1-C-induced activation of PRC1 promotes acquisition of the CSC state. In support of such a model, targeting MUC1-C in cancer cells results in downregulation of PRCs, reversal of the EMT phenotype and decreases in self-renewal capacity 68, 81, 89, 90.
MUC1-C is a target for inhibiting immune evasion in cancer
The EMT program has been linked to the induction of programmed death ligand 1 (PD-L1) expression and adverse clinical outcomes in patients with breast, lung and other types of cancers 91–93, suggesting that immune evasion promotes an invasive and metastatic phenotype. Immune evasion of tumors has also been associated with EZH2-mediated suppression of chemokines and effector T-cell recruitment 94, 95. Accordingly, the involvement of MUC1-C in inducing EMT and EZH2 invoked a possible role in immune evasion. Indeed, recent work has shown that MUC1-C→NF-κB signaling, which drives EZH2 35, EMT 68, and the CSC state of self-renewal 81, 89, 90, also induces the CD274 gene and PD-L1 expression 96. In addition, the MUC1-C→NF-κB→ZEB1 pathway represses effectors of the immune response, including IFNγ and GM-CSF 96. Inhibiting MUC1-C function with GO-203 thus results in downregulation of PD-L1, induction of IFNγ, and activation of anti-tumor immunity in an immune competent MUC1 transgenic mouse model 97. MUC1-C also protects cancer cells from killing by TRAIL, FAS ligand and perforin/granzyme B-mediated lysis 98, 99. These findings hold important implications for MUC1-C in the accumulating evidence that EMT and CSCs are associated with the induction of PD-L1 and other events linked to immune evasion 92, 100, 101.
MUC1-C confers anticancer drug resistance
The association between EMT and the CSC state has also been linked to development of drug resistance by mechanisms that have been largely unexplored 83, 102. MUC1-C contributes to the development of resistance to cytotoxic 62, 103 and targeted 47, 90, 104 agents. How MUC1-C has the capacity to promote such pleiotropic mechanisms for anticancer drug resistance is in part related to activation of multidrug resistance (MDR) genes, including ABCB1, which encodes the P-glycoprotein 105. MUC1-C-induced drug resistance could also be related to epigenetic regulation of genes that confer the resistant phenotype, and/or a previously unrecognized effect on genomic instability that selects for cell populations refractory to drug treatment. Of potential importance for this paradigm is the finding that the MUC1-C→EZH2 pathway suppresses expression of BRCA1, which functions in cell cycle checkpoint activation and DNA repair, and RAD51, which directs homologous strand exchange 35, 106. DSBs are repaired by HR or by the more error-prone NHEJ pathway. With a defective HR pathway, for example in the setting of MUC1-C→EZH2-mediated BRCA1 and RAD51 suppression, insufficient repair of DSBs could result in genomic instability and thereby the selection of cells with anticancer drug resistance.
The repair of DSBs is facilitated by BMI1-mediated ubiquitylation of H2A and γH2AX 107. BMI1-induced suppression of DSB-induced CHK1 and CHK2 activation has further implicated PRC1 in promoting genomic instability and transformation 107. Thus, the role of MUC1-C in activating BMI1 and suppressing CDKN2/p16INK4a expression could thereby enhance MUC1-C-mediated genomic instability 36. Additionally, RNA-seq studies have demonstrated that MUC1-C suppresses multiple effectors of DNA damage response (DDR) pathways, including HR, NHEJ, mismatch repair and transcription-coupled repair, among others 35. How MUC1-C regulates these additional genes and whether there is involvement of the MUC1-C→EZH2 or MUC1-C→BMI1 pathways is not presently known. Nonetheless, these observations highlight the potential importance of MUC1-C function in integrating histone modifications, genomic instability and the propensity for acquiring anticancer drug resistance.
MUC1-C is a highly promising target for cancer treatment
Emerging evidence has thus demonstrated that MUC1-C promotes hallmarks of the cancer cell, including epigenetic regulation, EMT, the CSC state, immune evasion and anticancer drug resistance. Accordingly, MUC1-C has emerged a promising target for cancer therapy. However, to date, there are no approved agents that target MUC1-C function. This situation derives in part from the undruggable nature of this target. The MUC1-C cytoplasmic domain is devoid of a kinase function and is an intrinsically disordered protein 108, a characteristic that provides for interactions with multiple signaling pathways 109, but represents a challenge for drug development. Despite this hurdle, a CQC motif in the MUC1-C cytoplasmic domain is necessary for MUC1-C homodimerization, nuclear localization and oncogenic function, and is the Achilles’ heel of this oncoprotein 37, 39. Thus, MUC1-C with a CQC→AQA mutation functions as a dominant-negative for transformation 110. In addition, blocking the MUC1-C CQC motif with the cell-penetrating GO-203 peptide inhibits MUC1-C function 39. Thus, in concert with targeting MUC1-C by genetic approaches, treatment with GO-203 inhibits MUC1-C-induced EZH2 and BMI1 expression 35, 36, EMT signaling 68, 81 and self-renewal capacity 81, 89, 90. Significantly, GO-203 treatment of cancer cells is also synergistic with cytotoxic drugs 111 and reverses acquired resistance to targeted agents 47, 90, 104, consistent with the premise that MUC1-C confers pleotropic drug-resistant phenotypes.
The findings that MUC1-C function is blocked by GO-203 provided the experimental basis for the clinical evaluation of this agent. A Phase I trial of GO-203 in patients with advanced carcinomas demonstrated an acceptable safety profile and early evidence of anti-tumor activity. Pharmacokinetic studies further demonstrated a circulating GO-203 half-life of 5–7 h that necessitated daily delivery to maintain drug levels in a therapeutic range. Accordingly, GO-203 has been developed in a nanoparticle formulation for less frequent and more sustained delivery in the clinic 112. In parallel and based on the findings that (i) MUC1 is expressed by AML stem-like cells 113, and (ii) GO-203 is highly synergistic with the DNA methylation inhibitor decitabine 78, a Phase II trial of GO-203 in combination with decitabine is now underway for the treatment of patients with relapsed/refractory AML (ClinicalTrials.gov Identifier: NCT02204085).
MUC1 has also emerged as an attractive target for the immunotherapy of cancer. In this regard, vaccines targeting MUC1 for the treatment of malignancies, including NSCLC and breast cancer, have been advanced to late-stage clinical trials, but have had limited success to date 114, 115. Indeed, a challenge for an effective anti-cancer MUC1 vaccine is overcoming tolerance to MUC1, which is widely expressed by normal epithelial cells. In the course of addressing this challenge, a dendritic cell (DC)-based vaccine was found to be effective in reversing tolerance to MUC1 in a MUC1 transgenic mouse background, and eradicating MUC1-positive tumors in the absence of autoimmunity against normal tissues 116. This DC-based vaccine is also effective in reversing tolerance to MUC1 in patients with solid tumors and the hematologic malignancies, multiple myeloma and AML 117, 118. In addition, the findings that induction of anti-MUC1 immunity is associated with anti-tumor activity 117, 118 provided the basis for advancing this vaccine to national multi-center Phase II trials (ClinicalTrials.gov Identifiers: NCT02728102 and NCT03059485). Moreover, the finding that targeting MUC1-C with GO-203 in cancer cells inhibits immune evasion also provides the experimental rationale for combining GO-203 treatment with the above DC-based or other anti-cancer vaccines.
Acknowledgments
This publication was supported by the US Department of Defense under award number BC151648 and the National Cancer Institute of the National Institutes of Health under award numbers R01CA097098, R01CA166480 and R21CA216553.
Abbreviations
- MUC1
mucin 1
- MUC1-C
MUC1 C-terminal subunit
- PcG
Polycomb Group
- PRC
Polycomb Repressive Complex
- EZH2
enhancer of zeste homolog 2
- SUZ12
the suppressor of zeste 12 homolog
- EED
embryonic ectoderm development
- BMI1
B-cell-specific Moloney murine leukemia virus integration site 1 protein
- HOX
homeobox
- DNMT
DNA methyltransferase
- TSG
tumor suppressor gene
- RTK
receptor tyrosine kinase
- EMT
epithelial-mesenchymal transition
- CSC
cancer stem cell
- CTCF
CCCTC-binding factor
- DDR
DNA damage response
- DSB
DNA double strand break
- HR
homologous recombination
- NHEJ
non-homologous end-joining
- IDR
intrinsically disordered region
Footnotes
Conflict of Interest
DK has ownership interest (including patents) and is a consultant/advisory board member of Genus Oncology. No potential conflicts of interest were disclosed by the other authors.
References
- 1.Sparmann A, van Lohuizen M. Polycomb silencers control cell fate, development and cancer. Nat Rev Cancer. 2006;6:846–856. doi: 10.1038/nrc1991. [DOI] [PubMed] [Google Scholar]
- 2.Bracken AP, Helin K. Polycomb group proteins: navigators of lineage pathways led astray in cancer. Nat Rev Cancer. 2009;9:773–784. doi: 10.1038/nrc2736. [DOI] [PubMed] [Google Scholar]
- 3.Koppens M, van Lohuizen M. Context-dependent actions of Polycomb repressors in cancer. Oncogene. 2016;35:1341–1352. doi: 10.1038/onc.2015.195. [DOI] [PubMed] [Google Scholar]
- 4.Blackledge NP, Rose NR, Klose RJ. Targeting Polycomb systems to regulate gene expression: modifications to a complex story. Nat Rev Mol Cell Biol. 2015;16:643–649. doi: 10.1038/nrm4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Riising EM, Comet I, Leblanc B, Wu X, Johansen JV, Helin K. Gene silencing triggers Polycomb Repressive Complex 2 recruitment to CpG islands genome wide. Mol Cell. 2014;55:347–360. doi: 10.1016/j.molcel.2014.06.005. [DOI] [PubMed] [Google Scholar]
- 6.Mills AA. Throwing the cancer switch: reciprocal roles of polycomb and trithorax proteins. Nat Rev Cancer. 2010;10:669–682. doi: 10.1038/nrc2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Comet I, Riising EM, Leblanc B, Helin K. Maintaining cell identity: PRC2-mediated regulation of transcription and cancer. Nat Rev Cancer. 2016 doi: 10.1038/nrc.2016.83. [DOI] [PubMed] [Google Scholar]
- 8.Siddique HR, Saleem M. Role of BMI1, a stem cell factor, in cancer recurrence and chemoresistance: preclinical and clinical evidences. Stem Cells. 2012;30:372–378. doi: 10.1002/stem.1035. [DOI] [PubMed] [Google Scholar]
- 9.Wang H, Wang L, Erdjument-Bromage H, Vidal M, Tempst P, Jones RS, et al. Role of histone H2A ubiquitination in Polycomb silencing. Nature. 2004;431:873–878. doi: 10.1038/nature02985. [DOI] [PubMed] [Google Scholar]
- 10.Cao R, Tsukada Y, Zhang Y. Role of Bmi-1 and Ring1A in H2A ubiquitylation and Hox gene silencing. Mol Cell. 2005;20:845–854. doi: 10.1016/j.molcel.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 11.Vire E, Brenner C, Deplus R, Blanchon L, Fraga M, Didelot C, et al. The Polycomb group protein EZH2 directly controls DNA methylation. Nature. 2006;439:871–874. doi: 10.1038/nature04431. [DOI] [PubMed] [Google Scholar]
- 12.Schlesinger Y, Straussman R, Keshet I, Farkash S, Hecht M, Zimmerman J, et al. Polycomb-mediated methylation on Lys27 of histone H3 pre-marks genes for de novo methylation in cancer. Nat Genet. 2007;39:232–236. doi: 10.1038/ng1950. [DOI] [PubMed] [Google Scholar]
- 13.Cedar H, Bergman Y. Linking DNA methylation and histone modification: patterns and paradigms. Nat Rev Genet. 2009;10:295–304. doi: 10.1038/nrg2540. [DOI] [PubMed] [Google Scholar]
- 14.Bracken AP, Pasini D, Capra M, Prosperini E, Colli E, Helin K. EZH2 is downstream of the pRB-E2F pathway, essential for proliferation and amplified in cancer. EMBO J. 2003;22:5323–5335. doi: 10.1093/emboj/cdg542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gong Y, Huo L, Liu P, Sneige N, Sun X, Ueno NT, et al. Polycomb group protein EZH2 is frequently expressed in inflammatory breast cancer and is predictive of worse clinical outcome. Cancer. 2011;117:5476–5484. doi: 10.1002/cncr.26179. [DOI] [PubMed] [Google Scholar]
- 16.Collett K, Eide GE, Arnes J, Stefansson IM, Eide J, Braaten A, et al. Expression of enhancer of zeste homologue 2 is significantly associated with increased tumor cell proliferation and is a marker of aggressive breast cancer. Clin Cancer Res. 2006;12:1168–1174. doi: 10.1158/1078-0432.CCR-05-1533. [DOI] [PubMed] [Google Scholar]
- 17.Kleer CG, Cao Q, Varambally S, Shen R, Ota I, Tomlins SA, et al. EZH2 is a marker of aggressive breast cancer and promotes neoplastic transformation of breast epithelial cells. Proc Natl Acad Sci U S A. 2003;100:11606–11611. doi: 10.1073/pnas.1933744100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jang SH, Lee JE, Oh MH, Lee JH, Cho HD, Kim KJ, et al. High EZH2 protein expression is associated with poor overall survival in patients with luminal A breast cancer. J Breast Cancer. 2016;19:53–60. doi: 10.4048/jbc.2016.19.1.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Inari H, Suganuma N, Kawachi K, Yoshida T, Yamanaka T, Nakamura Y, et al. Expression of enhancer of zeste homolog 2 correlates with survival outcome in patients with metastatic breast cancer: exploratory study using primary and paired metastatic lesions. BMC Cancer. 2017;17:160. doi: 10.1186/s12885-017-3154-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sauvageau M, Sauvageau G. Polycomb group proteins: multi-faceted regulators of somatic stem cells and cancer. Cell Stem Cell. 2010;7:299–313. doi: 10.1016/j.stem.2010.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Behrens C, Solis LM, Lin H, Yuan P, Tang X, Kadara H, et al. EZH2 protein expression associates with the early pathogenesis, tumor progression, and prognosis of non-small cell lung carcinoma. Clin Cancer Res. 2013;19:6556–6565. doi: 10.1158/1078-0432.CCR-12-3946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sato T, Kaneda A, Tsuji S, Isagawa T, Yamamoto S, Fujita T, et al. PRC2 overexpression and PRC2-target gene repression relating to poorer prognosis in small cell lung cancer. Sci Rep. 2013;3:1911. doi: 10.1038/srep01911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang X, Zhao H, Lv L, Bao L, Wang X, Han S. Prognostic significance of EZH2 expression in non-small cell lung cancer: A meta-analysis. Sci Rep. 2016;6:19239. doi: 10.1038/srep19239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Varambally S, Dhanasekaran SM, Zhou M, Barrette TR, Kumar-Sinha C, Sanda MG, et al. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature. 2002;419:624–629. doi: 10.1038/nature01075. [DOI] [PubMed] [Google Scholar]
- 25.Chen S, Huang L, Sun K, Wu D, Li M, Li M, et al. Enhancer of zeste homolog 2 as an independent prognostic marker for cancer: a meta-analysis. PLoS One. 2015;10:e0125480. doi: 10.1371/journal.pone.0125480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim KH, Roberts CW. Targeting EZH2 in cancer. Nat Med. 2016;22:128–134. doi: 10.1038/nm.4036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang Y, Zhe H, Ding Z, Gao P, Zhang N, Li G. Cancer stem cell marker Bmi-1 expression is associated with basal-like phenotype and poor survival in breast cancer. World J Surg. 2012;36:1189–1194. doi: 10.1007/s00268-012-1514-3. [DOI] [PubMed] [Google Scholar]
- 28.Crea F, Paolicchi E, Marquez VE, Danesi R. Polycomb genes and cancer: time for clinical application? Crit Rev Oncol Hematol. 2012;83:184–193. doi: 10.1016/j.critrevonc.2011.10.007. [DOI] [PubMed] [Google Scholar]
- 29.Vrzalikova K, Skarda J, Ehrmann J, Murray PG, Fridman E, Kopolovic J, et al. Prognostic value of Bmi-1 oncoprotein expression in NSCLC patients: a tissue microarray study. J Cancer Res Clin Oncol. 2008;134:1037–1042. doi: 10.1007/s00432-008-0361-y. [DOI] [PubMed] [Google Scholar]
- 30.Glinsky GV, Berezovska O, Glinskii AB. Microarray analysis identifies a death-from-cancer signature predicting therapy failure in patients with multiple types of cancer. J Clin Invest. 2005;115:1503–1521. doi: 10.1172/JCI23412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yan KS, Lin CY, Liao TW, Peng CM, Lee SC, Liu YJ, et al. EZH2 in cancer progression and potential application in cancer therapy: A friend or foe? Int J Mol Sci. 2017:18. doi: 10.3390/ijms18061172. [DOI] [PMC free article] [PubMed]
- 32.Kreso A, van Galen P, Pedley NM, Lima-Fernandes E, Frelin C, Davis T, et al. Self-renewal as a therapeutic target in human colorectal cancer. Nat Med. 2014;20:29–36. doi: 10.1038/nm.3418. [DOI] [PubMed] [Google Scholar]
- 33.Yong KJ, Basseres DS, Welner RS, Zhang WC, Yang H, Yan B, et al. Targeted BMI1 inhibition impairs tumor growth in lung adenocarcinomas with low CEBPalpha expression. Sci Transl Med. 2016;8:350ra104. doi: 10.1126/scitranslmed.aad6066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang R, Cheung NK, Vider J, Cheung IY, Gerald WL, Tickoo SK, et al. MYCN and MYC regulate tumor proliferation and tumorigenesis directly through BMI1 in human neuroblastomas. FASEB J. 2011;25:4138–4149. doi: 10.1096/fj.11-185033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rajabi H, Hiraki M, Tagde A, Alam M, Bouillez A, Christensen CL, et al. MUC1-C activates EZH2 expression and function in human cancer cells. Sci Rep. 2017 Aug 7;7 doi: 10.1038/s41598-017-07850-0. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hiraki M, Maeda T, Bouillez A, Alam M, Tagde A, Hinohara K, et al. MUC1-C activates BMI1 in human cancer cells. Oncogene. 2016 Nov 28; doi: 10.1038/onc.2016.439. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kufe D. Mucins in cancer: function, prognosis and therapy. Nat Rev Cancer. 2009;9:874–885. doi: 10.1038/nrc2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Duraisamy S, Kufe T, Ramasamy S, Kufe D. Evolution of the human MUC1 oncoprotein. Int J Oncology. 2007;31:671–677. [PubMed] [Google Scholar]
- 39.Kufe D. MUC1-C oncoprotein as a target in breast cancer: activation of signaling pathways and therapeutic approaches. Oncogene. 2013;32:1073–1081. doi: 10.1038/onc.2012.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li Y, Liu D, Chen D, Kharbanda S, Kufe D. Human DF3/MUC1 carcinoma-associated protein functions as an oncogene. Oncogene. 2003;22:6107–6110. doi: 10.1038/sj.onc.1206732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rajabi H, Kufe D. MUC1-C oncoprotein integrates a program of EMT, epigenetic reprogramming and immune evasion in human carcinomas. BBA Reviews on Cancer. 2017;1868:117–122. doi: 10.1016/j.bbcan.2017.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li Y, Ren J, Yu W, Li G, Kuwahara H, Yin L, et al. The EGF receptor regulates interaction of the human DF3/MUC1 carcinoma antigen with c-Src and β-catenin. J Biol Chem. 2001;276:35239–35242. doi: 10.1074/jbc.C100359200. [DOI] [PubMed] [Google Scholar]
- 43.Ramasamy S, Duraisamy S, Barbashov S, Kawano T, Kharbanda S, Kufe D. The MUC1 and galectin-3 oncoproteins function in a microRNA-dependent regulatory loop. Mol Cell. 2007;27:992–1004. doi: 10.1016/j.molcel.2007.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ren J, Raina D, Chen W, Li G, Huang L, Kufe D. MUC1 oncoprotein functions in activation of fibroblast growth factor receptor signaling. Mol Cancer Res. 2006;4:873–883. doi: 10.1158/1541-7786.MCR-06-0204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Singh PK, Wen Y, Swanson BJ, Shanmugam K, Kazlauskas A, Cerny RL, et al. Platelet-derived growth factor receptor beta-mediated phosphorylation of MUC1 enhances invasiveness in pancreatic adenocarcinoma cells. Cancer Res. 2007;67:5201–5210. doi: 10.1158/0008-5472.CAN-06-4647. [DOI] [PubMed] [Google Scholar]
- 46.Singh PK, Behrens ME, Eggers JP, Cerny RL, Bailey JM, Shanmugam K, et al. Phosphorylation of MUC1 by Met modulates interaction with p53 and MMP1 expression. J Biol Chem. 2008;283:26985–26995. doi: 10.1074/jbc.M805036200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Raina D, Uchida Y, Kharbanda A, Rajabi H, Panchamoorthy G, Jin C, et al. Targeting the MUC1-C oncoprotein downregulates HER2 activation and abrogates trastuzumab resistance in breast cancer cells. Oncogene. 2014;33:3422–3431. doi: 10.1038/onc.2013.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kosugi M, Ahmad R, Alam M, Uchida Y, Kufe D. MUC1-C oncoprotein regulates glycolysis and pyruvate kinase M2 activity in cancer cells. PLoS One. 2011;6:e28234. doi: 10.1371/journal.pone.0028234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yin L, Kosugi M, Kufe D. Inhibition of the MUC1-C oncoprotein induces multiple myeloma cell death by downregulating TIGAR expression and depleting NADPH. Blood. 2012;119:810–816. doi: 10.1182/blood-2011-07-369686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ahmad R, Alam M, Hasegawa M, Uchida Y, Al-Obaid O, Kharbanda S, et al. Targeting MUC1-C inhibits the AKT-S6K1-elF4A pathway regulating TIGAR translation in colorectal cancer. Molecular Cancer. 2017;16:33. doi: 10.1186/s12943-017-0608-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gunda V, Souchek J, Abrego J, Shukla SK, Goode GD, Vernucci E, et al. MUC1-mediated metabolic alterations regulate response to radiotherapy in pancreatic cancer. Clin Cancer Res. 2017 Jul 18; doi: 10.1158/1078-0432.CCR-17-1151. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hasegawa M, Takahashi H, Rajabi H, Alam M, Suzuki Y, Yin L, et al. Functional interactions of the cystine/glutamate antiporter, CD44v and MUC1-C oncoprotein in triple-negative breast cancer cells. Oncotarget. 2016;7:11756–11769. doi: 10.18632/oncotarget.7598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Goode G, Gunda V, Chaika NV, Purohit V, Yu F, Singh PK. MUC1 facilitates metabolomic reprogramming in triple-negative breast cancer. PLoS One. 2017;12:e0176820. doi: 10.1371/journal.pone.0176820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Leng Y, Cao C, Ren J, Huang L, Chen D, Ito M, et al. Nuclear import of the MUC1-C oncoprotein is mediated by nucleoporin Nup62. J Biol Chem. 2007;282:19321–19330. doi: 10.1074/jbc.M703222200. [DOI] [PubMed] [Google Scholar]
- 55.Yamamoto M, Bharti A, Li Y, Kufe D. Interaction of the DF3/MUC1 breast carcinoma-associated antigen and β-catenin in cell adhesion. J Biol Chem. 1997;272:12492–12494. doi: 10.1074/jbc.272.19.12492. [DOI] [PubMed] [Google Scholar]
- 56.Rajabi H, Ahmad R, Jin C, Kosugi M, Alam M, Joshi M, et al. MUC1-C oncoprotein induces TCF7L2 transcription factor activation and promotes cyclin D1 expression in human breast cancer cells. J Biol Chem. 2012;287:10703–10713. doi: 10.1074/jbc.M111.323311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wei X, Xu H, Kufe D. Human MUC1 oncoprotein regulates p53-responsive gene transcription in the genotoxic stress response. Cancer Cell. 2005;7:167–178. doi: 10.1016/j.ccr.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 58.Ahmad R, Raina D, Joshi MD, Kawano T, Kharbanda S, Kufe D. MUC1-C oncoprotein functions as a direct activator of the NF-κB p65 transcription factor. Cancer Res. 2009;69:7013–7021. doi: 10.1158/0008-5472.CAN-09-0523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Khodarev N, Ahmad R, Rajabi H, Pitroda S, Kufe T, McClary C, et al. Cooperativity of the MUC1 oncoprotein and STAT1 pathway in poor prognosis human breast cancer. Oncogene. 2010;29:920–929. doi: 10.1038/onc.2009.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ahmad R, Rajabi H, Kosugi M, Joshi M, Alam M, Vasir B, et al. MUC1-C oncoprotein promotes STAT3 activation in an auto-inductive regulatory loop. Sci Signal. 2011;4:ra9. doi: 10.1126/scisignal.2001426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chaika NV, Gebregiworgis T, Lewallen ME, Purohit V, Radhakrishnan P, Liu X, et al. MUC1 mucin stabilizes and activates hypoxia-inducible factor 1 alpha to regulate metabolism in pancreatic cancer. Proc Natl Acad Sci USA. 2012;109:13787–13792. doi: 10.1073/pnas.1203339109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shukla SK, Purohit V, Mehla K, Gunda V, Chaika NV, Vernucci E, et al. MUC1 and HIF-1alpha signaling crosstalk induces anabolic glucose metabolism to impart gemcitabine resistance to pancreatic cancer. Cancer Cell. 2017;32:71–87. e77. doi: 10.1016/j.ccell.2017.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Khodarev N, Pitroda S, Beckett M, MacDermed D, Huang L, Kufe D, et al. MUC1-induced transcriptional programs associated with tumorigenesis predict outcome in breast and lung cancer. Cancer Res. 2009;69:2833–2837. doi: 10.1158/0008-5472.CAN-08-4513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.MacDermed DM, Khodarev NN, Pitroda SP, Edwards DC, Pelizzari CA, Huang L, et al. MUC1-associated proliferation signature predicts outcomes in lung adenocarcinoma patients. BMC Medical Genomics. 2010;3:16. doi: 10.1186/1755-8794-3-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pitroda S, Khodarev N, Beckett M, Kufe D, Weichselbaum R. MUC1-induced alterations in a lipid metabolic gene network predict response of human breast cancers to tamoxifen treatment. Proc Natl Acad Sci USA. 2009;106:5837–5841. doi: 10.1073/pnas.0812029106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Takahashi H, Jin C, Rajabi H, Pitroda S, Alam M, Ahmad R, et al. MUC1-C activates the TAK1 inflammatory pathway in colon cancer. Oncogene. 2015;34:5187–5197. doi: 10.1038/onc.2014.442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ahmad R, Raina D, Trivedi V, Ren J, Rajabi H, Kharbanda S, et al. MUC1 oncoprotein activates the IκB kinase β complex and constitutive NF-κB signaling. Nat Cell Biol. 2007;9:1419–1427. doi: 10.1038/ncb1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rajabi H, Alam M, Takahashi H, Kharbanda A, Guha M, Ahmad R, et al. MUC1-C oncoprotein activates the ZEB1/miR-200c regulatory loop and epithelial-mesenchymal transition. Oncogene. 2014;33:1680–1689. doi: 10.1038/onc.2013.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bouillez A, Rajabi H, Pitroda S, Jin C, Alam M, Kharbanda A, et al. Inhibition of MUC1-C suppresses MYC expression and attenuates malignant growth in KRAS mutant lung adenocarcinomas. Cancer Res. 2016;76:1538–1548. doi: 10.1158/0008-5472.CAN-15-1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Min J, Zaslavsky A, Fedele G, McLaughlin SK, Reczek EE, De Raedt T, et al. An oncogene-tumor suppressor cascade drives metastatic prostate cancer by coordinately activating Ras and nuclear factor-kappaB. Nat Med. 2010;16:286–294. doi: 10.1038/nm.2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chang CJ, Yang JY, Xia W, Chen CT, Xie X, Chao CH, et al. EZH2 promotes expansion of breast tumor initiating cells through activation of RAF1-beta-catenin signaling. Cancer Cell. 2011;19:86–100. doi: 10.1016/j.ccr.2010.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Puppe J, Drost R, Liu X, Joosse SA, Evers B, Cornelissen-Steijger P, et al. BRCA1-deficient mammary tumor cells are dependent on EZH2 expression and sensitive to Polycomb Repressive Complex 2-inhibitor 3-deazaneplanocin A. Breast Cancer Res. 2009;11:R63. doi: 10.1186/bcr2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Li Y, Bharti A, Chen D, Gong J, Kufe D. Interaction of glycogen synthase kinase 3β with the DF3/MUC1 carcinoma-associated antigen and β-catenin. Mol Cell Biol. 1998;18:7216–7224. doi: 10.1128/mcb.18.12.7216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tagde A, Rajabi H, Bouillez A, Alam M, Gali R, Bailey S, et al. MUC1-C drives MYC in multiple myeloma. Blood. 2016;127:2587–2597. doi: 10.1182/blood-2015-07-659151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tagde A, Markert T, Rajabi H, Hiraki M, Alam M, Bouillez A, et al. Targeting MUC1-C suppresses Polycomb Repressive Complex 1 in multiple myeloma. Oncotarget. 2017 Jul; doi: 10.18632/oncotarget.20144. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hernandez-Munoz I, Taghavi P, Kuijl C, Neefjes J, van Lohuizen M. Association of BMI1 with polycomb bodies is dynamic and requires PRC2/EZH2 and the maintenance DNA methyltransferase DNMT1. Mol Cell Biol. 2005;25:11047–11058. doi: 10.1128/MCB.25.24.11047-11058.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rajabi H, Tagde A, Alam M, Bouillez A, Pitroda S, Suzuki Y, et al. DNA methylation by DNMT1 and DNMT3b methyltransferases is driven by the MUC1-C oncoprotein in human carcinoma cells. Oncogene. 2016;35:6439–6445. doi: 10.1038/onc.2016.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Tagde A, Rajabi H, Stroopinsky D, Gali R, Alam M, Bouillez A, et al. MUC1-C induces DNA methyltransferase 1 and represses tumor suppressor genes in acute myeloid leukemia. Oncotarget. 2016;7:38974–38987. doi: 10.18632/oncotarget.9777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tam WL, Weinberg RA. The epigenetics of epithelial-mesenchymal plasticity in cancer. Nat Med. 2013;19:1438–1449. doi: 10.1038/nm.3336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mody HR, Hung SW, AlSaggar M, Griffin J, Govindarajan R. Inhibition of S-Adenosylmethionine-dependent methyltransferase attenuates TGFbeta1-induced EMT and metastasis in pancreatic cancer: Putative roles of miR-663a and miR-4787-5p. Mol Cancer Res. 2016;14:1124–1135. doi: 10.1158/1541-7786.MCR-16-0083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Alam M, Bouillez A, Tagde A, Ahmad R, Rajabi H, Maeda T, et al. MUC1-C represses the Crumbs complex polarity factor CRB3 and downregulates the Hippo pathway. Mol Cancer Res. 2016;14:1266–1276. doi: 10.1158/1541-7786.MCR-16-0233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Polyak K, Weinberg RA. Transitions between epithelial and mesenchymal states: acquisition of malignant and stem cell traits. Nat Rev Cancer. 2009;9:265–273. doi: 10.1038/nrc2620. [DOI] [PubMed] [Google Scholar]
- 83.Shibue T, Weinberg RA. EMT, CSCs, and drug resistance: the mechanistic link and clinical implications. Nat Rev Clin Oncol. 2017 doi: 10.1038/nrclinonc.2017.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wen Y, Cai J, Hou Y, Huang Z, Wang Z. Role of EZH2 in cancer stem cells: from biological insight to a therapeutic target. Oncotarget. 2017;8:37974–37990. doi: 10.18632/oncotarget.16467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Park IK, Morrison SJ, Clarke MF. Bmi1, stem cells, and senescence regulation. J Clin Invest. 2004;113:175–179. doi: 10.1172/JCI20800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Richly H, Aloia L, Di Croce L. Roles of the Polycomb group proteins in stem cells and cancer. Cell Death Dis. 2011;2:e204. doi: 10.1038/cddis.2011.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Song LB, Li J, Liao WT, Feng Y, Yu CP, Hu LJ, et al. The polycomb group protein Bmi-1 represses the tumor suppressor PTEN and induces epithelial-mesenchymal transition in human nasopharyngeal epithelial cells. J Clin Invest. 2009;119:3626–3636. doi: 10.1172/JCI39374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Yang MH, Hsu DS, Wang HW, Wang HJ, Lan HY, Yang WH, et al. Bmi1 is essential in Twist1-induced epithelial-mesenchymal transition. Nat Cell Biol. 2010;12:982–992. doi: 10.1038/ncb2099. [DOI] [PubMed] [Google Scholar]
- 89.Alam M, Rajabi H, Ahmad R, Jin C, Kufe D. Targeting the MUC1-C oncoprotein inhibits self-renewal capacity of breast cancer cells. Oncotarget. 2014;5:2622–2634. doi: 10.18632/oncotarget.1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kharbanda A, Rajabi H, Jin C, Tchaicha J, Kikuchi E, Wong K, et al. Targeting the oncogenic MUC1-C protein inhibits mutant EGFR-mediated signaling and survival in non-small cell lung cancer cells. Clin Cancer Res. 2014;20:5423–5434. doi: 10.1158/1078-0432.CCR-13-3168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Alsuliman A, Colak D, Al-Harazi O, Fitwi H, Tulbah A, Al-Tweigeri T, et al. Bidirectional crosstalk between PD-L1 expression and epithelial to mesenchymal transition: significance in claudin-low breast cancer cells. Mol Cancer. 2015;14:149. doi: 10.1186/s12943-015-0421-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lou Y, Diao L, Parra Cuentas ER, Denning WL, Chen L, Fan YH, et al. Epithelial-mesenchymal transition is associated with a distinct tumor microenvironment including elevation of inflammatory signals and multiple immune checkpoints in lung adenocarcinoma. Clin Cancer Res. 2016;22:3630–3642. doi: 10.1158/1078-0432.CCR-15-1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Noman MZ, Janji B, Abdou A, Hasmim M, Terry S, Tan TZ, et al. The immune checkpoint ligand PD-L1 is upregulated in EMT-activated human breast cancer cells by a mechanism involving ZEB-1 and miR-200. Oncoimmunology. 2017;6:e1263412. doi: 10.1080/2162402X.2016.1263412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Peng D, Kryczek I, Nagarsheth N, Zhao L, Wei S, Wang W, et al. Epigenetic silencing of TH1-type chemokines shapes tumour immunity and immunotherapy. Nature. 2015;527:249–253. doi: 10.1038/nature15520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Nagarsheth N, Peng D, Kryczek I, Wu K, Li W, Zhao E, et al. PRC2 epigenetically silences Th1-Type chemokines to suppress effector T-Cell trafficking in colon cancer. Cancer Res. 2016;76:275–282. doi: 10.1158/0008-5472.CAN-15-1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Bouillez A, Rajabi H, Jin C, Samur M, Tagde A, Alam M, et al. MUC1-C integrates PD-L1 induction with repression of immune effectors in non-small cell lung cancer. Oncogene. 2017 Mar 13; doi: 10.1038/onc.2017.47. Epub ahead of print. [DOI] [PMC free article] [PubMed]
- 97.Bouillez A, Adeegbe D, Jin C, Hu X, Tagde A, Alam M, et al. MUC1-C promotes the suppressive immune microenvironment in non-small cell lung cancer. Oncoimmunology. 2017 doi: 10.1080/2162402X.2017.1338998. In press. [DOI] [PMC free article] [PubMed]
- 98.Agata N, Kawano T, Ahmad R, Raina D, Kharbanda S, Kufe D. MUC1 oncoprotein blocks death receptor-mediated apoptosis by inhibiting recruitment of caspase-8. Cancer Res. 2008;68:6136–6144. doi: 10.1158/0008-5472.CAN-08-0464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.David JM, Hamilton DH, Palena C. MUC1 upregulation promotes immune resistance in tumor cells undergoing brachyury-mediated epithelial-mesenchymal transition. Oncoimmunology. 2016;5:e1117738. doi: 10.1080/2162402X.2015.1117738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Mak MP, Tong P, Diao L, Cardnell RJ, Gibbons DL, William WN, et al. A patient-derived, pan-cancer EMT signature identifies global molecular alterations and immune target enrichment following epithelial-to-mesenchymal transition. Clin Cancer Res. 2016;22:609–620. doi: 10.1158/1078-0432.CCR-15-0876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Dongre A, Rashidian M, Reinhardt F, Bagnato A, Keckesova Z, Ploegh HL, et al. Epithelial-to-mesenchymal transition contributes to immunosuppression in breast carcinomas. Cancer Res. 2017;77:3982–3989. doi: 10.1158/0008-5472.CAN-16-3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Singh A, Settleman J. EMT, cancer stem cells and drug resistance: an emerging axis of evil in the war on cancer. Oncogene. 2010;29:4741–4751. doi: 10.1038/onc.2010.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ren J, Agata N, Chen D, Li Y, Yu W-H, Huang L, et al. Human MUC1 carcinoma-associated protein confers resistance to genotoxic anti-cancer agents. Cancer Cell. 2004;5:163–175. doi: 10.1016/s1535-6108(04)00020-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kharbanda A, Rajabi H, Jin C, Raina D, Kufe D. MUC1-C oncoprotein induces tamoxifen resistance in human breast cancer. Mol Cancer Res. 2013;11:714–723. doi: 10.1158/1541-7786.MCR-12-0668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Nath S, Daneshvar K, Roy LD, Grover P, Kidiyoor A, Mosley L, et al. MUC1 induces drug resistance in pancreatic cancer cells via upregulation of multidrug resistance genes. Oncogenesis. 2013;2:e51. doi: 10.1038/oncsis.2013.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Prakash R, Zhang Y, Feng W, Jasin M. Homologous recombination and human health: the roles of BRCA1, BRCA2, and associated proteins. Cold Spring Harbor perspectives in biology. 2015;7:a016600. doi: 10.1101/cshperspect.a016600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Lin X, Ojo D, Wei F, Wong N, Gu Y, Tang D. A novel aspect of tumorigenesis-BMI1 functions in regulating DNA damage response. Biomolecules. 2015;5:3396–3415. doi: 10.3390/biom5043396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Raina D, Agarwal P, Lee J, Bharti A, McKnight C, Sharma P, et al. Characterization of the MUC1-C cytoplasmic domain as a cancer target. PLoS One. 2015;10:e0135156. doi: 10.1371/journal.pone.0135156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Dyson HJ, Wright PE. Intrinsically unstructured proteins and their functions. Nat Rev Mol Cell Biol. 2005;6:197–208. doi: 10.1038/nrm1589. [DOI] [PubMed] [Google Scholar]
- 110.Kufe D. Functional targeting of the MUC1 oncogene in human cancers. Canc Bio Ther. 2009;8:1201–1207. doi: 10.4161/cbt.8.13.8844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Uchida Y, Raina D, Kharbanda S, Kufe D. Inhibition of the MUC1-C oncoprotein is synergistic with cytotoxic agents in treatment of breast cancer cells. Canc Bio Ther. 2013;14:127–134. doi: 10.4161/cbt.22634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Hasegawa M, Sinha RK, Kumar M, Alam M, Yin L, Raina D, et al. Intracellular targeting of the oncogenic MUC1-C protein with a novel GO-203 nanoparticle formulation. Clin Cancer Res. 2015;21:2338–2347. doi: 10.1158/1078-0432.CCR-14-3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Stroopinsky D, Rosenblatt J, Ito K, Mills H, Yin L, Rajabi H, et al. MUC1 is a potential target for the treatment of acute myeloid leukemia stem cells. Cancer Res. 2013;73:5569–5579. doi: 10.1158/0008-5472.CAN-13-0677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Butts C, Socinski MA, Mitchell PL, Thatcher N, Havel L, Krzakowski M, et al. Tecemotide (L-BLP25) versus placebo after chemoradiotherapy for stage III non-small-cell lung cancer (START): a randomised, double-blind, phase 3 trial. Lancet Oncol. 2014;15:59–68. doi: 10.1016/S1470-2045(13)70510-2. [DOI] [PubMed] [Google Scholar]
- 115.Quoix E, Lena H, Losonczy G, Forget F, Chouaid C, Papai Z, et al. TG4010 immunotherapy and first-line chemotherapy for advanced non-small-cell lung cancer (TIME): results from the phase 2b part of a randomised, double-blind, placebo-controlled, phase 2b/3 trial. Lancet Oncol. 2015;17:212–223. doi: 10.1016/S1470-2045(15)00483-0. [DOI] [PubMed] [Google Scholar]
- 116.Gong J, Chen D, Kashiwaba M, Li Y, Takeuchi H, Qu H, et al. Reversal of tolerance to human MUC1 antigen in MUC1 transgenic mice immunized with fusions of dendritic and carcinoma cells. Proc Natl Acad Sci USA. 1998;95:6279–6283. doi: 10.1073/pnas.95.11.6279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Rosenblatt J, Avivi I, Vasir B, Uhl L, Munshi NC, Katz T, et al. Vaccination with dendritic cell/tumor fusions following autologous stem cell transplant induces immunologic and clinical responses in multiple myeloma patients. Clin Cancer Res. 2013;19:3640–3648. doi: 10.1158/1078-0432.CCR-13-0282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Rosenblatt J, Stone R, Uhl L, Neuberg D, Joyce R, Levine J, et al. Individualized vaccination of AML patients in remission is associated with induction of antileukemia immunity and prolonged remissions. Sci Transl Med. 2016 doi: 10.1126/scitranslmed.aag1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Li LC, Dahiya R. MethPrimer: designing primers for methylation PCRs. Bioinformatics. 2002;18:1427–1431. doi: 10.1093/bioinformatics/18.11.1427. [DOI] [PubMed] [Google Scholar]
- 120.Muller H, Bracken AP, Vernell R, Moroni MC, Christians F, Grassilli E, et al. E2Fs regulate the expression of genes involved in differentiation, development, proliferation, and apoptosis. Genes Dev. 2001;15:267–285. doi: 10.1101/gad.864201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Dyson HJ, Wright PE. Role of intrinsic protein disorder in the Function and interactions of the transcriptional coactivators CREB-binding protein (CBP) and p300. J Biol Chem. 2016;291:6714–6722. doi: 10.1074/jbc.R115.692020. [DOI] [PMC free article] [PubMed] [Google Scholar]