Abstract
Angiosperms, the flowering plants, provide the essential resources for human life, such as food, energy, oxygen, and materials. They also promoted the evolution of human, animals, and the planet earth. Despite the numerous advances in genome reports or sequencing technologies, no review covers all the released angiosperm genomes and the genome databases for data sharing. Based on the rapid advances and innovations in the database reconstruction in the last few years, here we provide a comprehensive review for three major types of angiosperm genome databases, including databases for a single species, for a specific angiosperm clade, and for multiple angiosperm species. The scope, tools, and data of each type of databases and their features are concisely discussed. The genome databases for a single species or a clade of species are especially popular for specific group of researchers, while a timely-updated comprehensive database is more powerful for address of major scientific mysteries at the genome scale. Considering the low coverage of flowering plants in any available database, we propose construction of a comprehensive database to facilitate large-scale comparative studies of angiosperm genomes and to promote the collaborative studies of important questions in plant biology.
Keywords: angiosperm genomes, genome database, data sharing, big data, comparative genomics
Introduction
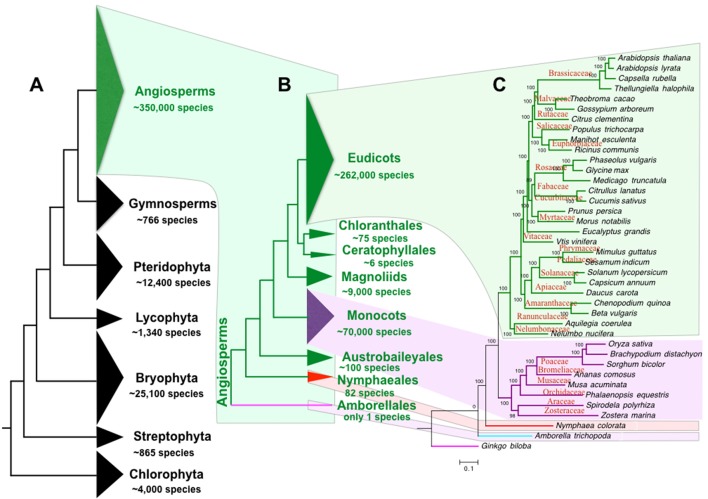
The “green lineage” or the plant kingdom comprises ~4,000 chlorophyta algae, 865 charophyta algae, 25,100 bryophytes, 1,340 lycophytes, 12,400 pteridophytes, 766 gymnosperms (Pryer et al., 2002), and ~350,000 angiosperms (or flowering plants, estimated by www.theplantlist.org). Therefore, angiosperm is by far the most diverse group among all clades of the green lineage (Figure 1A). Originated from a single ancestor at about 167–199 mya (Bell et al., 2010), angiosperms have diverged into 8 extant clades, including Amborellales, Nymphaeales, Austrobaileyales, monocots, Magnoliids, Ceratophyllales, Chloranthales, and Eudicots (Zeng et al., 2014). Only one species is found in the basal branch angiosperm clade Amborellales, whereas the largest angiosperm clade eudicot contains ~262,000 species (Zeng et al., 2014; Figures 1B,C). Compared to other green lineage clades, the angiosperms play the most important roles in our human life. Our food, health, energy, materials, and environment largely depend on angiosperms. In addition, human culture is tightly linked to the utilization of angiosperms (Raskin et al., 2002). For example, early human cultures were shaped by the agriculture (Balick and Cox, 1996) including food production by rice and wheat, fruit gathering, wine fermentation, tea plantation, and flower culturing. Furthermore, angiosperms play important roles in the evolution of animal vision (Osorio and Vorobyev, 2008), taste (Li and Zhang, 2014), and olfactory sense (Niimura, 2012). The angiosperms also contributed greatly to the evolution of planet earth in the atmospheric cycle, water cycle, and the carbon cycle.
Figure 1.
Phylogeny and species of green plants and angiosperms. (A) The tree of the plant life ranging from green algae to flowering plants. (B) The tree of angiosperm life. (C) Angiosperm species tree of representative references genomes. The angiosperm species number is estimated by www.theplantlist.org. Species number of other plant phylum is reported by Pryer et al. (2002). The number of Nymphaeales and other angiosperms species is summarized by Borsch et al. (2008) and Zeng et al. (2014), respectively.
The Science editorial “So much more to know” raised 100 scientific questions to be answered and several of them are angiosperm-related (Hubble, 2005), such as: (1) How does a single somatic cell become a whole plant? (2) Why are some genomes really big and others quite compact? (3) What is all that “junk” doing in our genomes? (4) How did flowers evolve? Moreover, other important questions include (1) the origin of important innovations of the flowers and fruits, (2) the evolution of C4 and CAM photosynthesis, (3) the mechanisms of life style changes such as the epiphytes and parasites, (4) the genetic changes responsible for various ecological adaptations. Comparative genomics may hold the keys to these questions. The genomic sequences, bioinformatics tools, databases, and computing resources are essential infrastructures for comparative genomics.
Rich information can be identified in the angiosperm genomes, which contain various elements, including the genes, repetitive elements, centromeres. All angiosperms are paleo-polyploids (Van de Peer et al., 2017) and some harbor sex chromosomes (Charlesworth, 2016). A genome database is designed to store and present all the information. With the rapid development of bioinformatics, genome database has evolved from mere data storage platform to a novel discipline. Two textbooks focused on the genome databases have been published: “Bioinformatics for Beginners: Genes, Genomes, Molecular Evolution, Databases and Analytical Tools” (Choudhuri, 2014) and “Genomes, Browsers and Databases: Data-Mining Tools for Integrated Genomic Databases” (Schattner, 2008). Furthermore, new journals focused on database have been launched and database articles becoming more popular in various journals. A journal named as Database: The Journal of Biological Databases and Curation was launched in 2009 centered in the biological database. The annual special issues of database published by two high-impact journals, Plant and Cell Physiology and Nucleic Acids Research, have built a good reputation and become influential in the biological research community. In addition, database articles are also frequently published in other leading journals of plant science, such as The Plant Cell and Molecular Plant.
The main function of genome databases has evolved from data storage to online analysis, to lead the jigsaw puzzle in genome sequencing and resequencing projects. For example, Genome Database for Rosaceae (GDR) aims to host the genomes of all rosaceae species although only a few of their genomes have been sequenced. GDR integrated all the released rosaceae genomes and it is expected that more genome resequencing data would be added into the GDR. In the XIX International Botanical Congress (Shenzhen China, 2017), as a key part of the Earth BioGenome Project (EBP), a “10KP plan” was announced with an aim to sequence more than 10,000 genomes representing every major clade of plants and eukaryotic microbes.
In this review, we dedicate to provide readers the latest advances of angiosperm genome projects and database constructions. We discussed the pros and cons of three types of genome databases. We advocate a genome database for all the sequenced angiosperms for prompting data sharing. We also suggest a suit of standards for genome database establishment to boost the development of future databases. The future challenges in facing the biological big data were also discussed. We believe this review will shed new light on the development of angiosperm genome database in the near future.
Genome and database overview
The first angiosperm genome database was launched in 2001 for the model plant Arabidopsis thaliana (Huala et al., 2001). Since 2001, various angiosperm genome databases have been developed synchronizing with the progress of sequencing projects of angiosperm genomes. The earliest angiosperm genome databases were designed as a repository of genome sequencing data. These databases have then evolved to serve as genome portals/hubs that integrate various genomic information, as well as web servers that provide online genomics analyses. These genome databases can be generally classified as three different types: single species database, comprehensive database, and clade-oriented database.
The sequenced angiosperm genomes
As of August 31, 2017, the genomes of 236 angiosperm species have been completely sequenced. The list of sequenced angiosperm genomes and genome databases are provided in Table 1. The 236 species are found in 31 of the 64 angiosperm orders, thus nearly 50% of angiosperm orders have at least one genome sequenced. Most of them are plants of high economic importance or their wild relatives. More effort should be done on genome sequencing of more species that are important for study of evolutionary history of angiosperms such as magnoliids and basal angiosperms.
Table 1.
A list of the public accessible plant genomes and their database construction status.
| Species | Order | Type | URL |
|---|---|---|---|
| Nymphaea colorata | Nymphaeales | Ornamental | www.angiosperms.org |
| Amborella trichopoda | Amborellales | Wild | phytozome.jgi.doe.gov |
| Elaeis guineensis | Arecales | Economic | chibba.agtec.uga.edu/duplication/ |
| Phoenix dactylifera | Arecales | Fruit | pgsb.helmholtz-muenchen.de |
| Elaeis oleifera | Arecales | Wild | |
| Brachypodium distachyon | Poales | Economic | plants.ensembl.org |
| Eragrostis tef | Poales | Economic | |
| Eleusine coracana | Poales | Food | |
| Hordeum vulgare | Poales | Food | phytozome.jgi.doe.gov |
| Oryza indica | Poales | Food | plants.ensembl.org |
| Oryza sativa | Poales | Food | rice.plantbiology.msu.edu/ |
| Setaria italica | Poales | Food | phytozome.jgi.doe.gov |
| Sorghum bicolor | Poales | Food | gramene.org/ |
| Triticum aestivum | Poales | Food | phytozome.jgi.doe.gov |
| Triticum turgidum | Poales | Food | gigadb.org |
| Zea mays | Poales | Food | plants.ensembl.org |
| Secale cereale | Poales | Food, | pgsb.helmholtz-muenchen.de |
| Ananas comosus | Poales | Fruit | phytozome.jgi.doe.gov |
| Lolium perenne | Poales | Ornamental | pgsb.helmholtz-muenchen.de |
| Zizania latifolia | Poales | Vegetable | |
| Oryza punctats | Poales | Weed | plants.ensembl.org |
| Oryza rufipogon | Poales | Weed | plants.ensembl.org |
| Aegilops tauschii | Poales | Wild | plants.ensembl.org |
| Brachypodium stacei | Poales | Wild | genome.jgi.doe.gov |
| Dichanthelium oligosanthes | Poales | Wild | genomevolution.org/CoGe |
| Hordeum pubiflorum | Poales | Wild | |
| Leersia perrieri | Poales | Wild | plants.ensembl.org |
| Oropetium thomaeum | Poales | Wild | genomevolution.orgauth.iplantc.org |
| Oryza barthii | Poales | Wild | plants.ensembl.org |
| Oryza brachyantha | Poales | Wild | plants.ensembl.org |
| Oryza glaberrima | Poales | Wild | plants.ensembl.org |
| Oryza glumaepatula | Poales | Wild | plants.ensembl.org |
| Oryza glumipatula | Poales | Wild | plants.ensembl.org |
| Oryza meridionalis | Poales | Wild | plants.ensembl.org |
| Oryza nivara | Poales | Wild | plants.ensembl.org |
| Oryza longistaminata | Poales | Wild | plants.ensembl.org |
| Panicum hallii | Poales | Wild | genomevolution.orgauth.iplantc.org |
| Panicum virgatum | Poales | Wild | genome.jgi.doe.gov |
| Setaria viridis | Poales | Wild | phytozome.jgi.doe.gov |
| Triticum uratu | Poales | Wild | gigadb.org |
| Camptotheca acuminata | Cornales | Ornamental, Economic | datadryad.org |
| Musa balbisiana | Zingiberales | Fruit | genomevolution.org |
| Musa itinerans | Zingiberales | Fruit | banana-genome-hub.southgreen.fr |
| Ensete ventricosum | Zingiberales | Ornamental | |
| Musa acuminata | Zingiberales | Ornamental | plants.ensembl.org |
| Dendrobium catenatum | Asparagales | Ornamental | |
| Phalaenopsis equestris | Asparagales | Ornamental | chibba.agtec.uga.edu/duplication/ |
| Xerophyta viscosa | Pandanales | Wild | |
| Lemna minor | Alismatales | Economic | genomevolution.org |
| Spirodela polyrhiza | Alismatales | Economic | phytozome.jgi.doe.gov |
| Zoysia japonica | Alismatales | Ornamental | zoysia.kazusa.or.jp |
| Zoysia matrella | Alismatales | Ornamental | zoysia.kazusa.or.jp |
| Zoysia pacifica | Alismatales | Ornamental | zoysia.kazusa.or.jp |
| Zostera marina | Alismatales | Wild | phytozome.jgi.doe.gov |
| Macleaya cordata | Ranunculales | Economic | |
| Aquilegia coerulea | Ranunculales | Wild | genome.jgi.doe.gov |
| Nelumbo nucifera | Proteales | Ornamental | chibba.agtec.uga.edu/duplication |
| Trifolium pratense | Fabales | Drink | phytozome.jgi.doe.gov |
| Lotus japonicus | Fabales | Economic | chibba.agtec.uga.edu/duplication/ |
| Glycine soja | Fabales | Food | soybase.org |
| Lupinus angustifolius | Fabales | Ornamental | |
| Cajanus cajan | Fabales | Vegetable | chibba.agtec.uga.edu/duplication/ |
| Cicer arietinum | Fabales | Vegetable | chibba.agtec.uga.edu/duplication/ |
| Glycine max | Fabales | Vegetable | plants.ensembl.org |
| Medicago truncatula | Fabales | Vegetable | www.medicagogenome.org/ |
| Phaseolus vulgaris | Fabales | Vegetable | phytozome.jgi.doe.gov |
| Vicia faba | Fabales | Vegetable | |
| Vigna angularis | Fabales | Vegetable | viggs.dna.affrc.go.jp |
| Vigna radiata | Fabales | Vegetable | genomevolution.org/CoGe |
| Vigna unguiculata | Fabales | Vegetable | phytozome.jgi.doe.gov |
| Arachis duranensis | Fabales | Wild | www.peanutbase.org/ |
| Arachis ipaensis | Fabales | Wild | www.peanutbase.org/ |
| Cicer reticulatum | Fabales | Wild | www.coolseasonFoodlegume.org |
| Trifolium subterraneum | Fabales | Wild | |
| Betula pendula | Fabales | Wood, Economic | genomevolution.org |
| Humulus lupulus | Rosales | Drink | hopbase.cgrb.oregonstate.edu |
| Cannabis sativa | Rosales | Economic | genome.ccbr.utoronto.ca/cgi-bin/hgGateway |
| Prunus avium | Rosales | Food | ftp://ftp.bioinfo.wsu.edu |
| Artocarpus camansi | Rosales | Fruit | datadryad.org |
| Ficus carica | Rosales | Fruit | |
| Fragaria nipponica | Rosales | Fruit | |
| Fragaria orientalis | Rosales | Fruit | |
| Fragaria vesca | Rosales | Fruit | phytozome.jgi.doe.gov |
| Fragaria × ananassa | Rosales | Fruit | |
| Malus domestica | Rosales | Fruit | phytozome.jgi.doe.gov |
| Morus notabilis | Rosales | Fruit | morus.swu.edu.cn/morusdb |
| Prunus persica | Rosales | Fruit | phytozome.jgi.doe.gov |
| Pyrus bretschneideri | Rosales | Fruit | peargenome.njau.edu.cn |
| Pyrus communis | Rosales | Fruit | genomevolution.org |
| Rubus occidentalis | Rosales | Fruit | ftp://ftp.bioinfo.wsu.edu |
| Ziziphus jujuba | Rosales | Fruit | jujube.genomics.cn |
| Prunus mume | Rosales | Ornamental | chibba.agtec.uga.edu/duplication |
| Rosa × damascena | Rosales | Ornamental | |
| Fragaria iinumae | Rosales | Wild | |
| Fragaria nubicola | Rosales | Wild | |
| Castanea mollissima | Fagales | Fruit | hardwoodgenomics.org |
| Juglans regia | Fagales | Fruit | |
| Betula nana | Fagales | Wild | |
| Quercus lobata | Fagales | Wood | valleyoak.ucla.edu/genomesequence |
| Cucumis melo | Cucurbitales | Fruit | bioinformatics.psb.ugent.be/plaza/ |
| Cucumis sativus | Cucurbitales | Vegetable | phytozome.jgi.doe.gov |
| Lagenaria siceraria | Cucurbitales | Vegetable | |
| Momordica charantia | Cucurbitales | Vegetable | |
| Cephalotus follicularis | Oxalidales | Economic | genomevolution.orgauth.iplantc.org |
| Hevea brasiliensis | Malpighiales | Economic | www4a.biotec.or.th/rubber |
| Jatropha curcas | Malpighiales | Economic | www.kazusa.or.jp/jatropha |
| Populus tremula | Malpighiales | Economic | plantgenie.org |
| Populus tremulax | Malpighiales | Economic | plantgenie.org |
| Populus trichocarpa | Malpighiales | Economic | phytozome.jgi.doe.gov |
| Ricinus communis | Malpighiales | Economic | phytozome.jgi.doe.gov |
| Linum usitatissimum | Malpighiales | Economic, Fiber | phytozome.jgi.doe.gov |
| Manihot esculenta | Malpighiales | Vegetable | phytozome.jgi.doe.gov |
| Populus tremuloides | Malpighiales | Wild | plantgenie.org |
| Populus euphratica | Malpighiales | Wild | |
| Salix purpurea | Malpighiales | Wild | phytozome.jgi.doe.gov |
| Populus pruinosa | Malpighiales | Wood | gigadb.org |
| Populus deltoides | Malpighiales | Wood | phytozome.jgi.doe.gov |
| Eucalyptus camaldulensis | Myrtales | Economic | www.kazusa.or.jp |
| Punica granatum | Myrtales | Food, Ornamental | |
| Metrosideros polymorpha | Myrtales | Wild | |
| Eucalyptus grandis | Myrtales | Wood | phytozome.jgi.doe.gov |
| Theobroma cacao | Malvales | Drink | www.cacaogenomedb.org/ |
| Gossypium raimondii | Malvales | Economic | chibba.agtec.uga.edu/duplication/ |
| Corchirus olitorius | Malvales | Fiber | |
| Corchorus capsularis | Malvales | Fiber | |
| Gossypium arboreum | Malvales | Fiber | www.cottongen.org |
| Gossypium barbadense | Malvales | Fiber | www.cottongen.org |
| Gossypium hirsutum | Malvales | Fiber | cgp.genomics.org.cn |
| Hibiscus syriacus | Malvales | Ornamental | hibiscus.kobic.re.kr/hibiscus.en |
| Aquilaria agallochum | Malvales | Wood | |
| Brassica nigra | Brassicales | Economic | brassicadb.org |
| Camelina sativa | Brassicales | Economic | |
| Capsella orientalis | Brassicales | Food | genomevolution.org |
| Carica papaya | Brassicales | Fruit | phytozome.jgi.doe.gov |
| Lepidium meyenii | Brassicales | Economic | ftp://202.203.187.112/genome/maca/ |
| Tarenaya hassleriana | Brassicales | Ornamental | genomevolution.org/coge |
| Barbarea vulgaris | Brassicales | Vegetable | 185.45.23.197:5080/Barbarea_data/ |
| Brassica juncea | Brassicales | Vegetable | brassicadb.org |
| Brassica napus | Brassicales | Vegetable | gramene.org/ |
| Brassica oleracea | Brassicales | Vegetable | plants.ensembl.org |
| Brassica rapa | Brassicales | Vegetable | brassicadb.org/brad/ |
| Capsella bursa-pastoris | Brassicales | Vegetable | |
| Capsella rubella | Brassicales | Vegetable | phytozome.jgi.doe.gov |
| Moringa oleifera | Brassicales | Vegetable | ftp://202.203.187.112/genome/lamu/ |
| Raphanus sativus | Brassicales | Vegetable | radish.kazusa.or.jp/index.html |
| Thlaspi arvense | Brassicales | Vegetable | |
| Aethionema arabicum | Brassicales | Wild | brassicadb.org |
| Arabidopsis halleri | Brassicales | Wild | phytozome.jgi.doe.gov |
| Arabidopsis lyrata | Brassicales | Wild | bioinformatics.psb.ugent.be/plaza/ |
| Arabidopsis thaliana | Brassicales | Wild | www.arabidopsis.org/index.jsp |
| Arabis alpina | Brassicales | Wild | arabis-alpina.org |
| Arabis montbretiana | Brassicales | Wild | |
| Arabis nordmanniana | Brassicales | Wild | |
| Boechera stricta | Brassicales | Wild | genome.jgi.doe.gov |
| Capsella grandiflora | Brassicales | Wild | genomevolution.org |
| Cardamine hirsuta | Brassicales | Wild | chi.mpipz.mpg.de |
| Eutrema salsugineum | Brassicales | Wild | phytozome.jgi.doe.gov |
| Leavenworthia alabamica | Brassicales | Wild | brassicadb.org |
| Raphanus raphanistrum | Brassicales | Wild | genomevolution.org/coge |
| Sisymbrium irio | Brassicales | Wild | brassicadb.org |
| Thellungiella parvula | Brassicales | Wild | thellungiella.org/data/ |
| Citrus grandis | Sapindales | Food, | citrus.hzau.edu.cn |
| Citrus ichangensis | Sapindales | Food, | citrus.hzau.edu.cn |
| Citrus medica | Sapindales | Food | citrus.hzau.edu.cn |
| Citrullus lanatus | Sapindales | Fruit | chibba.agtec.uga.edu/duplication |
| Citrus clementina | Sapindales | Fruit | phytozome.jgi.doe.gov |
| Citrus sinensis | Sapindales | Fruit | phytozome.jgi.doe.gov |
| Dimocarpus longan | Sapindales | Fruit | gigadb.org |
| Atalantia buxifolia | Sapindales | Wild | citrus.hzau.edu.cn |
| Azadirachta indica | Sapindales | Wild | |
| Vitis vinifera | Vitales | Fruit | plants.ensembl.org |
| Vitis aestivalis | Vitales | Fruit | |
| Vitis cinerea × Vitis riparia | Vitales | Fruit | |
| Rhodiola crenulata | Saxifragales | Economic | gigadb.org |
| Kalanchoe fedtschenkoi | Saxifragales | Ornamental | phytozome.jgi.doe.gov |
| Kalanchoe laxiflora | Saxifragales | Ornamental | phytozome.jgi.doe.gov |
| Kalanchoe marnieriana | Saxifragales | Ornamental | genomevolution.org |
| Chenopodium pallidicaule | Caryophyllales | Food | |
| Chenopodium quinoa | Caryophyllales | Food | phytozome.jgi.doe.gov |
| Fagopyrum esculentum | Caryophyllales | Food | buckwheat.kazusa.or.jp |
| Amaranthus hypochondriacus | Caryophyllales | Ornamental | phytozome.jgi.doe.gov |
| Dianthus caryophyllus | Caryophyllales | Ornamental | carnation.kazusa.or.jp |
| Drosera capensis | Caryophyllales | Ornamental | |
| Beta vulgaris | Caryophyllales | Vegetable | plants.ensembl.org |
| Spinacia oleracea | Caryophyllales | Vegetable | genomevolution.org |
| Amaranthus tuberculatus | Caryophyllales | Weed | |
| Chenopodium suecicum | Caryophyllales | Wild | |
| Silene latifolia subsp. alba | Caryophyllales | Wild | |
| Eichhornia paniculata | Commelinales | Wild | |
| Camellia sinensis | Ericales | Drink | www.plantkingdomgdb.com |
| Actinidia chinensis | Ericales | Fruit | bdg.hfut.edu.cn |
| Diospyros lotus | Ericales | Fruit | chibba.agtec.uga.edu/duplication/ |
| Vaccinum macrocarpon | Ericales | Fruit | |
| Primula veris | Ericales | Wild | |
| Helianthus annuus | Asterales | Economic | www.sunflowergenome.org |
| Silybum marianum | Asterales | Economic | |
| Carthamus tinctorius | Asterales | Oil | |
| Lactuca sativa | Asterales | Vegetable | phytozome.jgi.doe.gov |
| Conyza canadensis | Asterales | Weed | |
| Panax notoginseng | Apiales | Economic | ftp://202.203.187.112/genome/sanqi |
| Daucus carota | Apiales | Vegetable | phytozome.jgi.doe.gov |
| Nicotiana benthamiana | Solanales | Economic | genomevolution.org |
| Nicotiana tabacum | Solanales | Economic | solgenomics.net |
| Ipomoea nil | Solanales | Ornamental | |
| Nicotiana sylvestris | Solanales | Ornamental | solgenomics.net |
| Petunia inflata | Solanales | Ornamental | genomevolution.org |
| Petunia integrifolia | Solanales | Ornamental | genomevolution.org |
| Capsicum annuum | Solanales | Vegetable | chibba.agtec.uga.edu/duplication |
| Solanum lycopersicum | Solanales | Vegetable | phytozome.jgi.doe.gov |
| Solanum melongena | Solanales | Vegetable | genomevolution.org |
| Solanum pimpinellifolium | Solanales | Vegetable | solgenomics.net |
| Solanum tuberosum | Solanales | Vegetable | phytozome.jgi.doe.gov |
| Ipomoea trifida | Solanales | Wild | sweetpotato-garden.kazusa.or.jp |
| Nicotiana attenuata | Solanales | Wild | |
| Nicotiana otophora | Solanales | Wild | |
| Nicotiana tomentosiformis | Solanales | Wild | solgenomics.net |
| Petunia axilaris | Solanales | Wild | genomevolution.org |
| Solanum arcanum | Solanales | Wild | |
| Solanum commersonii | Solanales | Wild | solgenomics.net |
| Solanum habrochaites | Solanales | Wild | |
| Solanum pennellii | Solanales | Wild | solgenomics.net |
| Sesamum indicum | Lamiales | Economic | www.ocri-genomics.org/Sinbase/index.html |
| Dorcoceras hygrometricum | Lamiales | Economic | |
| Mentha longifolia | Lamiales | Economic | langelabtools.wsu.edu/mgr/organism/Mentha/longifolia |
| Ocimum tenuiflorum | Lamiales | Economic | |
| Pogostemon cablin | Lamiales | Economic | ftp://202.203.187.112 |
| Salvia miltiorrhiza | Lamiales | Economic | ftp://202.203.187.112 |
| Erythranthe guttata | Lamiales | Ornamental | |
| Fraxinus excelsior | Lamiales | Ornamental | www.ashgenome.org |
| Mimulus guttatus | Lamiales | Ornamental | phytozome.jgi.doe.gov |
| Antirrhinum majus | Lamiales | Ornamental, Economic | genomevolution.org |
| Genlisea aurea | Lamiales | Wild | |
| Utricularia gibba | Lamiales | Wild | chibba.agtec.uga.edu/duplication/ |
| Coffea canephora | Gentianales | Drink | genomevolution.org |
| Catharanthus roseus | Gentianales | Economic | plantgenomics.msu.edu |
| Rhazya stricta | Gentianales | Economic |
The blank link of 58 species suggest these genome information were stored in NCBI-Genome column.
After the completion of the genome sequencing, an urgent issue is to share the genome data with the research community immediately after the genome release. The importance of data sharing is well recognized because it expands the impact of these valuable sequence data and promotes collaboration. A good genome database should meet two criteria: (i) integration of various types of genomic data, and (ii) providing genome analysis tools.
Genome database for a single species
Among the 236 sequenced angiosperm genomes, only a few of them have a well-constructed customized database (Table 1) to host its various genome information. 58 genomes are only stored at NCBI Genome without a customized database. The genome databases of model plants Arabidopsis and rice (Oryza sativa) appear to be most well constructed. The most popular Arabidopsis database is the Arabidopsis Information Resource (TAIR, www.arabidopsis.org) (Garcia-Hernandez et al., 2002). TAIR provides updated genome sequence (currently V10) and various genomic information, including SNP, transposons, genes, gene families, gene annotations, gene names, proteins, and mutant orderings. Multiple web-integrated bioinformatics tools are also provided by TAIR. For examples, BLAST, WU-BLAST, FASTA, Gbrowse, Synteny Viewer, Seqviewer, Motif analysis, and Chromosome Map tool are powerful for visualization and comparative studies of genes and genome sequence at different scales. Pathway maps provide predicted gene interaction information, and has gained its popularity for the rapid development of metabolic and metabolomics researches. Other tools include Mapviewer, Metabolic Pathways, N-browse, Patmatch, VxInsight, Java Tree View, Bulk Data Retrieval, Gene Symbol Registry, and Textpresso Full Text. ARAPOT (www.araport.org) is also an important Arabidopsis genome database that provides updated genome sequence (currently V11), various gene information and protein interaction networks. However, another Arabidopsis database (Schoof et al., 2002) is no longer accessible. Among the web-integrated bioinformatics tools, such as those in TAIR as an example, BLAST, WU-BLAST, FASTA, Gbrowse, Synteny Viewer, Seqviewer, Motif analysis, and Chromosome Map tool were developed for visualize and compare genes and genome sequence at various scales. Patmatch and Bulk Data Retrieval tools help users to fetch data from servers. Pathway maps provide predicted gene interaction information, and are becoming popular nowadays for the rapid development of metabolic and metabolomics researches. Besides, the other tools are good complementary to various purposes.
However, unlike TAIR, most species-specific genome databases do not offer a rich collection of bioinformatic tools. For example, the pear (Pyrus bretschneideri.) genome database (peargenome.njau.edu.cn) only provide data download. The ash tree (Hymenoscyphus fraxinea and Agrilus planipennis) genome database (ashgenome.org/) includes the BLAST, Jbrowse tools, and data download service. The jujube (Ziziphus jujuba) genome database (jujube.genomics.cn/page/species/index.jsp) has tools such as gene search, Mapview and BLAST. The limited availability of bioinformatic tools may have reduced the popularity and usability of these databases.
Constrution a comprehensive database requires the knowledge of databases and a plethora of database and web programming languages such as Java, HTML, PHP, MySQL, Python, Perl et al., which are not the expertise of most experimental biologists. Most of none-model plant genomes are sequenced by experimental biologists, which is probably the main factor for the different levels of functionalities among species-specific genome databases. Three problems are constantly encountered for species-specific genome databases. First, these databases are often constructed by outsourcing companies, or by one of the bioinformatics graduate student/staff. The cost of such database is usually low and less time consuming. However, the content of these databases is usually rarely or never updated, probably due to the expiration of the service contract with outsourcing companies or departures of graduate students/post-doctoral scholars. For this reason, many species-specific genome database are unstable and eventually become inactivated. For instance, the databases of Mei and pineapple are no longer accessible (only accessible for several months after the release of genomes). Although accessing some databases is not convenient, a few species have more than one genome database. Second, the visual design of these databases usually does not match to those comprehensive databases. The findability and accessibility are usually limited because the users of a species-specific genome database are usually limited to those who work on the same species (Adam-Blondon et al., 2016). Third, with the rapid development of sequencing technology, many more genomes have been sequenced but no customized genome database was built for these genomes, although they may be economically or evolutionarily important.
Comprehensive databases for various angiosperm species
With the advances of next generation sequencing (NGS) and the latest third generation sequencing platforms, angiosperms with genome sequences are rapidly accumulating. The Pacific Biosciences (PacBio) company have developed a single-molecule real-time sequencing platform, which outputs long and unbiased reads with average length >10Kb, greatly facilitates the assembly of large and complex angiosperm genomes. The genome sequencing of a desiccation grass Oropetium thomaeum (VanBuren et al., 2015), sunflower (Badouin et al., 2017), and quinoa (Jarvis et al., 2017) all relied on PacBio and produced high-quality genome assembly. Comparative analysis of these genomic data allows scientists to answer many important questions of plant biology. Therefore, a high demand for comprehensive genome databases is expected. Currently, several comprehensive databases that include a large collection of plant genomes have been constructed (Figure 2A).
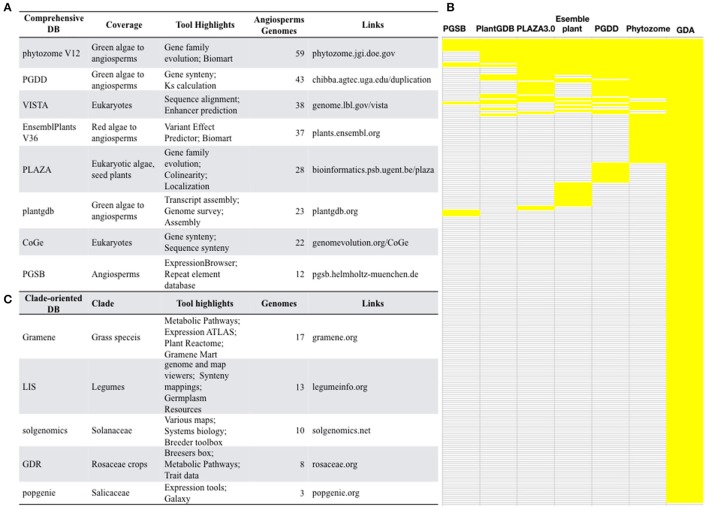
Figure 2.
Selected well-constructed genome databases for/covering angiosperms. (A) Comprehensive databases and their featured tools and indexed genomes. (B) Small-scale genome databases for specific clade of angiosperms. (C) A comparison of the comprehensive genome databases. PGSB, Plant Genome and Systems Biology; PlantGDB, Plant Genome Database; PGDD, Plant Genome Duplication Database; GDA, Genome Database for Angiosperms.
Phytozome (phytozome.jgi.doe.gov) is a large plant genomic portal sponsored by the USA Department of Energy (DOE). The current release of Phytozome (v12) hosts assembled and annotated genomes from 59 angiosperm species, as well as other green lineage species, such as algae, moss, liverworts, selaginella (Goodstein et al., 2012). In addition to BLAST and Gbrowse tools, Phytozome also provide Biomart which allow users to annotate plant gene families, to study the evolution of plant gene families, to display genes in the genomic context (Goodstein et al., 2012), which is valuable for a wide range of scientists who are interested in gene family evolution. However, considering that the genomes of 236 angiosperm species have been sequenced, <1 third of all sequenced angiosperms are included by Phytozome, suggesting the presence of a major gap in the availability of most angiosperm genomes at Phytozome.
The Plant Genome Duplication Database (PGDD) (chibba.agtec.uga.edu/duplication/) is a database currently hosting 43 angiosperm genomes, with tools to identify the intragenome and cross-genome synteny relationships. Synonymous substitutions of homologs inferred from syntenic alignments could be calculated from this database (Lee et al., 2012). By the synteny comparison, PGDD facilitates the identification of evolutionary analysis of gene and genome duplication (Lee et al., 2012).
Ensembl is well-known for developing bioinformatics tools and annotating various eukaryotic genomes (Kersey et al., 2014). The Ensembl Plants (plants.ensembl.org/index.html) provide a HMMER tool for homology searches of gene family members. However, it only covers 37 angiosperm genomes and does not include genome browsers for genomic context views thereby limits its readership.
VISTA (genome.lbl.gov/vista/index.shtml), which includes 38 angiosperm genomes, provides comprehensive tools for analyzing multiple genomes, such as tools for alignment of multiple sequences and large genomic sequences. VISTA has been extensively used by the biomedical community (Poliakov et al., 2014). However, some tools are not applicable to angiosperms as they are restricted to human and mouse data.
Other databases such as PLAZA (Proost et al., 2009), plantgdb (Duvick et al., 2008), CoGe (Lyons, 2008), PGSB (Spannagl et al., 2016) also provide valuable tools for angiosperm comparative genomics. However, none of these databases contains more than 30 plant genomes, which is less than one-eighth of total sequenced angiosperms.
Many of these comprehensive genome databases are well-maintained and are frequently updated with release of new versions, such as PlantGDB V187, Phytozome V12, and Gramene V36. Phytozome updates roughly each year, and PGDD and CoGe update timely upon availability of new genome data. Another feature is that these databases are empowered with various tools other than the open-sourced BLAST and browsers. They offer tools for gene family copies (Phytozome, PLAZA), gene/chromosome synteny (CoGe, PGDD, phytozome, PLAZA, PGSB), protein domains (PlantGDB, Phytozome, EnsemblPlant), gene expression (Phytozome, PGSB), biomart (Phytozome, EnsemblPlants), Intermine (Phytozome), GO annotation (Phytozome, EnsemblPlants, PLAZA, PGSB), alternative splicing (EnsemblPlants, plantgdb).
Although these comprehensive databases contain a large array of species, the largest one Phytozome only includes 59 angiosperms genomes, accounting for about ¼ of sequenced angiosperm genomes. We constructed the Genome Database for Angiosperms (GDA, www.angiosperms.org) to host all of the released angiosperms genome (Figures 2B, 3). GDA aims to updates all the recently sequenced angiosperm genomes by supplying a timeline (Figure 3A). Currently, all the 236 angiosperm genomes, CDSs, and proteins are provided and can be accessed via BLAST suits and download service.
Figure 3.
The proposed genome database for angiosperms (GDA, www.angiosperms.org). (A) The menu of the database. (B,C) Statistics of the sequenced genomes in all the angiosperm orders. (D) Statistics of the sequenced crop genomes. (E) Timeline for the genome updates and related hyperlinks. (F) Ongoing projects for specific angiosperms. (G) BLAST page for all the released 235 angiosperm genomes, CDS, and protein information.
Database for clade-oriented angiosperms species
The NGS techniques has significantly accelerated the decoding of genomes. For example, 10 rice species have been sequenced (Table 1). Comparative genomics is a powerful strategy to decode the genetic basis underlying trait evolution and the evolution of genes and genomes. We summarized the five well-constructed clade-oriented genome databases that have a clear goal (Figure 2).
The current version of Gramene (gramene.org) provides curated and integrated genomic information for plants, especially the 17 grass species. Gramene's bioinformatics platforms provide specific softwares for studying the grass traits. Gramene is an early adopter of BioMart (Smedley et al., 2009) and develops the GrameneMart that enables scientists to perform advanced querying, download, and online comparison of grass genomic data sources through a single portal (Tello-Ruiz et al., 2016). Besides the genome framework, Gramene hosts a pathway framework that integrates a plant reactome pathway, and the pathway tool platform “Cyc Pathways,” allowing the fast comparison of grass-specific pathways.
LIS (legumeinfo.org) is the genome information portal for 13 economically important legumes. LIS provides bioinformatics tools for genome and map viewers, and synteny mappings (Dash et al., 2016). LIS also supports the bridge between the genomic information and the crop improvement by supplying the Germplasm Resources. Likewise, Sol Genomics Network (solgenomics.net) is a Solanaceae-oriented database containing genome data, genomic tools, and breeders' tools.
Other clade-oriented genome databases host <10 genomes (Figure 2), such as the Rosaceae crop oriented GDR (Jung et al., 2014), poplar oriented PopGenIE (Sjödin et al., 2009), cool season food legume oriented CSFL (Main et al., 2013). These clade-oriented genome databases gather multiple species, often with economic importance from the same clade, and provide genome data as well as tools for traditional breeders. However, the integration of various genomes needs more frequent updates. The visibility of these databases is often limited to specific scientists, and will be time-consuming for plant kingdom-wide researchers to obtain these data.
These databases are clade-oriented and distinctive in data and tools compared to other databases. They include economically important crops and their related wild species, and contain genome data that are not included in those comprehensive genome database. They usually provide breeding markers such as molecular markers, various maps, breeder's toolbox, primer design, and germplasm resources. Other genomic tools such as BLAST and synteny mapping are useful to visualize and compare the genome data in various scales. Expression visualization often provides large quantity of expression datasets for fast comparison of various genes and gene families. Furthermore, re-sequencing genomes and biological pathway are also provided in these databases. However, the visibility of these databases needs to be improved. They have been frequently accessed by researchers from the same field, but they are less well-known throughout the plant research community. We recommended that a good genome database should be engaged in the alliance, such as Sol and root, tuber and banana (RTB) crops co-sponsored workshop, sharing and co-developing bioinformatics tools.
Outlook and challenges
A suit of standards required
First, the transparent operation of genome datasets or tools is required. For data changes in the database, updates need to be recorded, so that we can grasp the new information in a timely manner. This can be published by news and so on. The release of data or tools needs to be forecasted. Solgenomics (solgenomics.net) serves as a good example, as it provides very detailed recent changes to the database. Phytozome also provides genome update in a very conspicuous position.
Second, the genome databases, especially for a single species-oriented ones, require a series of minimum standard tools. Data should integrate the reference but not draft genome, CDS, protein, GFF, GO annotation, and the sequencing quality report. Tools should include the gene search, blast, browse, download.
Third, databases should be maintained for at least 3 years. Good maintenance secures a steady population of users whereas a bad one will only narrow its academic impact.
Tripal: a toolkit for genome database construction
Tripal is a member of the Generic Model Organism Database (GMOD) organization suite of genome tools. The first official version of Tripal was released in 2009 by Stephen Ficklin and Meg Staton at the Clemson University Genomics Institute (CUGI). Tripal incoperates several features: (1) Chado database and related modules for data storage and search; (2) Community-developed modules to help fasten site construction; (3) Provide an out-of-the-box setup for a genomics site for those who simply want to put new genome assemblies and annotations online; (4) Provide Application Programming Interfaces (APIs) for complete customization such that more advanced displays, look-and-feel, and new functionality can be supported; (5) Sites can be customized as desired or using theme packages from drupal. During the last several years, Tripal has been implemented in 12 genome projects, including both single genome centered and multiple genome databases: banana, cacao, citrus, cool season food legume, cotton, curcurbit, rosaceae, Vaccinium, hardwood genomics project, legume information system, Medicago truncatula, and peanut (Figure 4).
Figure 4.
The tripal toolkit for genome database construction. (A) The Tripal is a biological application of Drupal. (B) core modules of Tripal. (C) carious modules developed during the last several years. (D) The successful applications of tripal in 12 angiosperm plant projects.
The database facilitates the implementation of the toronto agreement
The papers that reporting genome data usually include (i) genetic and functional changes of the genome and various components such as genes, repetitive elements, and (ii) molecular mechanisms of important traits. However, much information is still not being studied and reported, so the sharing of genomic data raised more concerns. Toronto agreement was proposed in 2009 (Toronto International Data Release Workshop Authors, 2009), aimed to share the scientific data such as the genome sequences before the publication. Data sharing promotes collaboration and contributes to the efficient use of data. Unfortunately, rapid sharing of genome data is still an area that need improvement.
There are a variety of technical means through database to fulfill the Toronto agreement: (i) the establishment of data access thresholds, such as a detailed disclaimer, or the registration system to identify the academic institutions (e-mail address ending with .edu) to share information only to the academic staff; (ii) only provide BLAST and/or Jbrowse and other tools, and DO not provide (it depends and could be optional) download data for data sharing. If only BLAST tools are provided, providers only need to contribute the protein and CDS without the whole genome. Jbrowse could provide genetic information without protein sequence. The provision of data and data sharing on the integrated database is not a nonexistent behavior. The earlier exposed to the public, the earlier intellectual property is committed, and the more efficient it promotes scientific collaborations. At present, due to the requirements by DOE, more genomic data in Phytozome have been released prior to the publication of related genome paper, such as the genome of kalanchoe, monkey flower, and so on. However, the Toronto agreement is yet to be fully implemented and needs in-depth practice.
Because of the value of angiosperms, the large number of genomic data has attracted many scientists and still brings us great challenges: the immediacy, integrity and analytical ability of the data. We provide on GDA database the timeline to update each of the recently sequenced plants (Figure 3). At present, 1,001 Arabidopsis strains (Weigel and Mott, 2009), 2,489 millet varieties (db.cngb.org/millet/), and 3,000 rice genomes have completed genome sequencing (The 3,000 rice genomes project). The 10K orchid genome project (J-J Project, sinicaorchid.gzit.net) has been put forward. All these projects made a huge challenge to the current database.
The current database for the processing of such large-scale data also lacks large-capacity computing devices and bioinformatics tools. Visualization of large data also poses a major challenge. The variety of data quality also requires an evaluation system to ensure that low-quality data is filtered to speed up the analysis. In general, the plant genome database will become a new biological branch. The supercomputing equipment, bioinformatics algorithms, and tool development need to be introduced and upgraded. In addition, a user-developer interactive than user-friendly interface is required. Overall, the upgrade of the angiosperm database will greatly enhance our understanding of important issues related to angiosperms and greatly promote the crop breeding process.
Concluding remarks
The genome data of flowering plants are rapidly accumulating in quantity and complexity. With the big data concept more and more popular in solving big questions, there will be a strong demand to integrate all related data. What's more, bioinformatics tools are usually developed and built firstly in comprehensive or large databases because they attract more researchers. We reviewed and compared the pros and cons on the data, tools, special highlights from three types of genome databases that are mostly used. We also proposed that a comprehensive genome database to host the genomes of all released angiosperms to accelerate the research of major scientific questions at the genome scale.
Author contributions
LZ: designed the research; FC, WD, and LZ: collected and analyzed the data; FC, WD, JZ, XG, JC, ZW, ZL, HT, and LZ: wrote, revised, and approved the manuscript.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by the National Key Research and Development Program of China (2016YFD0100305) and National Natural Science Foundation of China (81502437), and a start-up fund from Fujian Agriculture and Forestry University to LZ, and FC is supported by a grant from State Key Laboratory of Ecological Pest Control for Fujian and Taiwan Crops (SKB2017004).
References
- Adam-Blondon A.-F., Alaux M., Pommier C., Cantu D., Cheng Z., Cramer G. R., et al. (2016). Towards an open grapevine information system. Hort. Res. 3:16056. 10.1038/hortres.2016.56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badouin H., Gouzy J., Grassa C. J., Murat F., Staton S. E., Cottret L., et al. (2017). The sunflower genome provides insights into oil metabolism, flowering and Asterid evolution. Nature 546, 148–152. 10.1038/nature22380 [DOI] [PubMed] [Google Scholar]
- Balick M. J., Cox P. A. (1996). Plants, People, and Culture: the Science of Ethnobotany. New York, NY: Scientific American Library. [Google Scholar]
- Bell C. D., Soltis D. E., Soltis P. S. (2010). The age and diversification of the angiosperms re-revisited. Am. J. Bot. 97, 1296–1303. 10.3732/ajb.0900346 [DOI] [PubMed] [Google Scholar]
- Borsch T., Löhne C., Wiersema J. (2008). Phylogeny and evolutionary patterns in Nymphaeales: integrating genes, genomes and morphology. Taxon 57, 1052–1081. Available online at: http://www.ingentaconnect.com/content/iapt/tax/2008/00000057/00000004/art00004 [Google Scholar]
- Charlesworth D. (2016). Plant sex chromosomes. Annu. Rev. Plant Biol. 67, 2.1–2.24. 10.1146/annurev-arplant-043015-111911 [DOI] [PubMed] [Google Scholar]
- Choudhuri S. (2014). Bioinformatics for Beginners: Genes, Genomes, Molecular Evolution, Databases and Analytical Tools. New York, NY: Academic Press. [Google Scholar]
- Dash S., Campbell J. D., Cannon E. K. S., Cleary A. M., Huang W., Kalberer S. R., et al. (2016). Legume information system (legumeinfo.org): a key component of a set of federated data resources for the legume family. Nucleic Acids Res. 44, D1181–D1188. 10.1093/nar/gkv1159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duvick J., Fu A., Muppirala U., Sabharwal M., Wilkerson M. D., Lawrence C. J., et al. (2008). PlantGDB: a resource for comparative plant genomics. Nucleic Acids Res. 36, 959–965. 10.1093/nar/gkm1041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Hernandez M., Berardini T. Z., Chen G., Crist D., Doyle A., Huala E., et al. (2002). TAIR: a resource for integrated Arabidopsis data. Funct. Integr. Genomics 2, 239–253. 10.1007/s10142-002-0077-z [DOI] [PubMed] [Google Scholar]
- Goodstein D. M., Shu S., Howson R., Neupane R., Hayes R. D., Fazo J., et al. (2012). Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 40, 1178–1186. 10.1093/nar/gkr944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huala E., Dickerman A. W., Garcia-hernandez M., Weems D., Reiser L., Lafond F., et al. (2001). The Arabidopsis Information Resource (TAIR): a comprehensive database and web-based information retrieval, analysis, and visualization system for a model plant. Nucleic Acids Res. 29, 102–105. 10.1093/nar/29.1.102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubble E. (2005). So much more to know. Science 309, 78–102. 10.1126/science.309.5731.78b [DOI] [PubMed] [Google Scholar]
- Jarvis D. E., Ho Y. S., Lightfoot D. J., Schmöckel S. M., Li B., Borm T. J. A., et al. (2017). The genome of Chenopodium quinoa. Nature 542, 307–312. 10.1038/nature21370 [DOI] [PubMed] [Google Scholar]
- Jung S., Ficklin S. P., Lee T., Cheng C. H., Blenda A., Zheng P., et al. (2014). The Genome Database for Rosaceae (GDR): year 10 update. Nucleic Acids Res. 42, 1237–1244. 10.1093/nar/gkt1012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kersey P. J., Allen J. E., Christensen M., Davis P., Falin L. J., Grabmueller C., et al. (2014). Ensembl Genomes 2013: scaling up access to genome-wide data. Nucleic Acids Res. 42, 546–552. 10.1093/nar/gkt979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T., Tang H., Wang X., Paterson A. H. (2012). PGDD: a database of gene and genome duplication in plants. Nucleic Acids Res. 41, D1152–D1158. 10.1093/nar/gks1104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D., Zhang J. (2014). Diet shapes the evolution of the vertebrate bitter taste receptor gene repertoire. Mol. Biol. Evol. 31, 303–309. 10.1093/molbev/mst219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons E. H. (2008). CoGe, a New Kind of Comparative Genomics Platform: Insights Into the Evolution of Plant Genomes. Ann Arbor, MI: Proquest, Umi Dissertation Publishing. [Google Scholar]
- Main D., Cheng C.-H., Ficklin S. P., Jung S., Zheng P., Coyne C. J., et al. (2013). The cool season food legume database: an integrated resource for basic, translational and applied research, in Plant and Animal Genome XXI Conference (San Diego, CA: ). [Google Scholar]
- Niimura Y. (2012). Olfactory receptor multigene family in vertebrates: from the viewpoint of evolutionary genomics. Curr. Genomics 13, 103–114. 10.2174/138920212799860706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osorio D., Vorobyev M. (2008). A review of the evolution of animal colour vision and visual communication signals. Vis. Res. 48, 2042–2051. 10.1016/j.visres.2008.06.018 [DOI] [PubMed] [Google Scholar]
- Poliakov A., Foong J., Brudno M., Dubchak I. (2014). GenomeVISTA — an integrated software package for whole-genome alignment and visualization. Bioinformatics 30, 2654–2655. 10.1093/bioinformatics/btu355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proost S., Van Bel M., Sterck L., Billiau K., Van Parys T., Van De Peer Y., et al. (2009). PLAZA : A comparative genomics resource to study gene and genome evolution in Plants. Plant Cell 21, 3718–3731. 10.1105/tpc.109.071506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pryer K. M., Schneider H., Zimmer E. A., Banks J. A. (2002). Deciding among green plants for whole genome studies. Trends Plant Sci. 1385, 550–554. 10.1016/S1360-1385(02)02375-0 [DOI] [PubMed] [Google Scholar]
- Raskin I., Ribnicky D. M., Komarnytsky S., Ilic N., Poulev A., Borisjuk N., et al. (2002). Plants and human health in the twenty-first century. Trends Biotechnol. 20, 522–531. 10.1016/S0167-7799(02)02080-2 [DOI] [PubMed] [Google Scholar]
- Schattner P. (2008). Genomes, Browsers and Databases: Data-Mining Tools for Integrated Genomic Databases. Cambridge: Cambridge University Press. [Google Scholar]
- Schoof H., Zaccaria P., Gundlach H., Lemcke K., Rudd S., Kolesov G., et al. (2002). MIPS Arabidopsis thaliana Database (MAtDB): an integrated biological knowledge resource based on the first complete plant genome. Nucleic Acids Res. 30, 91–93. 10.1093/nar/30.1.91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjödin A., Street N. R., Sandberg G., Gustafsson P., Jansson S. (2009). The Populus Genome Integrative Explorer (PopGenIE): a new resource for exploring the Populus genome. New Phytol. 182, 1013–1025. 10.1111/j.1469-8137.2009.02807.x [DOI] [PubMed] [Google Scholar]
- Smedley D., Haider S., Ballester B., Holland R., London D., Thorisson G., et al. (2009). BioMart-biological queries made easy. BMC Genomics 10:22. 10.1186/1471-2164-10-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spannagl M., Nussbaumer T., Bader K. C., Martis M. M., Seidel M., Kugler K. G., et al. (2016). PGSB PlantsDB : updates to the database framework for comparative plant genome research. Nucleic Acids Res. 44, 1141–1147. 10.1093/nar/gkv1130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tello-Ruiz M. K., Stein J., Wei S., Preece J., Olson A., Naithani S., et al. (2016). Gramene 2016: comparative plant genomics and pathway resources. Nucleic Acids Res. 44, D1133–D1140. 10.1093/nar/gkv1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toronto, International Data Release Workshop Authors (2009). Prepublication data sharing. Nature 461, 168–170. 10.1038/461168a [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanBuren R., Bryant D., Edger P. P., Tang H., Burgess D., Challabathula D., et al. (2015). Single-molecule sequencing of the desiccation-tolerant grass Oropetium thomaeum. Nature 527, 508–511. 10.1038/nature15714 [DOI] [PubMed] [Google Scholar]
- Van de Peer Y., Mizrachi E., Marchal K. (2017). The evolutionary significance of polyploidy. Nat. Rev. Genet. 18, 411–424. 10.1038/nrg.2017.26 [DOI] [PubMed] [Google Scholar]
- Weigel D., Mott R. (2009). The 1001 genomes project for Arabidopsis thaliana. Genome Biol. 10:107. 10.1186/gb-2009-10-5-107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng L., Zhang Q., Sun R., Kong H., Zhang N., Ma H. (2014). Resolution of deep angiosperm phylogeny using conserved nuclear genes and estimates of early divergence times. Nat. Commun. 5, 4956. 10.1038/ncomms5956 [DOI] [PMC free article] [PubMed] [Google Scholar]