Abstract
Targeted and inducible regulation of mammalian gene expression is a broadly important capability. We engineered drug-inducible catalytically inactive Cpf1 fused to transcriptional activation domains to tune the expression of endogenous genes in human cells. Leveraging the multiplex capability of the Cpf1 platform, we demonstrate both synergistic and combinatorial gene expression in human cells. Our work should enable the development of multiplex gene perturbation library screens for understanding complex cellular phenotypes.
Sequence-specific RNA-guided CRISPR-Cas nucleases are simple to program1, 2; the widely used Streptococcus pyogenes Cas9 (SpCas9) can be targeted to specific DNA sequences by an associated complementary guide RNA (gRNA), provided that a protospacer adjacent motif (PAM) of the form NGG is also present. Catalytically inactive SpCas9 (dSpCas9) has been fused to transcriptional activation or repression domains to alter the expression of individual genes or to perform genome-wide library screens in mammalian cells3. Both small molecule- and light-inducible dSpCas9-based fusions have been developed4–6, enabling the ability to regulate the activity of this platform. Recently described CRISPR-Cpf1 nucleases offer additional capabilities beyond those of SpCas9 including shorter length CRISPR RNAs (crRNAs) for guiding Cpf1 to targets, the ability to target T-rich PAMs7, 8, and RNase processing of multiple crRNAs from a single transcript by sequence-specific ribonuclease activity resident within Cpf1 itself9, 10. However, to our knowledge, “dead” Cpf1 (dCpf1)-based gene regulators have thus far only been shown to repress gene expression in bacteria11, 12 and plant (Arabidopsis)13.
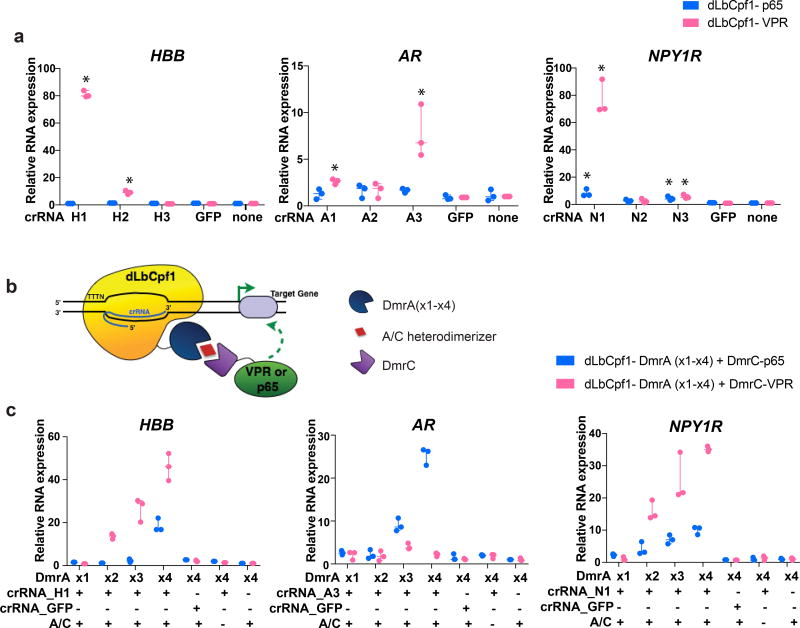
Based on preliminary experiments suggesting its higher activity (Supplementary Note 1 and Supplementary Fig. 1), we fused dCpf1 from Lachnospiraceae bacterium (dLbCpf1) to the strong synthetic VPR activator (Herpes Simplex Virus-derived VP16 activator, the human NF-KB p65 activation domain, and the Epstein-Barr Virus-derived R transactivator (Rta))14. We targeted these dLbCpf1-VPR fusions to the promoters of three different endogenous genes that are either epigenetically silenced according to ENCODE data (HBB) or expressed at low levels (AR and NPY1R) in HEK293 cells by designing three crRNAs for each promoter, located at various distances within 1 kb upstream of the transcription start site (TSS). At least one crRNA for each of the three target genes achieved robust transcriptional activation as assayed by real-time RT-PCR, whereas a dLbCpf1-p65 fusion alone showed little or no transcriptional activation of the target gene promoter (Fig. 1a), confirming previous observations with dSpCas914. We also tested a larger series of 32 crRNAs positioned within 1 kb upstream or 500 bp downstream of the TSSs of two additional endogenous genes, CD5 and CD22, which encode cell surface proteins. Most of the 32 crRNAs tested could significantly activate the target gene promoter when positioned between ~600 bp upstream and ~400 bp downstream of the TSSs (Supplementary Fig. 2), consistent with results obtained using dSpCas9 activators15. The levels of activation observed with dLbCpf1-based activators are comparable to what might be observed in naturally occurring biological systems and are similar to what has been reported previously for analogous dCas9-based activators16.
Figure 1. Targeted human endogenous gene regulation using individual crRNAs with dLbCpf1-based activators.
(a) Activities of dLbCpf1-p65 or dLbCpf1-VPR using single crRNAs at three endogenous human genes (HBB, AR, and NPY1R) in HEK293 cells. Relative mRNA expression is calculated by comparison to the control sample in which no crRNA is expressed. *, statistically significant difference compared to the control sample by Student t-test (two-tailed test assuming equal variance, p < 0.05).
(b) Schematic illustrating drug-dependent bi-partite dLbCpf1-based activator fusion proteins.
(c) Activities of drug-dependent bi-partite dLbCpf1-based activators using single crRNAs at three endogenous human genes (HBB, AR, and NPY1R) in HEK293 cells. The crRNA used for each promoters was the one that showed the highest activity in (a) above. Data shown in (a) and (c) represent three biological independent replicates.
To construct drug-regulated versions of dCpf1-VPR we used DmrA and DmrC domains, which interact only in the presence of a rapamycin analog known as the A/C heterodimerizer4, 17. We created split dLbCpf1 activators consisting of two fusions: dLbCpf1 fused to a DmrA domain and a DmrC domain fused to VPR or p65 (Fig. 1b), making activation dependent on the A/C drug. Although single crRNAs failed to reveal inducible activation of the HBB, AR or NPY1R genes (Fig. 1c), dLbCpf1 fusions harboring two, three or four tandem copies of the DmrA domain in the presence of DmrC-VPR and the A/C heterodimerizer drug increased gene activation at two of the three endogenous gene promoters (HBB and NPY1R; Fig. 1c). The degree of activation observed correlated with the number of DmrA domains, with maximum levels reaching approximately half of that observed with direct dLbCpf1-VPR fusions (Fig. 1c). Surprisingly, we found that using a DmrC-p65 fusion with dLbCpf1-DmrA fusions led to drug-dependent transcriptional upregulation from all three target gene promoters (Fig. 1c), an unexpected result given the lack of activation observed with dLbCpf1-p65 direct fusions and these same crRNAs (Fig. 1a).
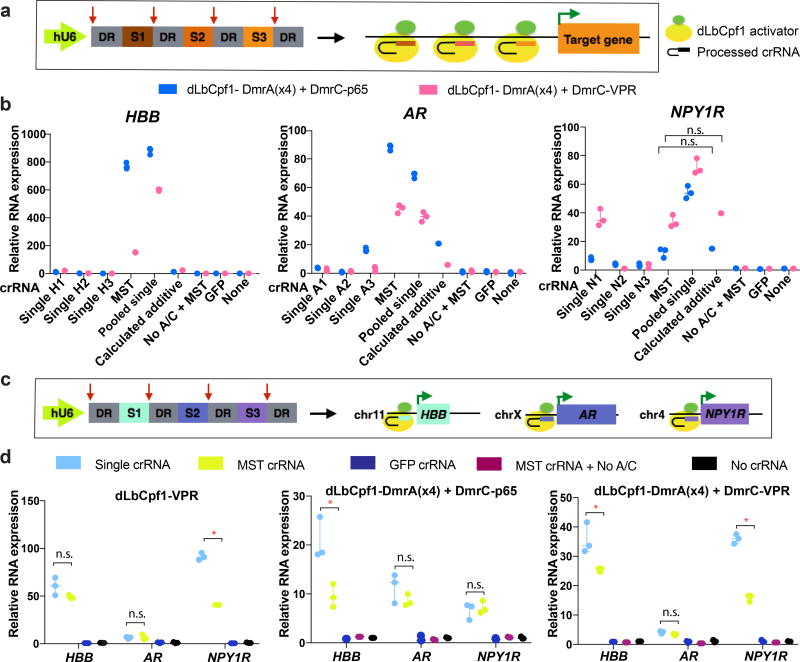
One major advantage of Cpf1 compared to Cas9 is the ability to encode two or more crRNAs in a multiplex single transcript (MST), with subsequent processing of individual crRNAs by the RNase processing activity of Cpf19, 10 (Fig. 2) potentially simplifying the implementation of synergistic and multiplex gene activation. We tested the use of MST crRNAs for synergistic activation by encoding sets of the 3 crRNAs targeting the promoters of HBB, AR or NPY1R genes on single transcripts (Fig. 2a). With dLbCpf1-VPR, we observed synergistic activation for HBB and AR but not for NPY1R (Supplementary Fig. 3). We also failed to observe synergistic activation of any of the three genes with dLbCpf1-p65, with either MST crRNAs or pooled single crRNAs (Supplementary Fig. 3). We also observed drug-dependent synergy with dLbCpf1-DmrA(x4) and DmrC-VPR at the HBB and AR promoters but again not at the NPY1R promoter (even though synergy was observed with pooled single crRNAs introduced together) (Fig. 2b). A similar pattern of results for these three gene promoters was observed with dLbCpf1-DmrA(x4) and DmrC-p65 (Fig. 2b) with higher activation observed on the HBB and AR genes relative to comparable experiments performed with DmrC-VPR (Fig. 2b). Interestingly, for the HBB gene, this higher activation was not seen with the single crRNA that worked the best (Fig. 1c). Although the reason for this difference is not known, it is possible that it relates to the number and configuration of transcriptional activation domains recruited to the promoter.
Figure 2. Multiplex and synergistic regulation of endogenous human genes by dLbCpf1-based activators.
(a) Schematic illustrating multiplex expression of three crRNAs each targeted to the same endogenous gene promoter in the same cell.
(b) Activities of dLbCpf1-DmrA(x4) and DmrC-VPR fusions (pink bars) or of dLbCpf1-DmrA(x4) and DmrC-p65 fusions (blue bars) with three single crRNAs, pooled sets of single crRNAs, and a MST encoding all three crRNAs on the HBB, AR, or NPY1R endogenous gene promoters. Synergistic effects of dLbCpf1-VPR/p65 fusions with MST crRNAs or individual pooled crRNAs are all statistically significant (Student t-test, two-tailed test assuming equal variance, p < 0.05) except for cases where n.s. (not significant) is indicated.
(c) Schematic illustrating multiplex expression of three crRNAs each targeted to a different endogenous gene promoter in a single cell.
(d) Simultaneous activation of three endogenous human genes using crRNAs expressed singly or from a MST together with dLbCpf1-VPR direct fusions (left panel), dLbCpf1-DmrA(x4) and DmrC-p65 fusions (middle panel), or dLbCpf1-DmrA(x4) and DmrC-VPR fusions (right panel). For the direct fusion experiment, a total of 250 ng of plasmids encoding the indicated crRNAs was used. For each drug-regulated fusion experiment, a total of 400ng of plasmids encoding the indicated crRNAs was used. Transcripts were measured in HEK293 cells using RT-qPCR with relative mRNA expression calculated by comparison to the control sample in which no crRNA is expressed. Representative data shown in (b) and (d) are of three biological independent replicates. hU6, human U6 Polymerase III promoter; DR; direct repeat sequence; n.s., not significant by Student t-test (p > 0.05); *, statistically significant by Student t-test (two-tailed test assuming equal variance, p< 0.05)
We also tested the use of MST crRNAs for multiplex activation of different gene promoters in the same cells (Fig. 2c). We found that MST crRNAs could be used together with (i) dLbCpf1-VPR, (ii) dLbCpf1-DmrA(x4) and DmrC-VPR fusions, or (iii) dLbCpf1-DmrA(x4) and DmrC-p65 fusions to simultaneously activate the endogenous HBB, AR, and NPY1R gene promoters in HEK293 cells (Fig. 2d). The activities of MST crRNAs were generally comparable relative to single individually expressed crRNAs with more than half of the gene/activator configurations we examined showing no statistically significant difference (Fig. 2d). For the cases where a difference was observed, the MST crRNAs generally showed only ~2-fold lower activation and the absolute level of fold-activation was still quite high, ranging from ~9-fold to ~40-fold. Attempts to further increase the activities of MST crRNAs did not yield substantial improvements (Supplementary Note 1 and Supplementary Fig. 4). To extend our findings to another human cell line, we also tested the direct VPR activator fusions and drug-regulated VPR and p65 activators in human U2OS cells, targeting the same genes (HBB, AR, and NPY1R) with MST crRNAs and single crRNAs (Supplementary Fig. 5). We observed that HBB and NPY1R were highly upregulated and that HBB activation with DmrC-p65 was even greater than in HEK293s. We did not observe activation of the AR gene but this is likely because its baseline expression is already highly elevated in U2OS cells (with a basal quantitative RT-PCR Ct value of ~28). Relative differences in the efficacies of crRNAs expressed singly or in MSTs were similar to what we observed in HEK293s.
In addition, we assessed the kinetics of activator effects to the addition and withdrawal of A/C heterodimerizer. We found that maximum activation of the HBB and AR genes was observed ~25 to 35 hours after the addition of drug (Supplementary Fig. 6a) and return of activated gene expression to baseline occurred ~35 to 45 hours after withdrawal of the drug (Supplementary Fig. 6b). We envision that drug-inducibility could be easily extended to other orthologues and we have successfully used these same strategies to regulate and tune dSpCas9-based activators (Supplementary Fig. 7) as have others who published similar drug-regulated dCas9-based activators while this work was in progress5.
To quantify the ability of Cpf1 to complement the targeting range of SpCas9, we performed an informatics analysis of sequences located between 600 bp upstream and 400 bp downstream of 32,696 TSSs in the human genome for sites with TTTV, TYCV and TATV PAMs7, 8 (Online Methods). We identified “desert” regions of ≥20 bps within these sequences in which no SpCas9 target sites could be found and summed these for each TSS. We then determined for each TSS, what percentage of these summed desert regions could be targeted by wild-type Cpf1 only (Supplementary Fig. 8a) and the two Cpf1 PAM recognition variants with wild-type Cpf1 (Supplementary Fig. 8b). Wild-type Cpf1 permits the targeting of 66% of the sum of all desert regions while the two PAM variants together with wild-type Cpf1 permit the targeting of 88% of these regions. As might be expected, the targeting capability increases as the size of the desert increases (Supplementary Figs. 8a and 8b).
Overall, our findings should enable and motivate the use of dCpf1-based gene regulatory proteins for performing both focused and genome-wide gene perturbation screens in mammalian cells. More broadly, our results should also motivate the engineering of dCpf1-based direct and drug-regulated fusions to other heterologous regulatory domains (e.g., p300, DNMT, TET1) as has been done previously with dSpCas918–20. Successful development of such fusions could enable recruitment of two different effector domains in a controlled manner using dCas9 and dCpf1 scaffolds targeted to the same promoter.
ONLINE METHODS
Life Sciences Reporting Summary
A checklist summarizing experimental design, software used, and characterization of materials and reagents used is attached.
Plasmids and oligonucleotides
Diagram of constructs and a list of plasmids and related sequences used in this study are found in Supplementary Note 2; LbCpf1 target sites and multiplex crRNA sequences, and SpCas9 target sites are found Supplementary Table 1. Plasmids dLbCpf1-p65 (JG1202) and dLbCpf1-VPR (JG1211) were constructed by cloning p65 and VPR domains into dLbCpf1(D832A) (MMW1578) using BstZ17I and Not I sites through isothermal assembly. VPR was amplified from dSpCas9-VPR which was a gift from George Church (Addgene plasmid # 63798)14. Plasmids encoding dLbCpf1-DmrA(x1), dLbCpf1-DmrA(x2), dLbCpf1-DmrA(x3), and dLbCpf1-DmrA(x4) (JG674, JG676, JG693, and YET1000, respectively) were generated by subcloning dLbCpf1(D832A) into AgeI and XhoI digested constructs that have different numbers of DmrA domains (BPK1019, BPK1033, BPK1140, BPK1179 for dSpCas9-DmrA(x1) to dSpCas9-DmrA(X4), respectively) using isothermal assembly. Plasmids encoding dSpCas9(D10A/H840A) effector fusions to VP64, p65, or DmrA were cloned via isothermal assembly (for a complete list of plasmids, please see Supplementary Note 2). The DmrC entry vector was digested with NruI, and p65 or VPR were inserted via isothermal assembly to generate DmrC-p65 (BPK1169) and DmrC-VPR (MMW948). Single LbCpf1 crRNA expression plasmids were constructed by ligating annealed oligo duplexes into BsmBI-digested BPK3082 (Addgene #78742)21. MST LbCpf1 crRNA plasmids were assembled by annealing, phosphorylating, and ligating three pairs of oligonucleotides into BsmBI and HindIII-digested BPK3082. Sequences for all oligo pairs are listed in Supplementary Table 1. The NF-KB p65 domain encoded in JG1202, BPK1169, BPK1160, and BPK1163 encompasses amino acids 285 to 551 of the native protein while domain encoded in constructs containing VPR encompass amino acids 428 to 546.
Human cell culture and transfection
HEK293 cells (Invitrogen) and HEK293T cells (ATCC) were grown at 37° C, in 5% CO2 in Dulbecco’s Modified Eagle Medium (DMEM) with 10% heat-inactivated fetal bovine serum (FBS) and 1% penicillin and streptomycin. U2OS cells (obtained from Toni Cathomen, Freiburg) were grown at 37° C, in 5% CO2 in Advanced DMEM supplemented with 10% heat-inactivated FBS, 2 mM GlutaMax, and 1% penicillin and streptomycin. Media supernatant was analyzed biweekly for the presence of Mycoplasma. For the GFP activation assays in Supplementary Fig. 1, 500 ng of plasmid DNA expressing dCas9-VPR-BFP (JKNp44), dAsCpf1-VPR-BFP (JKNp49) or dLbCpf1-VPR-BFP (JKNp47) and 500 ng of crRNA plasmid were co-transfected into HEK293T cells that stably encode GFP under a Tet-On 3G doxycycline-inducible promoter (Takara Clontech). Both the dLbCpf1 and dAsCpf1 protein coding sequences were codon-optimized for expression in human cells. Median GFP fluorescence was measured 3 days post-transfection using a LSR II Flow Cytometer (BD) and values were normalized to HEK293T-GFP cells with no plasmids transfected. For LbCpf1 direct fusion experiments on endogenous genes, 750 ng of dLbCpf1-p65 or dLbCpf1-VPR plasmids with 250 ng of LbCpf1 crRNA plasmids were co-transfected using a 3ul of TransIT®-LT1 Transfection Reagent (Mirus, cat# MIR2300) into HEK293 cells in a 12-well plate. For experiments with dLbCpf1 fusions to DmrA domains, 400 ng of dLbCpf1-DmrA fusions plasmids, 200 ng of DmrC-p65 or DmrC-VPR plasmids, and 400 ng of LbCpf1 crRNA plasmids were co-transfected using 3 ul of LT1 into HEK293 cells in a 12-well plate. A complete media containing 500 µM A/C heterodimerizer (Takara Clontech) was used at the time of transfection. For LbCpf1 experiments in U2OS cells, same amount of plasmids used in HEK293 cells were co-transfected by nucleofection using the DN-100 program on a Lonza 4-D Nucleofector with the SE Cell Line Kit (Lonza). For experiments in Supplementary Fig. 6, 1000 ng of dLbCpf1-DmrA fusions plasmids, 500 ng of DmrC-p65 plasmid, and 1000 ng of LbCpf1 MST crRNA plasmids targeting HBB promoter or AR promoter were co-transfected using 7.5 ul of LT1 into HEK293 cells in a 6-well plate. After 34 hours of post-transfection, cells were split into 24-well plates to harvest at different time points.
Quantitative reverse transcription PCR
Total RNA was extracted from the transfected cells 72 hours post-transfection using the NucleoSpin® RNA Plus (Clontech, cat# 740984.250), and 250 ng of purified RNA was used for cDNA synthesis using High-Capacity RNA-cDNA kit (ThermoFisher, cat# 4387406). cDNA was diluted 1:20 and 3 ul of cDNA was used for quantitative PCR (qPCR). qPCR reaction samples were prepared using cDNA, SYBR (ThermoFisher, cat# 4385612), and primers detecting each target transcript. Primer sequences are listed in Supplementary Table 2. qPCR was performed using Roche LightCycler480 with the following cycling protocols (Supplementary Table 2). When Ct values are over 35, we considered them as 35, because Ct values fluctuate for very low expressed transcripts. Samples that were transfected with LbCpf1 crRNA backbone plasmid, BPK3082 were used as negative controls, and the levels of fold activation over negative controls were normalized to the expression of HPRT1.
Flow Cytometry
Cells were washed with cell staining buffer (Biolegends, #420201) after 72 hours post-transfection and incubated with CD22 (Biolegends, #363511) or CD5 antibody (Biolegends, #364008) for 15 minutes, followed by twice wash with cell staining buffer. PE/Cy-7 for CD5 positive cells and BV421 for CD22 positive cells were measured by a LSR Fortessa X-20 flow cytometer (BD).
VEGFA ELISA
HEK293 cells were seeded in 24-well plates roughly 20 hours prior to transfection using Lipofectamine 3000 (Thermo Fisher Scientific). Unless otherwise indicated, 250 ng of dCas9-DmrA plasmid was co-transfected with 250 ng of sgRNA plasmid (single VEGFA site 3 sgRNA or pooled VEGFA sites 1, 2, and 3 sgRNAs22) and 125 ng of DmrC-effector plasmid. A complete media exchange was performed approximately 20 hours post-transfection, with the exchanged media containing 500 µM A/C heterodimerizer (Takara Clontech), unless otherwise noted. Approximately 42 hours post-transfection, supernatant media was removed and clarified prior to analysis using a Human VEGF Quantikine ELISA kit (R&D Systems). Optical density of stopped ELISA reactions was determined using a Model 860 Microplate reader (Bio-Rad).
Western Blot
HEK293T cells were transfected in 6-well plates with 3 µg of plasmids encoding VPR-fused dCas9, dAsCpf1, dLbCpf1 and 3 µg of guide RNA-encoding plasmid. Cells were harvested 48 hours post-transfection with PBS and lysed in RIPA buffer (Thermo Fisher Scientific) supplemented with protease inhibitors (Sigma-Aldrich). Soluble lysates were quantified by BCA protein assay (Thermo Scientific) and 40 µg of each sample was loaded into a 4–12% Bis-Tris protein gel (Thermo Scientific). HA-tagged VPR fusions were detected with rat anti-HA antibody (Sigma-Aldrich). GAPDH was detected with rabbit anti-GAPDH antibody (Cell Signaling Technology). Blots were imaged by LI-COR Odyssey® CLx.
Computational analysis of Cpf1-targetable sites in the human genome
Cpf1 targeting range was characterized by analyzing sequences −600 bps and +400 bps relative to the TSS of 32,696 genes in the human genome (GRCh37/hg19). To assess the complementarity between the targeting ranges of Cpf1 and SpCas9, “desert” regions of ≥20 bps that are not targetable by SpCas9 (20 bp spacer + NGG PAM) were enumerated and summed for each gene. These desert regions were then analyzed to quantify the proportion that could be targetable by wild-type Cpf1 alone or with the two Cpf1 PAM recognition variants (TYCV and TATV)8 together with wild-type Cpf1. The desert region sums for each gene were binned into the following groups to assess the distributions: 20 – 100 bps, 101 – 200 bps, 201 – 300 bps, 301 – 400 bps, 401 – 500 bps, and 501 – 1000 bps. Codes for the analysis performed are available from the corresponding author upon reasonable request.
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Supplementary Material
Acknowledgments
We thank Moira Welch and Alexander Sousa for technical assistance with constructing certain plasmids. B.P.K. acknowledges support from Banting (Natural Sciences and Engineering Research Council of Canada) and Charles A. King Trust Postdoctoral Fellowships. This work was supported by the National Institutes of Health R35 GM118158 (J.K.J.), NIH R01 GM107427 (J.K.J.), R01 DA036858 (J.S.W.) and U01 CA168370 (J.S.W.), the Howard Hughes Medical Institute (J.S.W.), and the Desmond and Ann Heathwood Massachusetts General Hospital Research Scholar Award (J.K.J.).
Footnotes
AUTHOR CONTRIBUTIONS
Y.E.T., B.P.K., J.K.N., J.Y.H., J.S.W., and J.K.J. conceived of and designed experiments. Y.E.T., B.P.K., J.K.N., J.Y.H., J.E.H., and J.G. performed experiments. Y.E.T., B.P.K., J.K.N., J.Y.H., J.S.W., and J.K.J. wrote the manuscript.
COMPETING FINANCIAL INTERESTS STATEMENT
B.P.K. is a consultant for Avectas. J.S.W. is a founder and scientific advisory board member of KSQ Therapeutics. J.K.J. has financial interests in Beacon Genomics, Beam Therapeutics, Editas Medicine, Pairwise Plants, Poseida Therapeutics, and Transposagen Biopharmaceuticals. J.K.J.’s interests were reviewed and are managed by Massachusetts General Hospital and Partners HealthCare in accordance with their conflict of interest policies.
References
- 1.Komor AC, Badran AH, Liu DR. CRISPR-Based Technologies for the Manipulation of Eukaryotic Genomes. Cell. 2017;168:20–36. doi: 10.1016/j.cell.2016.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wright AV, Nunez JK, Doudna JA. Biology and Applications of CRISPR Systems: Harnessing Nature's Toolbox for Genome Engineering. Cell. 2016;164:29–44. doi: 10.1016/j.cell.2015.12.035. [DOI] [PubMed] [Google Scholar]
- 3.Dominguez AA, Lim WA, Qi LS. Beyond editing: repurposing CRISPR-Cas9 for precision genome regulation and interrogation. Nat Rev Mol Cell Biol. 2016;17:5–15. doi: 10.1038/nrm.2015.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zetsche B, Volz SE, Zhang F. A split-Cas9 architecture for inducible genome editing and transcription modulation. Nature biotechnology. 2015;33:139–142. doi: 10.1038/nbt.3149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bao Z, Jain S, Jaroenpuntaruk V, Zhao H. Orthogonal Genetic Regulation in Human Cells Using Chemically Induced CRISPR/Cas9 Activators. ACS synthetic biology. 2017 doi: 10.1021/acssynbio.6b00313. [DOI] [PubMed] [Google Scholar]
- 6.Polstein LR, Gersbach CA. A light-inducible CRISPR-Cas9 system for control of endogenous gene activation. Nature chemical biology. 2015;11:198–200. doi: 10.1038/nchembio.1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zetsche B, et al. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell. 2015;163:759–771. doi: 10.1016/j.cell.2015.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gao L, et al. Engineered Cpf1 variants with altered PAM specificities. Nat Biotechnol. 2017 doi: 10.1038/nbt.3900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fonfara I, Richter H, Bratovic M, Le Rhun A, Charpentier E. The CRISPR-associated DNA-cleaving enzyme Cpf1 also processes precursor CRISPR RNA. Nature. 2016;532:517–521. doi: 10.1038/nature17945. [DOI] [PubMed] [Google Scholar]
- 10.Zetsche B, et al. Multiplex gene editing by CRISPR-Cpf1 using a single crRNA array. Nature biotechnology. 2017;35:31–34. doi: 10.1038/nbt.3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim SK, et al. Efficient Transcriptional Gene Repression by Type V-A CRISPR-Cpf1 from Eubacterium eligens. ACS synthetic biology. 2017 doi: 10.1021/acssynbio.6b00368. [DOI] [PubMed] [Google Scholar]
- 12.Zhang X, et al. Multiplex gene regulation by CRISPR-ddCpf1. Cell Discov. 2017;3:17018. doi: 10.1038/celldisc.2017.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tang X, et al. A CRISPR-Cpf1 system for efficient genome editing and transcriptional repression in plants. Nature plants. 2017;3:17018. doi: 10.1038/nplants.2017.18. [DOI] [PubMed] [Google Scholar]
- 14.Chavez A, et al. Highly efficient Cas9-mediated transcriptional programming. Nature methods. 2015;12:326–328. doi: 10.1038/nmeth.3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gilbert LA, et al. Genome-Scale CRISPR-Mediated Control of Gene Repression and Activation. Cell. 2014;159:647–661. doi: 10.1016/j.cell.2014.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chavez A, et al. Comparison of Cas9 activators in multiple species. Nature methods. 2016;13:563–567. doi: 10.1038/nmeth.3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rivera VM, Berk L, Clackson T. Dimerizer-mediated regulation of gene expression in vivo. Cold Spring Harbor protocols. 2012;2012:821–824. doi: 10.1101/pdb.prot070144. [DOI] [PubMed] [Google Scholar]
- 18.Hilton IB, et al. Epigenome editing by a CRISPR-Cas9-based acetyltransferase activates genes from promoters and enhancers. Nature biotechnology. 2015;33:510–517. doi: 10.1038/nbt.3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu XS, et al. Editing DNA Methylation in the Mammalian Genome. Cell. 2016;167:233–247.e217. doi: 10.1016/j.cell.2016.08.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Amabile A, et al. Inheritable Silencing of Endogenous Genes by Hit-and-Run Targeted Epigenetic Editing. Cell. 2016;167:219–232.e214. doi: 10.1016/j.cell.2016.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kleinstiver BP, et al. Genome-wide specificities of CRISPR-Cas Cpf1 nucleases in human cells. Nature biotechnology. 2016;34:869–874. doi: 10.1038/nbt.3620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maeder ML, et al. CRISPR RNA-guided activation of endogenous human genes. Nat Methods. 2013;10:977–979. doi: 10.1038/nmeth.2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.