Abstract
Activated carbon (AC) is an increasingly attractive remediation alternative for the sequestration of dioxins at contaminated sites globally. However, the potential for AC to reduce the bioavailability of dioxins in mammals and the residing gut microbiota has received less attention. This question was partially answered in a recent study examining 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD)-induced hallmark toxic responses in mice administered with TCDD sequestered by AC or freely available in corn oil by oral gavage. Results from that study support the use of AC to significantly reduce the bioavailability of TCDD to the host. Herein, we examined the bioavailability of TCDD sequestered to AC on a key murine gut commensal and the influence of AC on the community structure of the gut microbiota. The analysis included qPCR to quantify the expression of segmented filamentous bacteria (SFB) in the mouse ileum, which has responded to TCDD-induced host toxicity in previous studies and community structure via sequencing the 16S ribosomal RNA (rRNA) gene. The expression of SFB 16S rRNA gene and functional genes significantly increased with TCDD administered with corn oil vehicle. Such a response was absent when TCDD was sequestered by AC. In addition, AC appeared to have a minimal influence on murine gut community structure and diversity, affecting only the relative abundance of Lactobacillaceae and two other groups. Results of this study further support the remedial use of AC for eliminating bioavailability of TCDD to host and subsequent influence on the gut microbiome.
Keywords: TCDD, Gut microbiome, Segmented filamentous bacteria, Dysbiosis, Activated carbon sequestration, Corn oil, Remediation, Dioxin
Introduction
Polychlorinated dibenzo-p-dioxins and furans, or simply dioxins, are a group of persistent organic pollutants that are widespread in the environment (Ferrario et al. 2000; Gadomski et al. 2004). Dioxin exposure, typically from food such as fish or dairy, is partly the result of persistence and accumulation in river sediment and floodplains, necessitating ongoing remedial efforts (Yamashita et al. 2000; Ghosh et al. 2011). Compared to dredging or isolation with walls and capping, alternative remediation methods such as in situ activated carbon (AC) sorbent amendments are an economically viable control mechanism (Bridges et al. 2010; E.P.A. 2013). Thus, AC has recently been implemented in multiple contaminated sites throughout the world (Patmont et al. 2015). Despite increasing use and demonstrated potential for AC to reduce the bioavailability of dioxins in aquatic invertebrates and earthworms (Fagervold et al. 2010; Josefsson et al. 2012) influence on mammals, their residing gut microbiota has received less attention.
This question was addressed, in part, in a recent study that examined the availability of 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) amended to AC (AC-TCDD) in a murine model (Boyd et al. 2017). Classic markers of TCDD-induced toxicity were measured including modulation of the IgM antibody-forming cell (AFC) and the dioxin-inducible cytochrome P450 family cyp1a1 messenger RNA (mRNA) gene expression. Results from that study supported the use of AC to eliminate bioavailability of dioxins in mammals in that TCDD-induced host responses were observed only when administered with corn oil (CO-TCDD). Responses were not observed with equivalent TCDD doses administered with AC.
The goals of this study were to (1) examine the bioavailability of TCDD sequestered to AC (AC-TCDD) on a key gut commensal in the murine model and (2) characterize the influence of AC on the gut microbiota. This is an important question as the gut microbiota plays a crucial role in host health and immune competence (Littman and Pamer 2011; Honda and Littman 2016). Previous studies have observed TCDD-induced dysbiosis of gut commensals including Candidatus Savagella also known as segmented filamentous bacteria (SFB), Firmicutes, and Enterobacteriaceae (Bhaduri 2015; Lefever et al. 2016; Stedtfeld et al. 2017, Modulatory influence of segmented filamentous bacteria on transcriptomic response of gnotobiotic mice exposed to TCDD, submitted). Experiments performed by our group and others suggest that gut commensal response is the result of TCDD-induced toxicity to the host, mediated in part, through the aryl hydrocarbon receptor (AhR), and not the direct influence of TCDD on bacteria (Lefever et al. 2016; Stedtfeld et al. 2017). Given the recently demonstrated limited bioavailability of AC-TCDD to hallmark responses in mice (Boyd et al. 2017), we hypothesized that the response of key gut commensals would also be muted. To test this hypothesis, the transcriptomic response of SFB was examined, which has previously been shown to respond to the toxicity induced by TCDD (Bhaduri 2015; Stedtfeld et al. 2017) and other AhR ligands (Zhang et al. 2015).
A secondary question was to examine the influence of food grade AC on the gut microbiome. Along with the potential for entering the food chain by way of ingestion, AC has also been used to remove residual levels of antibiotics from the colon (Khoder et al. 2010), or as a food additive to reduce bioavailability of contaminants in animal feed (Avantaggiato et al. 2004)—a potential alternative to antibiotics in animal husbandry (Chu et al. 2013). AC is also used as an emergency treatment for accidental dosing of poisonous substances or overdosing (Holt and Holz 1963). It is increasingly used as a remedy or supplement for intestinal disorders and detoxification. However, the potential for AC to interfere with the gut microbiome and subsequently host health has not been well documented. This was examined here via 16S ribosomal RNA (rRNA) gene sequencing using DNA extracted from ileal tissues of mice exposed to AC-TCDD.
Materials and methods
Animals and experimental design
Pathogen-free female B6C3F1 mice that were 5 to 8 weeks old were purchased from Charles River Breeding Laboratories (Portage, MI, USA) and handled as previously described (Boyd et al. 2017). Briefly, 45 mice were randomly separated, five per cage into nine separate cages, one for each experimental group. The experimental design (Fig. S1) included mice dosed by oral gavage with corn oil vehicle (CO) with 0, 0.1, 1.0, and 10 μg/kg TCDD and mice dosed by oral gavage with AC vehicle with 0, 0.1, 1.0, and 10 μg/kg TCDD. A naïve group (n = 5) with no oral exposure was also used. Five mice were placed in each group for replication. Food grade WPC coconut AC (Calgon Carbon Corp) was characterized and adsorbed with TCDD as described previously (Boyd et al. 2017). Briefly, for AC-TCDD, 156.25 mg of AC loaded with TCDD via the incipient wetness method (Boyd et al. 2011; Kaplan et al. 2011) and confirmed through analysis using pyrolysis gas chromatography mass spectrometry was suspended in 5 ml deionized water prior to dosing. Mice were dosed by oral gavage 200 μl once per day for four consecutive days, resulting in 6.25 mg of AC-TCDD per day. Three days after the fourth and final exposure with TCDD (7 days into the experiment), mice were sacrificed and tissues were collected for analysis. During all treatments, mice had access to water and food ad libitum. The entire experiment was replicated and included three naïve mice as a no treatment control. All animals received humane care in compliance with the animal use protocol approved by Michigan State University.
Isolation of tissue and transcriptomic response with qPCR
RNA was extracted from the ileum of all mice to evaluate the transcriptome response of SFB in response to TCDD. At the time of sacrifice, a 0.25-cm segment proximal to the caecum was removed from the same spot for all animals and was used for subsequent analysis. Intestinal tissue samples were added to RNA/DNA stabilizer and stored at − 80 °C until further use. The PureLink RNA Mini Kit with Trizol (12183018A, Ambion/Thermo Fisher Scientific, Waltham, MA) was used to extract and purify RNA from ileal tissue and content. RNA was processed further to remove DNA using a two-step procedure that included DNAse I (Invitrogen/Thermo Fisher Scientific, Waltham, MA) and Turbo DNAse I kit (Life Technologies/Thermo Fisher Scientific, Waltham, MA). After extraction, isolated RNA was quantified using a Qubit (Life Technologies) and assessed for purity using the Nanodrop ND-1000 UV-Vis spectrophotometer (Nanodrop Products, Wilmington, DE). Complementary DNA was synthesized via random primers using the High-Capacity cDNA Reverse Transcription Kit (Thermo Fisher Scientific, Waltham, MA), with a starting amount of 1.0 μg of RNA.
For mRNA expression analysis of bacteria, qPCR primers were designed from functional gene sequences available for SFB (Table S1). Genes targeting SFB function were selected based on potential interaction with the host and Th17 cell induction (Pamp et al. 2012). Primers were designed as previously described (Stedtfeld et al. 2008) using default parameters of Primer Express version 2.0.
QPCR was performed using a custom SmartChip™ (Wafergen Biosystems, Fremont, CA) with SFB-specific 16S rRNA gene and functional gene primers (Table S1). Briefly, Wafergen’s custom SmartChip™ array permits 5184 qPCR reactions with 100 nl volumes to be run in parallel. Sample and primers were dispensed into the SmartChip™ using a Multi-sample Nano-dispenser (Wafergen Biosystems, Fremont, CA). PCR cycling conditions and initial data processing were performed as previously described (Wang et al. 2014; Stedtfeld et al. 2016). Amplification reactions on the SmartChip™ consisted of 1×LightCycler 480 SYBR® Green I Master Mix (Roche Inc., USA), nuclease-free PCR-grade water, 10 ng/μl cDNA template per sample in the qPCR reaction (0.5 ng per reaction well), and 0.5 μM of each forward and reverse primer. Each reaction was tested in triplicate, and a no template control was also run. The mean genomic copies per reaction were calculated as previously described (Looft et al. 2012). Genes detected in only one of the three technical replicates in each sample were considered false positives and were filtered from the analysis.
Analysis of 16S rRNA genes using DNA extracted from ileum
For 16S rRNA gene sequence analysis, community DNA was extracted from ileal tissue and content using the Power Soil Extraction kit (MoBio). Amplicon sequencing libraries of the bacterial 16S V4 hypervariable region were made as previously described (Kozich et al. 2013). Following PCR, reaction outputs were normalized using Invitrogen SequalPrep DNA Normalization plates and pooled. The library pool was quantified using a combination of Qubit dsDNA HS, Caliper LabChipGX HS DNA, and Kapa Illumina Library Quantification qPCR assays. The pool was loaded on an Illumina MiSeq v2 standard flow cell for sequencing, which was performed in a 2 × 250 bp paired-end format with a v2, 500 cycle reagent cartridge. Sequencing primers specific for the V4 region (515f/806r) were added to the appropriate wells of the reagent cartridge as described in (Kozich et al. 2013). Base calling was done by Illumina Real Time Analysis (RTA) v1.18.64, and output of RTA was demultiplexed and converted to a FastQ format with Illumina Bcl2fastq v1.8.4. Raw sequence reads were submitted to NCBI (BioProject Accession No. PRJNA381198). Sequences were analyzed using default parameters of Qiime (Caporaso et al. 2010). Analysis of structural shifts was performed at the family level. Chao and Shannon diversity indices were calculated using the same sequencing depth (6472 sequences per sample).
Statistical analysis and SFB gene quantification
QPCR was analyzed for absolute abundance based on the mass of tissue and content used to extract RNA and extraction/digestion yield. Expression analysis of SFB functional genes included normalizing to SFB-specific 16S rRNA gene primer set (calculating relative expression) and examining analysis of fold change between the group administered 10 μg/kg TCDD with corn oil (CO_10TCDD) and all other groups. The ROUT method with a Q = 1% was used to identify and remove outliers (Motulsky and Brown 2006). A one-way ANOVA and a multiple comparison Sidak tests were performed to compare all groups with the naïve control and to compare the 10 μg/kg CO-TCDD group with the CO group without TCDD and the 10 μg/kg AC-TCDD group. For comparison of just two groups, a two-tailed unpaired t test was used. All statistical analysis and some plots were generated using Prism (version 7 for Windows, GraphPad Software, San Diego, CA, USA). Volcano plots were made using Excel. Shifts in abundance were deemed significantly different if fold change > |1.5| and p < 0.05.
Results
Effect of AC-TCDD on SFB
ATCDD-elicited host response significantly influenced the absolute abundance of SFB solely in the group dosed with 10 μg/kg CO-TCDD compared to the naïve control (Fig. 1a). A TCDD-induced response in SFB was not observed with any of the doses of AC-TCDD. Overall, a 5.0-, 8.5-, and 3.7-fold higher level of SFB was observed in the group dosed with 10 μg/kg CO-TCDD compared to naïve, the CO without TCDD, and 10 μg/kg AC-TCDD group, respectively. There was no measurable difference between the 10 μg/kg AC-TCDD and AC control (fold change = 0.92), and the naïve group (fold change = 2.28, p = 0.98). A similar result was observed with the replicate experiment, in which a significant increase in SFB was only observed in the group that received the highest dose of CO-TCDD (Fig. S2). Concordant with previously described studies, TCDD administered on AC eliminated both the toxic response in mice (Boyd et al. 2017) and the subsequent effect on a key member of the gut microbiome. However, a concentration-dependent dose response that was previously observed with host bioassays was not observed with SFB.
Fig. 1.
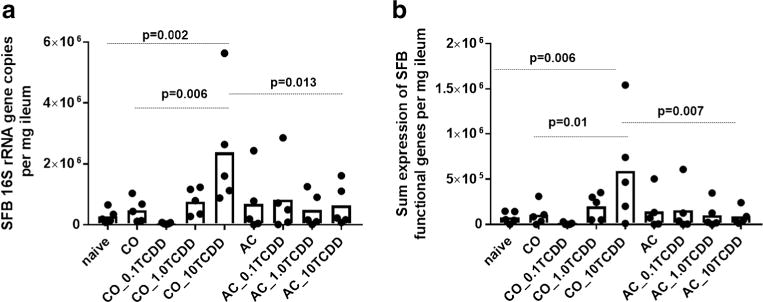
QPCR analysis of SFB gene expression from ileal tissue in response to 0, 0.1, 1.0, or 10 μg/kg TCDD administered with corn oil vehicle (CO_TCDD) or adsorbed onto activated carbon (AC_TCDD): a expression of SFB-specific 16S rRNA gene primer set and b sum expression of SFB functional genes (n = 21 primer sets). p values are listed from an ANOVA with a multiple mean comparison Sidak test between naïve, CO with no TCDD (CO), and 10 μg/kg TCDD administered on AC (AC_10TCDD) versus 10 μg/kg TCDD administered on CO (CO_10TCDD). Numbers 0.1, 1.0, and 10 indicate microgram per kilogram dose of TCDD
To further verify TCDD-induced dysbiosis of SFB solely in groups dosed with CO, the expression of select functional genes in ileal tissues was also analyzed via qPCR. Functional SFB genes selected for analysis included factors putative to host interaction (Pamp et al. 2012). The number of functional genes detected with 10 μg/kg of CO-TCDD was higher (11.8 ± 6.2 number of genes detected) than other groups (4.3 ± 4.0 number of genes detected). The summed expression of all tested functional genes was also significantly higher for the 10 μg/kg CO-TCDD group of mice (Fig. 1b). All groups administered with AC-TCDD did not show a significant increase in SFB functional gene expression.
The relative expression of functional genes (normalized to SFB-specific 16S rRNA gene) tended to decrease in the 10 μg/kg of CO-TCDD group compared to all other groups (Fig. 2a). However, this was only significant in the putative D-ala-D-ala-dipeptidase gene (Fig. 2b) and tended to be lower (p = 0.06) in the putative fibronectin-binding protein. The relative expression analysis was only performed on genes that were detected in a majority of replicates within a given group; thus, only 11 genes are displayed (Fig. 2a). The mean expression of all genes detected shows that while the SFB 16S rRNA gene increased in the group dosed with 10 μg/kg of CO-TCDD, the level of expression varied among functional genes (Fig. S3). Taken together, the expression of SFB functional genes further verifies that TCDD was not bioavailable when adsorbed on to AC.
Fig. 2.
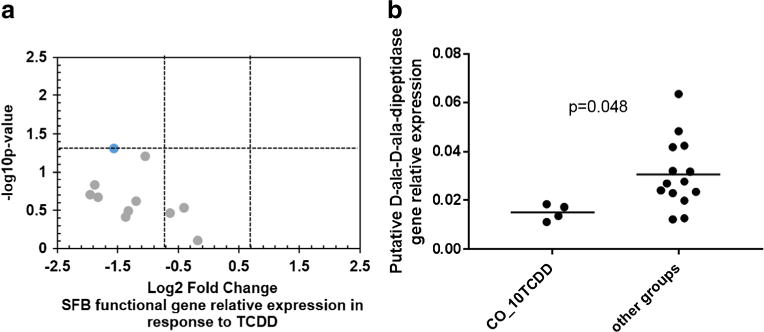
QPCR analysis of SFB functional gene relative expression in the ileum. a Volcano plot showing −Log10 p value and Log2 fold change comparing the relative functional gene expression of the group dosed with 10 μg/kg TCDD administered with corn oil (CO_10TCDD) with all other groups and b the relative expression of the putative D-ala-D-ala dipeptidase gene. The functional gene relative expression was normalized to SFB-specific 16S rRNA gene
Effect of AC and CO on gut microbiome
The 16S rRNA gene was sequenced to examine the influence of AC and CO on gut microbiota in the ileum. Results showed that AC significantly increased the amount of Lactobacillaceae in the ileum (Fig. 3b). Two separate bacterial groups significantly decreased in response to AC including Ruminococcaceae and Rikenellaceae (Fig. 3c, d). Chao and Shannon diversity indices were also examined, and no significant shift in diversity was observed due to AC, CO, or TCDD (Fig. S4). While the Shannon diversity indices tended to be lower with groups that were administered with AC, this was not significant (p = 0.114).
Fig. 3.
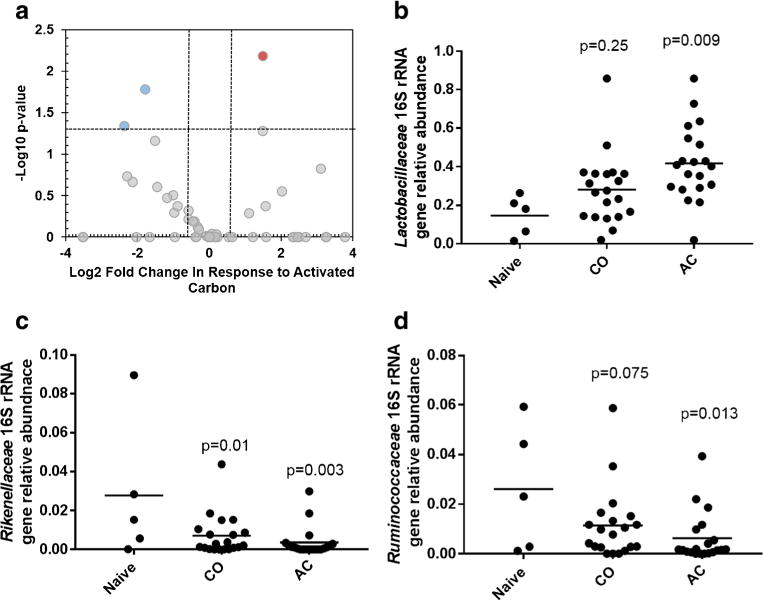
16S rRNA gene sequence analysis using DNA extracted from ileum comparing naïve mice with groups dosed with corn oil (CO) or dosed with activated carbon (AC). a Volcano plot showing −Log10 p value and Log2 fold change for bacterial sequences classified at the family level between naïve mice and mice dosed with AC and b–d the taxonomic families significantly influenced with AC
CO, a common vehicle for administering TCDD and other AhR ligands in murine studies (Fernandez-Salguero et al. 1996; Bruner-Tran and Osteen 2011), also influenced the gut microbiota (Fig. S5b), but to a lesser extent than previously observed with sesame oil. In detail, only three bacterial families including Methanosaetaceae, Erysipelotrichaceae, and a group from clostridia were significantly reduced (p < 0.05) in mice dosed with CO. In our previous studies using sesame oil—a common vehicle (Brown et al. 1998; Kopec et al. 2008), we observed a response that opposed TCDD in terms of shifting SFB, Firmicutes, and Enterobacteriaceae (Bhaduri 2015; Stedtfeld et al. 2017, submitted). For example, compared to the relative abundance 1 day before dosing, Enterobacteriaceae in fecal pellets decreased ~ 10-fold and 300-fold, 3 and 7 days following an initial dose of sesame oil, respectively. However, Enterobacteriaceae was not influenced with CO compared to the naive control in the ileum or fecal pellets measured five days after the initial dosing (Fig. S6). Collectively, CO may be a more suitable vehicle compared to sesame oil for investigating the influence of host toxicity and gut modulation with persistent organic pollutants.
Discussion
SFB was selected as a marker because it interacts with host immunity (Ivanov et al. 2009) and responds to TCDD in the murine model (Stedtfeld et al. 2017; Bhaduri 2015). Previous studies have surmised that shift in the gut microbiome, including SFB, is an indirect response to TCDD-induced modulation of the immune system (Zhang et al. 2015; Lefever et al. 2016; Stedtfeld et al. 2017). Indeed, analysis of TCDD effect on immune function in these mice, evaluated with anti-sheep red blood cell immunoglobulin M antibody-forming cells, induced significant suppression of humoral activity at doses down to 0.1 μg/kg CO-TCDD (Boyd et al. 2017). Measured in mice feces, previous studies have also found that mice deficient in mucosal immunity maintainers displayed a 10-fold increase in SFB abundance (Upadhyay et al. 2012). Furthermore, in vitro studies have also shown that TCDD did not directly influence gut commensals (Lefever et al. 2016; Stedtfeld et al. 2017). Taken together, these results further verify that bacterial dysbiosis is due to TCDD-induced changes in the host. Additional analysis is warranted to differentiate if dysbiosis is the result of TCDD-induced changes to host luminal metabolites or immune suppression.
The significantly higher number of SFB with 10 μg/kg CO-TCDD compared to CO was only observed in RNA and not in DNA. Measured via 16S rRNA gene sequencing with DNA, SFB only tended to be higher in response to TCDD (fold change = 2.2, p = 0.15). Other bacterial groups (e.g., Enterobacteriacea; Fig. S6) also responded to TCDD as described in previous studies (Stedtfeld et al. 2017, submitted; Lefever et al. 2016). Doses less than 10 μg/kg TCDD did not significantly influence SFB in this study, which had been observed previously (Bhaduri 2015; Fader et al. 2015). In detail, a significant shift in SFB abundance for TCDD doses ranging from 1 to 30 μg/kg administered every 4 days for 28 days was previously observed (Bhaduri 2015). We suspect that the shorter duration of this experiment, compared to previous studies (28 to 90 days), influenced overall bacterial response. Assuming a TCDD half-life of 11 days in mice (Gasiewicz et al. 1983; Birnbaum 1986), a theoretical analysis showed that the level of TCDD in mice from both experiments was similar at the time of measurement, but varied in duration of exposure (Fig. S7). As previously described (Boyd et al. 2017), analytical determination of TCDD concentrations in murine gut feces was not determined due to the challenge of accurately measuring low levels of exposure used in this study; further complicated by the use of AC.
Previous studies testing AC as an alternative to antibiotics in swine also observed a higher level of lactic acid bacteria counts via culture analysis (Chu et al. 2013). The study by Chu and coauthors also observed significantly lower levels of coliforms and lactic acid. In general, lactic acid bacteria are considered probiotics due to the creation of organic acids, providing an acidic environment that inhibits pathogenic organisms. Therefore, the short-term ingestion of AC does not appear to have a threatening influence on the structure of the gut microbiome.
Results of this study support the remedial use of AC to mute TCDD-induced responses to the gut microbiome of a murine host. In detail, SFB functional genes were only expressed with the highest dose of TCDD administered with CO. This most plausibly resulted from the elimination of bio-availability to the host due to its sequestration by AC as previously demonstrated (Boyd et al. 2017). As measured by 16S rRNA gene analysis of ileal content, AC also had a minimal influence on gut community structure and diversity. Three members were significantly shifted with AC, including a slight increase in Lactobacillaceae, generally considered a probiotic. CO vehicle influenced the gut microbiota to a lesser extent compared to previously used sesame oil vehicle. Overall, the response of a key member of the gut microbiome was absent when TCDD was administered on AC.
Supplementary Material
Acknowledgments
The authors would like to thank Premachandra Gnanasiri for the assistance in the preparation of AC.
Funding
This study was funded by the National Institute of Environmental Health Sciences Superfund Research Program (NIEHS SRP P42ES004911).
Footnotes
Compliance with ethical standards
Conflict of interest The authors declare that they have no conflict of interest.
Ethical approval All applicable international, national, and/or institutional guidelines for the care and use of animals were followed. This article does not contain any studies with human participants performed by any of the authors.
Electronic supplementary material The online version of this article (doi:10.1007/s00253-017-8460-9) contains supplementary material, which is available to authorized users.
References
- Avantaggiato G, Havenaar R, Visconti A. Evaluation of the intestinal absorption of deoxynivalenol and nivalenol by an in vitro gastrointestinal model, and the binding efficacy of activated carbon and other adsorbent materials. Food Clin Toxicol. 2004;42:817–824. doi: 10.1016/j.fct.2004.01.004. [DOI] [PubMed] [Google Scholar]
- Bhaduri P. Thesis. Michigan State University. Civil and Environmental Engineering; 2015. Exposure to 2, 3, 7, 8-tetrachlorodibenzo-p-dioxin differentially impacts key members of mice gut microbiome. [Google Scholar]
- Birnbaum LS. Distribution and excretion of 2,3,7,8-tetrachlorodibenzo-p-dioxin in congenic strains of mice which differ at the Ah locus. Drug Metab Dispos. 1986;14:34–40. [PubMed] [Google Scholar]
- Boyd SA, Sallach JB, Zhang Y, Crawford RB, Li H, Johnston CT, Teppen BJ, Kaminski NE. Sequestration of TCDD by activated carbon eliminates bioavailability and the suppression of immune function in mice. Environ Toxicol Chem. 2017 doi: 10.1002/etc.3815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd SA, Johnston CT, Pinnavaia TJ, Kaminski NE, Teppen BJ, Li H, Khan B, Crawford RB, Kovalova N, Kim SS, Shao H, Gu C, Kaplan BLF. Suppression of humoral immune responses by 2,3,7,8-tetrachlorodibenzo-p-dioxin intercalated in smectite clay. Environ Toxicol Chem. 2011;30:2748–2755. doi: 10.1002/etc.701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridges TS, Gustavson KE, Schroeder P, Ells SJ, Hayes D, Nadeau SC, Palermo MR, Patmont C. Dredging processes and remedy effectiveness: relationship to the 4 Rs of environmental dredging review. Integr Environ Assess Manag. 2010;6:619–630. doi: 10.1002/ieam.71. [DOI] [PubMed] [Google Scholar]
- Brown NM, Manzolillo PA, Zhang JX, Wang J, Lamartiniere CA. Prenatal TCDD and predisposition to mammary cancer in the rat. Carcinogenesis. 1998;19:1623–1629. doi: 10.1093/carcin/19.9.1623. [DOI] [PubMed] [Google Scholar]
- Bruner-Tran KL, Osteen KG. Developmental exposure to TCDD reduces fertility and negatively affects pregnancy outcomes across multiple generations. Reprod Toxicol. 2011;31:344–350. doi: 10.1016/j.reprotox.2010.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, Huttley GA, Kelly ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu GM, Jung CK, Kim HY, Ha JH, Kim JH, Jung MS, Lee SJ, Song Y, Ismael R, Ibrahim H, Cho JH, Lee SS, Song YM. Effects of bamboo charcoal and bamboo vinegar as antibiotic alternatives on growth performance, immune responses and fecal microflora population in fattening pigs. Anim Sci J. 2013;84:113–120. doi: 10.1111/j.1740-0929.2012.01045.x. [DOI] [PubMed] [Google Scholar]
- E.P.A. U. Use of amendments for in situ remediation at superfund sediment sites 2013 [Google Scholar]
- Fader KA, Nault R, Ammendolia DA, Harkema JR, Williams KJ, Crawford RB, Kaminski NE, Potter D, Sharratt B, Zacharewski TR. 2, 3, 7, 8-Tetrachlorodibenzo-p-dioxin (TCDD) alters lipid metabolism and depletes immune cell populations in the jejunum of C57BL/6 mice. Toxicol Sci. 2015;148:567–580. doi: 10.1093/toxsci/kfv206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagervold SK, Chai Y, Davis JW, Wilken M, Cornelissen G, Ghosh U. Bioaccumulation of polychlorinated dibenzo-p-dioxins/dibenzofurans in E. fetida from floodplain soils and the effect of activated carbon amendment. Environ Sci Technol. 2010;44:5546–5552. doi: 10.1021/es9027138. [DOI] [PubMed] [Google Scholar]
- Fernandez-Salguero PM, Hilbert DM, Rudikoff S, Ward JM, Gonzalez FJ. Aryl-hydrocarbon receptor-deficient mice are resistant to 2, 3, 7, 8-tetrachlorodibenzo-p-dioxin-induced toxicity. Toxicol Appl Pharmacol. 1996;140:173–179. doi: 10.1006/taap.1996.0210. [DOI] [PubMed] [Google Scholar]
- Ferrario JB, Byrne CJ, Cleverly DH. 2,3,7,8-Dibenzo-p-dioxins in mined clay products from the United States: evidence for possible natural origin. Environ Sci Technol. 2000;34:4524–4532. doi: 10.1021/es001052r. [DOI] [Google Scholar]
- Gadomski D, Tysklind M, Irvine RL, Burns PC, Andersson R. Investigations into the vertical distribution of PCDDs and mineralogy in three ball clay from the United States exhibiting the natural formation pattern. Environ Sci Technol. 2004;38:4956–4963. doi: 10.1021/es049579h. [DOI] [PubMed] [Google Scholar]
- Gasiewicz T, Geiger L, Rucc G, Neal R. Distribution, excretion, and metabolism of 2, 3, 7, 8-tetrachlorodibenzo-p-dioxin in C57BL/6J, DBA/2J, and B6D2F1/J mice. Drug Metab Dispos. 1983;11:397–403. [PubMed] [Google Scholar]
- Ghosh U, Luthy R, Cornelissen G, Werner D, Menzie C. In-situ sorbent amendments: a new direction in contaminated sediment management. Env Sci Technol. 2011;45:1163–1168. doi: 10.1021/es102694h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt L, Holz P. The black bottle: a consideration of the role of charcoal in the treatment of poisoning in children. J Pediatr. 1963;63:306.14. doi: 10.1016/s0022-3476(63)80344-3. [DOI] [PubMed] [Google Scholar]
- Honda K, Littman DR. The microbiota in adaptive immune homeostasis and disease. Nature. 2016;535:75–84. doi: 10.1038/nature18848. [DOI] [PubMed] [Google Scholar]
- Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, Wei D, Goldfarb KC, Santee CA, Lynch SV. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139:485–498. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Josefsson S, Schaanning M, Samuelsson GS, Gunnarsson JS, Olofsson I, Eek E, Wiberg K. Capping efficiency of various carbonaceous and mineral materials for in situ remediation of polychlorinated dibenzo-p-dioxin and dibenzofuran contaminated marine sediments: sediment-to-water fluxes and bioaccumulation in boxcosm tests. Environ Sci Technol. 2012;46:3343–3351. doi: 10.1021/es203528v. [DOI] [PubMed] [Google Scholar]
- Kaplan BLF, Crawford RB, Kovalova N, Arencibia A, Kim SS, Pinnavaia TJ, Boyd SA, Teppen BJ, Kaminski NE. TCDD adsorbed on silica as a model for TCDD contaminated soils: evidence for suppression of humoral immunity in mice. Toxicology. 2011;282:82–87. doi: 10.1016/j.tox.2011.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoder M, Tsapis N, Domergue-dupont V, Gueutin C, Fattal E. Removal of residual colonic ciprofloxacin in the rat by activated charcoal entrapped within zinc-pectinate beads. Eur J Pharm Sci. 2010;41:281–288. doi: 10.1016/j.ejps.2010.06.018. [DOI] [PubMed] [Google Scholar]
- Kopec AK, Boverhof DR, Burgoon LD, Ibrahim-aibo D, Harkema JR, Tashiro C, Chittim B, Zacharewski TR. Comparative toxicogenomic examination of the hepatic effects of PCB126 and TCDD in immature, ovariectomized C57BL/6 mice. Toxicol Sci. 2008;102:61–75. doi: 10.1093/toxsci/kfm289. [DOI] [PubMed] [Google Scholar]
- Kozich JJ, Westcott SL, Baxter NT, Highlander SK, Schloss PD. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl Env Microbiol. 2013;79:5112–5120. doi: 10.1128/AEM.01043-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefever DE, Xu J, Chen Y, Huang G, Tamas N, Guo TL. TCDD modulation of gut microbiome correlated with liver and immune toxicity in streptozotocin (STZ)-induced hyperglycemic mice. Toxicol Appl Pharmacol. 2016;304:48–58. doi: 10.1016/j.taap.2016.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Littman DR, Pamer EG. Role of the commensal microbiota in normal and pathogenic host immune responses. Cell Host Microbe. 2011;10:311–323. doi: 10.1016/j.chom.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Looft T, Johnson TA, Allen HK, Bayles DO, Alt DP, Stedtfeld RD, Sul WJ, Stedtfeld TM, Chai B, Cole JR. In-feed antibiotic effects on the swine intestinal microbiome. Proc Natl Acad Sci U S A. 2012;109:1691–1696. doi: 10.1073/pnas.1120238109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motulsky HJ, Brown RE. Detecting outliers when fitting data with nonlinear regression–a new method based on robust nonlinear regression and the false discovery rate. BMC Bioinformatics. 2006;7:123. doi: 10.1186/1471-2105-7-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pamp SJ, Harrington ED, Quake SR, Relman DA, Blainey PC. Single-cell sequencing provides clues about the host interactions of segmented filamentous bacteria (SFB) Genome Res. 2012;22:1107–1119. doi: 10.1101/gr.131482.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patmont CR, Ghosh U, LaRosa P, Menzie CA, Luthy RG, Greenberg MS, Cornelissen G, Eek E, Collins J, Hull J, Hjartland T, Glaza E, Bleiler J, Quadrini J. In situ sediment treatment using activated carbon: a demonstrated sediment cleanup technology. Integr Environ Assessand Manag. 2015;11:195–207. doi: 10.1002/ieam. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stedtfeld RD, Baushke SW, Tourlousse DM, Miller SM, Stedtfeld TM, Gulari E, Tiedje JM, Hashsham SA. Development and experimental validation of a predictive threshold cycle equation for quantification of virulence and marker genes by high-throughput nanoliter-volume PCR on the OpenArray platform. Appl Env Microbiol. 2008;74:3831–3838. doi: 10.1128/AEM.02743-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stedtfeld RD, Williams MR, Fakher U, Johnson TA, Stedtfeld TM, Wang F, Khalife WT, Hughes M, Etchebarne BE, Tiedje JM. Antimicrobial resistance dashboard application for mapping environmental occurrence and resistant pathogens. FEMS Microbiol Ecol. 2016;92:fiw020. doi: 10.1093/femsec/fiw020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stedtfeld R, Stedtfeld T, Fader K, Williams M, Quensen J, Zacharewski T, Tiedje J, Hashsham S. TCDD influences reservoir of antibiotic resistance genes in murine gut microbiome. FEMS Microbiol Ecol. 2017;93:fix058. doi: 10.1093/femsec/fix058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upadhyay V, Poroyko V, Kim T, Devkota S, Fu S, Liu D, Tumanov AV, Koroleva EP, Deng L, Nagler C. Lymphotoxin regulates com-mensal responses to enable diet-induced obesity. Nat Immunol. 2012;13:947–953. doi: 10.1038/ni.2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F-H, Qiao M, Su J-Q, Chen Z, Zhou X, Zhu Y-G. High throughput profiling of antibiotic resistance genes in urban park soils with reclaimed water irrigation. Env Sci Technol. 2014;48:9079–9085. doi: 10.1021/es502615e. [DOI] [PubMed] [Google Scholar]
- Yamashita N, Imagawa T, Villeneuve DL, Hashimoto S, Miyazaki A, Giesy JP. Vertical profile of polychlorinated naphthalenes, biphenyls, polycyclic aromatic hydrocarbons, and alkylphenols in a sediment core from Tokyo Bay, Japan. Env SCI Technol. 2000;34:3560–3567. doi: 10.1021/es001043i. [DOI] [Google Scholar]
- Zhang L, Nichols RG, Correll J, Murray IA, Tanaka N, Smith PB, Hubbard TD, Sebastian A, Albert I, Hatzakis E. Persistent organic pollutants modify gut microbiota-host metabolic homeostasis in mice through aryl hydrocarbon receptor activation. Env Heal Perspect. 2015;123:679–688. doi: 10.1289/ehp.1409055. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


