Abstract
Farnesyl pyrophosphate synthase (FPS) catalyzes the synthesis of farnesyl pyrophosphate, a key intermediate in sterol and sesquiterpene biosynthesis. Using a polymerase chain reaction-based approach, we have characterized LeFPS1, a tomato (Lycoperscion esculentum cv Wva 106) fruit cDNA, which encodes a functional FPS. We demonstrate that tomato FPSs are encoded by a small multigenic family with genes located on chromosomes 10 and 12. Consistent with farnesyl pyrophosphate requirement in sterol biosynthesis, FPS genes are ubiquitously expressed in tomato plants. Using an LeFPS1 specific probe, we show that the corresponding gene can account for most of FPS mRNA in most plant organs, but not during young seedling development, indicating a differential regulation of FPS genes in tomato. FPS gene expression is also under strict developmental control: FPS mRNA was mainly abundant in young organs and decreased as organs matured with the exception of fruits that presented a biphasic accumulation pattern. In this latter case in situ hybridization studies have shown that FPS mRNA is similarly abundant in all tissues of young fruit. Taken together our results suggest that several FPS isoforms are involved in tomato farnesyl pyrophosphate metabolism and that FPS genes are mostly expressed in relation to cell division and enlargement.
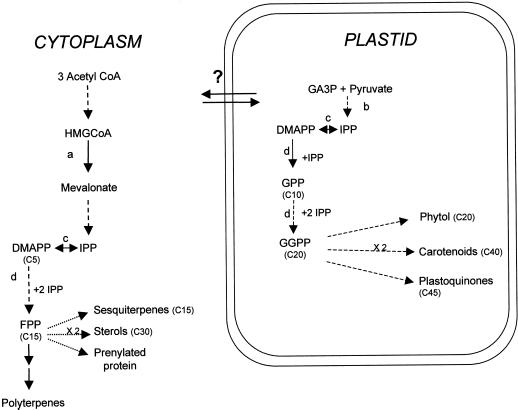
Isoprenoids constitute a widespread family of chemical compounds. More than 22,000 different molecules have been identified in plants. They include sterols, gibberellins, carotenoids, phytol chains, and prenyl groups that are involved in membrane stability, cell growth, and proliferation. They are also essential for respiration, photosynthesis, photoprotection, and plant environment interactions (for review, see Chappell, 1995). Isoprenoids derive from prenyl precursors produced by the sequential condensation of isopentenyl pyrophosphate (IPP) on different allylic acceptors. In plants it is now clear that IPP synthesis occurs following two different pathways located, respectively, in the cytoplasm/endoplasmic reticulum compartment (McGarvey and Croteau, 1995) and in the plastids (Lichtenthaler et al., 1997). The cytoplasmic pathway (or mevalonate pathway) involves the synthesis of mevalonate from acetyl-Co-A by the enzyme 3-hydroxy-3-methylglutaryl coenzyme A reductase (HMGR), whereas in plastids IPP is produced from pyruvate and glyceraldehyde 3-phosphate following the Rohmer pathway. Both IPP pools presumably lead to different isoprenoid end products, sterols being produced in the cytoplasm/endoplasmic reticulum compartment and carotenoids, phytols, or gibberellins in the plastids.
Tomato (Lycoperscion esculentum) fruit is commonly used as a model to study the function of isoprenoids during development. Tomato fruit development follows a complex process that consists of three main steps followed by fruit ripening: (a) fruit set, which involves ovary development and the decision to abort or to proceed further; (b) a cell division step, occurring during early fruit development; and (c) a cell expansion step responsible for further growth until the fruit reaches its final size at the mature green stage (Gillaspy et al., 1993). Several lines of evidence indicate that early disruption of the cytosolic isoprenoid biosynthesis pathway affects fruit organogenesis and maturation (Narita and Gruissem, 1989; Rodriguez-Concepcion and Gruissem, 1999). Hence, isoprenoids including sterols, hormones (notably gibberellins), and prenyl groups, are essential for fruit development and ripening. However the precise role of the different isoprenoid compounds has not been determined. Among the isoprenoid intermediates, farnesyl pyrophosphate (FPP) plays a central function (Fig. 1). In plants, FPP is a precursor of phytosterols, sesquiterpenoids, phytoalexins, and is involved in protein farnesylation, which plays an essential role in cell cycle progression (Chappell, 1995). Thus limitation in FPP synthesis may affect the abundance of compounds essential in fruit growth and metabolism.
Figure 1.
Depiction of terpenoid biosynthesis in plants. Broken arrows indicate multiple steps or reactions. The number of carbon molecules is indicated in brackets. Question mark indicates putative exchanges of isoprenoids between the cytosol and the plastidic compartment. AcetylCoA, Acetyl coenzyme A; HMGCoA, 3-hydroxy-3 methylglutaryl coenzyme A; DMAPP, dimethylallyl pyrophosphate; GGPP, geranylgeranyl pyrophosphate. a, HMGCoA reductase. b, Plastidic 1-deoxy-d-xylulose-5-phosphate pathway. c, Isopentenyl pyrophosphate isomerase. d, Prenyl transferase (adapted from Lichtenthaler [1999]).
FPP synthase (FPS) synthesizes FPP in two separate steps (Chappell, 1995). FPS genes and cDNAs have first been characterized in vertebrates and yeast (Anderson et al., 1989; Ashby and Edwards, 1989) and more recently in various plant species including Arabidopsis (Delourme et al., 1994), white lupine (Attucci et al., 1995), maize (Li and Larkins, 1996), rubber tree (Adiwilaga and Kush, 1996), and rice (Sanmiya et al., 1997). Both in rice (Sanmiya et al., 1999) and in Arabidopsis (Cunillera et al., 1996), two genes or cDNAs have been cloned indicating that FPS is encoded by small multigenic families. Regulation of FPS genes appears to be strictly controlled during plant organ development and depends on environmental conditions. Differential expression of the two genes characterized in Arabidopsis has been demonstrated: AtFPS1 is highly expressed in roots and flowers, whereas AtFPS2 transcription occurs mainly in flowers (Cunillera et al., 1996). In rice FPPS1 transcript accumulation is induced by blue light in germinating seedlings, and in leaves and FPPS2 mRNA is detected only in roots (Sanmiya et al., 1999). Tissue-specific expression of FPS genes has also been reported in maize endosperm (Li and Larkins, 1996). However, little is known about FPS gene expression during fruit development (Hugueney et al., 1996).
To gain a better understanding of FPS physiological function in tomato fruits and plants, we have cloned and characterized LeFPS1, a cDNA encoding a functional tomato FPS. We show that the tomato genome contains two LeFPS1 hybridizing loci. FPS transcript accumulation was analyzed in young seedlings and during leaf, flower and fruit development. We also studied the tissue distribution of FPS transcripts in young tomato fruits and seeds by in situ hybridization. Our results show a differential regulation of FPS genes in tomato plants and suggest important functions of FPSs in early fruit development and in the development of other plant organs when cell division and growth occur.
RESULTS
Isolation and Characterization of LeFPS cDNAs from Tomato Fruit cDNA Libraries
Two sets of nested degenerate oligonucleotides (FPex and FPin) designed from conserved regions II and V of FPS (Cunillera et al., 1997) were used to amplify a 280-bp long cDNA fragment from a young tomato fruit cDNA library. This fragment, which shared a high similarity with known plant FPS sequences, hybridized with tomato genomic DNA (data not shown). We therefore used this fragment as a probe to screen the two tomato fruit cDNA libraries described in “Materials and Methods.” Five independent cDNA clones were isolated from the young fruit cDNA library and were shown to contain the same 1.26-kb long insert (LeFPS1). Screening of a mature fruit cDNA library allowed us to isolate six truncated forms of LeFPS1.
Sequence Analysis and Comparison
LeFPS1 hybridized to the tomato's transcripts of approximately 1.3 kb as estimated after northern analysis, which suggested that the cDNA was nearly complete. A putative ATG initiation codon was found at position 33 (Fig. 2A) surrounded by the sequence AAAAATGGC highly homologous to the plant consensus translation initiation site (Joshi et al., 1987). This ATG starts a putative open reading frame (ORF) encoding a 342-amino acid polypeptide with a predicted molecular mass of 39.3 kD. This is in good agreement with the size of the polypeptide produced by in vitro transcription/translation of LeFPS1 (Fig. 2B) and by overexpressing the LeFPS1 protein in Escherichia coli (Fig. 3A). In Figure 2B we also observed a second polypeptide with a molecular mass of 30 kD, which is likely to be due to an internal translation initiation event as may happen when in vitro translation is performed (see Promega's instructions). A sequence identical to the consensus polyadenylation site (AATAAA) is found at position 1,221, 162 bp downstream of the stop codon (Fig. 2A). We also noticed a small ORF of 153 bp encoding a 51-amino acid peptide located just downstream of the main ORF. Small ORFs in the 3′-untranslated region (UTR) are present in several other FPS plant cDNAs including peppers FPS and Arabidopsis FPS2. In all cases they correspond to hydrophobic peptides of unknown function. Under our in vitro transcription/translation conditions, this ORF was not translated either alone or as a fusion with the main ORF (Fig. 2B).
Figure 2.
Characteristics of the LeFPS1 cDNA sequence. A, Nucleotide and deduced amino acid sequence of the LeFPS1 cDNA. Gray boxes correspond to the highly conserved regions of FPS (see text). Stop codons are indicated with asterisks and the putative polyadenylation site is written in italics. Underlined nucleotides denote a small ORF of unknown function. B, Autoradiography of in vitro transcription-translation products of LePFS1. LeFPS1 cloned in pBS was in vitro transcribed and translated in the presence of [35S]Met as described in “Materials and Methods.” Radiolabeled products were separated on a 12% (w/v) SDS-PAGE and autoradiographed. “No RNA” and “LeFPS1” represent the transcription-translation respectively products in the absence and presence of LeFPS1. Numbers on the left indicate the molecular mass standards expressed in kD (New England Biolabs, Beverly, MA).
Figure 3.
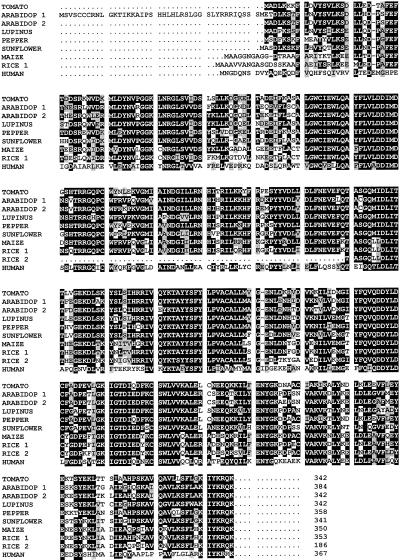
Alignment of the predicted amino acid sequences of tomato LeFPS1 (AF048747) with those from other origins: Arabidopsis (Arabidop1, U80605; Arabidop2, L46349), white lupine (Lupinus, U20771), peppers (Pepper, X84695), maize (Maize, L39789), sunflower (Sunflower, AF019892), rice (Rice 1, AB021747; Rice 2, AB021979), and human (Human, J05262). Black and gray shading correspond respectively to identical and similar amino acid residues. Dots indicate gaps introduced to allow optimal alignment of the sequence.
Protein sequence comparison indicated that LeFPS1 is closely related (88.3% identity) to the FPS of peppers, another Solanaceae. The identity level remains high with other dicotylenous plants ranging between 76% and 80%. As expected, monocotyledonous FPSs are more distantly related since rice and maize FPSs only share about 68% identity with LeFPS1. Yeast and animal FPSs show even greater divergence, although significant similarity (65%) and identity (47%) levels are still found between human FPS and LeFPS1.
LeFPS Encodes an Active FPS
The conservation of the five characteristic domains found in all FPSs characterized so far (Fig. 3) suggests that we cloned a cDNA encoding an active FPS polypeptide. These domains include the two type I FPS repeated motifs YFX1VX2DDX3X4D involved in allylic substrate recognition (Ohnuma et al., 1996). The Tyr residue, which determines the allylic product chain length, is located five amino acids upstream of the first Asp-rich domain as already described for other eukaryotic FPSs (Ohnuma et al., 1996).
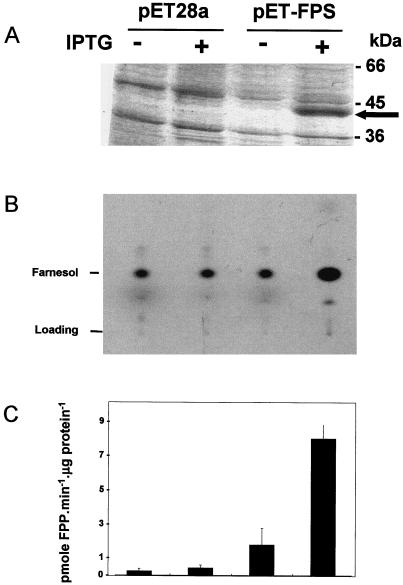
To demonstrate the prenyl transferase activity of LeFPS1, we expressed the recombinant protein in E. coli. The pET-FPS plasmid was obtained by inserting LeFPS1 cDNA in pET28a(+) downstream of the His tag sequence. isopropylthio-β-galactoside (IPTG) induction of bacteria transformed with pET-FPS led to the production of a recombinant protein of 39 kD (Fig. 4A). Crude protein extracts prepared from induced or non induced bacteria were incubated with geranyl pyrophosphate (GPP) and [14C]IPP as described in “Materials and Methods.” Accumulation of [14C]farnesol was detected in all cases after either gas-liquid chromatography (GLC) (data non shown) or thin layer chromatography analysis of the dephosphorylated products(Fig. 4B). Overexpression of LeFPS1 led to a more than 18-fold increase in FPS-specific activity as compared with the control transformed with the non-recombinant pET28 plasmid (Fig. 4C). Low synthesis of farnesol in control extracts was probably due to endogenous E. coli FPS activity. These results indicate that after IPTG induction of protein synthesis, E. coli cells transformed with pET-FPS accumulate a 39-kD protein, which correlates with a strong increase in FPS specific activity.
Figure 4.
Expression and activity of LeFPS1 in recombinant E. coli. A, SDS-PAGE analysis of 50 μg of total protein extracts from bacteria transformed with the non-recombinant pET28a vector or with the pET-LeFPS plasmid with (+) or without (−) IPTG induction. After Coomassie Blue staining, a new polypeptide (approximately 40 kD) was detected in IPTG-induced bacteria containing the pET-LeFPS plasmid. B, Sonicated bacterial extracts were incubated with [14C]IPP and GPP. Isoprenoids were extracted, dephosphorylated, and separated on thin layer chromatography prior to autoradiography. Radiolabeled products were identified after GLC analysis. Lanes are as indicated in A. C, Quantification of [14C]farnesol produced by recombinant bacteria protein extracts. After thin layer chromatographic analysis and autoradiography, farnesol spots were collected and radioactivity was estimated. Results are the average of three independent experiments and are expressed in picomoles of FPP produced per microgram of total bacterial protein. Bars indicate the sd. Lanes are as indicated in A.
Southern Analysis and FPS Gene Mapping
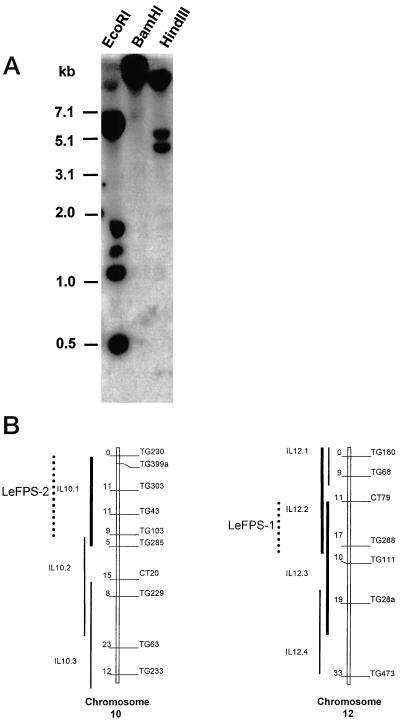
The full length LeFPS1 cDNA probe hybridizes with six EcoRI and three HindIII fragments on Southern blots using tomato genomic DNA (Fig. 5A). However LeFPS1 cDNA 3′-UTR hybridized only with the larger HindIII fragment, suggesting that it can be used as a gene specific probe (data not shown). These results indicated that FPS is encoded by a small gene family and/or that multiple introns are present in the LeFPS1 gene as already shown for Arabidopsis AtFPS1 and AtFPS2 genes (Cunillera et al., 1996). A similar hybridization pattern is found in other tomato cultivars such as Marmande and “beef heart” (data not shown).
Figure 5.
Genomic organization of tomato LeFPS genes. A, Thirty micrograms of tomato genomic DNA was digested with the indicated restriction endonuclease and subjected to DNA gel-blot analysis using LeFPS1 cDNA as a hybridization probe. The blot was exposed for 4 d. Numbers on the left indicate size in kb from the markers (1-kb ladder, Gibco BRL, Cleveland). B, Partial map of the chromosomes 10 and 12. The putative location of the two loci (thick line) was deduced from the introgression line showing a polymorphism (dotted lines). LeFPS1 and LeFPS2 were mapped using the 1.3-kb LeFPS1 cDNA insert as a hybridization probe. The 0.18-kb cDNA fragment corresponding to the LeFPS1 3′-UTR was used to locate LeFPS1 on chromosome 12.
To determine the number of loci hybridizing with LeFPS1 in the tomato genome, gene mapping was performed using the population of introgressed lines from Lycopersicon pennellii developed by Eshed and Zamir (1994). As a low-copy pattern was observed with most of the restriction enzymes, we mapped all the polymorphic bands with only two enzymes, EcoRI and HindIII. In both cases two loci were mapped, respectively, at the top of chromosome 10 (polymorphism on the introgressed fragment, 10.1), and on the common part of chromosome fragments, 12.2 and 12.3 (Fig. 5B). Even when high-stringency washes (0.1× SSC and 0.1% [w/v] SDS, 65°C) were performed, two loci were revealed, indicating that both genes share a high level of similarity. Since the LeFPS1 3′-UTR hybridized only with the HindIII fragment that was mapped on chromosome 12, we concluded that the gene LeFPS1 is located on this chromosome (Fig. 5B).
Three other loci giving a low intensity hybridization signal were detected on the common part of introgressed fragments 3.2, and 3.3, and on fragments 4.3 and 9.2 (data not shown).
Differential and Developmental Accumulation of FPS mRNA in Tomato Plant Organs
Mapping experiments indicated that tomato FPSs are encoded by genes located at least at two different LeFPS1 hybridizing loci. Hence, the full length LeFPS1 cDNA was used as a probe to study total FPS transcript accumulation, and LeFPS1 gene expression was investigated using the 3′-UTR of the LeFPS1 cDNA as a gene specific probe.
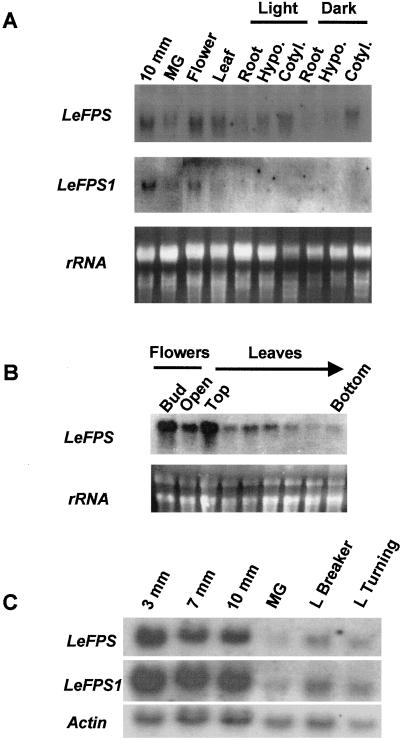
As shown in Figure 6A, the full-length probe hybridizes with a 1.3-kb mRNA in all tissues analyzed. The highest expression levels were found in young fruits, but the signals were still intense in leaves, flowers, and young seedling cotyledons (Fig. 6A). Weaker signals are observed in young seedling roots, hypocotyls, and mature green fruits. When the same blot was hybridized with the LeFPS1 gene specific probe, hybridization signals were much weaker due to smaller probe size, and blots were exposed for longer periodes (Fig. 6A). Similar mRNA accumulation patterns were observed in leaves, flowers, and fruits. However, with the shorter probe, we could not detect any signal in young seedling cotyledons, even after a prolonged exposure time (Fig. 6A).
Figure 6.
Analysis of FPS mRNA accumulation during the development of tomato plant organs. A, Total RNA (15 μg per sample) prepared from fruits, leaves, flowers, and light or dark young seedlings (as indicated) were subjected to northern analysis. The blot was successively hybridized with a 182-bp cDNA fragment corresponding to the 3′-UTR of LeFPS1 (LeFPS1) and with the full-length LeFPS1 cDNA probe (LeFPS). Blots were respectively exposed for 12 (LeFPS1) and 2 d (LeFPS). Ribosomal RNA stained with ethidium bromide was used as a loading control. B, Total RNA (15 μg) extracted from flowers, and tomato leaves collected along the stem from the apex to the base of the plant were subjected to northern analysis using the full-length LeFPS1 cDNA as a probe. Ribosomal RNA stained with ethidium bromide was used as a loading control. The blot was exposed for 5 d. C, Total RNA (15 μg) prepared from tomato fruits harvested at different stages of development and ripening as indicated were subjected to northern analysis using the two probes described in A and with an actin probe (Germain et al., 1997), which was used as a loading control. Exposure times are as described in A. MG, Mature green; Hypo, hypocotyl; Cotyl, cotyledon; L breaker, late breaker; L turning, late turning.
The effect of light on FPS mRNA accumulation during tomato seedlings development was also studied using both probes. No clear differences were found between dark- or light-grown seedlings thus giving no evidence of light regulation of LeFPS1 related genes (Fig. 6A).
Total FPS mRNA accumulation was further investigated in leaves and flowers harvested at different developmental stages (Fig. 6B). The steady-state level of FPS transcript was higher in young leaves in the apical part of the plant and in unopened flowers, whereas a noticeable decrease was noted during leaf development and in opened flowers. FPS mRNA was still detected in leaves harvested in the lower part of the plant. We concluded from these experiments that FPS mRNA accumulation occurs ubiquitously in tomato plants and is developmentally regulated.
FPS mRNA Accumulates in All Tissues of Tomato Fruits
To analyze FPS mRNA accumulation during fruit development, total RNA extracts were prepared from fruits with a diameter of 3 mm (including the seeds), and from fruit pericarp and columella for larger fruits. As shown in Figure 6C total FPS mRNAs are abundant in young tomato fruits. As fruits mature, the abundance of FPS transcript decreased slightly with a minimum at the mature green stage. During the ripening process, FPS mRNA amount increased again, but to a lesser extent compared with young fruits. A similar accumulation pattern was obtained when the LeFPS1 gene specific probe was used (Fig. 6C), showing that this gene is expressed at all fruit developmental stages.
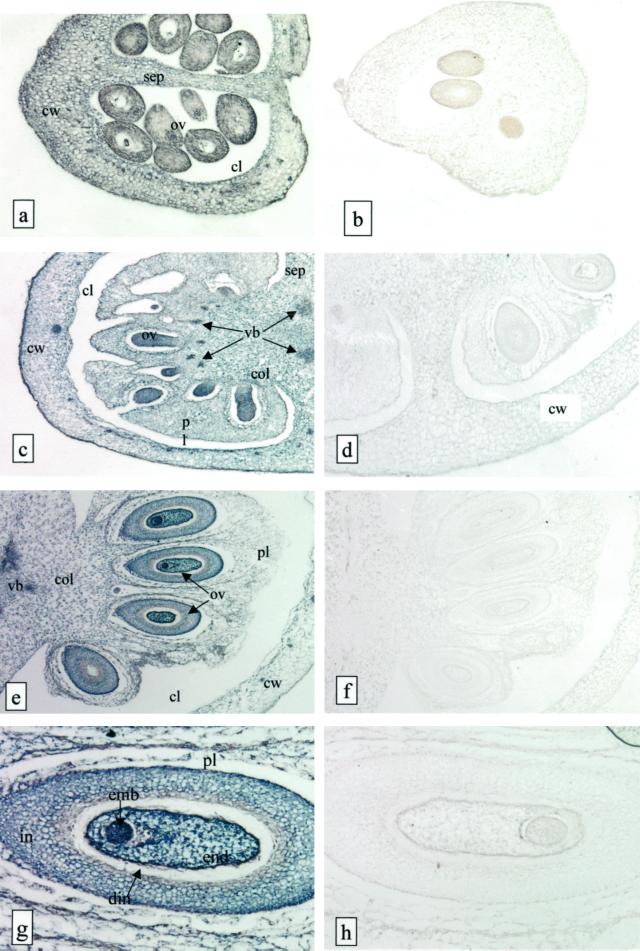
We examined the tissue distribution of FPS mRNA in developing tomato fruits using in situ hybridization with full length digoxigenin-labeled sense or antisense LeFPS1 RNA probes. An intense staining was observed when the antisense probe was used with sections prepared from fruits of a diameter of 4 (Fig. 7A), 6 (Fig. 7C), and 8 mm (Fig. 7E), whereas almost no staining was visible with the sense probe (Fig. 7, B, D, and F) or in the control without probe (not shown). The FPS transcripts were detected in fruit pericarp, columella, and placenta at all developmental stages analyzed (Fig. 7, A, C, and E). The signal appeared homogenous in all pericarp cell types with the exception of vascular bundles and epidermal cells, which showed a more intense staining. This effect may simply be due to smaller cell size and lower vacuolization of the cytoplasm of these cells compared with cells from other fruit tissues. We observed a slight decrease in staining intensity in all pericarp cells during fruit development (Fig. 7, A, C, and E), but no clear variations among tissues.
Figure 7.
In situ hybridization of LeFPS to young tomato fruit sections. Bright field micrographs of 7-μm tissue sections from 3.5-mm diameter fruits (a and b), 6-mm (c and d), and 8-mm (e and f) large fruits are shown. g and h, Higher magnification of e and f showing the concentration of labeling in the developing seeds. Sections were hybridized either with a sense (a, c, e, and f) or an antisense (b, d, f, and h) LeFPS1 DIG-labeled RNA probe. The hybridization signal appears as a dark-blue staining and is localized in cells from all fruit tissues. Cw, Carpel wall; sep, septum; ov, ovules; pl, placenta; col, columella; vb, vascular bundles; emb, embryo; end, endosperm; in, integument; din, disintegrating portion of integument. a and b, ×60; c, ×30; d, ×70; e and f, ×20; g and h, ×80.
Since seeds were eliminated during northern experiment (see above), we analyzed FPS mRNA distribution in maturing seeds. Embryos and endosperm showed an intense blue color at all developmental stages analyzed (Fig. 7F). Ovule teguments presented a high labeling intensity as well, with the exception of the two most internal cell layers that correspond to the disintegrating part of the teguments described by Smith (1935). Thus the expression of FPS genes appears very high in young developing seeds, which is in agreement with the high accumulation of FPS mRNA in maize endosperm (Li and Larkins, 1996).
To summarize, the FPS transcripts were detected in all tissues during fruit development and growth. Variations in FPS mRNA amount during fruit growth were not cell type-specific, but appeared homogenous in all fruit tissues.
DISCUSSION
In plants and animals FPS plays an essential role in isoprenoid metabolism. FPS enzymes have been located in different cellular compartments, namely the cytosol (Hugueney et al., 1996), the mitochondria (Cunillera et al., 1997), and the plastids (Sanmiya et al., 1999) in plants and the peroxisomes in animals (Biardi and Krisans, 1996). It is now widely accepted that the FPP produced in the cytosol is dedicated to the biosynthesis of sterols (Biardi and Krisans, 1996), although it is also used for farnesylation and sesquiterpenoid biosynthesis (for review, see Chappell, 1995). In this context it is noteworthy that overexpression of a yeast FPS in tobacco plants led to an apparent increase of both sterols and carotenoids (Daudonnet et al., 1997). Whereas the increase in sterols can be easily interpreted, the effect on the accumulation of carotenoids is unexpected and lays open to question the precise involvement of FPSs in various isoprenoid biosynthetic pathways during plant development.
In tomato fruits sterols accumulate during both early fruit development and ripening (Gillaspy et al., 1993; Whitaker, 1984), whereas carotenoids, mainly lycopene, are actively produced during the ripening phase (Fraser et al., 1994). To get a better understanding of the physiological functions of FPS during these processes, we have isolated and characterized LeFPS1, a cDNA clone that encodes an active isoform of FPS. We found very high similarities between LeFPS1 and other plant FPSs both at the amino acid and at the nucleic acid level, which can be correlated with the genetic distance between plant families. We could not find any evidence of chloroplast or mitochondrial targeting signals, which suggests that LeFPS1 probably encodes a cytosolic FPS isoform.
Little information is available concerning FPS plant gene family complexity. There is evidence that FPS is encoded by at least two genes in several plant species including Arabidopsis (Cunillera et al., 1996), rice (Sanmiya et al., 1999) and Parthenium argentatum (Pan et al., 1996). Here we show that the tomato genome contains two LeFPS1 hybridizing loci, on chromosome 12 and on chromosome 10. Taking advantage of the specific hybridization of the LeFPS1 3′-UTR to a single HindIII restriction fragment, we deduced that the corresponding gene is located on chromosome 12. A second gene that we propose to name LeFPS2 is likely to be located on chromosome 10.
We also identified a second group of loci located on chromosomes 3, 4, and 9, which only weakly hybridized with LeFPS1. In rice cDNAs encoding, respectively, a cytosolic and a chloroplastic isoform, do not cross-hybridize, suggesting that they have diverged significantly (Sanmiya et al., 1999). It is possible that a similar situation has occurred in other plants since a chloroplastic isoform was identified by the same authors in wheat and tobacco. Hence the 3 loci described above could correspond to tomato FPS genes having a low homology level with LeFPS1. Alternatively, weak cross hybridization with unrelated genes cannot be completely ruled out.
Consistent with the requirement of FPP in sterol biosynthesis, it has been shown that FPS genes are ubiquitously expressed in plants (Cunillera et al., 1996; Sanmiya et al., 1997). Similarly, total FPS mRNA was present in all tomato organs we tested. We then investigated a possible organ specific regulation of LeFPS1 using a gene specific probe. Our results have shown that LeFPS1 is expressed in most tomato organs with the exception of young seedling cotyledons where no LeFPS1 specific signal was detected. Though it does not rule out a weak expression of LeFPS1, accumulation of FPS mRNA in cotyledons must be due to the expression of another gene, possibly LeFPS2 since it cross-hybridized with the full length LeFPS1 cDNA. Hence, as already shown in Arabidopsis and rice (Cunillera et al., 1996; Sanmiya at al., 1999), tomato FPS genes are differentially regulated. We cannot exclude the possibility that LeFPS2 is also expressed in other parts of the plant. In this case it would either be regulated similarly to LeFPS1 or expressed at a very low level since there was little difference between the hybridization pattern obtained with LeFPS1 gene-specific and non-specific probes (Fig. 6, A and C). As far as fruit is concerned, LeFPS1 is probably the major gene expressed since all the cDNA clones isolated from the young fruit and the ripened fruit cDNA libraries correspond to the same cDNA species (see “Results”).
Light induction of FPS mRNA and protein accumulation was demonstrated in rice and concerns only FPPS1, a chloroplastic FPS isoform. FPPS2, which encodes a rice cytosolic FPS isoform, is unaffected by light and specifically expressed in roots (Sanmiya et al., 1997, 1999). In our case neither total FPS mRNA nor LeFPS1 mRNA accumulation was dependent on light exposure during seedling development (Fig. 6A). Similarly Arabidopsis genes that encode cytosolic or mitochondrial FPS isoforms are not light inducible (Cunillera et al., 1996, 1997). Hence, light induction of FPS genes might only concern isoforms targeted to the chloroplast and involved in the synthesis of the photosynthetic machinery (Sanmiya et al., 1999).
Terpenoids are essential compounds during tomato fruit development, as demonstrated by altering HMGR activity in young tomato fruits (Narita and Gruissem, 1989; Rodriguez-Concepcion and Gruissem, 1999). The blocking of fruit development that these authors observed was suggested to be caused by inhibition of sterol biosynthesis (Gillaspy et al., 1993), although the synthesis of many other essential isoprenoid end products may also be affected. Since FPP is located at a central branch point in isoprenoid biosynthesis, its function may also be essential for correct fruit development. We first addressed this question by analyzing FPS gene expression at different fruit developmental stages.
Our results showed a very high expression level during early fruit development and a slight decrease during fruit growth. A basal level of FPS mRNA expression was attained as fruit reached their mature size. In young fruits, cell division occurs in most pericarp cell layers and continues mainly in subepidermal cell layers as fruits develop. Further fruit development proceeds via enlargement of mitotically arrested cells (Gillaspy et al., 1993; J.-P. Carde, unpublished observations). In in situ hybridization experiments, no labeling enhancement was noticed in areas containing dividing cells as compared with other pericarp parts, showing that FPS mRNAs are abundant both in dividing and elongating cells. Thus the decrease observed in FPS mRNA level during fruit growth might simply reflect a decrease in FPP requirement due to a progressive cessation of both cell division and elongation. In this context it is noteworthy that tomato FPS genes are highly expressed in young plant organs when cell division and cell elongation events occur, whereas FPS transcripts are barely detectable in mature organs. This situation parallels the demonstration by Jelesko et al. (1999) that HMG1 is primarily transcribed in dividing and elongating cells during the development of tomato plants. Since HMGR catalyzes the synthesis of mevalonate, an essential step in sterol biosynthesis, genes encoding enzymes involved in sterol biosynthesis might be coregulated during plant development.
Tomato fruit ripening is characterized by an intense accumulation of carotenoids, mainly due to a 500-fold increase in lycopene concentration from the mature green to the red ripe stage (Fraser et al., 1994). Carotenoid biosynthesis occurs in plastids (for review, see Camara et al., 1995; Cunningham and Gantt, 1998) and seems to proceed independently from the cytosolic isoprenoid biosynthetic pathway (Rodriguez-Concepcion and Gruissem, 1999). Thus we found it surprising that the level of LeFPS1 mRNA increased again during tomato fruit ripening. This pattern of accumulation correlates with tomato HMG2 gene expression (Daraselia et al., 1996). Rodriguez-Concepcion and Gruissem (1999) proposed that expression of HMG2 might be part of a general defense mechanism activated during fruit ripening. Le FPS1 could participate in such a mechanism providing precursors for the synthesis of sesquiterpenoids produced in tomato fruits. Alternatively, Whitaker (1984) has shown that sterols accumulate significantly during tomato fruit ripening, which might explain the expression of genes encoding enzymes involved in this pathway. We are now developing sense and antisense strategies to analyze the physiological function of FPS during tomato fruit development and ripening.
MATERIALS AND METHODS
Plant Material
Tomato (Lycopersicon esculentum cv Wva 106) plants were germinated and grown in soil in greenhouse conditions under natural light. Experiments with tomato plantlets were performed with 10-d-old seedlings germinated on moist absorbent paper in darkness or 16 h of light, 8 h of darkness. Plant and fruit samples were frozen in liquid nitrogen and stored at −80°C until use. Tomato fruits up to 3 mm in diameter were directly frozen. Pericarp and columella from larger fruits were separated from seeds and locular jelly before storage.
Library Screening and cDNA Clone Analysis
Tomato fruit cDNA libraries prepared from poly(A+) RNA from either young fruits (Joubès et al., 1999) or red ripe tomato fruits (Kausch et al., 1997) were used for PCR amplification and library screening. Two sets of nested degenerate oligonucleotides were designed from conserved regions II and V of FPS (Cunillera et al., 1997). External degenerate primers were FPexd (5′TTYYTIGTIYTIGAYGAYATIATGG3′) and FPexr (5′CYTCIAIRTCIGTICCIATYTICC3′). Internal degenerate primers were FPind (5′GGGAATTCACIMGIGGICARCCIGYTGG3′) and FPinr (5′GGGATCCRTCIARRTARTCRTCYTGIACYTG3′). Dilutions of young and red ripe tomato fruit cDNA libraries were used as templates for PCR amplifications. PCR reactions were performed in a volume of 50 μL containing 25 pmol of each FPex primer. After 5 min at 95°C, 1 unit of Taq polymerase (Appligene Oncor, Illkirch, France) was added. Thirty-five amplification cycles (at 94°C, 50°C, and 72°C for 40 s each) were performed followed by a 10-min incubation at 72°C. An aliquot of 4 μL of the first PCR reaction was amplified a second time using the two FPin primers. Thirty-five cycles were performed in the conditions described above, except for the annealing temperature, which was increased to 51°C. PCR products were cloned in the pGEM-T vector (Promega, Madison, WI) and electroporated in the Escherichia coli strain, DH5α.
For library screening, about 250,000 plaque-forming units from each tomato fruit library were plated. Duplicate nitrocellulose filters (Hybond C, Amersham, Buckinghamshire, UK) were probed with the [32P]dCTP-radiolabeled FPS PCR product (Megaprime labeling kit, Amersham). After hybridization, membranes were washed under low- stringency conditions (2× SSC and 0.1% [w/v] SDS, three times for 15 min at 65°C and 1× SSC and 0.1% [w/v] SDS, three times for 10 min at 65°C) prior to autoradiography. Phages from independent plaques (five for the young tomato fruit and six for the red ripe tomato fruit cDNA library) were isolated, purified, and in vivo excised according to the library manufacturer's instructions (Stratagene, La Jolla, CA).
DNA was sequenced by the dideoxy method (Sanger and Coulson 1975). Sequence comparisons and alignments were performed with the ENTREZ software from National Center for Biotechnology Information and DNASIS and PROSIS (Hitachi, San Bruno, CA).
DNA and RNA Gel-Blot Hybridization
Genomic DNA from tomato leaves was extracted as previously described (Tieman et al., 1992). Total genomic DNA (30 μg) was digested with the indicated restriction endonucleases, separated by electrophoresis on an 0.8% (w/v) agarose Tris-Borate EDTA gel, denatured, and transferred to Hybond C membrane (Amersham) with 20× SSC.
Total RNA was extracted from various plant organs using a hot phenol extraction procedure (Hernould et al., 1992). For northern analysis, total RNA was fractionated on a 1.2% (w/v) agarose, 6% (w/v) formaldehyde gel as described by Sambrook et al. (1989) and transferred to Hybond N+ membrane (Amersham).
Hybridizations at 65°C were performed using either the full-length LeFPS1 cDNA fragment, or the 180-bp long LeFPS1 3′-UTR. Washing was normally performed in 2× SSC and 0.1% (w/v) SDS at 65°C. Where indicated the filters were washed at higher stringency. DNA probes were labeled with [32P]dCTP using a Ready-to-go labeling kit (Amersham Pharmacia Biotech, Orsay, France) and nonincorporated nucleotides were removed by spin chromatography using the probe columns (Quant G-50 Micro, Amersham Pharmacia Biotech). The size of FPS transcripts was estimated using an RNA size marker as standard (Boehringer Mannheim, Basel).
Expression of LeFPS1 in Bacteria and Measurement of Recombinant Prenyl Transferase Activity
LeFPS1 was subcloned in the expression vector pET28(a)+ (Novagen, Madison, WI) between the BamHI and XhoI sites. A BamHI site was inserted upstream of the ATG codon in LeFPS1 by PCR using the Expand High fidelity DNA polymerase (Boehringer Mannheim). The direct primer, 5′CCGGATCCATGGCTGATCTGAAGAAG3′, overlaps the ATG and contains a 5′ extension with a BamHI recognition sequence. The T7 primer was used as reverse primer. LeFPS1 cloned in pBluescript SK was used as a template. Hot start PCR was performed in a 50-μL reaction mixture containing 20 pmol of each primer and 40 ng of template. Thirty PCR amplification cycles, at 94°C for 30 s, 44°C for 30 s, and 72°C for 2 min were followed by a 5 min extension step at 72°C. PCR products were inserted in pET28a(+) generating pET-FPS. Both plasmids were introduced in the E. coli strain BL21(DE3).
A 5-mL Luria-Bertani (125 mg/L kanamycin) liquid culture was inoculated with 0.5 mL of an overnight culture of BL 21 transformed with pET-FPS or pET28a in BL21 (DE3). After 2 h, production of recombinant protein was induced during 30 min at 37°C by addition of 1 mm IPTG. One milliliter of culture was then resuspended in 100 μL of the extraction buffer (10 mm Mg2+ in 25 mm Tris (Tris[hydroxymethyl]-aminomethane)Cl, pH 7.5) and sonicated. Bacterial extracts were incubated with 5 mm [14C] IPP (50 mCi/mmol, Amersham), 50 mm GPP, 10 mm Mg2+ in 25 mm TrisCl, pH 7.5, at 30°C for 30 min prior to pentane extraction. The aqueous phase was dephosphorylated overnight at 37°C with 1 mg/mL alkaline phosphatase (Sigma, St. Louis) in a Gly buffer (50 mm Gly, pH 9.5). Alcohols were extracted with hexane:ether (1:1) prior to scintillation counting. 30,000 cpm were separated and analyzed on GLC (INTERSMAT IGC 121 FL) linked to a radiometer (model 894, Packard, Meriden, CT). In parallel, reaction products (equivalent to 30,000 cpm) were separated using thin layer chromatography (Silica gel SE60, Merck, Lyon, France) with benzene:methanol (9:1) as solvent and autoradiographied. Radiolabeled farnesol was eluted and counted.
Protein concentration was estimated using the Bradford microassay (Bio-Rad, Hercules, CA) with bovine serum albumin as standard. SDS-PAGE analysis of the total bacterial proteins were performed on 10% (w/v) acrylamide gels. Proteins were detected using a Coomassie Blue staining procedure.
In Vitro Transcription/Translation
LeFPS1 cDNA cloned in pBluescript (Stratagene) was digested with EcoRV, XhoI and inserted, downstream to the T7 RNA polymerase promoter, in pCDNA3A (Invitrogen, Carlsbad, CA) digested with SmaI and XhoI. In vitro transcription/translation was performed using the TnT/T7 system (Promega) according to the manufacturer's instructions using [35S]-Met (400 Ci/mmol, ICN) as substrate. The reaction products were analyzed by SDS-PAGE analysis as described above. TnT reaction products were detected after autoradiography of the dried gels.
In Situ Hybridization
Tomato fruits harvested from plants grown in a growth chamber (16 h of light, 8 h of dark) were collected, measured, cut, and immediately immersed in saline formaldehyde/acetic/acid ethanol prior to dehydration and embedding essentially as described in Cox and Goldberg (1988). In situ hybridization was performed according to the Boehringer Mannheim protocol.
To synthesize LeFPS1 DIG-labeled RNA probes, LeFPS1 cDNA was cloned in pBS-SK. The plasmid was linearized with Xho1 or BamH1 and transcribed respectively with T7 (antisense probe) or T3 (sense probe) RNA polymerases using DIG-labeled UTP according to the supplier's instructions.
Mapping FPS Loci
FPS loci were mapped using the population of introgressed lines from Lycopersicon pennelli developed by Eshed and Zamir (1994). Each line contains a single homozygous fragment covering a portion of the genome. Polymorphism was first checked between the parental lines L. esculentum M82 and L. pennellii LA716 with five restriction enzymes. Mapping on the 50 lines was then performed with HindIII and EcoRI using the LeFPS1 cDNA as a probe.
ACKNOWLEDGMENTS
We thank Avtar K. Handa (Horticulture Department, Purdue University, West Lafayette, IN) for the gift of the red ripe tomato fruit cDNA library and Christian Chevalier (Institut National de la Recherche Agronomique [INRA], Grande Ferrade, France) for the gift of the young tomato fruit cDNA library. The authors are indebted to Rachel Cowling-Carol (University of Lyon, France), Armand Mouras (University of Bordeaux II, France), Philippe Raymond (INRA, Grande Ferrade, France), Michel Herzog, Pierre Carol, G. Langenkamper (University of Grenoble, France), and Marcel Kuntz (Centre National de la Recherche Scientifique, Grenoble, France) for critical reading of the manuscript.
Footnotes
J.G. received a post-doctoral fellowship from the région Aquitaine.
LITERATURE CITED
- Adiwilaga K, Kush A. Cloning and characterization of cDNA encoding farnesyl diphosphate synthase from rubber tree (Hevea brasiliensis) Plant Mol Biol. 1996;30:935–946. doi: 10.1007/BF00020805. [DOI] [PubMed] [Google Scholar]
- Anderson MS, Yarger JG, Burck CL, Poulter CD. Farnesyl diphosphate synthetase: molecular cloning, sequence, and expression of an essential gene from Saccharomyces cerevisiae. J Biol Chem. 1989;264:19176–19184. [PubMed] [Google Scholar]
- Ashby MN, Edwards PA. Identification and regulation of a rat liver cDNA encoding farnesyl pyrophosphate synthetase. J Biol Chem. 1989;264:635–640. [PubMed] [Google Scholar]
- Attucci S, Aitken SM, Gulick PJ, Ibrahim RK. Farnesyl pyrophosphate synthase from white lupin: molecular cloning, expression and purification of the expressed protein. Arch Biochem Biophys. 1995;20:493–500. doi: 10.1006/abbi.1995.1422. [DOI] [PubMed] [Google Scholar]
- Biardi L, Krisans SK. Compartmentalization of cholesterol biosynthesis: conversion of mevalonate to farnesyl diphosphate occurs in the peroxisomes. J Biol Chem. 1996;271:1784–1788. doi: 10.1074/jbc.271.3.1784. [DOI] [PubMed] [Google Scholar]
- Camara B, Hugueney P, Bouvier F, Kuntz M, Moneger R. Biochemistry and molecular biology of chromoplast development. Int Rev Cytol. 1995;163:175–247. doi: 10.1016/s0074-7696(08)62211-1. [DOI] [PubMed] [Google Scholar]
- Chappell J. Biochemistry and molecular biology of the isoprenoid biosynthetic pathway in plants. Annu Rev Plant Physiol Plant Mol Biol. 1995;46:521–547. [Google Scholar]
- Cox KH, Goldberg RB. Analysis of plant gene expression. In: Shaw CH, editor. Plant Molecular Biology: A Practical Approach. Oxford: IRL Press; 1988. pp. 1–35. [Google Scholar]
- Cunillera N, Arro M, Delourme D, Karst F, Boronat A, Ferrer A. Arabidopsis thaliana contains two differentially expressed farnesyl-diphosphate synthase genes. J Biol Chem. 1996;271:7774–7780. doi: 10.1074/jbc.271.13.7774. [DOI] [PubMed] [Google Scholar]
- Cunillera N, Boronat A, Ferrer A. The Arabidopsis thaliana FPS1 gene generates a novel mRNA that encodes a mitochondrial farnesyl-diphosphate isoform. J Biol Chem. 1997;272:15381–15388. doi: 10.1074/jbc.272.24.15381. [DOI] [PubMed] [Google Scholar]
- Cunningham FX, Gantt E. Genes and enzymes of carotenoid biosynthesis in plants. Annu Rev Plant Phys Plant Mol Biol. 1998;49:557–583. doi: 10.1146/annurev.arplant.49.1.557. [DOI] [PubMed] [Google Scholar]
- Daraselia ND, Tarchevskaya S, Narita JO. The promoter for tomato 3-hydroxy-3-methylglutaryl coenzyme A reductase gene 2 has unusual regulatory elements that direct high-level expression. Plant Physiol. 1996;112:727–733. doi: 10.1104/pp.112.2.727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daudonnet S, Karst F, Tourte Y. Expression of the farnesyldiphosphate synthase of Saccharomyces cerevisiae in tobacco. Mol Breed. 1997;3:137–145. [Google Scholar]
- Delourme D, Lacroute F, Karst F. Cloning of an Arabidopsis thaliana cDNA encoding for farnesyl diphosphate synthase by functional complementation in yeast. Plant Mol Biol. 1994;26:1867–1873. doi: 10.1007/BF00019499. [DOI] [PubMed] [Google Scholar]
- Eshed Y, Zamir D. Introgressions from Lycopersicon pennellii can improve the soluble-solids yield of tomato hybrids. Theor Appl Genet. 1994;88:891–897. doi: 10.1007/BF01254002. [DOI] [PubMed] [Google Scholar]
- Fraser PD, Truesdale MR, Bird CR, Schuch W, Bramley PM. Carotenoid biosynthesis during tomato fruit development. Plant Physiol. 1994;105:405–413. doi: 10.1104/pp.105.1.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Germain V, Raymond P, Ricard B. Differential expression of two tomato lactate dehydrogenase genes in response to oxygen deficit. Plant Mol Biol. 1997;35:711–721. doi: 10.1023/a:1005854002969. [DOI] [PubMed] [Google Scholar]
- Gillaspy G, Ben-David H, Gruissem W. Fruits: a developmental perspective. Plant Cell. 1993;5:1439–1451. doi: 10.1105/tpc.5.10.1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernould M, Mouras A, Litvak S, Araya A. RNA editing of the mitochondrial atp9 transcript from tobacco. Nucleic Acids Res. 1992;199:1809. doi: 10.1093/nar/20.7.1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugueney P, Bouvier F, Badillo A, Quennemet J, d'Harlingue A, Camara B. Developmental and stress regulation of gene expression for plastid and cytosolic isoprenoid pathway in pepper fruits. Plant Physiol. 1996;111:619–626. doi: 10.1104/pp.111.2.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jelesko JG, Jenkins SM, Rodriguez-Concepcion M, Gruissem W. Regulation of tomato HMG1 during cell proliferation and growth. Planta. 1999;208:310–318. [Google Scholar]
- Joshi CP. An inspection of the domain between putative TATA box and translation start site in 79 plant genes. Nucleic Acids Res. 1987;15:6643–6653. doi: 10.1093/nar/15.16.6643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joubès J, Phan T-H, Rothan C, Bergounioux C, Ratmond P, Chevalier C. Molecular and biochemical characterization of the involvement of cyclin-dependant kinase A during the early development of tomato fruit. Plant Physiol. 1999;121:857–869. doi: 10.1104/pp.121.3.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kausch KD, Handa AK. Molecular cloning of a ripening-specific lipoxygenase and its expression during wild-type and mutant tomato fruit development. Plant Physiol. 1997;113:1041–1050. doi: 10.1104/pp.113.4.1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li CP, Larkins BA. Identification of a maize endosperm-specific cDNA encoding farnesyl pyrophosphate synthetase. Gene. 1996;171:193–196. doi: 10.1016/0378-1119(95)00880-2. [DOI] [PubMed] [Google Scholar]
- Lichtenthaler HK. The 1-deoxy-d-xylulose-5-phosphate pathway of isoprenoid biosynthesis in plants. Annu Rev Plant Physiol Plant Mol Biol. 1999;50:47–65. doi: 10.1146/annurev.arplant.50.1.47. [DOI] [PubMed] [Google Scholar]
- Lichtenthaler HK, Schwender J, Disch A, Rohmer M. Biosynthesis of isoprenoids in higher plant chloroplasts proceeds via a mevalonate-independent pathway. FEBS Lett. 1997;400:271–274. doi: 10.1016/s0014-5793(96)01404-4. [DOI] [PubMed] [Google Scholar]
- McGarvey DJ, Croteau R. Terpenoid metabolism. Plant Cell. 1995;7:1015–1026. doi: 10.1105/tpc.7.7.1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narita JO, Gruissem W. Tomato hydroxymethylglutaryl-CoA reductase is required early in fruit development but not during ripening. Plant Cell. 1989;1:181–190. doi: 10.1105/tpc.1.2.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnuma S, Nakazawa T, Hemmi H, Hallberg AM, Koyama T, Ogura K, Nishino T. Conversion from farnesyl diphosphate synthase to geranylgeranyldiphosphate synthase by random chemical mutagenesis. J Biol Chem. 1996;271:10087–10095. doi: 10.1074/jbc.271.17.10087. [DOI] [PubMed] [Google Scholar]
- Pan Z, Herickhoff L, Backhaus RA. Cloning, characterization, and heterologous expression of cDNAs for farnesyl diphosphate synthase from the guayule rubber plant reveals that this prenyltransferase occurs in rubber particles. Arch Biochem Biophys. 1996;332:196–204. doi: 10.1006/abbi.1996.0333. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Concepcion M, Gruissem W. Arachidonic acid alters tomato HMG expression in fruit growth and induces 3-hydroxy-3-methylglutaryl coenzyme A reductase-independent lycopene accumulation. Plant Physiol. 1999;119:41–48. doi: 10.1104/pp.119.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Ed 2. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Sanger F, Coulson AR. A rapid method for determining sequences in DNA by primed synthesis with DNA polymerase. J Mol Biol. 1975;94:441–448. doi: 10.1016/0022-2836(75)90213-2. [DOI] [PubMed] [Google Scholar]
- Sanmiya K, Iwasaki T, Matsuoka M, Miyao M, Yamamoto N. Cloning of a cDNA that encodes farnesyl diphosphate synthase and the blue light-induced expression of the corresponding gene in the leaves of rice plants. Biochim Biophys Acta. 1997;1350:240–246. doi: 10.1016/s0167-4781(96)00231-x. [DOI] [PubMed] [Google Scholar]
- Sanmiya K, Ueno O, Matsuoka M, Yamamoto N. Localization of farnesyl diphosphate synthase in chloroplasts. Plant Cell Physiol. 1999;40:348–354. doi: 10.1093/oxfordjournals.pcp.a029549. [DOI] [PubMed] [Google Scholar]
- Smith O. Pollination and life history studies of the tomato (Lycopersicon esculentum Mill.) Memoirs of the Cornell University Agricultural Experimental Station. 1935;184:3–16. [Google Scholar]
- Tieman DM, Harriman RW, Ramamohan G, Handa AK. An antisense pectin methylesterase gene alters pectin chemistry and soluble solids in tomato fruit. Plant Cell. 1992;4:667–679. doi: 10.1105/tpc.4.6.667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitaker BD. Changes in the steryl lipid content and composition of tomato fruit ripening. Phytochemistry. 1984;27:34111–34116. [Google Scholar]