Abstract
Breast cancer is a heterogeneous disease and patients are managed clinically based on ER, PR, HER2 expression, and key risk factors. The use of gene expression assays for early stage disease is already common practice. These tests have found a place in risk stratifying the heterogeneous group of stage I–II breast cancers for recurrence, for predicting chemotherapy response, and for predicting breast cancer-related mortality. Most guidelines for hormone receptor (HR)–positive early breast cancer recommend addition of adjuvant chemotherapy for most women, leading to overtreatment, which causes considerable morbidity and cost. Expert oncologist discussed about strategies of gene expression assays and aid in chemotherapy recommendations for treatment of HR + ve EBC and the expert group used data from published literature, practical experience and opinion of a large group of academic oncologists to arrive at this practical consensus recommendations for the benefit of community oncologists.
Keywords: ki67, mammaprint, oncotype dx, predictive test, prosigna, taxane
Introduction
Breast cancer is one of the leading causes of cancer-related morbidity worldwide.[1] Approximately 20% of women diagnosed with EBC will experience recurrence at a distant site within 10 years.[2] One key challenge is that breast cancer is a heterogeneous disease that is categorized clinically by immunohistochemical (IHC) staining of the three receptors; estrogen receptor (ER), progesterone receptor (PR), and the human epidermal growth factor receptor-2 (ERBB2/HER2).[3] Seminal studies in the early 2000s demonstrated that gene expression signatures could classify breast cancers into distinct and reproducible molecular subgroups.[4,5,6,7] In essence, breast cancer can be molecularly classified into luminal A and luminal B subgroups that are mostly comprised of hormone-receptor positive (HR+) breast cancers; a basal-like subgroup that is mostly comprised of triple-negative breast cancers (TNBC); a HER2-enriched subgroup that is mostly comprised of HER2 + breast cancers and a normal-like subgroup that has been proposed to be mostly comprised of the contaminating tumor-surrounding stroma.[8] PAM50 predicted subtypes within a defined IHC subgroup have prognostic implications, in that the luminal A subgroup has a better prognosis than the luminal B subtype.
Traditionally, adjuvant chemotherapy was recommended based on tumor features such as stage (tumor size, regional nodal involvement), grade, expression of hormone receptors (estrogen receptor [ER] and progesterone receptor [PR]) and human epidermal growth factor receptor-2 (HER2), and patient features (age, menopausal status). However, this approach is not accurate enough to guide individualized treatment recommendations, which are based on the risk for recurrence and the reduction in this risk that can be achieved with various systemic treatments.
In particular, there are individuals with low-risk HR-positive, HER2-negative breast cancers who could be spared the toxicities of cytotoxic chemotherapies without compromising the prognosis. Beyond chemotherapy, endocrine therapies also have risks, especially when given for extended durations. Recently, extended endocrine therapy has been shown to prevent late recurrences of HR-positive breast cancers.
Expert group of oncologist meet in the update in oncology-X-2017 to discuss on how to Manage HR + ve early breast cancer and role of Genomics in diagnosis as well as in treatment of early breast cancer.
The update in oncology-X-2017 was organized by Sir Ganga Ram Hospital group met to discuss and arrive at a consensus statement to provide community oncologists practical guidelines for challenging common case scenarios in Breast Cancer out of these we are discus about how to Manage HR + ve early breast cancer and role of Genomics in this chapter. While the discussions will take the scenario as exists in India as a representative country with limited resources, the final manuscript is applicable globally.[9,10] The discussion was based on domain expertise of the National as well as international faculty, published evidence and practical experience in real life management of breast cancer patients. Opinion of the 250 oncologist including medical oncologist, radiation oncologist, surgical oncologist, molecular oncologist and radiologist are present in the update in oncology-X-2017 was taken into consideration by the expert panel.
The expert group was chaired by Dr. D.C. Doval and Dr. Rajeshwar Singh whereas the discussions were moderated by Dr. Ashok Vaid and Dr. Anita Ramesh. The core expert group consists Dr. Samit Purohit, Dr. Bhawan Avasthi, Dr. Sumant Gupta, Dr. Vivek Kaushal, Dr. Shad Salim and Dr. Stephen C Malamud. Consensus answers were used as the basis of formulating the consensus statement providing community oncologists with ready-to-use practical recommendations. The survey answers were used as the basis for formulating the consensus statement so that community oncologists have a ready-to-use Fertility Prevention in Breast cancer patients.
As part of the background work, the best existing evidence was compiled and provided to the expert group panel members for review in preparation of the expert group meeting.[11,12,13] The national and international experts invited to this meeting were also provided the data on the voting by the audience delegates from the update in oncology-X-2017. Members of the panel were also allowed to share their ersonal experiences, make comments and record dissent while voting for the consensus statements. Total of Seven broad question categories were part of the expert group discussions [Table 1].
Table 1.
Question categories addressed by the update in oncology-X-2017
In order to have a more concrete understanding of the risks and benefits of adjuvant chemotherapy, several gene expression assays have been developed to better stratify this group of diverse patients. The assays evaluate varying numbers of genes in the breast tumor, to quantify their expression levels, and output a score that correlates with risk of recurrence. These tests are commercially available now days are being used in clinical practice to assist with prognostication and often to aid decision making regarding adjuvant chemotherapy.
Genomic Profiling-Tables 2–8
Table 2.
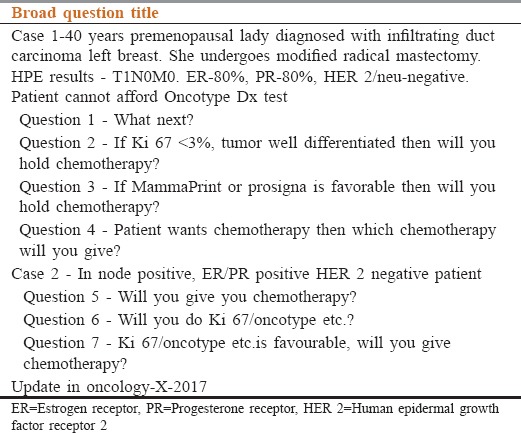
Question 1-40 years premenopausal lady diagnosed with infiltrating duct carcinoma left breast has undergone modified radical mastectomy. Final diagnosis is infiltrating duct carcinoma pT1N0M0. Estrogen receptor-80%, progesterone receptor-80%, human epidermal growth factor receptor 2/neu negative. She is not willing for Oncotype Dx test. What will you recommend next?

Table 8.
Question 7 - Ki 67/oncotype Dx result is favourable, will you give chemotherapy?

Table 3.
Question 2 - If Ki 67 <3%, tumors well differentiated then will you withhold chemotherapy?

Table 4.
Question 3 - If MammaPrint or prosigna is favorable then will you withhold chemotherapy?

Table 5.
Question 4 - Patient wants chemotherapy then which chemotherapy will you give?

Table 6.
Question 5 - In a node positive, estrogen receptor/progesterone receptor positive human epidermal growth factor receptor 2 negative patient, will you give you chemotherapy?

Table 7.
Question 6 - Will you do Ki 67/oncotype Dx etc?

Gene expression profiling by microarray was initially used to identify unique subtypes of breast cancer, but these subtypes also have strong prognostic implications. For example, patients with luminal A tumors have consistently been shown to have a better prognosis than all other subtypes, including the luminal B tumors, which are also ER-positive.[8] There are several assays that clinicians are currently using in their practices to assess the molecular profile of a tumor prior to making recommendations regarding adjuvant systemic therapy.
Ki-67 Assays, Including IHC4 and PEPI
Chronic proliferation is a hallmark of cancer cells.[14] Ki-67, a nuclear nonhistone protein whose expression varies in intensity throughout the cell cycle, has been used as a measurement of tumor cell proliferation.[15] Two large meta-analyses have demonstrated that high Ki-67 expression in breast tumors is independently associated with worse disease-free and overall survival rates.[16,17] Ki-67 expression has also been used to classify HR-positive tumors as luminal A or B. After classifying tumor subtypes based on intrinsic gene expression profiling, Cheang et al. determined that a Ki-67 cut point of 13.25% differentiated luminal A and B tumors.[18] However, the ideal cut point for Ki-67 remains unclear, as the sensitivity and specificity in this study was 77% and 78%, respectively. Others have combined Ki-67 with standard ER, PR, and HER2 testing. This IHC4 score, which weighs each of these variables, was validated in postmenopausal patients from the ATAC (Arimidex, Tamoxifen, Alone or in Combination) trial who had ER-positive tumors and did not receive chemotherapy.[19] The prognostic informati on from the IHC4 was similar to that seen with the 21-gene recurrence score (Oncotype DX), which is discussed later in this article. The key challenge with Ki-67 testing currently is the lack of a validated test methodology, and intraobserver variability in interpreting the Ki-67 results.[20] Recent series have suggested that Ki-67 be considered as a continuous marker rather than a set cut point.[21] These issues continue to impact the clinical utility of Ki-67 for decision making for adjuvant chemotherapy.
Ki-67 and the preoperative endocrine prognostic index (PEPI) score have been explored in the neoadjuvant setting to separate postmenopausal women with endocrine-sensitive versus intrinsically resistant disease and identify patients at risk for recurrent disease.[22] Patients with low pathological stage (0 or 1) and a favorable biomarker profile (PEPI score 0) at surgery had the best prognosis in the absence of chemotherapy.
On the other hand, higher pathological stage at surgery and a poor biomarker profile with loss of ER positivity or persistently elevated Ki-67 (PEPI score of 3) identified de novo endocrine resistant tumors which are at higher risk for early relapse.[23]
Oncotype DX
This 21-gene assay developed by Genomic Health (Redwood City, CA, http://www.genomichealth.com) is the most frequently used test in clinical practice in the U.S.[23] Based on quantitative reverse transcription polymerase chain reaction (PCR) expression levels of 5 reference genes and 16 selected genes related mostly to the estrogen receptor (ER), HER2, proliferation, and invasion, the assay determines a recurrence score (RS) that assigns patients into a low-, intermediate-, or high-risk category.
Originally, the 21-gene recurrence score assay was analyzed as a prognostic biomarker tool in a prospective-retrospective biomarker substudy of the National Surgical Adjuvant Breast and Bowel Project (NSABP) B-14 clinical trial in which patients with node-negative, ER-positive tumors were randomly assigned to receive tamoxifen or placebo without chemotherapy.[24] Using the standard reported values of low risk (<18), intermediate risk (18–30), or high risk (≥31) for recurrence, among the tamoxifen-treated patients, cancers with a high-risk recurrence score had a significantly worse rate of distant recurrence and overall survival.[25] Inferior breast cancer survival with a high recurrence score was also confirmed in other series of endocrine-treated patients with node-negative and node-positive disease.[26,27,28]
PAM50 (Breast Cancer Prognostic Gene Signature)
Using microarray and quantitative reverse transcriptase PCR (RT-PCR) on formalin-fixed paraffin-embedded (FFPE) tissues, the Breast Cancer Prognostic Gene Signature (PAM50) assay was initially developed to identify intrinsic breast cancer subtypes, including luminal A, luminal B, HER2-enriched, and basal-like.[4,8] Based on the prediction analysis of microarray (PAM) method, the assay measures the expression levels of 50 genes, provides a risk category (low, intermediate, and high), and generates a numerical risk of recurrence score (ROR). The intrinsic subtype and ROR have been shown to add significant prognostic value to the clinicopathological characteristics of tumors. Clinical validity of PAM50 was evaluated in postmenopausal women with HR-positive, early-stage breast cancer treated in the prospective ATAC and ABCSG-8 (Austrian Breast and Colorectal Cancer Study Group 8) trials.[29,30] PAM50 has been designed to be carried out in any qualified pathology laboratory. Moreover, the ROR score provides additional prognostic information about risk of late recurrence in breast cancer.
70-Gene Breast Cancer Recurrence Assay (MammaPrint)
MammaPrint is a 70-gene assay that was initially developed using an unsupervised, hierarchical clustering algorithm on whole-genome expression arrays with early-stage breast cancer.
Among 295 consecutive patients who had Mamma Print testing, those classified with a good-prognosis tumor signature (n = 115) had an excellent 10-year survival rate (94.5%) compared to those with a poor prognosis signature (54.5%), and the signature remained prognostic upon multivariate analysis.[7] Subsequently, a pooled analysis comparing outcomes by Mamma Print score in patients with node-negative or 1 to 3 node-positive breast cancers treated as per discretion of their medical team with either adjuvant chemotherapy plus endocrine therapy or endocrine therapy alone reported that only those patients with a high-risk score benefited from chemotherapy.[31] Recently, a prospective phase 3 study (MINDACT [Microarray In Node negative Disease may Avoid ChemoTherapy]) evaluating the utility of Mamma-Print for adjuvant chemotherapy decision-making reported results.[32]
EndoPredict
EndoPredict (EP) is another quantitative RT-PCR–based assay which uses FFPE tissues to calculate a risk score based on 8 cancer-related and 3 reference genes. The score is combined with clinicopathological factors including tumor size and nodal status to make a comprehensive risk score (EPclin). EPclin is used to dichotomize patients into EP low- and EP high-risk groups. EP has been validated in 2 cohorts of patients enrolled in separate randomized studies, ABCSG-6 and ABCSG-8. EP provided prognostic information beyond clinicopathological variables to predict distant recurrence in patients with HR-positive, HER2-negative early breast cancer.[33]
Endo Predict is the first multi gene expression assay that could be routinely performed in decentral molecular pathological laboratories with a short turnaround time.[34]
Breast Cancer Index
The BCI is a RT-PCR–based gene expression assay that consists of 2 gene expression biomarkers: molecular grade index (MGI) and HOXB13/IL17BR (H/I). The BCI was developed as a prognostic test to assess risk for breast cancer recurrence using a cohort of ER-positive patients (n = 588) treated with adjuvant tamoxifen versus observation from the prospective randomized Stockholm trial.[35] The prognostic and predictive values of the BCI have been validated in other large, randomized studies and in patients with both node-negative and node-positive disease.[36,37] The predictive value of the endocrine-response biomarker, the H/I ratio, has been demonstrated in randomized studies. In the MA.17 trial, a high H/I ratio was associated with increased risk for late recurrence in the absence of letrozole. However, extended endocrine therapy with letrozole in patients with high H/I ratios predicted benefit from therapy and decreased the probability of late disease recurrence.[38]
Adjuvant Treatment Options
Luminal subtypes
The luminal A and B subtypes are both characterized by HR expression, and 5 years of adjuvant anti-estrogen therapy became the standard of care based upon results from multiple trials.[39] The addition of aromatase inhibitors in the adjuvant setting for postmenopausal women has improved disease-free survival compared with tamoxifen alone. Aromatase inhibitors can be used as upfront continuous treatment for 5 years,[40,41] as sequential therapy after 2 to 3 years of tamoxifen,[42,43] or as extended adjuvant therapy after 5 years of tamoxifen.[44]
Patients with HR-positive breast cancer continue to have relapse rates of 1% to 4% per year between 5 and 15 years from diagnosis, and the optimal duration of adjuvant hormonal therapy remains an important clinical question.[45,46] The use of gene expression profiling can help to identify key genes that can then be exploited therapeutically.
Basal-like subtype
When patients were stratified by breast tumor subtype and analyzed for time to distant metastasis and overall survival, those with the basal subtype had the worst clinical outcome.[47] This likely reflects both the aggressive nature of basal-subtype breast tumors and the lack of targeted therapies, since these tumors do not express the ER and do not overexpress HER2. Conventional anthracycline- and taxane-based regimens are currently used to treat patients with the basal-like subtype of breast cancer.
HER2-enriched subtype
The HER2-enriched subtype is characterized by high expression of HER2, most commonly due to amplification of the HER2 gene. Genes such as GRB7 and TOP2A, which are located in close proximity to the HER2 gene on chromosome 17, are often co-amplified.[48] Multiple studies have been performed to correlate TOP2A gene status, topo2a expression levels, and response to anthracyclines.[49,50,51,52,53,54] The role of TOP2A amplification was examined in the Breast Cancer International Research Group (BCIRG) 006 trial in which early-stage, HER2-positive patients were randomized between three arms: standard anthracycline- and taxane-based chemotherapy with or without trastuzumab, and a third non–anthracycline-containing regimen of docetaxel, carboplatin, and trastuzumab. Patients without co-amplification derived greater benefit from the addition of trastuzumab. In patients with co-amplification of TOP2A and HER2, minimal incremental benefit was seen with the addition of trastuzumab; however, the long-term toxicity profile favored the non–anthracycline-containing regimen.[53]
Conclusion
Reduction in breast cancer mortality is mainly the result of improved systemic treatments. With advances in breast cancer screening tools in recent years, the rate of cancer detection has increased. This has raised concerns regarding over diagnosis. To prevent unwanted toxicities associated with overtreatment, better treatment decision tools are needed. Several genomic assays are currently available and widely used to provide prognostic and predictive information and aid in decisions regarding appropriate use of adjuvant chemotherapy in HR-positive/HER2-negative early-stage breast cancer. Gene expression assays have the potential to fill the gap where clinicopathologic criteria fall short.
Summary of our discussion is based on the individual patients' and their unique tumor biology will likely result in better outcomes overall by making sure the right patient receives the right therapy. The aim of our discussion is to improved treatment selection (with or without chemotherapy), there will be improved quality of life, more efficient use of resources, and reduced direct and indirect costs of treatment.
Take Home Message

Financial support and sponsorship
Nil.
Conflicts of interest
There are no conflicts of interest.
References
- 1.Goss PE, Lee BL, Badovinac-Crnjevic T, Strasser-Weippl K, Chavarri-Guerra Y, St. Louis J, et al. Planning cancer control in Latin America and the Caribbean. Lancet Oncol. 2013;14:391–436. doi: 10.1016/S1470-2045(13)70048-2. [DOI] [PubMed] [Google Scholar]
- 2.Early Breast Cancer Trialists' Collaborative Group (EBCTCG) Effects of chemotherapy and hormonal therapy for early breast cancer on recurrence and 15-year survival: An overview of the randomised trials. Lancet. 2005;365:1687–717. doi: 10.1016/S0140-6736(05)66544-0. [DOI] [PubMed] [Google Scholar]
- 3.Hammond ME, Hayes DF, Dowsett M, Allred DC, Hagerty KL, Badve S, et al. American Society of Clinical Oncology/College of American Pathologists guideline recommendations for immunohistochemical testing of estrogen and progesterone receptors in breast cancer. J Clin Oncol. 2010;28:2784–95. doi: 10.1200/JCO.2009.25.6529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Perou CM, Sørlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–52. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 5.Sørlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A. 2001;98:10869–74. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van 't Veer LJ, Dai H, van de Vijver MJ, He YD, Hart AA, Mao M, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–6. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 7.van de Vijver MJ, He YD, van't Veer LJ, Dai H, Hart AA, Voskuil DW, et al. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med. 2002;347:1999–2009. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- 8.Parker JS, Mullins M, Cheang MC, Leung S, Voduc D, Vickery T, et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. J Clin Oncol. 2009;27:1160–7. doi: 10.1200/JCO.2008.18.1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.National Cancer Registry Programme, Indian Council of Medical Research. In: Consolidated Report of Population Based Cancer Registries 2001-2004, Incidence and Distribution of Cancer. Bangalore: Coordinating Unit, National Cancer Registry Programme (ICMR) 2006. Leading sites of cancer; pp. 8–30. [Google Scholar]
- 10.Badwe RA, Gangawal S, Mitra I, Desai PB. Clinico-pathological featuresand prognosis of breast cancer in different religious communities in India. Indian J Cancer. 1990;27:220–8. [PubMed] [Google Scholar]
- 11.Altekruse SF, Kosary CL, Krapcho M, editors. National Cancer Institute. 1975-2007. In: SEER Cancer Statistics Review. [Google Scholar]
- 12.National Cancer Registry Program. Ten Year Consolidated Report of the Hospital Based Cancer Registries, 1984–1993, An Assessment of the Burden and Care of Cancer Patients. New Delhi: Indian Council of Medical Research; 2001. [Google Scholar]
- 13.Agarwal G, Pradeep PV, Aggarwal V, Yip CH, Cheung PS. Spectrum of breast cancer in Asian women. World J Surg. 2007;31:1031–40. doi: 10.1007/s00268-005-0585-9. [DOI] [PubMed] [Google Scholar]
- 14.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 15.de Azambuja E, Cardoso F, de Castro G, Jr, Colozza M, Mano MS, Durbecq V, et al. Ki-67 as prognostic marker in early breast cancer: A meta-analysis of published studies involving 12,155 patients. Br J Cancer. 2007;96:1504–13. doi: 10.1038/sj.bjc.6603756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Petrelli F, Viale G, Cabiddu M, Barni S. Prognostic value of different cut-off levels of Ki-67 in breast cancer: A systematic review and meta-analysis of 64,196 patients. Breast Cancer Res Treat. 2015;153:477–91. doi: 10.1007/s10549-015-3559-0. [DOI] [PubMed] [Google Scholar]
- 17.Cheang MC, Chia SK, Voduc D, Gao D, Leung S, Snider J, et al. Ki67 index, HER2 status, and prognosis of patients with luminal B breast cancer. J Natl Cancer Inst. 2009;101:736–50. doi: 10.1093/jnci/djp082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cuzick J, Dowsett M, Pineda S, Wale C, Salter J, Quinn E, et al. Prognostic value of a combined estrogen receptor, progesterone receptor, Ki-67, and human epidermal growth factor receptor 2 immunohistochemical score and comparison with the Genomic Health Recurrence score in early breast cancer. J Clin Oncol. 2011;29:4273–8. doi: 10.1200/JCO.2010.31.2835. [DOI] [PubMed] [Google Scholar]
- 19.Pathmanathan N, Balleine RL. Ki67 and proliferation in breast cancer. J Clin Pathol. 2013;66:512–6. doi: 10.1136/jclinpath-2012-201085. [DOI] [PubMed] [Google Scholar]
- 20.Denkert C, Budczies J, von Minckwitz G, Wienert S, Loibl S, Klauschen F, et al. Strategies for developing Ki67 as a useful biomarker in breast cancer. Breast. 2015;24(Suppl 2):S67–72. doi: 10.1016/j.breast.2015.07.017. [DOI] [PubMed] [Google Scholar]
- 21.Ma CX, Bose R, Ellis MJ. Prognostic and predictive biomarkers of endocrine responsiveness for estrogen receptor positive breast cancer. Adv Exp Med Biol. 2016;882:125–54. doi: 10.1007/978-3-319-22909-6_5. [DOI] [PubMed] [Google Scholar]
- 22.Ellis MJ, Tao Y, Luo J, A'Hern R, Evans DB, Bhatnagar AS, et al. Outcome prediction for estrogen receptor-positive breast cancer based on postneoadjuvant endocrine therapy tumor characteristics. J Natl Cancer Inst. 2008;100:1380–8. doi: 10.1093/jnci/djn309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang M, Rajan S, Issa AM. Cost effectiveness of gene expression profiling for early stage breast cancer: A decision-analytic model. Cancer. 2012;118:5163–70. doi: 10.1002/cncr.27443. [DOI] [PubMed] [Google Scholar]
- 24.Fisher B, Jeong JH, Bryant J, Anderson S, Dignam J, Fisher ER, et al. Treatment of lymph-node-negative, oestrogen-receptor-positive breast cancer: Long-term findings from national surgical adjuvant breast and bowel project randomised clinical trials. Lancet. 2004;364:858–68. doi: 10.1016/S0140-6736(04)16981-X. [DOI] [PubMed] [Google Scholar]
- 25.Paik S, Shak S, Tang G, Kim C, Baker J, Cronin M, et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med. 2004;351:2817–26. doi: 10.1056/NEJMoa041588. [DOI] [PubMed] [Google Scholar]
- 26.Habel LA, Shak S, Jacobs MK, Capra A, Alexander C, Pho M, et al. A population-based study of tumor gene expression and risk of breast cancer death among lymph node-negative patients. Breast Cancer Res. 2006;8:R25. doi: 10.1186/bcr1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Albain KS, Barlow WE, Shak S, Hortobagyi GN, Livingston RB, Yeh IT, et al. Prognostic and predictive value of the 21-gene recurrence score assay in postmenopausal women with node-positive, oestrogen-receptor-positive breast cancer on chemotherapy: A retrospective analysis of a randomised trial. Lancet Oncol. 2010;11:55–65. doi: 10.1016/S1470-2045(09)70314-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dowsett M, Cuzick J, Wale C, Forbes J, Mallon EA, Salter J, et al. Prediction of risk of distant recurrence using the 21-gene recurrence score in node-negative and node-positive postmenopausal patients with breast cancer treated with anastrozole or tamoxifen: A TransATAC study. J Clin Oncol. 2010;28:1829–34. doi: 10.1200/JCO.2009.24.4798. [DOI] [PubMed] [Google Scholar]
- 29.Dowsett M, Sestak I, Lopez-Knowles E, Sidhu K, Dunbier AK, Cowens JW, et al. Comparison of PAM50 risk of recurrence score with oncotype DX and IHC4 for predicting risk of distant recurrence after endocrine therapy. J Clin Oncol. 2013;31:2783–90. doi: 10.1200/JCO.2012.46.1558. [DOI] [PubMed] [Google Scholar]
- 30.Gnant M, Filipits M, Greil R, Stoeger H, Rudas M, Bago-Horvath Z, et al. Predicting distant recurrence in receptor-positive breast cancer patients with limited clinicopathological risk: Using the PAM50 Risk of Recurrence score in 1478 postmenopausal patients of the ABCSG-8 trial treated with adjuvant endocrine therapy alone. Ann Oncol. 2014;25:339–45. doi: 10.1093/annonc/mdt494. [DOI] [PubMed] [Google Scholar]
- 31.Knauer M, Mook S, Rutgers EJ, Bender RA, Hauptmann M, van de Vijver MJ, et al. The predictive value of the 70-gene signature for adjuvant chemotherapy in early breast cancer. Breast Cancer Res Treat. 2010;120:655–61. doi: 10.1007/s10549-010-0814-2. [DOI] [PubMed] [Google Scholar]
- 32.Cardoso F, van't Veer LJ, Bogaerts J, Slaets L, Viale G, Delaloge S, et al. 70-gene signature as an aid to treatment decisions in early-stage breast cancer. N Engl J Med. 2016;375:717–29. doi: 10.1056/NEJMoa1602253. [DOI] [PubMed] [Google Scholar]
- 33.Filipits M, Rudas M, Jakesz R, Dubsky P, Fitzal F, Singer CF, et al. A new molecular predictor of distant recurrence in ER-positive, HER2-negative breast cancer adds independent information to conventional clinical risk factors. Clin Cancer Res. 2011;17:6012–20. doi: 10.1158/1078-0432.CCR-11-0926. [DOI] [PubMed] [Google Scholar]
- 34.Müller BM, Keil E, Lehmann A, Winzer KJ, Richter-Ehrenstein C, Prinzler J, et al. The endoPredict gene-expression assay in clinical practice – Performance and impact on clinical decisions. PLoS One. 2013;8:e68252. doi: 10.1371/journal.pone.0068252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jerevall PL, Ma XJ, Li H, Salunga R, Kesty NC, Erlander MG, et al. Prognostic utility of HOXB13:IL17BR and molecular grade index in early-stage breast cancer patients from the Stockholm trial. Br J Cancer. 2011;104:1762–9. doi: 10.1038/bjc.2011.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sgroi DC, Chapman JA, Badovinac-Crnjevic T, Zarella E, Binns S, Zhang Y, et al. Assessment of the prognostic and predictive utility of the breast cancer index (BCI): An NCIC CTG MA. 14 study. Breast Cancer Res. 2016;18:1. doi: 10.1186/s13058-015-0660-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang Y, Schnabel CA, Schroeder BE, Jerevall PL, Jankowitz RC, Fornander T, et al. Breast cancer index identifies early-stage estrogen receptor-positive breast cancer patients at risk for early- and late-distant recurrence. Clin Cancer Res. 2013;19:4196–205. doi: 10.1158/1078-0432.CCR-13-0804. [DOI] [PubMed] [Google Scholar]
- 38.Sgroi DC, Carney E, Zarrella E, Steffel L, Binns SN, Finkelstein DM, et al. Prediction of late disease recurrence and extended adjuvant letrozole benefit by the HOXB13/IL17BR biomarker. J Natl Cancer Inst. 2013;105:1036–42. doi: 10.1093/jnci/djt146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Early Breast Cancer Trialists' Collaborative Group (EBCTCG) Davies C, Godwin J, Gray R, Clarke M, Cutter D, et al. Relevance of breast cancer hormone receptors and other factors to the efficacy of adjuvant tamoxifen: Patient-level meta-analysis of randomised trials. Lancet. 2011;378:771–84. doi: 10.1016/S0140-6736(11)60993-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cuzick J, Sestak I, Baum M, Buzdar A, Howell A, Dowsett M, et al. Effect of anastrozole and tamoxifen as adjuvant treatment for early-stage breast cancer: 10-year analysis of the ATAC trial. Lancet Oncol. 2010;11:1135–41. doi: 10.1016/S1470-2045(10)70257-6. [DOI] [PubMed] [Google Scholar]
- 41.Regan MM, Neven P, Giobbie-Hurder A, Goldhirsch A, Ejlertsen B, Mauriac L, et al. Assessment of letrozole and tamoxifen alone and in sequence for postmenopausal women with steroid hormone receptor-positive breast cancer: The BIG 1-98 randomised clinical trial at 8·1 years median follow-up. Lancet Oncol. 2011;12:1101–8. doi: 10.1016/S1470-2045(11)70270-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dubsky PC, Jakesz R, Mlineritsch B, Pöstlberger S, Samonigg H, Kwasny W, et al. Tamoxifen and anastrozole as a sequencing strategy: A randomized controlled trial in postmenopausal patients with endocrine-responsive early breast cancer from the Austrian Breast and Colorectal Cancer Study Group. J Clin Oncol. 2012;30:722–8. doi: 10.1200/JCO.2011.36.8993. [DOI] [PubMed] [Google Scholar]
- 43.Bliss JM, Kilburn LS, Coleman RE, Forbes JF, Coates AS, Jones SE, et al. Disease-related outcomes with long-term follow-up: An updated analysis of the intergroup exemestane study. J Clin Oncol. 2012;30:709–17. doi: 10.1200/JCO.2010.33.7899. [DOI] [PubMed] [Google Scholar]
- 44.Jin H, Tu D, Zhao N, Shepherd LE, Goss PE. Longer-term outcomes of letrozole versus placebo after 5 years of tamoxifen in the NCIC CTG MA. 17 trial: Analyses adjusting for treatment crossover. J Clin Oncol. 2012;30:718–21. doi: 10.1200/JCO.2010.34.4010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Early Breast Cancer Trialists' Collaborative Group. Tamoxifen for early breast cancer. Cochrane Database Syst Rev. 2001 CD000486. [Google Scholar]
- 46.Saphner T, Tormey DC, Gray R. Annual hazard rates of recurrence for breast cancer after primary therapy. J Clin Oncol. 1996;14:2738–46. doi: 10.1200/JCO.1996.14.10.2738. [DOI] [PubMed] [Google Scholar]
- 47.Sorlie T, Tibshirani R, Parker J, Hastie T, Marron JS, Nobel A, et al. Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc Natl Acad Sci U S A. 2003;100:8418–23. doi: 10.1073/pnas.0932692100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Slamon DJ, Press MF. Alterations in the TOP2A and HER2 genes: Association with adjuvant anthracycline sensitivity in human breast cancers. J Natl Cancer Inst. 2009;101:615–8. doi: 10.1093/jnci/djp092. [DOI] [PubMed] [Google Scholar]
- 49.Arriola E, Moreno A, Varela M, Serra JM, Falo C, Benito E, et al. Predictive value of HER-2 and topoisomerase IIalpha in response to primary doxorubicin in breast cancer. Eur J Cancer. 2006;42:2954–60. doi: 10.1016/j.ejca.2006.06.013. [DOI] [PubMed] [Google Scholar]
- 50.Pritchard KI. Are HER2 and TOP2A useful as prognostic or predictive biomarkers for anthracycline-based adjuvant chemotherapy for breast cancer? J Clin Oncol. 2009;27:3875–6. doi: 10.1200/JCO.2009.22.8361. [DOI] [PubMed] [Google Scholar]
- 51.Gianni L, Valagussa P. Anthracyclines and early breast cancer: The end of an era? J Clin Oncol. 2009;27:1155–7. doi: 10.1200/JCO.2008.20.1640. [DOI] [PubMed] [Google Scholar]
- 52.Buzdar AU. Topoisomerase IIalpha gene amplification and response to anthracycline-containing adjuvant chemotherapy in breast cancer. J Clin Oncol. 2006;24:2409–11. doi: 10.1200/JCO.2006.05.9113. [DOI] [PubMed] [Google Scholar]
- 53.Slamon D, Eiermann W, Robert N, Pienkowski T, Martin M, Press M, et al. Adjuvant trastuzumab in HER2-positive breast cancer. N Engl J Med. 2011;365:1273–83. doi: 10.1056/NEJMoa0910383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Martin M, Romero A, Cheang MC, López García-Asenjo JA, García-Saenz JA, Oliva B, et al. Genomic predictors of response to doxorubicin versus docetaxel in primary breast cancer. Breast Cancer Res Treat. 2011;128:127–36. doi: 10.1007/s10549-011-1461-y. [DOI] [PubMed] [Google Scholar]


