Abstract
Mutation of SMARCA4 (BRG1), the ATPase of BAF (mSWI/SNF) and PBAF complexes, contributes to a range of malignancies and neurologic disorders. Unfortunately, the effects of SMARCA4 missense mutations have remained uncertain. Here we show that SMARCA4 cancer missense mutations target conserved ATPase surfaces and disrupt the mechanochemical cycle of remodeling. We find that heterozygous expression of mutants alters the open chromatin landscape at thousands of sites across the genome. Loss of DNA accessibility does not directly overlap with Polycomb accumulation, but is enriched in “A compartments” at active enhancers, which lose H3K27ac but not H3K4me1. Affected positions include hundreds of sites identified as superenhancers in many tissues. Dominant-negative mutation induced pro-oncogenic expression changes, including increased expression of Myc and its target genes. Together, our data suggest that disruption of enhancer accessibility represents a key source of altered function in SMARCA4-mutated disorders in a wide variety of tissues.
Keywords: Cancer, chromatin remodeling, epigenomics, enhancers
Introduction
Epigenetic deregulation is widely regarded as a hallmark of cancer and certain neurologic disorders. Among the most frequently mutated chromatin regulatory complexes are polymorphic BAF (mSWI/SNF) and PBAF complexes1,2. These ATP-dependent chromatin remodelers are mutated in ~20% of cancers3,4, with mutations in specific subunits contributing to specific cancers1. For cancers such as malignant rhabdoid tumors5,6 and synovial sarcoma7, tissue-specific alterations of individual BAF subunits arise in ~100% of cases, and repair of these defects in cell-culture models leads to loss of both transformation and proliferation. Moreover, animal models and epigenomic studies of human patients have provided extensive evidence that these complexes are major tumor suppressors8 whose mutations contribute to transformation in many cell types9.
In malignant rhabdoid tumors (MRTs) and synovial sarcoma (SS), the ability of BAF complexes to oppose Polycomb repression proximal to transcription start sites (TSSs) is central to their role in these malignancies. In MRTs, biallelic loss of SMARCB1 induces abnormal subunit composition10,11, and impairs opposition to Polycomb silencing at the tumor-suppressor gene p16 (INK4A, refs. 12–14). Silencing of p16 in turn impairs cell-cycle control, thereby inducing rapid transformation and proliferation without genomic instability15,16. In SS, the protein product of the SS18-SSX gene fusion17 incorporates into BAF complexes, leading to reduction of the Polycomb-associated silencing mark H3K27me3 near the TSSs of oncogenic genes such as EGR1 (ref. 18) and SOX2 (ref. 7). For both MRTs and SS, changes of Polycomb occupancy at TSSs across the genome coincide with transformation, and in both cases, these changes are reversible by exogenous expression or repair of the underlying BAF defect7,19,20.
Recent studies in BAF-deficient cancer models have revealed alterations at enhancers distal to TSSs, which contain transcription factor binding sites. In MRT cells, loss of SMARCB1 induces characteristic changes of the permissive mark H3K27ac at tissue-restricted enhancers, but largely preserves the function of superenhancers11. Moreover, in animal models of colon cancer, loss of the BAF subunit Arid1a leads to impaired enhancer function, with deregulation of APC–beta-catenin signaling pathways21. Similar results have been observed in mouse embryonic fibroblasts22. In mouse embryonic stem cells (mESCs), topoisomerase inhibition leads to impairment of BAF’s ability to generate accessible sites over the genome, indicating these factors work in tandem to generate accessibility23. Thus, emerging evidence suggests that altered function at enhancers arises upon loss of BAF activity, in addition to Polycomb deregulation.
SMARCA4 (BRG1), a central ATPase of BAF complexes, is the most frequently mutated Snf2-like gene in human cancer1, and has been identified as a major tumor suppressor in pan-cancer studies1,8. Unlike the tissue-restricted alterations of SMARCB1 and SS18, loss-of-function mutations of SMARCA4 are enriched in diverse cancer types, contributing to a range of cancers including those of the lung24–26, ovaries27–29, skin4,30, thoracic sarcomas31, and lymphomas32,33. SMARCA4 inactivation is especially common in small cell carcinoma of the ovary, hypercalcemic type (SCCOHT)27,28. Dual biallelic inactivation of SMARCA4 and the related ATPase SMARCA2 are highly specific to SCCOHT29, indicating that complete loss of BAF and PBAF ATP-dependent chromatin remodeling activity is an essential feature of this malignancy.
In contrast to the precise, reproducible genetic changes observed in SCCOHT, the impacts of diverse heterozygous SMARCA4 mutations distributed across many cancer types have remained uncertain and controversial. Recently, we showed that deletion and heterozygous cancer mutations of the SMARCA4 ATPase induce genome-wide accumulation of Polycomb Repressive Complexes 1 and 2 (PRC1 and PRC2) at bivalent TSSs across the genome34. However, it has remained uncertain how these mutations impair SMARCA4’s fundamental mechanisms to promote malignancy. Here we show that these mutations induce divergent effects on SMARCA4 dynamics in living cells, but the impairment of normal function through distinct mechanisms results in convergent changes of the DNA accessibility landscape at enhancers and superenhancers used in many cell types.
Results
Heterozygous mutation of SMARCA4 is a common feature of many cancers
To assess the frequency of heterozygous SMARCA4 missense mutations in cancer, we performed a pan-cancer analysis of annotated human tumor sequencing studies, including The Cancer Genome Atlas (TCGA) whole-exome studies35 and MSK-IMPACT36 targeted sequencing studies where SMARCA4 mutations and deletions were observed. We eliminated samples with only silent mutations and no other changes, and furthermore restricted our analysis to samples where copy number analysis had also been performed, leaving 927 human tumor samples. Analysis of the overall genetic state of SMARCA4 in these samples (Figure 1a) shows that most samples (56.5%, 524/927) contain a single allele affected by missense mutations without other aberrations. In contrast, other major tumor suppressors, such as ARID1A (20.9%, 370/1773, p<2.2e-16) and RB1 (14.5%, 155/1066, p<2.2e-16), have significantly lower fractions of samples with only single missense mutations (Figure 1b).
Figure 1. Heterozygous SMARCA4 mutations contribute to many cancers.
(a) Heat map illustrating the genomic states of 927 human tumor samples from different tissues with SMARCA4 mutations and deletions. Samples without genetic changes to SMARCA4 or with only silent mutations are not shown. (b) SMARCA4 has disproportionately more missense mutations than other tumor suppressors like ARID1A and RB1. Pie chart showing the proportion of tumor samples showing patterns of genetic aberrations. Fisher exact test comparing proportion of missense mutations compared to SMARCA4, *** p<2.2e-16. (c) Expression of SMARCA4 in tumor samples bearing missense mutations show comparable levels of SMARCA4, consistent with heterozygous expression.
To assess whether samples with missense mutations had comparable levels of expression as non-mutated samples, we analyzed RNA-seq data from TCGA studies. We found that samples with single missense mutations had comparable levels of SMARCA4 expression (Figure 1c), consistent with heterozygous mutation. Our analysis shows that heterozygous mutation is the most common mode of disruption to many different cancers where SMARCA4 is mutated.
Cancer mutations of SMARCA4 induce distinct dynamic defects of remodeling in living cells
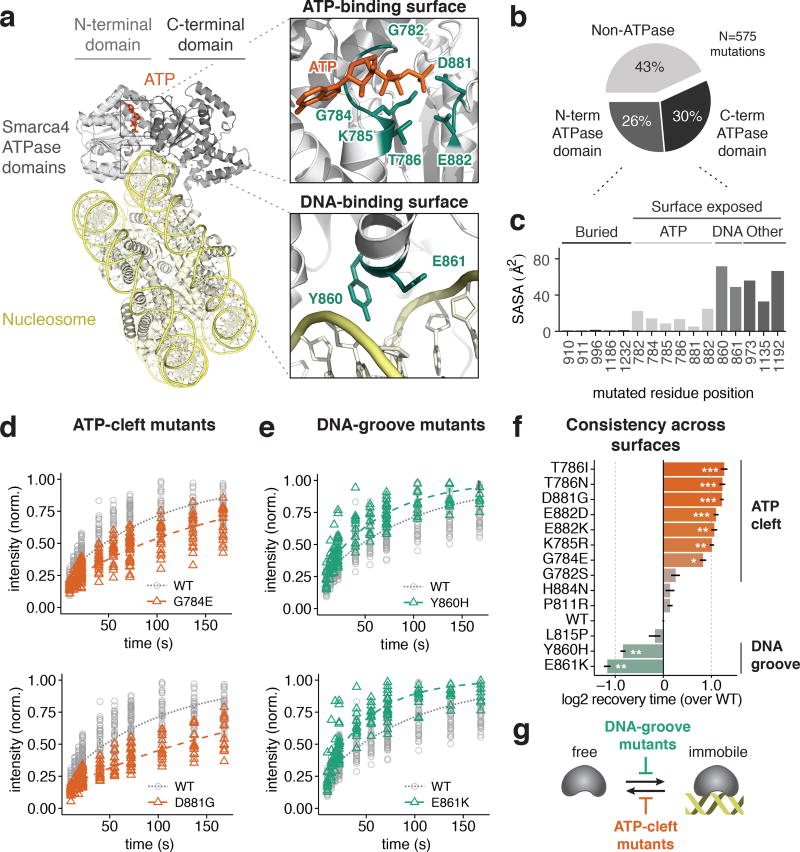
Frequent cancer mutations within the SMARCA4 ATPase domain have provided a rich source of alleles to probe the mechanisms of these proteins. To gain insight into their mechanisms, we generated a homology model of SMARCA4 bound to a nucleosome based on the high degree of sequence homology with structures of related Snf2 homologues from diverse organisms, including Saccharomyces cerevisiae Snf2 (PDB accession code 5X0X)37, Thermothelomyces thermophila Snf2 (PDB 5HZR)38, and Saccharomyces cerevisiae Chd1 (PDB 3MWY)39. Full details of the construction of this homology model are provided in the Methods section, and three-dimensional coordinates of mouse and human SMARCA4 homology models are provided in PDB format as supplemental data. Our model revealed that frequently mutated positions of SMARCA4 lie in functionally important surfaces, such as the DNA groove between the N- and C-terminal ATPase domains, and the ATP cleft (Figure 2a, Figure S1).
Figure 2. SMARCA4 cancer mutations affect functional surfaces and induce distinct dynamic defects.
(a) Homology model of SMARCA4 ATPase domains bound to a nucleosome, derived from Snf2 and Snf2-like family member structures from other organisms (details in main text and Methods). Three-dimensional coordinates of the SMARCA4 homology models are provided in PDB format as supplemental data. (b) Distribution of missense mutations of SMARCA4 across all cancers compiled by cBioPortal. 56% of mutations target the N- and C-terminal ATPase domain. (c) Solvent accessible surface area (SASA) for frequently mutated residue positions of SMARCA4 in human cancers. Buried and surface mutations are frequent sites of mutations, including the ATP-binding cleft, DNA-binding groove, and other sites with unknown function. (d) FRAP recovery curves for SMARCA4-GFP mutants of the ATP-binding cleft show slow photobleaching recovery. (e) FRAP recovery curves for SMARCA4-GFP mutants of the DNA-binding groove show fast photobleaching recovery. Curves for WT controls (dotted) are the same for all plots in panels (d) and (e). (f) Positions found within the ATP-binding cleft or DNA-binding groove show similar FRAP recovery kinetics. Two-sample KS test compared to WT, * p<0.05; ** p<0.01; *** p<0.001. Plotted values are mean t1/2 recovery times, error bars are SEM from n=10 cells. (g) Model of how SMARCA4 surface mutations alter dynamic engagement of chromatin. DNA-binding groove mutants impair engagement with DNA, while ATP-binding cleft mutants impair ATP hydrolysis, which may be required for transition to a mobile state.
We analyzed the positions of all cancer missense mutations on cBioPortal40; we found that most (56%, N=575) SMARCA4 mutations affected the bipartite ATPase domain (Figure 2b). By calculating the solvent-accessible surface area for each position, we found that frequently mutated positions in cancer are distributed in structurally diverse locations of the domain. Some mutation hotspots, such as T910, are expected to affect residues that are buried or have little exposed surface area (Figure 2c). As disruption of folding or thermodynamic stability would be expected to impair function in a non-specific manner, we turned our focus to residues that were frequently mutated at important surfaces, specifically, the ATP cleft and DNA groove (Figure S1).
We hypothesized that mutations of these positions might induce distinct dynamic defects, since they occur at surfaces with unique molecular functions. We therefore generated a targeted library of mutations at these sites, including p.G782S, p.G784E, p.K785R, p.T786I, p.T786N, p.P811R, p.L815P, p.Y860H, p.E861K, p.D881G, p.E882D, p.E882K, and p.H884N (numbered according to the human protein product; UniProt, P51532). We expressed these as C-terminal GFP fusions, and performed a screen of these for dynamic defects using fluorescence recovery after photobleaching (FRAP) following lentiviral transduction in 293T cells (Figure 2d–f). Compared to wild-type SMARCA4-GFP, we found that mutants located at the ATP cleft consistently resulted in slower FRAP recovery (Figure 2f). In contrast, mutants located at the DNA groove consistently resulted in fast recovery times compared to wild-type (Figure 2f). These data are consistent with a model whereby DNA-groove mutants disrupt engagement of immobile chromatin, but ATP-cleft mutants disrupt ATP-dependent release from immobile chromatin (Figure 2g). This model is supported by previous observations that the ATPase activity of SMARCA4 and orthologs is DNA-stimulated41,42, as ATP hydrolysis requires prior interaction with nucleic acid substrate.
To validate the observed dynamics from this screen, we generated a transgenic mouse expressing SMARCA4 fused to the fluorescent protein Dendra2 (ref. 43), comparable to GFP (Figure S2). This fusion did not affect protein function, and homozygous SMARCA4-Dendra2 mice have been bred and deposited at Jackson Laboratories for cryopreservation (stock #27901). In mESCs, SMARCA4-Dendra2 displays a high degree of stability (with stability half-life comparable to the cell doubling time), and shows FRAP dynamic properties identical to SMARCA4-GFP expressed via lentiviral transduction (Figure S3), confirming the validity of our approach. Interestingly, we find that the delayed FRAP recovery of ATPase mutants arises during interphase but not mitosis, where BAF is largely excluded from condensed mitotic chromosomes (Figure S3–S4) consistent with previous observations that SMARCA4 is phosphorylated during mitosis44. Our results reveal a convergence between tumor sequencing studies, molecular structure, and live-cell dynamics.
Cancer mutants induce convergent effects on the chromatin landscape
Because ATP-cleft and DNA-groove mutants induced opposing dynamic effects, we anticipated that they might give rise to distinct effects on the chromatin landscape, even in a heterozygous setting frequently observed in cancer. To probe the functional impacts of commonly mutated positions, we used mESCs as a convenient model to investigate the conserved effects following SMARCA4 disruption. These cells are advantageous for fundamental studies due to the clonal nature of their growth, lack of genetic heterogeneity, negligible mutation load, and the availability of genetic tools such as conditional knockouts. ES cells also do not express the alternate BAF ATPase SMARCA2 (BRM), permitting us to exclude confounding effects from this homologous gene. Using a conditional SMARCA4 knockout mESC line45, we rescued SMARCA4 deletion with lentiviral expression of wild-type SMARCA4-GFP fusion and either wild-type or mutant SMARCA4 tagged with V5 (SMARCA4-V5). Resulting cells had expression levels of SMARCA4 equivalent to those of endogenous protein, as previously described (Figure S4 in ref. 34). As a control, we compared results from these cells to those from similarly prepared cells expressing wild-type SMARCA4-V5 instead of a mutant. This strategy allowed us to observe the effects of mutation in a setting that mimics heterozygous mutation present in human cancers.
We performed ATAC-seq46 to identify locations where SMARCA4 mutants altered genomic DNA accessibility. To our surprise, we discovered that heterozygous mutants gave rise to highly-correlated changes across the genome compared to wild-type cells (Figure 3a–c). Accessibility losses associated with mutants were generally stronger and more frequent than sites that gained accessibility. We assigned sites into one of three categories: decreased, unchanged, and increased, using the definitions found in the Methods section. We analyzed the genomic annotations of sites that decreased accessibility and discovered that relative to unchanged sites, these were highly enriched at non-coding regions, particularly introns and intergenic regions (Figure 3d).
Figure 3. Convergent effects of SMARCA4 cancer mutants on the DNA accessibility landscape.
(a) Genome tracks showing loss of accessibility at an annotated enhancer downstream of Bcl11b. Heterozygous mutant SMARCA4 cells but not wild-type cells show similar loss of enhancer accessibility. (b) Heat map of Pearson correlation coefficients (PCC) of reads densities in peaks shows correlated genome-wide changes induced by heterozygous SMARCA4 mutants. Changes are uncorrelated with changes between wild-type cell-culture replicates. (c) Heat map showing the 2,000 genome-wide sites with the highest variance across datasets. All mutants show a similar trend of altered accessibility across these sites. (d) Genomic annotation enrichment datasets for decreased sites across mutant ATAC-seq. Decreased sites are enriched at non-coding regions such as introns and intergenic regions. (e) Lasso multivariate regression identifies a consistent set of features associated with positive or negative changes of accessibility in mutants compared to wild-type. Features associated with altered accessibility include several factors and marks associated with enhancers. (f) Characterization of ATAC-seq in sites with decreased accessibility in mutant cells, along with ChIP-seq of Smarcc1, Lsd1, and H3K4me1 in wild-type cells. (g) Characterization of ATAC-seq in sites with unchanged accessibility in mutant cells, along with ChIP-seq of Smarcc1, Lsd1, and H3K4me1 in wild-type cells. (h) Characterization of ATAC-seq in sites with increased accessibility in mutant cells, along with ChIP-seq of Smarcc1, Lsd1, and H3K4me1 in wild-type cells.
We furthermore analyzed a dataset of 111 public ChIP-seq studies in wild-type mESCs, which permitted us to investigate the association of factors with gains or losses of accessibility in our current study. Lasso multivariate regression for all our mutant ATAC-seq datasets revealed a robust and consistent association with several factors related to enhancers in mESCs, including Lsd1, Smc1, H3K27ac, Oct4, and Brd4 (Figure 3e).
Because these changes were highly correlated across all mutants, we conclude that these mutants surprisingly have convergent effects on the DNA accessibility landscape. We therefore focused the remainder of our analysis on G784E, which is located at the ATP-binding cleft in the Walker A motif and makes direct contact with ATP (Figure 2a). We find that ATAC accessibility sites were enriched in wild-type cells with ChIP density of Smarcc1, a core subunit of BAF complexes (Figure 3f–h). Furthermore, sites with decreased accessibility following expression of G784E SMARCA4 (n=8,805) were highly enriched with H3K4me1 in wild-type cells (Figure 3f), a mark frequently found at enhancers. Sites that increased accessibility (n=2,887) typically had low levels of Lsd1 (Figure 3h), an H3K4 demethylase involved in enhancer decommissioning47. These results suggest that the gains of accessibility following expression of mutant SMARCA4 depend on the absence of Lsd1. Unchanged sites (n=24,656) generally contained mixed amounts of these factors, and include many regulatory sites such as housekeeping TSSs.
Polycomb accumulation does not reduce local accessibility
Polycomb repression is thought to be mediated by altering DNA accessibility48. We investigated the relationship between changes of accessibility and Polycomb accumulation that occurs over the genome. We examined direct overlap between changes of Polycomb occupancy (reflected by ChIP-seq of Ring1b, a core catalytic subunit of PRC1), and DNA accessibility (reflected by ATAC-seq), in two settings: SMARCA4 conditional biallelic deletion23,34, and heterozygous ATPase mutation (G784E). Although both conditional knockout and heterozygous mutation result in strong and characteristic accumulation of PRC1 and PRC2 at bivalent TSSs over the genome34, we observed no direct relationship between increases of Polycomb and altered accessibility (Figure 4a–d). This observation did not arise due to non-overlap of peaks, as ATAC-seq and Ring1b ChIP peaks had high enrichment at both TSSs and enhancers (Figure S5), and changes at individual sites were highly reproducible (Figure S6).
Figure 4. Accessibility losses and PRC1 changes do not directly overlap.
(a) ATAC accessibility sites ranked by altered ATAC-seq read density in SMARCA4 knockout (left) and in hetereozygous G784E SMARCA4 (right). Genomic sites lacking both ATAC and Ring1b changes are not shown. Corresponding changes of Ring1b ChIP-seq read density at these same sites show that Ring1b is broadly unchanged at these sites, and shows no clear relationship to accessibility changes measured by ATAC. (b) Example genome track showing representative accessibility but not Ring1b changes. (c) Ring1b ChIP sites ranked by altered ATAC-seq read density in SMARCA4 knockout (left) and in heterozygous G784E SMARCA4 (right). Corresponding changes of Ring1b ChIP-seq read density at these same sites show that Ring1b is broadly unchanged at these sites, and shows no clear relationship to accessibility changes measured by ATAC. Genomic sites lacking both ATAC and Ring1b changes are not shown. (d) Example genome track showing representative Ring1b but not accessibility changes. (e) Genomic fold changes of enhancers (Enh) and transcription start sites (TSS) using ATAC-seq upon conditional Ring1b knockout. TSSs show few changes while enhancers show significantly greater variability (KS test p<2.2e-16). Center line of box plot is mean, box limits are 1st and 3rd quartiles, whiskers are limits + 1.5*IQR (inter-quartile range), points are actual values of outliers.
We confirmed this result by performing ATAC-seq following dual conditional knockout of Ring1a and Ring1b (ref. 49), both catalytic subunits of PRC1, and found that global accessibility at TSSs showed few changes. Instead, altered accessibility at enhancers was far more apparent (Figure 4e, p<2.2e-16). Therefore, the increased occupancy of PRC1 we previously described at TSSs following SMARCA4 inactivation34 does not directly coincide with altered accessibility. We conclude that DNA accessibility changes are distinct from accumulation of Polycomb complexes at bivalent TSSs. Our results challenge longstanding in vitro models of the repressive mechanisms associated with Polycomb factors.
Epigenomic alterations to enhancer identity
We next sought to compare the effects of heterozygous SMARCA4 mutation on accessibility at enhancers and TSSs. Accessibility was broadly unaffected at TSSs, but was reduced at annotated mESC enhancers50 (Figure 5a–b, additional examples in Figure S7). Accessibility at TSSs and enhancers was also broadly mirrored by altered ChIP read density of RNAP2 (Figure S8), suggesting that altered accessibility has functional importance for regulation of transcription.
Figure 5. Heterozygous SMARCA4 mutation induces loss of accessibility and H3K27ac at active enhancers and superenhancers from many tissues.
(a) Mean ATAC-seq read density at TSSs and enhancers in wild-type and heterozygous mutant cells. (b) Example genome track showing representative loss of accessibility and H3K27ac but preservation of H3K4me1 at an enhancer. (c) Relationship between H3K4me1 ChIP and ATAC fold changes from wild-type and mutant cells shows significant correlation with a small effect size (m). (d) Relationship between RNAP2 ChIP and ATAC fold changes between wild-type and mutant cells shows significant correlation with a small effect size. (e) Relationship between H3K27ac ChIP and ATAC fold changes between wild-type and mutant cells shows significant correlation with a large effect size. (f) Abundance of H3K4me1 and H3K27ac in wild-type cells, classified by whether site increased, decreased accessibility, or was unchanged. Sites with decreased accessibility cluster have elevated levels of H3K4me1 and H3K27ac, consistent with active enhancers. (g) Mean accessibility changes at the Oct4-Sox2-Tcf-Nanog combination transcription factor motif. (h) Heat map of 18 transcription factor motifs associated with reduced accessibility across cell-culture replicates in heterozygous G784E SMARCA4 cells. (i) Cumulative distribution of ATAC-seq fold change, classified by genomic annotation. TSSs show few changes, while enhancers and superenhancers show biased losses. Superenhancers show distributions of fold changes comparable to enhancers. (j) Analysis of sites with decreased accessibility that overlap with superenhancers from multiple tissue types. Color in heat map shows fractional overlap with known annotated superenhancers from tissues obtained from dbSUPER. (k) Distribution of the number of tissue types associated with each superenhancer site analyzed in (j). (l) Pie chart of the proportion of tissue-restricted superenhancers compared with superenhancers that identified in >1 tissue type. RPM, reads per million; Enh, enhancer; SE, superenhancer.
To identify other chromatin features accompanying accessibility changes, we focused on three marks and factors: H3K4me1, RNAP2, and H3K27ac. We performed ChIP-seq for these factors in both WT/WT and WT/G784E mESCs to measure their changes at sites with altered accessibility. Despite significant correlation (R=0.334, p<2.22e-16), the effect size of altered H3K4me1 was low (m=0.104), showing that sites with decreased accessibility generally maintained high levels of H3K4me1 (Figure 5c). RNAP2 also showed significant correlation (R=0.230, p<2.2e-16), with a modest effect size (m=0.114). However, RNAP2 ChIP revealed that a small number of decreased sites also showed losses of RNAP2, suggesting that RNAP2 loading may only be affected when certain combinations of other changes occur (Figure 5d). On the other hand, H3K27ac showed robust effects (R=0.358, p<2.2e-16) with a prominent effect size (m=0.370), indicating a stronger relationship between H3K27ac and DNA accessibility (Figure 5e). Hence, these sites transition from being “active” enhancers (with high H3K27ac and high accessibility) to “poised” enhancers (with maintained H3K4me1, but reduced H3K27ac and reduced accessibility).
Consistently, sites that lost accessibility (N=2,692) had the highest levels of H3K27ac in wild-type cells (Figure 5f). In contrast, the sites that showed increased accessibility (N=1,040) generally had lower levels of both H3K4me1 and H3K27ac in wild-type cells. Since most of these did not take on new elevated levels of H3K4me1 in mutant cells (Figure 5c), we speculate that these sites may have taken on other unknown regulatory roles in mutant cells.
By exploring average accessibility changes over transcription factor sequence motifs, ATAC-seq permitted us to identify a set of transcription factors whose cognate sequences show reduced accessibility. One of these was the combination motif for Oct4-Sox2-Tcf-Nanog, which undergoes a ~2-fold reduction of nucleosome-free ATAC-seq reads over the motif (Figure 5g). By scanning for reproducible changes, we obtained a list of 18 transcription factor motifs, with recurrent losses of accessibility following expression of G784E SMARCA4 (Figure 5h). Our results show that mutant SMARCA4 induces alteration of the enhancer landscape affecting a diverse set of factors, rather than targeting a small subset of transcription factors. These changes in the enhancer network arise without expression of SMARCA2 (BRM, see Figure S9), allowing us to attribute our observations to SMARCA4 mutants.
Sites with lost accessibility in ES cells include hundreds of superenhancers from many tissue types
In malignant rhabdoid tumors, loss of SMARCB1 affects active enhancers, but leaves superenhancers relatively protected11. We therefore sought to assess whether heterozygous SMARCA4 mutations would have similar effects. By classifying sites into genomic annotations for mESC transcription start sites (TSS), overall enhancers (Enh), or superenhancers (SE), we find that superenhancers show distributions of accessibility changes comparable with overall enhancers (Figure 5i), both of which are significantly tilted towards losses compared to TSSs (p<2.2e-16). Therefore, superenhancers are significantly impacted by heterozygous SMARCA4 mutation, unlike biallelic loss of SMARCB1.
Chromatin regulatory interactions in embryonic stem cells anticipate cell-type-specific architecture that arises upon lineage commitment51. We therefore hypothesized that many sites we observed with reduced accessibility may correspond to sites with regulatory roles in differentiated cells. To assess whether affected sites correspond to important regulatory sites in multiple cell types, we examined overlap with an ensemble of annotated superenhancers across a variety of differentiated tissues, including those from lung, intestines, hematological, and neuronal lineages. By comparing to sites in the dbSUPER database of superenhancers52, we found 940 sites (34.9% of decreased sites overall) are significantly enriched for direct overlap with previously annotated superenhancer sites from at least one tissue (Figure 5j–k, p<2.2e-16). Plotting the distribution of associated tissue types for each superenhancer (Figure 5l) reveals that that most of these sites (51.5%, 484/940) are superenhancers in more than a single tissue type. Therefore, heterozygous mutations of SMARCA4 induce accessibility losses at sites identified to have essential roles in maintaining cell identity across multiple tissue types. Our findings furthermore show that BAF-dependent accessibility anticipates lineage specification in several lineages.
Effects on genomic accessibility are dominant negative
We next sought to investigate whether the observed changes reflected SMARCA4 haploinsufficiency, or were instead a dominant-negative effect53. To measure the impact of loss from a single allele, we derived a line of mESCs for conditional deletion of only one allele of SMARCA4. This actin::CreER SMARCA4(+/fl) line expresses Cre recombinase fused to the estrogen receptor (CreER), and encodes a single floxed allele of SMARCA4, with the remaining allele wild-type. This permitted us to conditionally delete a single allele in the presence of 4-hydroxytamoxifen (Tam), and compare against mock treatment with ethanol (EtOH). Western blotting showed reduction of half of SMARCA4 protein levels (Figure 6a–b) after tamoxifen-induced deletion. However, we did not observe reproducible changes in chromatin accessibility using ATAC-seq following loss of the single allele, and only a single site met the criteria for a decreased site out of 41,267 ATAC-seq sites across the genome (Figure 6c). Notably, we did not detect changes at enhancers or TSSs as we observed for the ATPase mutants (Figure 6d–e). Importantly, our results leave open the possibility of changes at sites of lower accessibility or in other cell types, or of other haploinsufficient roles (e.g., in DNA repair).
Figure 6. Chromatin organization influences dominant-negative effects of SMARCA4 mutants on the open chromatin landscape.
(a) Demonstration of a line of heterozygous conditional deletion SMARCA4 cells. A single allele of SMARCA4 is converted to a null allele in the presence of 4-hydroxytamoxifen (Tam) but not ethanol control (EtOH). (b) Quantification of bands in (a), showing loss of half the SMARCA4 present caused by incubation with Tam. (c) Plot of genome-wide fold changes for all ATAC-seq sites following acute deletion of one allele of SMARCA4. (d) Mean profiles of ATAC-seq read density at TSSs and (e) enhancers following acute deletion of one allele of SMARCA4. The absence of changes demonstrates that the changes we observe in Figure 4 are not due to haploinsufficiency, but are a dominant-negative effect. (f) Heterozygous G784E SMARCA4 but not heterozygous null cells show clusters of reduced accessibility in chromosomal A compartments (described in more detail in the main text and Methods). (g) Quantification of genome-wide ATAC-seq site changes based on presence in A or B compartments in heterozygous null cells. (h) In heterozygous G784E SMARCA4 cells, ATAC-seq sites in A compartments have reduced accessibility compared to sites in B compartments. CDF, cumulative distribution function.
In contrast to loss of a single allele, G784E SMARCA4 resulted in accessibility losses that were correlated in position over the body of the chromosome (Figure 6f). Analysis of Hi-C data (see Methods section) permitted us to partition each chromosome into large multi-megabase-scale compartments termed “A” and “B” compartments54,55. We found that the more transcriptionally active A compartments tended to have more acute accessibility losses than the more repressed B compartments (Figure 6f). Focal accessibility changes within A compartments were significantly tilted towards losses compared to B compartments (Figure 6g–h, p<2.2e-16). Thus, enhancers in A compartments are differentially sensitive to the expression of ATPase mutant SMARCA4. However, these same sites are unaffected by loss of expression from a single allele of SMARCA4. Therefore, the strong effects following heterozygous SMARCA4 ATPase mutation reflect genetic dominance rather than haploinsufficiency.
Pro-oncogenic changes of gene expression include Polycomb and non-Polycomb targets
To discover the effects of the altered enhancer landscape on transcription, we compared wild-type to heterozygous G784E SMARCA4 cells using RNA-seq. Despite the thousands of sites of altered accessibility described above, we found a modest set of changes to the transcriptional profiles of cells, with a small number of genes showing increased (n=49) or decreased (n=191) expression, using the criteria in the Methods section (Figure 7a). Genes with changes included previously annotated Polycomb targets56 (n=86), which were significantly enriched among all altered genes (odds ratio=1.74, p=7.7e-5). We also observed altered expression of genes that are not Polycomb targets (n=154; Figure 7b).
Figure 7. SMARCA4 ATPase mutants can induce pro-oncogenic gene expression changes.
(a) Summary of genome-wide RNA expression changes by RNA-seq. Each point is a gene, colored by whether it is increased, decreased, or unchanged, using the criteria described in the Methods section. (b) Heat map of genes with altered expression based on whether they are annotated to be promoters targeted by Polycomb Repressive Complexes (PRCs) in mESCs56, or are not annotated to have PRC-target promoters. (c) Expression of pluripotency factors remains elevated in heterozygous G784E SMARCA4 cells. Myc is expressed ~2-fold higher in mutant cells compared to wild-type cells. Plotted values are mean normalized expression values (a.u.), error bars are 95% confidence intervals from independent cell-culture replicates (n=2). (d) Gene set enrichment analysis shows significant enrichment of downstream targets of Myc (p<2.2e-16). (e) Model of dominant-negative effects following inactivation of SMARCA4 ATP hydrolysis or DNA/nucleosome binding. (f) Effect of SMARCA4 heterozygous mutation on the epigenetic landscape. Mutation and the ensuing compensatory changes shifts the epigenetic-phenotypic landscape, resulting in stabilization of a new phenotypic state that may adopt pathogenic patterns of gene expression.
Interestingly, despite reduced accessibility of their target sites, we do not observe loss of expression of the core pluripotency network, including Oct4 (Pou5f1), Sox2, Nanog, or Klf4. Expression of these factors is maintained or even enhanced in heterozygous mutant cells, as previously reported for complete knockout45. In contrast, we observe a significant ~2-fold increase in the expression of the proto-oncogene Myc (Figure 7a,c; log2 fold change=0.98, FDR-adjusted p=0.01). Gene set enrichment analysis also revealed a significant enrichment of Myc target genes (FDR-adjusted p<2.2e-16), one of the hallmarks of many malignancies (Figure 7d). Altogether, our results reveal that cell identity may be maintained through compensating mechanisms, despite a dramatic reshuffling of the enhancer and superenhancer landscape. In ES cells, these compensating mechanisms include upregulation of Myc, a hallmark of cancer that may be considered a pro-oncogenic response. Interestingly, biallelic deletion of SMARCA4 in the same cells leads to cell death45, thus dominant-negative mutations in cancer settings may similarly confer changes distinct from biallelic loss.
Discussion
Here we analyzed human tumor sequencing data to identify the major role played by heterozygous mutation of SMARCA4 (Figure 1). We have analyzed the available mutation frequencies obtained through tumor sequencing studies in the context of structural models of SMARCA4 we derived from recent discoveries in structural biology37–39. By doing so, we discovered that frequently mutated positions of SMARCA4 lie on biologically significant surfaces (Figure 2a). This clustering is consistent with earlier observations that mutations of key bioactive surfaces are significantly enriched across several cancer types57. These mutations induce characteristic dynamic defects, consistent with a model where binding of SMARCA4 to DNA/nucleosomes promotes immobilization, and release from this immobile state is catalyzed by ATP hydrolysis (Figure 2g).
Despite divergent dynamic effects, mutations in either the ATP cleft or DNA groove give rise to similar changes across the chromatin landscape (Figure 3). The convergence of these defects indicates that cancer positively selects for functional inactivation rather than disruption of individual surfaces per se. Our results show that frequently occurring disease mutations inactivate SMARCA4 by diverse modes, for example, by impairing ATP hydrolysis, by disrupting DNA binding, and likely through other mechanisms. Our ATAC-seq results confirm that the effects of these mutations converge on the open chromatin landscape; we find consistent changes to DNA accessibility at thousands of sites over the genome, including at a large number of superenhancers from diverse cell types, regardless of how SMARCA4 is disrupted. In contrast to biallelic loss of other BAF subunits, which largely arise in lineage-restricted settings, heterozygous mutations of SMARCA4 contribute to malignancies from diverse tissue types. We propose that this broad pattern largely reflects disruption of the enhancer accessibility landscape, including superenhancer sites used by many tissue types.
We find that a set of active enhancers in wild-type mESCs is particularly predisposed to lose accessibility (Figure 5). These changes lead to losses of accessibility at an array of transcription factor motifs, and concomitant losses of H3K27ac modification, while in most cases preserving H3K4me1 levels. Hence, enhancer identity (as defined by H3K4me1 marking) is largely preserved, but active enhancers with high H3K27ac are particularly sensitive to SMARCA4 ATPase mutation. These effects are particularly pronounced in active A compartments, but do not arise upon loss of a single allele of SMARCA4 (Figure 5), indicating dominant-negative effects for the missense mutants. Hence, we find that chromosomal organization heavily influences the dominant-negative effects of SMARCA4 disease mutants. Our data strongly argue that in addition to its direct effects on PRC1 at bivalent genes (ref. 34), SMARCA4 dynamically acts to generate accessibility in transcriptionally active compartments of the genome, notably at existing superenhancers and latent/primordial superenhancers.
Despite the thousands of changes in the open chromatin landscape, these alterations induce a relatively small number of significant transcriptional changes (Figure 7a). Altered genes include both Polycomb-target genes and non-Polycomb-targets, indicating that regulation of the open chromatin landscape at enhancers constitutes a separate layer of regulation in addition to the mechanisms previously described for PRC1 (ref. 34). Of particular interest is the up-regulation of the proto-oncogene Myc as well as its downstream targets (Figure 7). Myc enhances the stability of the pluripotency factors at their cognate sites on chromatin58, and relieves transcriptional pausing59,60. Therefore, it is appealing to speculate that heterozygous expression of mutant SMARCA4 in mESCs leads to loss of transcription factor accessibility or paused polymerases, and increased expression of Myc may compensate by stabilizing the interaction of transcription factors at their target sites or by relieving polymerase pausing following SMARCA4 mutation.
Altogether, our findings show that heterozygous SMARCA4 mutations induce loss of H3K27ac and accessibility at enhancers and superenhancers. Because ATP-cleft and DNA-groove mutants both induce these effects, we propose that SMARCA4’s dominant-negative role at enhancers arises via sequestration of binding partners at unproductive transient binding sites, or in the nucleoplasm (Figure 7e). Our results reveal that unlike biallelic loss of SMARCB1 or SMARCA4, heterozygous SMARCA4 missense mutations bias the epigenetic landscape by altering the accessibility of superenhancers used in a variety of different tissues. These changes permit exploration of other sites in phenotypic space, which may promote pro-oncogenic changes of gene expression (Figure 7f). Given our findings, additional studies in human models of cancer and neurologic disorders are warranted to fully test and examine the mechanisms we describe here. Taken together, the distinct effects of dominant-negative mutations suggest that therapeutic strategies targeting the large number of missense SMARCA4 mutations in cancer and neurologic disorders may require distinct approaches from those targeting biallelic inactivation.
Methods
Culture of animal and human cells
Mouse ES cells used in this study were derived from pregnant female mice in the Crabtree lab, and authenticated by examination of morphology and expression of embryonic stem cell markers. These cells were cultured using standard conditions. Briefly, ES culture media containing Dulbecco’s Modified Eagle’s Medium (Cat# 10829018; Life Technologies), 15% FBS (Cat# ASM-5007; Applied StemCell), Penicillin-Streptomycin (Cat# 15140122; Life Technologies), Glutamax (Cat# 35050061; Life Technologies), HEPES buffer (Cat# 15630080; Life Technologies), 2-mercaptoethanol (Cat# 21985023; Life Technologies), MEM-NEA (Cat# 11140050; Life Technologies), and LIF supplement61, was replaced daily, and ES cells were passaged every 48 hours. For inducible deletions, Smarca4flox/flox actin–CreER ES cells45 and Smarca4+/flox actin–CreER ES cells, which previously tested negative for mycoplasma contamination using PCR testing, were plated onto irradiated feeder mouse embryonic fibroblasts, treated with 0.8 uM 4-hydroxytamoxifen (Tam) or ethanol (EtOH) for 48 h, and harvested for further experiments after trypsin dissociation at 72 h.
Western blotting
Cells were lysed for at least 30 min at 4 °C in RIPA buffer (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 0.1% SDS, 0.5% sodium deoxycholate, 1% NP-40). Lysates were centrifuged for 30 min at 14,000g, and the supernatant was flash-frozen. The total protein concentration was measured by Bradford assay (Bio-Rad). We boiled 10 µg of protein in gel loading buffer (Life Technologies) with 50 mM of dithiothreitol and loaded it onto 4–10% BisTris NuPage gels (Life Technologies). Bands were transferred to PVDF membranes (Bio-Rad), and then membranes were blocked in 5% BSA/TBST. Blots were then incubated with primary antibodies for 1 h at room temperature. Proteins were detected with the LI-COR detection system.
Construction and breeding of Smarca4-Dendra2 transgenic mice
Mutation was obtained using a targeting sequence derived from BAC clone bMQ-242m22 (Source Bioscience). The Dendra2 sequence was inserted using homologous recombination with a floxed neomycin resistance cassette, This resistance cassette was later excised by breeding to a mouse expressing ubiquitous Cre recombinase, leaving behind a single loxP site downstream of Dendra2. Validation of successful construction was performed with Southern and Western blots, as well as fluorescence microscopy. Homozygous mice were obtained with no apparent phenotype, and bred through filial crossing.
Genotyping of Smarca4-Dendra2 mice
Standard PCR conditions were used to genotype Smarca4-Dendra2 transgenic mice. PCR amplification conditions were as follows: 55 °C annealing temperature, 72 °C extension temperature for 30 seconds, 28 cycles. Wild-type alleles are specifically amplified using forward primer: CCC TAC CAT GGT GCA GGG CA, and reverse primer: AAT GTC TGG TTC AGT CTT CCT CA, to yield a 429-bp amplicon product. Smarca4-Dendra2 alleles are specifically amplified using forward primer: GCC CAG CCA GGT GTG GTG AGC CCA ATT CCG ATC ATA TTC A, and reverse primer: GGG CCT GTG AGC CTC CTG GA, to yield a 469-bp amplicon product. Homozygous mice amplify only the wild-type or only the Smarca4-Dendra2 product, while heterozygous mice amplify both products.
Lentiviral preparation and infection
Lentivirus was produced in Lenti-X 293T cells (Clontech) via polyethylenimine transfection. Media were changed 24 h after transfection, and after another 48 h media were collected and centrifuged for 2 h at 20,000 rpm. Viral pellets were resuspended in PBS and used to infect cells. Cells were selected with 2 µg/ml puromycin, as appropriate beginning 48 h after infection.
Fluorescence recovery after photobleaching (FRAP)
Cells were maintained at 37 °C under humidified 5% CO2 using chambered µ-Dish 35mm coverslips (Cat# 81156, Ibidi) in an enclosed inverted Leica SP2 laser scanning confocal microscope. Using Leica software, half of the nucleus of each cell was photobleached using 100 mW 405-nm laser excitation. Nuclear fluorescence was monitored as a time course, and each image saved as a TIFF file. The degree of fluorescence recovery within the region of interest (ROI, the photobleached portion of the nucleus) was monitored within each image. Image masks were made using MATLAB to identify the full nucleus (visualized prior to photobleaching), and the ROI only. The ratio of fluorescence intensity within the ROI over the intensity throughout the entire nucleus was plotted and fit to exponential curves using non-linear least square fitting, to obtain the recovery rate. Recovery half-lives derived from this fit were collected for each condition. P-values to compare the populations of half-lives were obtained using the two-sample Kolmogorov-Smirnov (KS) test.
Deconvolution fluorescence microscopy
Deconvolution microscopy was performed on fixed cells using the OMX Blaze V4 microscope platform.
Assay of transposase-accessible chromatin (ATAC) library preparation
ATAC libraries were independently prepared from separately cultured samples in duplicate, using methods previously described46. Briefly, we obtained nuclei by resuspending cells in 0.5 ml of lysis buffer (0.1% Tween-20 in RSB buffer) and incubated them for 10 min on ice. Nuclei were pelleted by centrifugation for 10 min at 500g, resuspended in 50 µl of transposition mix [1× Tagmentation DNA buffer, 2.5 µl Tagment DNA enzyme (Illumina)], and incubated for 30 min at 37 °C. DNA was purified with a MinElute PCR purification kit (Qiagen), and libraries were amplified by PCR with barcoded Nextera primers (Illumina). For sequencing, libraries were size-selected with Agencourt AMPure XP (Beckman Coulter) for fragments between ~50 and 1,000 bp in length according to the manufacturer's instructions. Sequencing was performed using paired-end reads on the Illumina HiSeq2000 sequencer.
Processing of ATAC-seq data
Analysis of high-throughput sequencing data was not performed blindly, but the following techniques were applied uniformly to all datasets. Paired-end reads were processed by mapping to the mm9 reference mouse genome using Bowtie 2.1.0 (ref. 62). Duplicate fragments and reads with mapping quality < 10 were discarded, leaving only high-quality unique reads. For each fragment, two pseudofragments were generated, which occupied 200 bp centered at the true fragment ends. Peak calling was performed by MACS 2.1.1 (ref. 63) based on the density of pseudofragments.
For differential analysis, all peaks from control and treatment datasets within ±1 kb were merged, and peaks below a threshold of 10 RPM in at least one dataset were discarded to remove low-quality peak calls. Background density of ATAC-seq signal was obtained by calculating the mean read density in the 100-bp window 8-kb away from each peak. This background density was subtracted from the overall read density within each peak in each dataset. For each dataset, the total number of background-adjusted reads overlapping each of the resulting peaks was compared for differential peak calling. Differential peak calls were made using DESeq2 (ref. 64), using the summed number of background-adjusted reads at the top 5% of highest read-density sites as size factors. DESeq2 accounts for individual site variances across all replicates to make differential peak calls. Log2 fold changes were calculated by the default use of maximum a posteriori estimation using a zero-mean normal prior (Tikhonov-Ridge regularization). FDR-corrected P-values were calculated using the Benjamini-Hochberg procedure. Differential calls were made by requiring fold changes of >1.5-fold in either direction and FDR-corrected p < 0.10. RPM values in genome tracks are the mean values across both replicates from each condition. Mean density profiles were computed using bwtool65 by calculating the mean basepair coverage across all replicates for a given condition. Overlap of peaks was performed using Bedtools66.
ChIP-seq library preparation
ChIP libraries were independently prepared from separately cultured samples in duplicate, according to previously described protocols67–69. Briefly, we fixed 10–30 million cells in suspension for 15 min in 1% formaldehyde. Excess formaldehyde was quenched by the addition of glycine to 100 mM. Fixed cells were washed, pelleted, and flash-frozen. Pellets were thawed, resuspended in NP Rinse 1 buffer (50 mM HEPES-KOH, pH 8.0, 140 mM NaCl, 1 mM EDTA, pH 8.0, 10% glycerol, 0.5% NP-40, 0.25% Triton X-100) and incubated for 10 min on ice to isolate nuclei. Nuclei were washed once with NP Rinse 2 buffer (10 mM Tris-HCl, pH 8.0, 1 mM EDTA, pH 8.0, 0.5 mM EGTA, pH 8.0, 200 mM NaCl) and then twice with shearing buffer (0.1% SDS, 1 mM EDTA, pH 8.0, 10 mM Tris-HCl, pH 8.0). Pellets were resuspended in 900 µl of shearing buffer with protease inhibitors and sonicated in a Covaris E220 sonicator for 10–12 min to generate DNA fragments between approximately 200 and 1,000 bp in length. Chromatin was then immunoprecipitated overnight at 4 °C with antibodies bound to Protein G Dynabeads. (Life Technologies) in ChIP buffer (50 mM HEPES-KOH, pH 7.5, 300 mM NaCl, 1 mM EDTA, pH 8.0, 1% Triton X-100, 0.1% DOC, 0.1% SDS). The chromatin-bead slurry was washed with a magnetic rack four times with ChIP buffer, once with DOC buffer (10 mM Tris-HCl, pH 8.0, 250 mM LiCl, 0.5% NP-40, 0.5% DOC, 1 mM EDTA, pH 8.0), and once with TE buffer. Chromatin was eluted with Q7 elution solution (0.1 M NaHCO3, 1% SDS). After RNase A (Life Technologies) and proteinase K (New England BioLabs) digestion and de-crosslinking at 65 °C overnight, DNA was extracted with phenol-chloroform and precipitated with ethanol. Size selection was performed by extracting 200–400 bp DNA fragments on a 2% agarose E-gel (Invitrogen) before PCR amplification, then extracted using MinElute cleanup kits (Qiagen). PCR amplification was performed using ≤ 14 cycles, and the resulting DNA quantified by Qubit fluorometric quantitation. Sequencing was performed using single-end reads on the Illumina HiSeq2000 sequencer. Antibodies used in these studies include rabbit anti-H3K4me1 (Abcam, Cat# ab8895, Lot number: GR251663-1), rabbit anti-H3K27ac (Abcam, Cat# ab4729, Lot number: GR104852-1), and mouse monoclonal anti-RNAP2 (Covance clone 8WG16, Cat# MMS-126R).
Processing of ChIP-seq data
Analysis of high-throughput sequencing data was not performed blindly, but the following techniques were applied uniformly to all datasets. Single-end ChIP seq reads were processed by mapping to the mm9 reference mouse genome using Bowtie 2.1.0 (ref. 62), rejecting reads that contain more than a single mismatch. Duplicate reads were discarded, leaving only unique reads. For all analyses, peak calling was performed by MACS 2.1.1 (ref. 63) by comparing to input samples for each cell type.
All peaks from control and treatment datasets within ±1 kb were merged, and peaks below a threshold of 30 RPM (in at least one dataset) were discarded to remove low-quality peak calls. For each dataset, the total number of reads overlapping each of the resulting peaks was compared for differential peak calling. Differential peak calls were made using DESeq2 (ref. 64), using the summed number of reads at all sites above the 95th-percentile as size factors to avoid the influence of background ChIP-seq reads. DESeq2 accounts for individual site variances across all replicates to make differential peak calls. Log2 fold changes were calculated by the default use of maximum a posteriori estimation using a zero-mean normal prior (Tikhonov-Ridge regularization). FDR-corrected P-values were calculated using the Benjamini-Hochberg procedure. Differential calls were made by requiring fold changes of >1.5-fold in either direction and FDR-corrected p < 0.10. RPM values in genome tracks are the mean values across both replicates from each condition. Calculation of mean genome track densities was performed using bwtool65 and browser tracks were prepared using Gviz70. For presentation in heat maps, ChIP-seq data were ordered by the amount of signal in the middle 5% of an 8-kb window centered around each peak.
RNA-seq library preparation
RNA was isolated with QIAzol and extracted with phenol-chloroform. Using the Ovation V2 RNA-seq system, RNA-DNA hybrids were formed using primers with conjugated poly(dT) sequences fused to random DNA-hybridization sequences. Following first-strand synthesis and cDNA amplification, cDNA was sheared with Diagenode sonication (25-s pulse, followed by 30-s recovery at 4 °C for 2×10 cycles). Library preparation was performed as previously reported69 and sequencing was performed using single-end reads on the Illumina HiSeq2000 sequencer.
Processing of RNA-seq data
Reads within coding regions were counted using htseq71, with gene definitions obtained from the refGene of RefSeq genes in the UCSC Browser. The of all counts was processed using DESeq2, using default parameters. Log2 fold changes were calculated using maximum a posteriori estimation using a zero-mean normal prior (Tikhonov-Ridge regularization). FDR-corrected P-values were calculated using the Benjamini-Hochberg procedure. Differential calls were made by requiring fold changes of >1.5-fold in either direction and FDR-corrected p < 0.10.
Public datasets analyzed in this study
Read densities were obtained from publicly available as NCBI GEO datasets (Table S1). For each of these chromatin feature datasets, read fragments were extended to 200 bp from the 3’ end, and basepair coverage was determined using Bedtools66.
Gene set enrichment analysis (GSEA)
Gene set enrichment analysis was performed by preparing a list of Refseq transcripts based on the output of DESeq2 from RNA-seq data. RefSeq genes were ranked by log2 fold change and this pre-ranked set was imported into the GSEA application (Broad Institute). Enrichments and p-values were obtained directly from the GSEA application.
Genome annotation enrichment
Enrichment for overlap with genomic annotations was performed by comparing how frequently each peak fell into basic genomic annotations, separately determined for each class of differential peak call described above. Enriched GO terms and log enrichment of genomic annotation overlap was calculated for each class of site using HOMER72.
Statistics
All differential calls for genomic datasets (ChIP-seq and RNA-seq) were made using DESeq2, where P-values are calculated using the Wald test, as described in the DESeq2 documentation64. FDR-corrected P-values were calculated using the Benjamini-Hochberg procedure. Analysis of correlation was performed using the Pearson correlation test. Two-sample Kolmogorov-Smirnov (KS) tests were performed as two-sided tests. All the above statistical tests were performed using R. The hypergeometric test for genomic annotation overrepresentation was performed as a one-sided test using HOMER72. When calculated p-values are smaller than the 64-bit double precision machine epsilon (2-52= 2.22e-16), p-values are reported as p<2.2e-16.
Lasso multivariate regression
Lasso multivariate regression73 was performed essentially the same as previously described34. Briefly, we related the fold-change in ATAC-seq read density induced by Smarca4 ATPase mutants (fc) to a linear combination of 111 individual chromatin features:
At each ATAC peak, the number of reads within ±3 kb from the center of the peak was summed for each of 111 features, using previously published datasets. The summed read count at each site was log10 transformed, and scaled to unit variance across all sites (xi). Lasso regression was performed using the R package “glmnet” 74, with the default mixing penalty parameter α=1. Values for the restricted parameter
were obtained for each Smarca4 mutant by 10-fold cross-validation, with the minimal value selected that provided the lowest mean cross-validated error.
Homology modeling
Models of the three-dimensional structures of murine and human SMARCA4 (aa 754–1335) were created in the YASARA v16.7.22.L.64 (ref. 75) homology modeling module. The target sequence was PSI-BLASTed against UniRef90 to build a position-specific scoring matrix from related sequences. This profile was used to search the PDB for potential modeling templates. The identified templates were ranked based on the alignment score, the structural quality according to WHAT_CHECK (ref. 76) obtained from the PDBFinder2 database77. The template search resulted in identification of possible templates out of which 2 templates (PDB IDs 5HZR and 5X0X) were further used for homology modeling purposes.
Multiple sequence alignments anchored on structural alignments were created by aligning the target sequence against UniRef90 and YASARA's PSSP database using SSALN scoring matrices78. This approach provided 10 different homology modeling templates. An indexed version of the PDB was used to determine the optimal loop anchor points and collect possible loop conformations for the unassigned parts of the sequence in each template. The side-chain rotamers of these sequences were optimized in implicit solvent so that water molecules did not block the search. The side-chain rotamers of the whole molecule were then fine-tuned considering electrostatic and knowledge-based packing interactions as well as solvation effects. To relieve steric clashes and to fix disordered geometry, the models were subjected to a high-resolution energy minimization with a shell of explicit solvent molecules, using the knowledge-based YASARA force field. The stereochemical properties of each model were evaluated based on the per-residue quality Z-score. To increase the accuracy, YASARA combined the highest scoring parts of the 10 models to obtain a hybrid model (overall Z-score ~ −0.81).
The structure of SMARCA4 in the hybrid model resembles the structure of the Swi2/Snf2 core from Saccharomyces cerevisiae (PDB ID 5X0X) and represents the conformation in a nucleosome/DNA-bound state. The interface region of SMARCA4 and ATP was defined based on the crystal structure of ATPγS in complex with Chd1 (PDB ID 3MWY), a Snf2-like ATPase with a high degree of homology to mammalian SMARCA4. The model of full complex was subjected to side-chain rotamer optimization and refined in an explicit solvent knowledge-based YASARA force field. Three-dimensional coordinates of mouse and human SMARCA4 homology models are provided in PDB format as supplemental data.
Molecular visualization
The Smarca4 homology model was rendered using The PyMOL Molecular Graphics System, Version 1.8 (Schrödinger, LLC).
Calculation of solvent accessible surface area
Solvent accessible surface area was calculated based on the resulting homology model for each residue position using PyMOL (v1.8). Values were calculated using the get_area() function with parameters dot_solvent=1, and dot_density=3.
Analysis of public tumor datasets
Figures and results reviewed here are based in part upon data generated by the TCGA Research Network and hosted on cBioPortal40. Briefly, data was downloaded using the cBioPortal web API, then processed to characterize the abundance of mutations at each residue position of Smarca4.
Calculation of A/B compartments
Determination of chromatin compartments was performed essentially as previously reported54. Briefly, publicly available Hi-C data from mESCs79 was downsampled to 200-kb resolution, and expected contact frequency between contact pairs (i,j) was calculated based on the distance from position i to j for all pairs. Observed-over-expected ratios for each contact pair (i,j) was calculated by normalizing pair read counts to expected pair read counts calculated as described above. The matrix of observed-over-expected ratios constituted an apparent two-state “checkerboard” matrix, which was clarified further by calculating the pairwise correlation matrix of the pairwise observed-over-expected matrix. The top principal component of the correlation matrix permitted straightforward assignment into A and B compartments based on the position of the principal component plot above or below the mean and the transcriptional output of the contained genes obtained from RNA-seq.
Densitometry and analysis
All densitometry of Western blot bands was performed by calculating integrated band intensities with LiCor ImageStudio 5.2.5.
Data presentation
All browser tracks were prepared using Gviz70 based on bigWig files. Heatmaps were prepared using heatmap.2() from the ‘gplots’ R package, and the ‘pheatmap’ R package. MA plots were prepared using heatscatter() from the ‘LSD’ R package. Color combinations were heavily influenced by the “Paired” and “Set1” palettes from the ‘RColorBrewer’ R package.
Analysis of overlap with superenhancers in different cell types
Direct overlap with superenhancers from multiple cell types was performed by downloading the current version of the dbSUPER database of superenhancers52 . Briefly, the database of superenhancers was downloaded in BED format for each cell type and fractional overlap was assessed for each peak of ATAC-seq decreased sites using Bedtools. Homology to sequences in the human genome was determined using the liftOver tool from the UCSC Genome Browser to map sequence positions from the mouse mm9 genome build to the human hg19 genome build.
Data availability
Sequencing data sets generated and analyzed in this study are available in the Gene Expression Omnibus (GEO) repository under accession numbers GSE88968, GSE94041, and GSE98605. Source data for Figure 1 are available with the paper online. Three-dimensional coordinates of the mouse SMARCA4 homology model are provided in PDB format as Supplementary Dataset 1. Three-dimensional coordinates of the human SMARCA4 homology model are provided in PDB format as Supplementary Dataset 2. Other data and materials are available from the authors upon request. A Life Sciences Reporting Summary for this article is available.
Supplementary Material
Highlights.
Heterozygous SMARCA4 missense mutations are a major but overlooked class of BAF subunit disruptions that contribute to many different cancers
SMARCA4 cancer mutations result in distinct classes of dynamic defects in living cells that disrupt important enzymological sub-steps
Using ATAC-seq, we find that SMARCA4 mutations with distinct dynamic defects lead to convergent effects on DNA accessibility at enhancers
The effects on the epigenomic landscape reflect a dominant-negative behavior at superenhancers from many tissues, revealing how SMARCA4 mutations contribute to diverse cancer types
Acknowledgments
We thank C. Weber, E. Chory, K. Cui, G. Hu, and E. Son for helpful discussions, and dedicate this manuscript to the lasting memory of Joseph P. Calarco. We gratefully acknowledge the assistance of the DNA Sequencing and Genomics Core facility of NHLBI, the Stanford Cell Sciences Imaging Facility (S10OD01227601), and the Stanford BioX3 cluster (S10RR02664701). Ring1a−/−;Ring1bfl/fl mESCs were a generous gift from H. Koseki (RIKEN, Japan). This work was supported by the Simons Foundation Autism Research Initiative (G.R.C.), NIH grants R37NS046789 (G.R.C.), R01CA163915 (G.R.C.), T32CA009151 (J.G.K.), R00CA187565 (H.C.H.), the Division of Intramural Research of the NHLBI/NIH (K.Z.)., the Czech Science Foundation GACR 16-06357S (V.V.), the Cancer Prevention & Research Institute of Texas grant RR170036 (H.C.H.), and the Howard Hughes Medical Institute (G.R.C.).
Footnotes
Author Contributions
H.C.H. and B.Z.S. conceived of and performed experiments, and wrote the paper; K.C., H.C.H., and V.V. developed the homology model; H.C.H, C.-Y.C., and E.L.M performed analyses; B.Z.S., H.C.H., C.-Y.C., J.G.K., and W.L.K. prepared materials; K.Z. and G.R.C. designed experiments and wrote the paper.
Competing financial interests
The authors declare no competing financial interests.
Research animals
All studies involving laboratory animals were performed in accordance with animal welfare regulations established by the Stanford University Administrative Panel on Laboratory Animal Care (APLAC). All protocols used in this study were approved by the Stanford APLAC.
References
- 1.Hodges C, Kirkland JG, Crabtree GR. The Many Roles of BAF (mSWI/SNF) and PBAF Complexes in Cancer. Cold Spring Harb Perspect Med. 2016;6 doi: 10.1101/cshperspect.a026930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Son EY, Crabtree GR. The role of BAF (mSWI/SNF) complexes in mammalian neural development. Am J Med Genet C Semin Med Genet. 2014;166c:333–49. doi: 10.1002/ajmg.c.31416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kadoch C, et al. Proteomic and bioinformatic analysis of mammalian SWI/SNF complexes identifies extensive roles in human malignancy. Nat Genet. 2013;45:592–601. doi: 10.1038/ng.2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shain AH, Pollack JR. The spectrum of SWI/SNF mutations, ubiquitous in human cancers. PLoS One. 2013;8:e55119. doi: 10.1371/journal.pone.0055119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Versteege I, et al. Truncating mutations of hSNF5/INI1 in aggressive paediatric cancer. Nature. 1998;394:203–6. doi: 10.1038/28212. [DOI] [PubMed] [Google Scholar]
- 6.Biegel JA, et al. Germ-line and acquired mutations of INI1 in atypical teratoid and rhabdoid tumors. Cancer Res. 1999;59:74–9. [PubMed] [Google Scholar]
- 7.Kadoch C, Crabtree GR. Reversible disruption of mSWI/SNF (BAF) complexes by the SS18-SSX oncogenic fusion in synovial sarcoma. Cell. 2013;153:71–85. doi: 10.1016/j.cell.2013.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davoli T, et al. Cumulative haploinsufficiency and triplosensitivity drive aneuploidy patterns and shape the cancer genome. Cell. 2013;155:948–62. doi: 10.1016/j.cell.2013.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wilson BG, Roberts CW. SWI/SNF nucleosome remodellers and cancer. Nat Rev Cancer. 2011;11:481–92. doi: 10.1038/nrc3068. [DOI] [PubMed] [Google Scholar]
- 10.Wei D, et al. SNF5/INI1 deficiency redefines chromatin remodeling complex composition during tumor development. Mol Cancer Res. 2014;12:1574–85. doi: 10.1158/1541-7786.MCR-14-0005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang X, et al. SMARCB1-mediated SWI/SNF complex function is essential for enhancer regulation. Nat Genet. 2017;49:289–295. doi: 10.1038/ng.3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Oruetxebarria I, et al. P16INK4a is required for hSNF5 chromatin remodeler-induced cellular senescence in malignant rhabdoid tumor cells. J Biol Chem. 2004;279:3807–16. doi: 10.1074/jbc.M309333200. [DOI] [PubMed] [Google Scholar]
- 13.Kia SK, Gorski MM, Giannakopoulos S, Verrijzer CP. SWI/SNF mediates polycomb eviction and epigenetic reprogramming of the INK4b-ARF-INK4a locus. Mol Cell Biol. 2008;28:3457–64. doi: 10.1128/MCB.02019-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wilson BG, et al. Epigenetic antagonism between polycomb and SWI/SNF complexes during oncogenic transformation. Cancer Cell. 2010;18:316–28. doi: 10.1016/j.ccr.2010.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee RS, et al. A remarkably simple genome underlies highly malignant pediatric rhabdoid cancers. J Clin Invest. 2012;122:2983–8. doi: 10.1172/JCI64400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lawrence MS, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013;499:214–8. doi: 10.1038/nature12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.de Bruijn DR, et al. The synovial-sarcoma-associated SS18-SSX2 fusion protein induces epigenetic gene (de)regulation. Cancer Res. 2006;66:9474–82. doi: 10.1158/0008-5472.CAN-05-3726. [DOI] [PubMed] [Google Scholar]
- 18.Lubieniecka JM, et al. Histone deacetylase inhibitors reverse SS18-SSX-mediated polycomb silencing of the tumor suppressor early growth response 1 in synovial sarcoma. Cancer Res. 2008;68:4303–10. doi: 10.1158/0008-5472.CAN-08-0092. [DOI] [PubMed] [Google Scholar]
- 19.Zhang ZK, et al. Cell cycle arrest and repression of cyclin D1 transcription by INI1/hSNF5. Mol Cell Biol. 2002;22:5975–88. doi: 10.1128/MCB.22.16.5975-5988.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang X, et al. Oncogenesis caused by loss of the SNF5 tumor suppressor is dependent on activity of BRG1, the ATPase of the SWI/SNF chromatin remodeling complex. Cancer Res. 2009;69:8094–101. doi: 10.1158/0008-5472.CAN-09-0733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mathur R, et al. ARID1A loss impairs enhancer-mediated gene regulation and drives colon cancer in mice. Nat Genet. 2017;49:296–302. doi: 10.1038/ng.3744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Alver BH, et al. The SWI/SNF chromatin remodelling complex is required for maintenance of lineage specific enhancers. Nat Commun. 2017;8:14648. doi: 10.1038/ncomms14648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miller EL, et al. TOP2 synergizes with BAF chromatin remodeling for both resolution and formation of facultative heterochromatin. Nat Struct Mol Biol. 2017;24:344–352. doi: 10.1038/nsmb.3384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fillmore CM, et al. EZH2 inhibition sensitizes BRG1 and EGFR mutant lung tumours to TopoII inhibitors. Nature. 2015;520:239–42. doi: 10.1038/nature14122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Imielinski M, et al. Mapping the hallmarks of lung adenocarcinoma with massively parallel sequencing. Cell. 2012;150:1107–20. doi: 10.1016/j.cell.2012.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Network TCGAR. Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;511:543–50. doi: 10.1038/nature13385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Witkowski L, et al. Germline and somatic SMARCA4 mutations characterize small cell carcinoma of the ovary, hypercalcemic type. Nat Genet. 2014;46:438–43. doi: 10.1038/ng.2931. [DOI] [PubMed] [Google Scholar]
- 28.Jelinic P, et al. Recurrent SMARCA4 mutations in small cell carcinoma of the ovary. Nat Genet. 2014;46:424–6. doi: 10.1038/ng.2922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Karnezis AN, et al. Dual loss of the SWI/SNF complex ATPases SMARCA4/BRG1 and SMARCA2/BRM is highly sensitive and specific for small cell carcinoma of the ovary, hypercalcaemic type. J Pathol. 2016;238:389–400. doi: 10.1002/path.4633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hodis E, et al. A landscape of driver mutations in melanoma. Cell. 2012;150:251–63. doi: 10.1016/j.cell.2012.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Le Loarer F, et al. SMARCA4 inactivation defines a group of undifferentiated thoracic malignancies transcriptionally related to BAF-deficient sarcomas. Nat Genet. 2015;47:1200–5. doi: 10.1038/ng.3399. [DOI] [PubMed] [Google Scholar]
- 32.Love C, et al. The genetic landscape of mutations in Burkitt lymphoma. Nat Genet. 2012;44:1321–5. doi: 10.1038/ng.2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lunning MA, Green MR. Mutation of chromatin modifiers; an emerging hallmark of germinal center B-cell lymphomas. Blood Cancer J. 2015;5:e361. doi: 10.1038/bcj.2015.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stanton BZ, et al. Smarca4 ATPase mutations disrupt direct eviction of PRC1 from chromatin. Nat Genet. 2017;49:282–288. doi: 10.1038/ng.3735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Weinstein JN, et al. The Cancer Genome Atlas Pan-Cancer analysis project. Nat Genet. 2013;45:1113–20. doi: 10.1038/ng.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zehir A, et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat Med. 2017;23:703–713. doi: 10.1038/nm.4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu X, Li M, Xia X, Li X, Chen Z. Mechanism of chromatin remodelling revealed by the Snf2-nucleosome structure. Nature. 2017;544:440–445. doi: 10.1038/nature22036. [DOI] [PubMed] [Google Scholar]
- 38.Xia X, Liu X, Li T, Fang X, Chen Z. Structure of chromatin remodeler Swi2/Snf2 in the resting state. Nat Struct Mol Biol. 2016;23:722–9. doi: 10.1038/nsmb.3259. [DOI] [PubMed] [Google Scholar]
- 39.Hauk G, McKnight JN, Nodelman IM, Bowman GD. The chromodomains of the Chd1 chromatin remodeler regulate DNA access to the ATPase motor. Mol Cell. 2010;39:711–23. doi: 10.1016/j.molcel.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gao J, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Laurent BC, Treich I, Carlson M. The yeast SNF2/SWI2 protein has DNA-stimulated ATPase activity required for transcriptional activation. Genes Dev. 1993;7:583–91. doi: 10.1101/gad.7.4.583. [DOI] [PubMed] [Google Scholar]
- 42.Johnson TA, Elbi C, Parekh BS, Hager GL, John S. Chromatin remodeling complexes interact dynamically with a glucocorticoid receptor-regulated promoter. Mol Biol Cell. 2008;19:3308–22. doi: 10.1091/mbc.E08-02-0123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gurskaya NG, et al. Engineering of a monomeric green-to-red photoactivatable fluorescent protein induced by blue light. Nat Biotechnol. 2006;24:461–5. doi: 10.1038/nbt1191. [DOI] [PubMed] [Google Scholar]
- 44.Sif S, Stukenberg PT, Kirschner MW, Kingston RE. Mitotic inactivation of a human SWI/SNF chromatin remodeling complex. Genes Dev. 1998;12:2842–51. doi: 10.1101/gad.12.18.2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ho L, et al. esBAF facilitates pluripotency by conditioning the genome for LIF/STAT3 signalling and by regulating polycomb function. Nat Cell Biol. 2011;13:903–13. doi: 10.1038/ncb2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods. 2013;10:1213–8. doi: 10.1038/nmeth.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Whyte WA, et al. Enhancer decommissioning by LSD1 during embryonic stem cell differentiation. Nature. 2012;482:221–5. doi: 10.1038/nature10805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Francis NJ, Kingston RE, Woodcock CL. Chromatin compaction by a polycomb group protein complex. Science. 2004;306:1574–7. doi: 10.1126/science.1100576. [DOI] [PubMed] [Google Scholar]
- 49.Endoh M, et al. Histone H2A mono-ubiquitination is a crucial step to mediate PRC1-dependent repression of developmental genes to maintain ES cell identity. PLoS Genet. 2012;8:e1002774. doi: 10.1371/journal.pgen.1002774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Whyte WA, et al. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013;153:307–19. doi: 10.1016/j.cell.2013.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ha M, Hong S. Gene-regulatory interactions in embryonic stem cells represent cell-type specific gene regulatory programs. Nucleic Acids Research. 2017 doi: 10.1093/nar/gkx752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Khan A, Zhang X. dbSUPER: a database of super-enhancers in mouse and human genome. Nucleic Acids Res. 2016;44:D164–71. doi: 10.1093/nar/gkv1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bultman S, et al. A Brg1 null mutation in the mouse reveals functional differences among mammalian SWI/SNF complexes. Mol Cell. 2000;6:1287–95. doi: 10.1016/s1097-2765(00)00127-1. [DOI] [PubMed] [Google Scholar]
- 54.Lieberman-Aiden E, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–93. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dekker J, Marti-Renom MA, Mirny LA. Exploring the three-dimensional organization of genomes: interpreting chromatin interaction data. Nat Rev Genet. 2013;14:390–403. doi: 10.1038/nrg3454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Brookes E, et al. Polycomb associates genome-wide with a specific RNA polymerase II variant, and regulates metabolic genes in ESCs. Cell Stem Cell. 2012;10:157–70. doi: 10.1016/j.stem.2011.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Araya CL, et al. Identification of significantly mutated regions across cancer types highlights a rich landscape of functional molecular alterations. Nat Genet. 2016;48:117–25. doi: 10.1038/ng.3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Soufi A, Donahue G, Zaret KS. Facilitators and impediments of the pluripotency reprogramming factors' initial engagement with the genome. Cell. 2012;151:994–1004. doi: 10.1016/j.cell.2012.09.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rahl PB, et al. c-Myc regulates transcriptional pause release. Cell. 2010;141:432–45. doi: 10.1016/j.cell.2010.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lin CY, et al. Transcriptional amplification in tumor cells with elevated c-Myc. Cell. 2012;151:56–67. doi: 10.1016/j.cell.2012.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ho L, et al. An embryonic stem cell chromatin remodeling complex, esBAF, is essential for embryonic stem cell self-renewal and pluripotency. Proc Natl Acad Sci U S A. 2009;106:5181–6. doi: 10.1073/pnas.0812889106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang Y, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pohl A, Beato M. bwtool: a tool for bigWig files. Bioinformatics. 2014;30:1618–9. doi: 10.1093/bioinformatics/btu056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–2. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jin W, et al. Genome-wide detection of DNase I hypersensitive sites in single cells and FFPE tissue samples. Nature. 2015;528:142–6. doi: 10.1038/nature15740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Barski A, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–37. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 69.Kidder BL, Zhao K. Efficient library preparation for next-generation sequencing analysis of genome-wide epigenetic and transcriptional landscapes in embryonic stem cells. Methods Mol Biol. 2014;1150:3–20. doi: 10.1007/978-1-4939-0512-6_1. [DOI] [PubMed] [Google Scholar]
- 70.Hahne F, Ivanek R. Visualizing Genomic Data Using Gviz and Bioconductor. Methods Mol Biol. 2016;1418:335–51. doi: 10.1007/978-1-4939-3578-9_16. [DOI] [PubMed] [Google Scholar]
- 71.Anders S, Pyl PT, Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–9. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Heinz S, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38:576–89. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tibshirani R. Regression Shrinkage and Selection via the Lasso. Journal of the Royal Statistical Society. Series B (Methodological) 1996;58:267–288. [Google Scholar]
- 74.Friedman J, Hastie T, Tibshirani R. Regularization paths for generalized linear models via coordinate descent. Journal of statistical software. 2010;33:1. [PMC free article] [PubMed] [Google Scholar]
- 75.Krieger E, Vriend G. YASARA View - molecular graphics for all devices - from smartphones to workstations. Bioinformatics. 2014;30:2981–2. doi: 10.1093/bioinformatics/btu426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hooft RW, Vriend G, Sander C, Abola EE. Errors in protein structures. Nature. 1996;381:272. doi: 10.1038/381272a0. [DOI] [PubMed] [Google Scholar]
- 77.Hooft RW, Sander C, Scharf M, Vriend G. The PDBFINDER database: a summary of PDB, DSSP and HSSP information with added value. Comput Appl Biosci. 1996;12:525–9. doi: 10.1093/bioinformatics/12.6.525. [DOI] [PubMed] [Google Scholar]
- 78.Qiu J, Elber R. SSALN: an alignment algorithm using structure-dependent substitution matrices and gap penalties learned from structurally aligned protein pairs. Proteins. 2006;62:881–91. doi: 10.1002/prot.20854. [DOI] [PubMed] [Google Scholar]
- 79.Dixon JR, et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–80. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequencing data sets generated and analyzed in this study are available in the Gene Expression Omnibus (GEO) repository under accession numbers GSE88968, GSE94041, and GSE98605. Source data for Figure 1 are available with the paper online. Three-dimensional coordinates of the mouse SMARCA4 homology model are provided in PDB format as Supplementary Dataset 1. Three-dimensional coordinates of the human SMARCA4 homology model are provided in PDB format as Supplementary Dataset 2. Other data and materials are available from the authors upon request. A Life Sciences Reporting Summary for this article is available.