Abstract
Reactive oxygen species (ROS) are formed in mitochondria during electron transport and energy generation. Elevated levels of ROS lead to increased amounts of mitochondrial DNA (mtDNA) damage. We report that levels of M1dG, a major endogenous peroxidation-derived DNA adduct, are 50–100-fold higher in mtDNA than in nuclear DNA in several different human cell lines. Treatment of cells with agents that either increase or decrease mitochondrial superoxide levels leads to increased or decreased levels of M1dG in mtDNA, respectively. Sequence analysis of adducted mtDNA suggests that M1dG residues are randomly distributed throughout the mitochondrial genome. Basal levels of M1dG in mtDNA from pulmonary microvascular endothelial cells (PMVECs) from transgenic bone morphogenetic protein receptor 2 mutant mice (BMPR2R899X) (four adducts per 106 dG) are twice as high as adduct levels in wild-type cells. A similar increase was observed in mtDNA from heterozygous null (BMPR2+/−) compared to wild-type PMVECs. Pulmonary arterial hypertension is observed in the presence of BMPR2 signaling disruptions, which are also associated with mitochondrial dysfunction and oxidant injury to endothelial tissue. Persistence of M1dG adducts in mtDNA could have implications for mutagenesis and mitochondrial gene expression, thereby contributing to the role of mitochondrial dysfunction in diseases.
INTRODUCTION
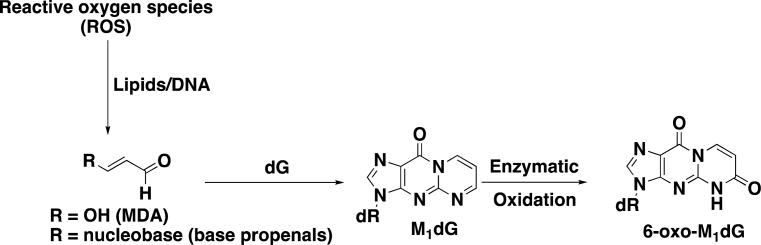
Reactive oxygen species (ROS) play a major role in physiological and pathophysiological processes (1–3). Mitochondria are particularly susceptible to oxidative damage since electrons that leak from the electron transport chain in the inner mitochondrial membrane react with oxygen to produce superoxide anion, O2.− (4). The formation of O2.− accounts for ∼0.15% of mitochondrial oxygen consumption (4). Superoxide anion is unstable and cannot traverse membranes; however, it is quickly converted to membrane-permeable hydrogen peroxide that can then undergo Fenton chemistry to yield the very reactive hydroxyl radical in the mitochondrial matrix. Reaction of the hydroxyl radical or other reactive species with mitochondrial DNA (mtDNA) or lipids can lead to the formation of base propenals and malondialdehyde (MDA) respectively (Figure 1) (5–10).
Figure 1.
Formation of M1dG from MDA and base propenals and the oxidation of M1dG to 6-oxo-M1dG.
Both base propenal and MDA are electrophiles that react with DNA to generate a variety of adducts, including the most abundant species, 3-(2-deoxy-β-d-erythro-pentofuranosyl)pyrimido[1,2-α]purin-10(3H)-one (M1dG) (Figure 1) (9,11). M1dG is mutagenic through its ability to induce base-pair substitutions and frameshifts (5,12–14). Earlier work suggests that it is removed by nucleotide excision repair (NER) and that the free mononucleoside is oxidized to generate a single metabolite, 6-oxo-M1dG (Figure 1) (15). Very recently, we demonstrated that M1dG is also oxidized to 6-oxo-M1dG in genomic DNA of intact cells and that M1dG is preferentially oxidized rather than removed by NER, at least in some cell lines (16).
In this study, we report that mtDNA isolated from several distinct cell types, including human embryonic kidney (HEK293), human liver carcinoma (HepG2), and human colon cancer (RKO) cells, exhibit basal levels of M1dG that are substantially higher than those observed in genomic DNA. Adduct levels increased with base propenal exposure and were strongly correlated with levels of oxidative stress. M1dG was distributed throughout the mitochondrial genome with no sequence specificity. 6-Oxo-M1dG was not observed in mtDNA from cells exposed to any conditions examined. Finally, we demonstrate a relationship between disruptions in the bone morphogenetic protein (BMPR2) signaling pathway and M1dG levels in mtDNA. These results provide new insight into the relationship between oxidative stress and mtDNA damage that may lead to mutations associated with aging and degenerative diseases (17–26).
MATERIALS AND METHODS
General procedures
All chemicals were obtained from commercial sources and used without further purification unless otherwise noted. Anhydrous solvents were purchased from Sigma Aldrich, St. Louis, MO, USA. All 1H and 13C NMR spectra were referenced to internal tetramethylsilane (TMS) at 0.0 ppm. The spin multiplicities are indicated by the symbols s (singlet), d (doublet), dd (doublet of doublets), t (triplet), q (quartet), m (multiplet) and br (broad). Reactions were monitored by thin-layer chromatography (TLC). Column chromatography was performed using commercial silica gel and eluted with the indicated solvent system. Yields refer to chromatographically and spectroscopically (1H and 13C NMR) homogeneous materials.
Preparation of adenine propenal
Adenine propenal was prepared as previously described (8,16). However, the pure compound was isolated by column chromatography eluted with 5% methanol in ethyl acetate.
Preparation of stably labeled M1dG and 6-oxo-M1dG internal standards
[15N5]-6-oxo-M1dG was synthesized as described previously by substituting [15N5]-dG (Cambridge Isotope Laboratories, Andover, MA, USA) for dG (15). Similarly, [13C, 15N2]-M1dG was synthesized as described previously by substituting [13C, 15N2]-dG (Cambridge Isotope Laboratories, Andover, MA, USA) for dG (27).
Cell culture and treatment of cells with adenine propenal
RKO, HEK293, or HepG2 cells (5 × 106) were grown as described previously (16). After 24 h, the medium was removed, and fresh medium without serum but containing adenine propenal (400 μM) was added for 1 h. The electrophile was removed, and fresh medium without serum was added. Then the cells were incubated for 0, 1, 2, 3, 6, 9, 12 or 24 h. Following incubation, the cells were harvested and lysed as described previously (16). The nuclei were isolated at 1000 × g at 4°C for 5 min, and the supernatants were removed as the mitochondrial fraction. The mitochondria were pelleted at 10 000 × g at 4°C for 10 min. They were then washed with 1 ml of a solution containing 250 mM sucrose, 1 mM EDTA, 20 mM HEPES (pH 7.5) and 0.2% protease inhibitor cocktail and pelleted at 10 000 × g at 4°C for 10 min. The mitochondria were then assessed for purity by western blot analysis probing for Atp5a following lysis of an aliquot of the organelle. DNA from the mitochondria was isolated, purified, and quantified as described previously (16).
Quantification of adducts in mtDNA
Prior to digestion, a portion of each sample was diluted 1:1000 in order to quantify dG levels. Internal standards, [15N5]-6-oxo-M1dG and [15N2,13C]-M1dG (5 or 10 pmol), were then added to the original samples, and [15N5]-dG (1 nmol) was added to the diluted samples. For all samples, the DNA was digested using standard conditions as described previously (16) with appropriate adjustments of enzyme concentrations for the diluted samples. The digested samples were desalted using Oasis HLB 1cc (30 mg) extraction cartridges (Waters Corporation). The cartridges were activated with two column volumes of methanol and then washed with five column volumes of water. The samples were loaded on individual cartridges and then washed with three column volumes of water, after which the nucleosides were eluted with two column volumes of methanol. The eluents were evaporated using a TurboVap LV evaporator giving a residue that was dissolved in water. The samples were then analyzed by liquid chromatography-tandem mass spectrometry (LC-MS/MS) using the conditions described previously (16).
Effect of TEMPO on M1dG levels during cell lysis and DNA isolation
RKO cells were grown as previously described (16) except that no electrophile or other treatments were added. The cells were lysed as described with the exception that 10 mM TEMPO was added to the cocktail used for the lysis. TEMPO was also added to cocktails used for the lysis of the mitochondria as well as the isolation of mtDNA. Digestion and analysis of mtDNA proceeded as described above.
M1dG levels in genomic mtDNA in RAW264.7 macrophages
RAW264.7 macrophages (5 × 106) were grown in DMEM + Glutamax medium (Invitrogen) with 10% fetal bovine serum at 37°C with 5% CO2 on plates 150 mm in diameter. After 24 h of incubation, the medium containing fetal bovine serum was replaced with serum-free medium for 24 h. The cells were then harvested at 0, 1, 2, 3, 6, 9, 12 or 24 h. DNA was isolated from the mitochondria and analyzed by LC–MS/MS as described above.
Effect of oxidative stress on M1dG levels in genomic mtDNA
RKO, HEK293, and HepG2 cells (5 × 106) were grown in RPMI 1640 medium with 10% fetal bovine serum at 37°C with 5% CO2 on plates 150 mm in diameter. RAW264.7 macrophages (5 × 106) were grown in DMEM + Glutamax medium with 10% fetal bovine serum at 37°C with 5% CO2 on plates 150 mm in diameter. After 24 h, the medium was removed, and fresh medium without serum was added for 24 h, after which, in separate experiments, the cells were treated with rotenone (100 nM, final concentration), TEMPOL (10 μM, final concentration), mitoTEMPO (10 μM, final concentration), or antimycin A (10 μM, final concentration). After 24 h of incubation, the cells were harvested, and mtDNA was isolated and analyzed as described above.
M1dG levels in genomic mtDNA in endothelial cells from BMPR2 mutant and heterozygous mice
Pulmonary microvascular endothelial cells (PMVEC) were isolated from wild-type, BMPR2 heterozygous null (BMPR2+/−) and transgenic BMPR2 mutant (BMPR2R899X) mice that had been crossed onto the Immortomouse background, as previously described (28–31). The conditionally immortalized lines were maintained at 33°C in the presence of murine interferon-gamma (IFN-γ) to maintain immortalization. Prior to all experiments, cells were transitioned to 37°C and IFN-γ removed from the media for at least 72 h prior to allow for transition back to primary phenotype. For the wild-type and the BMPR2R899X cells, doxycycline (300 ng/ml) was added to the medium to induce expression of the BMPR2 transgene (the construct being Rosa26-rtTA x TetO7-BMPR2R899X) at the time of transition to 37°C. At near confluency, cells were harvested, mitochondria isolated, and DNA was extracted, digested, and analyzed as described above.
DNA sequencing analysis
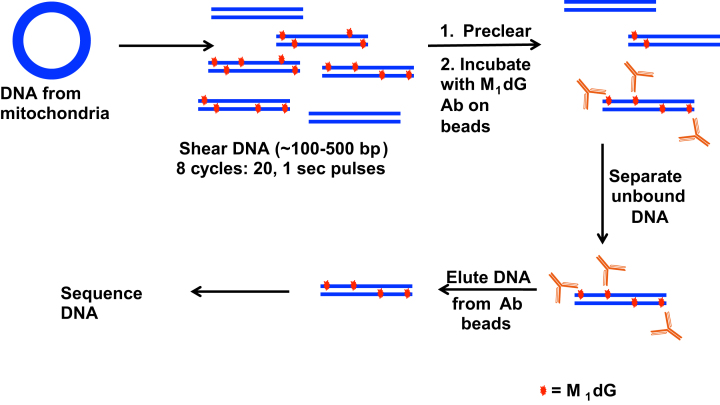
RKO cells were synchronized as described above and were then treated with or without 400 μM adenine propenal for 1 h. DNA was isolated from the mitochondria as described earlier. The DNA was then sheared by sonication (8 cycles of 20, 1 s pulses) to produce fragments between 100 and 500 base pairs in length (verified by agarose gel electrophoresis). M1dG enrichment in samples of DNA was performed as previously described using M1dG antibodies attached to sepharose beads (32–35). Following enrichment, samples of mtDNA from untreated cells (control) and adenine propenal-treated cells were submitted to HudsonAlpha (Huntsville, AL, USA) for ChIP library generation with the TruSeq ChIP Library Generation Kit (Illumina, San Diego, CA) and sequencing. Libraries were sequenced on the Illumina HiSeq 2500 with single-end 50 bp reads at a sequencing depth of 25M reads/sample. Fastq files were downloaded from HudsonAlpha and concatenated to single files. Paired-end reads were aligned to the hg19 build using Bowtie2 and converted to BAM files, sorted and indexed using Samtools1.2. Sorted BAM files were used as input for MACS2 using the narrow and broad options. Sequence alignments were viewed in Integrated Genome Viewer.
Statistical Analysis
Statistical analyses and generation of graphs were performed using Graph Pad Prism 6.0c (Graph Pad Software, San Diego, CA, USA). Differences in adduct levels between controls and treatments in triplicate experiments were determined using a one-way ANOVA and Tukey's post hoc analysis or another appropriate comparative test. Differences were considered significant if P < 0.05.
RESULTS
M1dG levels in mtDNA
We recently reported that M1dG levels in nuclear DNA increase on exposure to adenine propenal and that M1dG is oxidized to 6-oxo-M1dG at a faster rate than it is removed by NER in several human cell lines (16). Since mitochondria generate high levels of oxidative stress, we sought to determine the levels of M1dG in mtDNA under both basal conditions and following electrophile treatment. We observed basal M1dG levels in RKO mtDNA to be two adducts per 106 dG whereas the levels were 2.3 adducts and two adducts per 106 dG in HEK293 and HepG2 cells, respectively. Given that human cancer cells have ∼5000 mtDNA molecules, each containing 16 569 bp (36), a basal level of ∼2 M1dG adducts per 106 dG equates to ∼82 M1dG adducts per cell. It is important to note that this value only includes basal M1dG adducts in mtDNA and does not include contributions from nuclear DNA. The levels of M1dG in mtDNA were 2 orders of magnitude higher than those found in nuclear DNA of the same cell lines (∼1.5 adducts per 108 dG).
The comparatively high levels of M1dG in mtDNA, might be due to exposure to electrophiles generated during the cell lysis and DNA extraction procedure. To address this possibility, an experiment was conducted in which TEMPO, an antioxidant scavenger, was included during all phases of sample processing according to an approach previously reported by Swenberg et al. (37). The results indicated that there was no significant difference in M1dG levels in mtDNA isolated from control cells versus cells that were processed in the presence of TEMPO (Supplemental Figure S1). These results suggest that M1dG is not formed in mtDNA by exposure to electrophiles or oxidants during sample workup.
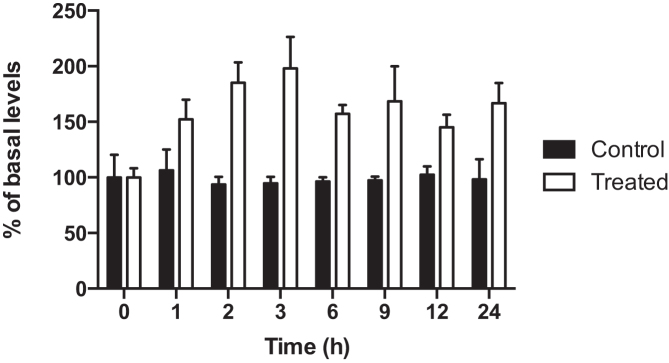
M1dG levels in mtDNA increased significantly with adenine propenal treatment, as exemplified in RKO cells, and they remained constant for up to 24 h thereafter (Figure 2). This is in contrast to M1dG levels in nuclear DNA that increase with adenine propenal treatment but then decrease over time after the electrophile is removed (16). The decrease in nuclear DNA M1dG levels correlates closely with the appearance of 6-oxo-M1dG (16). Remarkably, oxidation of M1dG to 6-oxo-M1dG was not observed in the mtDNA from any of the cell types investigated (Supplemental Figure S2). MtDNA from RAW264.7 macrophages was also isolated and analyzed. The RAW264.7 cell line is a well-studied system for investigating inflammation and associated metabolic processes (38–40). We found that the basal level of M1dG in mtDNA of RAW264.7 macrophages (∼1 adduct per 105 dG) was surprisingly high, and consistent with the other cell lines, it was two orders of magnitude above the nuclear DNA level (∼1 adduct per 107 dG). Again, no fluctuation in the mtDNA M1dG levels or conversion of mtDNA M1dG to 6-oxo-M1dG was observed.
Figure 2.
M1dG levels in mtDNA in RKO cells after treatment with adenine propenal (400 μM) for 1 h. The electrophile was removed, and cells were lysed at the indicated times for mtDNA isolation and analysis. Data represent the mean ± S.D. of triplicate determinations.
Effect of oxidative stress on M1dG levels in mtDNA
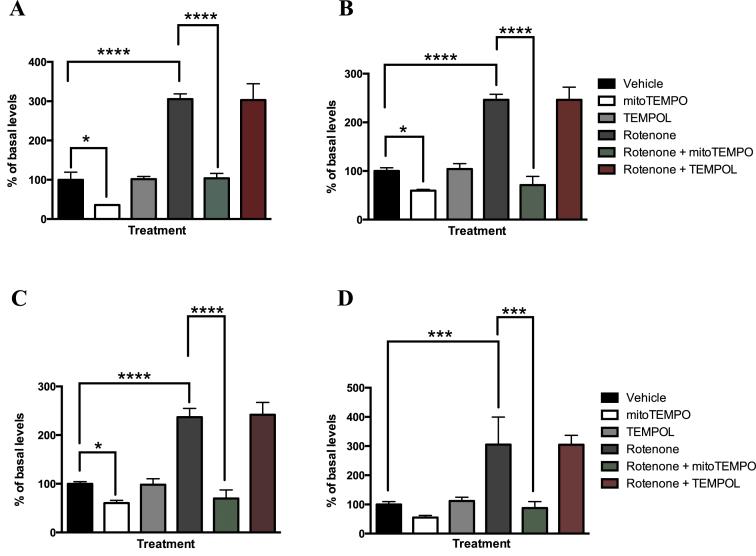
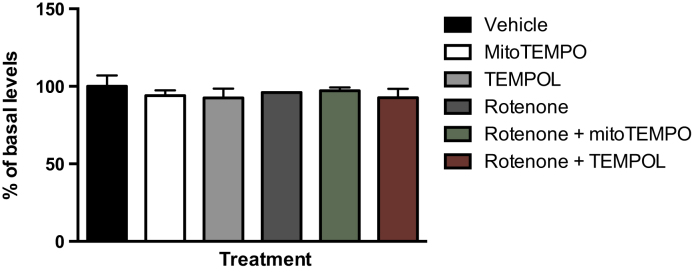
To investigate the effect of oxidative stress on levels of M1dG in mtDNA, all the cell types were treated with various agents that modulate levels of superoxide in the mitochondria. Rotenone is a known mitochondrial toxicant that inhibits complex I of the electron transport chain, resulting in more electron leakage and an increase in superoxide levels (41–43). We found that in the RKO cells, M1dG levels in mtDNA were increased by 200% compared to basal levels following rotenone treatment (Figure 3A). Treatment of RKO cells with antimycin A, a complex III inhibitor that also results in superoxide formation, produced similar results (Supplemental Figure S3). MitoTEMPO is a superoxide scavenger that decreases levels of superoxide selectively in mitochondria (44,45). Treatment of RKO cells with mitoTEMPO at 10 μM for 24 h, significantly decreased, by 65%, mtDNA M1dG levels compared to those from untreated cells (Figure 3A). However, treatment with TEMPOL, a ubiquitously dispersed superoxide scavenger (44), at 10 μM for 24 h, did not decrease M1dG levels in mtDNA (Figure 3). MitoTEMPO also significantly decreased the mtDNA M1dG levels of cells treated with rotenone, resulting in levels comparable to those seen in cells treated with vehicle alone. In contrast, TEMPOL had no effect on rotenone-mediated increases in M1dG levels in mtDNA. Importantly, in all the treatments described above, M1dG levels in nuclear DNA were unchanged compared to those in vehicle-treated cells (Figure 4). The same treatments were conducted in HEK293, HepG2 and RAW264.7 macrophages. In all of these cell types, similar trends were observed to those seen with the RKO cells (Figure 3B–D).
Figure 3.
M1dG levels in the mtDNA of RKO cells (panel A), HEK293 cells (panel B), HepG2 cells (panel C) and RAW264.7 cells (panel D) after treatment with rotenone (100 nM), and/or mitoTEMPO (10 μM), and/or TEMPOL (10 μM) for 24 h. Basal levels of M1dG in mtDNA were determined to be two adducts per 106 dG, 2.3 adducts per 106 dG, two adducts per 106 dG and one adduct per 105 dG in RKO cells, HEK293 cells, HepG2 cells and RAW 264.7 macrophages respectively. Data represent the mean ± S.D. of triplicate determinations.
Figure 4.
M1dG levels in nuclear DNA in RKO cells after a 24-h treatment with rotenone (100 nM), mitoTEMPO (10 μM), or TEMPOL (10 μM) alone or with rotenone (100 nM) and either mitoTEMPO (10 μM) or TEMPOL (10 μM). Data represent the mean ± S.D. of triplicate determinations. In RKO cells, basal levels of M1dG in genomic DNA was determined to be 1.5 adducts per 108 dG.
M1dG levels and BMP signaling
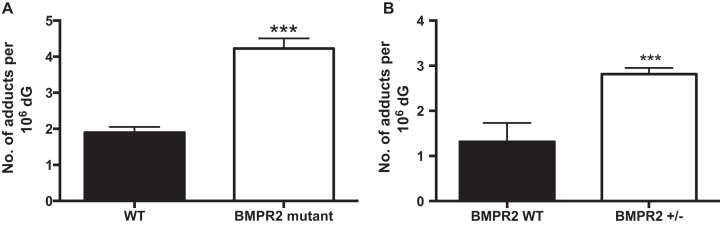
Pulmonary arterial hypertension (PAH) is a rare, progressive, often fatal disease resulting from endothelial dysfunction and vascular remodeling of small pulmonary arteries (46). A familial form of PAH results from a germline mutation of the gene encoding the bone morphogenetic protein receptor type II (BMPR2). BMPR2 is a transmembrane receptor consisting of an extracellular ligand-binding domain, a short transmembrane domain, a kinase domain, and a long cytoplasmic tail. Reduced expression of BMPR2 is also observed in many patients with idiopathic PAH and is associated with impaired mitochondrial function, oxidative stress, and increased mtDNA damage (47,48). Thus, we hypothesized that PAH-associated aberrant mitochondrial function could be correlated with an alteration in M1dG levels in mtDNA. To test this hypothesis, we employed BMPR2 heterozygous null (BMPR2+/−), and transgenic BMPR2 mutant (BMPR2R899X) mice that had been crossed onto the Immortomouse background as a model of familial PAH. We isolated mtDNA from PMVECs from wild-type, BMPR2+/− and BMPR2R899X mice and analyzed it for levels of M1dG. Consistent with our hypothesis, we found that M1dG levels in the mtDNA from BMPR2R899X and BMPR2+/− PMVECs were ∼2-fold higher than those in the corresponding wild-type mice (Figure 5).
Figure 5.
(A) Mitochondrial M1dG levels in mtDNA in PMVECs isolated from transgenic BMPR2 (bone morphogenetic protein receptor 2) mutant mice (BMPR2 R899X) and wild type mice. Data represent the mean ± S.D. of quadruplicate determinations. (B) Mitochondrial M1dG levels in mtDNA in PMVECs isolated from BMPR2 heterozygous null (BMPR2+/−) cells compared to wild-type. Data represent the mean ± S.D. of quadruplicate determinations.
DNA sequencing analysis
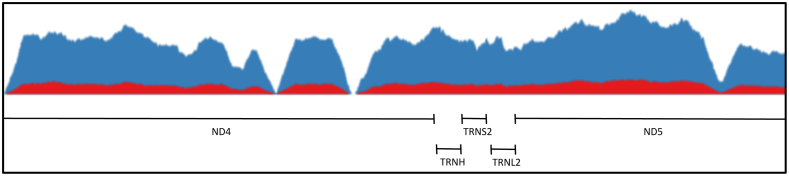
To determine if formation of M1dG in the mitochondrial genome occurs in a sequence-specific manner, we isolated mtDNA from RKO cells with or without prior adenine propenal treatment. Following isolation, the DNA was sheared by sonication, and M1dG-containing sequences were enriched using sepharose beads coated with anti-M1dG antibody (Figure 6) (32–35). Samples of the enriched mtDNA from both adenine propenal-treated and control untreated cells were subjected to Illumina sequencing. No regions of enrichment in the mtDNA were found, regardless of pretreatment, suggesting that M1dG is present throughout the mitochondrial genome (Figure 7).
Figure 6.
Workflow for the preparation of mtDNA from RKO cells for sequencing analysis following treatment with adenine propenal (400 μM) for 1 h.
Figure 7.
Representative coverage of a 2 kb region of the mitochondrial genome. The signal corresponding to mtDNA from cells treated with adenine propenal and enriched with the M1dG antibody (blue) shows increased sequence reads but does not show enrichment of distinct regions when compared to the signal corresponding to mtDNA from cells not treated with adenine propenal and enriched (red).
DISCUSSION
M1dG is an endogenous lesion that is detectable in the genomic DNA of humans and rodents (32,49–53). M1dG is repaired by NER, and the free nucleotide is oxidized to 6-oxo-M1dG by cytosolic xanthine oxidase and aldehyde oxidase (15,54). Very recently, we have shown that M1dG in genomic DNA of intact cells is oxidized to 6-oxo-M1dG across a variety of cell types by a process that appears to be enzymatic (16). In most cells that have been studied, basal levels of M1dG in genomic DNA are low, suggesting efficient repair/removal in the face of presumed ongoing production due to continuous exposure to MDA and/or base propenal (Figure 1). Under these conditions, repair of M1dG likely occurs by NER. Oxidation of M1dG might also occur, but if so, the resulting 6-oxo-M1dG must be removed or repaired very quickly such that its levels remain undetectable. When cells are exposed to adenine propenal, however, levels of M1dG in genomic DNA rise, indicating that the rate of formation of the adduct exceeds that of normal repair mechanisms. Under these conditions, conversion of M1dG to 6-oxo-M1dG becomes the primary route for the observed subsequent decrease in M1dG. Additionally, the rate of formation of 6-oxo-M1dG must exceed its rate of repair, as conversion of M1dG to 6-oxo-M1dG appears to be almost quantitative (16). Unlike RKO, HEK293, or HepG2 cells, 6-oxo-M1dG is detected in genomic DNA of RAW264.7 cells under basal conditions, presumably because higher levels of oxidative stress in these cells (38–40) lead to rates of M1dG formation that exceed the capacity of DNA repair mechanisms. The result is both a high steady state level of M1dG and detectable 6-oxo-M1dG (16).
In mtDNA, basal levels of M1dG are much higher than those found in genomic DNA, and we surmised that this might be due to the higher levels of ROS in mitochondria and/or the fact that mitochondria lack the ability to perform NER (55–57). Studies have revealed that mitochondria possess a rather robust base-excision repair (BER) pathway, including single-nucleotide base excision repair (SN-BER), long-patch BER (LP-BER), and single-strand break repair (SSBR) (58–62). However, M1dG does not appear to be repaired by any of these mechanisms. Furthermore, in mtDNA, no formation of 6-oxo-M1dG was observed, even after adenine propenal treatment that raised M1dG levels or in the case of RAW264.7 cells that exhibited high basal levels of M1dG. In fact, after adenine propenal exposure, the mitochondria maintained the elevated steady state level of M1dG for an extended period of time. The failure to detect 6-oxo-M1dG in mtDNA suggests that the putative enzyme that catalyzes M1dG oxidation is absent in the mitochondrion. However, we cannot completely rule out formation of 6-oxo-M1dG in mtDNA. It is possible that it occurs, but the rate of 6-oxo-M1dG repair/removal is so rapid that no detectable steady state levels are observed even when M1dG levels are very high. The apparent absence of M1dG repair mechanisms in mitochondria suggests that removal of M1dG might require total degradation of highly damaged mtDNA or destruction of the entire mitochondrion.
We hypothesized that the higher levels of M1dG in mtDNA than genomic DNA could be due to the leak of electrons from complex I and III of the mitochondrial electron transport chain leading to local production of superoxide and other ROS (63,64). In support of this hypothesis, we found that agents that increase electron leakage (rotenone and antimycin A) (41–43) resulted in an increase in M1dG in mtDNA (Figure 3). A recent report has shown that abasic sites and single strand breaks are the predominant forms of mtDNA damage produced by rotenone-induced stress in cultured cells (65). The C4′-oxidation of an abasic sugar results in the formation of base propenal (66,67), a direct precursor to M1dG (Figure 1). Our hypothesis was further validated by the reduction in both basal and rotenone-induced mtDNA M1dG levels obtained upon treatment with mitoTEMPO, a mitochondrially specific superoxide scavenger but not TEMPOL, a ubiquitously dispersed superoxide scavenger (44,45). It is important to note that none of the treatments described above affects M1dG levels in nuclear DNA. Therefore, these results imply that mitochondrial oxidant production is not the cause of this form of genomic DNA damage.
Relatively little is currently known about the potential contribution of mtDNA adducts to specific disease states. Consequently, we investigated the levels of M1dG in a mouse model of PAH, a disease associated with mitochondrial dysfunction and oxidant injury that has been associated with mutations in the BMPR2 gene (68,69). Recent work has revealed a direct link between BMPR2 mutation and impaired mitochondrial function accompanied by an increase in DNA damage (48). Specifically, mutation in the BMPR2 gene led to the induction of a large 4977-bp deletion in mtDNA (ΔmtDNA4977) (48), a lesion associated with the formation of the oxidation product 8-oxo-dG in mtDNA (70). In the present study, we found that the endogenous level of M1dG was 2-fold higher in mtDNA from PMVEC obtained from mice with a mutated BMPR2 gene than from wild-type mice. A similar trend was observed in mtDNA from PMVEC recovered from heterozygous null (BMPR2+/−) mice. The heterozygous null mutation is the same as is found in most hereditary PAH patients (29). The finding of elevated mtDNA M1dG levels in these mouse models of PAH is consistent with a recent report that there is an increased production of ROS, likely of mitochondrial origin, accompanied by evidence of lipid peroxidation in cultured cells, mice, and humans in the presence of a BMPR2 mutation (28). Numerous prior reports highlight a correlation between elevated levels of M1dG in various pathological states (49–52,71,72); however, it is unknown in these cases whether the adducted DNA was genomic or mitochondrial. Thus, the data presented here provide the first clear evidence of a correlation between mtDNA M1dG levels and a disease state.
The human mitochondrial genome has been sequenced in its entirety, and the individual genes have been identified and characterized (73,74). Additionally, several mutations in mtDNA have been correlated with specific disease states (18). Consequently, we sought to investigate whether M1dG accumulated in any particular mtDNA gene(s) or sequence contexts. The finding that M1dG is present at roughly equal levels across the mitochondrial genome (Figure 7) suggests that the adduct is not likely responsible for any specific known mutations; however, recent studies showing that M1dG arrests human mitochondrial RNA polymerase (POLRMT), support the hypothesis that it might impair mitochondrial gene expression (75).
The observation of M1dG in mtDNA is not unprecedented, as Jeong et al. reported an approximately two-fold higher level of M1dG in mtDNA than genomic DNA from rat liver (37). Thus, that report agrees with the current finding that adduct levels are higher in mtDNA than genomic DNA, but there appears to be a difference in the magnitude of that difference (2-fold versus 100-fold). It is possible that immortalized cells in culture experience higher levels of mitochondrial oxidative stress than in an intact organ. Alternatively, it is possible that post-mortem changes lead to degradation of the adduct in the mitochondria or during tissue processing (e.g. oxidation of M1dG to 6-oxo-M1dG). Another source of the difference might be the methodology used. In the earlier report, adduct levels were determined by LC–MS/MS analysis of the pentafluorobenzylhydrazine (PFBH) conjugate with the M1dG nucleobase whereas the present methodology involves direct quantification of M1dG residues by LC/MS–MS (37).
A review of the literature reveals that M1dG is not the only oxidative stress-related DNA adduct that is elevated in mtDNA compared to nuclear DNA. For example, 8-oxo-dG, one of the most studied oxidative lesions, has been found to be elevated in the mitochondria (76–78). Additionally, in an animal model of Wilson's disease, etheno-DNA adducts [1,N6-ethenodeoxyadenosine (ϵdA) and 3,N4-ethenodeoxycytidine (ϵdC)] were found to be elevated in mtDNA as compared to nuclear DNA (79). Also, in a study looking at the relationship between oxidative stress and Alzheimer's disease, oxidized bases in different lobes of the brain and cerebellum were isolated and quantified. The study identified the presence of 8-oxo-dG as well as 8-oxo-dA, 5-hydroxy-2′-deoxyuracil, 2,6-diamino-4-hydroxy-5-formamidopyrimidine (FapyG) and 4,6-diamino-5-formamidopyrimidine (FapyA), and demonstrated that levels of these adducts in the DNA from some brain regions were higher in Alzheimer's disease patients than in controls. Significantly, mtDNA possessed 10-fold higher levels of oxidized bases than nuclear DNA in both experimental and control groups (77).
The somatic mutation rate of mtDNA is 10–20 times higher than that of nuclear DNA (80). In our present studies, we have conducted investigations to study the relationship between oxidative stress and levels of M1dG in the mitochondrion. Our data provide strong evidence that increases in oxidative stress lead to a direct increase in levels of M1dG in mtDNA. The persistence of high levels of M1dG in mtDNA, could both promote mutation and have deleterious consequences for the transcription of mitochondrial genes. Importantly, the increase in mtDNA M1dG levels when BMP signaling is impaired provides a link between M1dG levels in mtDNA and pulmonary arterial hypertension, a mitochondrially related disease state. These results provide important new insight into the formation of a major endogenous adduct in the mitochondrion and suggest a possible role in pathological processes associated with mitochondrial dysfunction. Furthermore, as the levels of M1dG in mtDNA appear to be constant over time, it might serve as a biomarker for disease states, especially those associated with mitochondrial oxidative stress.
Supplementary Material
ACKNOWLEDGEMENTS
The authors are grateful to Carol Rouzer for critical reading and editing of this manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health [R37 CA87819 to L.J.M., HL121174 to J.P.F]. Funding for open access charge: National Institutes of Health.
Conflict of interest statement. None declared.
REFERENCES
- 1. Coussens L.M., Werb Z.. Inflammation and cancer. Nature. 2002; 420:860–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Elinav E., Nowarski R., Thaiss C.A., Hu B., Jin C., Flavell R.A.. Inflammation-induced cancer: crosstalk between tumours, immune cells and microorganisms. Nat. Rev. Cancer. 2013; 13:759–771. [DOI] [PubMed] [Google Scholar]
- 3. Karin M. NF-kappaB as a critical link between inflammation and cancer. Cold Spring Harb. Perspect. Biol. 2009; 1:a000141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. St-Pierre J., Buckingham J.A., Roebuck S.J., Brand M.D.. Topology of superoxide production from different sites in the mitochondrial electron transport chain. J. Biol. Chem. 2002; 277:44784–44790. [DOI] [PubMed] [Google Scholar]
- 5. Marnett L.J. Oxyradicals and DNA damage. Carcinogenesis. 2000; 21:361–370. [DOI] [PubMed] [Google Scholar]
- 6. Bigey P., Pratviel G., Meunier B.. Cleavage of double-stranded DNA by ‘metalloporphyrin-linker-oligonucleotide’ molecules: influence of the linker. Nucleic Acids Res. 1995; 23:3894–3900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Grollman J.H., Jr Cardiac radiology in Southern California: is there a future for cardiac radiologists. AJR Am. J. Roentgenol. 1985; 144:1083–1085. [DOI] [PubMed] [Google Scholar]
- 8. Johnson F., Pillai K.M., Grollman A.P., Tseng L., Takeshita M.. Synthesis and biological activity of a new class of cytotoxic agents: N-(3-oxoprop-1-enyl)-substituted pyrimidines and purines. J. Med. Chem. 1984; 27:954–958. [DOI] [PubMed] [Google Scholar]
- 9. Plastaras J.P., Riggins J.N., Otteneder M., Marnett L.J.. Reactivity and mutagenicity of endogenous DNA oxopropenylating agents: base propenals, malondialdehyde, and N(epsilon)-oxopropenyllysine. Chem. Res. Toxicol. 2000; 13:1235–1242. [DOI] [PubMed] [Google Scholar]
- 10. Halliwell B., Gutteridge J.M.. Free radicals, lipid peroxidation, and cell damage. Lancet. 1984; 2:1095. [DOI] [PubMed] [Google Scholar]
- 11. Plastaras J.P., Dedon P.C., Marnett L.J.. Effects of DNA structure on oxopropenylation by the endogenous mutagens malondialdehyde and base propenal. Biochemistry. 2002; 41:5033–5042. [DOI] [PubMed] [Google Scholar]
- 12. Dedon P.C., Plastaras J.P., Rouzer C.A., Marnett L.J.. Indirect mutagenesis by oxidative DNA damage: formation of the pyrimidopurinone adduct of deoxyguanosine by base propenal. Proc. Natl. Acad. Sci. U.S.A. 1998; 95:11113–11116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. VanderVeen L.A., Hashim M.F., Shyr Y., Marnett L.J.. Induction of frameshift and base pair substitution mutations by the major DNA adduct of the endogenous carcinogen malondialdehyde. Proc. Natl. Acad. Sci. U.S.A. 2003; 100:14247–14252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fink S.P., Reddy G.R., Marnett L.J.. Mutagenicity in Escherichia coli of the major DNA adduct derived from the endogenous mutagen malondialdehyde. Proc. Natl. Acad. Sci. U.S.A. 1997; 94:8652–8657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Otteneder M.B., Knutson C.G., Daniels J.S., Hashim M., Crews B.C., Remmel R.P., Wang H., Rizzo C., Marnett L.J.. In vivo oxidative metabolism of a major peroxidation-derived DNA adduct, M1dG. Proc. Natl. Acad. Sci. U.S.A. 2006; 103:6665–6669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wauchope O.R., Beavers W.N., Galligan J.J., Mitchener M.M., Kingsley P.J., Marnett L.J.. Nuclear oxidation of a major peroxidation DNA adduct, M1dG, in the genome. Chem. Res. Toxicol. 2015; 28:2334–2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wallace D.C., Singh G., Lott M.T., Hodge J.A., Schurr T.G., Lezza A.M., Elsas L.J. 2nd, Nikoskelainen E.K.. Mitochondrial DNA mutation associated with Leber's hereditary optic neuropathy. Science. 1988; 242:1427–1430. [DOI] [PubMed] [Google Scholar]
- 18. Chatterjee A., Mambo E., Sidransky D.. Mitochondrial DNA mutations in human cancer. Oncogene. 2006; 25:4663–4674. [DOI] [PubMed] [Google Scholar]
- 19. Ohta S. Contribution of somatic mutations in the mitochondrial genome to the development of cancer and tolerance against anticancer drugs. Oncogene. 2006; 25:4768–4776. [DOI] [PubMed] [Google Scholar]
- 20. Wallace D.C., Lott M.T., Shoffner J.M., Ballinger S.. Mitochondrial DNA mutations in epilepsy and neurological disease. Epilepsia. 1994; 35(Suppl. 1):S43–S50. [DOI] [PubMed] [Google Scholar]
- 21. Krishnan K.J., Reeve A.K., Turnbull D.M.. Do mitochondrial DNA mutations have a role in neurodegenerative disease. Biochem. Soc. Trans. 2007; 35:1232–1235. [DOI] [PubMed] [Google Scholar]
- 22. Newsholme P., Haber E.P., Hirabara S.M., Rebelato E.L., Procopio J., Morgan D., Oliveira-Emilio H.C., Carpinelli A.R., Curi R.. Diabetes associated cell stress and dysfunction: role of mitochondrial and non-mitochondrial ROS production and activity. J. Physiol. 2007; 583:9–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Rotig A., Bonnefont J.P., Munnich A.. Mitochondrial diabetes mellitus. Diabetes Metab. 1996; 22:291–298. [PubMed] [Google Scholar]
- 24. Ballinger S.W., Shoffner J.M., Hedaya E.V., Trounce I., Polak M.A., Koontz D.A., Wallace D.C.. Maternally transmitted diabetes and deafness associated with a 10.4 kb mitochondrial DNA deletion. Nat. Genet. 1992; 1:11–15. [DOI] [PubMed] [Google Scholar]
- 25. Alexeyev M.F., Ledoux S.P., Wilson G.L.. Mitochondrial DNA and aging. Clin. Sci. (Lond.). 2004; 107:355–364. [DOI] [PubMed] [Google Scholar]
- 26. Kujoth G.C., Bradshaw P.C., Haroon S., Prolla T.A.. The role of mitochondrial DNA mutations in mammalian aging. PLoS Genet. 2007; 3:e24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Szekely J., Wang H., Peplowski K.M., Knutson C.G., Marnett L.J., Rizzo C.J.. “One-pot” syntheses of malondialdehyde adducts of nucleosides. Nucleosides Nucleotides Nucleic Acids. 2008; 27:103–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lane K.L., Talati M., Austin E., Hemnes A.R., Johnson J.A., Fessel J.P., Blackwell T., Mernaugh R.L., Robinson L., Fike C. et al. Oxidative injury is a common consequence of BMPR2 mutations. Pulm Circ. 2011; 1:72–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Prewitt A.R., Ghose S., Frump A.L., Datta A., Austin E.D., Kenworthy A.K., de Caestecker M.P.. Heterozygous null bone morphogenetic protein receptor type 2 mutations promote SRC kinase-dependent caveolar trafficking defects and endothelial dysfunction in pulmonary arterial hypertension. J. Biol. Chem. 2015; 290:960–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chen X., Talati M., Fessel J.P., Hemnes A.R., Gladson S., French J., Shay S., Trammell A., Phillips J.A., Hamid R. et al. Estrogen metabolite 16alpha-hydroxyestrone exacerbates bone morphogenetic protein receptor type II-associated pulmonary arterial hypertension through microRNA-29-mediated modulation of cellular metabolism. Circulation. 2016; 133:82–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Fessel J.P., Chen X., Frump A., Gladson S., Blackwell T., Kang C., Johnson J., Loyd J.E., Hemnes A., Austin E. et al. Interaction between bone morphogenetic protein receptor type 2 and estrogenic compounds in pulmonary arterial hypertension. Pulm. Circ. 2013; 3:564–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Rouzer C.A., Chaudhary A.K., Nokubo M., Ferguson D.M., Reddy G.R., Blair I.A., Marnett L.J.. Analysis of the malondialdehyde-2′-deoxyguanosine adduct pyrimidopurinone in human leukocyte DNA by gas chromatography/electron capture/negative chemical ionization/mass spectrometry. Chem. Res. Toxicol. 1997; 10:181–188. [DOI] [PubMed] [Google Scholar]
- 33. Sevilla C.L., Mahle N.H., Eliezer N., Uzieblo A., O’Hara S.M., Nokubo M., Miller R., Rouzer C.A., Marnett L.J.. Development of monoclonal antibodies to the malondialdehyde-deoxyguanosine adduct, pyrimidopurinone. Chem. Res. Toxicol. 1997; 10:172–180. [DOI] [PubMed] [Google Scholar]
- 34. Hoberg A.M., Otteneder M., Marnett L.J., Poulsen H.E.. Measurement of the malondialdehyde-2′-deoxyguanosine adduct in human urine by immuno-extraction and liquid chromatography/atmospheric pressure chemical ionization tandem mass spectrometry. J. Mass Spectrom. 2004; 39:38–42. [DOI] [PubMed] [Google Scholar]
- 35. Akingbade D., Kingsley P.J., Shuck S.C., Cooper T., Carnahan R., Szekely J., Marnett L.J.. Selection of monoclonal antibodies against 6-oxo-M(1)dG and their use in an LC-MS/MS assay for the presence of 6-oxo-M(1)dG in vivo. Chem. Res. Toxicol. 2012; 25:454–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bogenhagen D.F. Mitochondrial DNA nucleoid structure. Biochim. Biophys. Acta. 2012; 1819:914–920. [DOI] [PubMed] [Google Scholar]
- 37. Jeong Y.C., Nakamura J., Upton P.B., Swenberg J.A.. Pyrimido[1,2-a]-purin-10(3H)-one, M1G, is less prone to artifact than base oxidation. Nucleic Acids Res. 2005; 33:6426–6434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Dennis E.A., Deems R.A., Harkewicz R., Quehenberger O., Brown H.A., Milne S.B., Myers D.S., Glass C.K., Hardiman G., Reichart D. et al. A mouse macrophage lipidome. J. Biol. Chem. 2010; 285:39976–39985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Raschke W.C., Baird S., Ralph P., Nakoinz I.. Functional macrophage cell lines transformed by Abelson leukemia virus. Cell. 1978; 15:261–267. [DOI] [PubMed] [Google Scholar]
- 40. Jiang W., Reich I.C., Pisetsky D.S.. Mechanisms of activation of the RAW264.7 macrophage cell line by transfected mammalian DNA. Cell. Immunol. 2004; 229:31–40. [DOI] [PubMed] [Google Scholar]
- 41. Li N., Ragheb K., Lawler G., Sturgis J., Rajwa B., Melendez J.A., Robinson J.P.. Mitochondrial complex I inhibitor rotenone induces apoptosis through enhancing mitochondrial reactive oxygen species production. J. Biol. Chem. 2003; 278:8516–8525. [DOI] [PubMed] [Google Scholar]
- 42. Verkaart S., Koopman W.J., van Emst-de Vries S.E., Nijtmans L.G., van den Heuvel L.W., Smeitink J.A., Willems P.H.. Superoxide production is inversely related to complex I activity in inherited complex I deficiency. Biochim. Biophys. Acta. 2007; 1772:373–381. [DOI] [PubMed] [Google Scholar]
- 43. Aluri H.S., Simpson D.C., Allegood J.C., Hu Y., Szczepanek K., Gronert S., Chen Q., Lesnefsky E.J.. Electron flow into cytochrome c coupled with reactive oxygen species from the electron transport chain converts cytochrome c to a cardiolipin peroxidase: role during ischemia-reperfusion. Biochim. Biophys. Acta. 2014; 1840:3199–3207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Dikalov S. Cross talk between mitochondria and NADPH oxidases. Free Radic. Biol. Med. 2011; 51:1289–1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Dikalova A.E., Bikineyeva A.T., Budzyn K., Nazarewicz R.R., McCann L., Lewis W., Harrison D.G., Dikalov S.I.. Therapeutic targeting of mitochondrial superoxide in hypertension. Circ. Res. 2010; 107:106–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lai Y.C., Potoka K.C., Champion H.C., Mora A.L., Gladwin M.T.. Pulmonary arterial hypertension: the clinical syndrome. Circ. Res. 2014; 115:115–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Atkinson C., Stewart S., Upton P.D., Machado R., Thomson J.R., Trembath R.C., Morrell N.W.. Primary pulmonary hypertension is associated with reduced pulmonary vascular expression of type II bone morphogenetic protein receptor. Circulation. 2002; 105:1672–1678. [DOI] [PubMed] [Google Scholar]
- 48. Diebold I., Hennigs J.K., Miyagawa K., Li C.G., Nickel N.P., Kaschwich M., Cao A., Wang L., Reddy S., Chen P.I. et al. BMPR2 preserves mitochondrial function and DNA during reoxygenation to promote endothelial cell survival and reverse pulmonary hypertension. Cell Metab. 2015; 21:596–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Singh R., Leuratti C., Josyula S., Sipowicz M.A., Diwan B.A., Kasprzak K.S., Schut H.A., Marnett L.J., Anderson L.M., Shuker D.E.. Lobe-specific increases in malondialdehyde DNA adduct formation in the livers of mice following infection with Helicobacter hepaticus. Carcinogenesis. 2001; 22:1281–1287. [DOI] [PubMed] [Google Scholar]
- 50. Jeong Y.C., Walker N.J., Burgin D.E., Kissling G., Gupta M., Kupper L., Birnbaum L.S., Swenberg J.A.. Accumulation of M1dG DNA adducts after chronic exposure to PCBs, but not from acute exposure to polychlorinated aromatic hydrocarbons. Free Radic. Biol. Med. 2008; 45:585–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kadlubar F.F., Anderson K.E., Haussermann S., Lang N.P., Barone G.W., Thompson P.A., MacLeod S.L., Chou M.W., Mikhailova M., Plastaras J. et al. Comparison of DNA adduct levels associated with oxidative stress in human pancreas. Mutat. Res. 1998; 405:125–133. [DOI] [PubMed] [Google Scholar]
- 52. Ma B., Villalta P.W., Balbo S., Stepanov I.. Analysis of a malondialdehyde-deoxyguanosine adduct in human leukocyte DNA by liquid chromatography nanoelectrospray-high-resolution tandem mass spectrometry. Chem. Res. Toxicol. 2014; 27:1829–1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Chaudhary A.K., Nokubo M., Reddy G.R., Yeola S.N., Morrow J.D., Blair I.A., Marnett L.J.. Detection of endogenous malondialdehyde-deoxyguanosine adducts in human liver. Science. 1994; 265:1580–1582. [DOI] [PubMed] [Google Scholar]
- 54. Knutson C.G., Wang H., Rizzo C.J., Marnett L.J.. Metabolism and elimination of the endogenous DNA adduct, 3-(2-deoxy-beta-D-erythropentofuranosyl)-pyrimido[1,2-alpha]purine-10(3H)-one, in the rat. J. Biol. Chem. 2007; 282:36257–36264. [DOI] [PubMed] [Google Scholar]
- 55. Pascucci B., Versteegh A., van Hoffen A., van Zeeland A.A., Mullenders L.H., Dogliotti E.. DNA repair of UV photoproducts and mutagenesis in human mitochondrial DNA. J. Mol. Biol. 1997; 273:417–427. [DOI] [PubMed] [Google Scholar]
- 56. Clayton D.A., Doda J.N., Friedberg E.C.. The absence of a pyrimidine dimer repair mechanism in mammalian mitochondria. Proc. Natl. Acad. Sci. U.S.A. 1974; 71:2777–2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Wallace D.C. Mitochondrial genetics: a paradigm for aging and degenerative diseases. Science. 1992; 256:628–632. [DOI] [PubMed] [Google Scholar]
- 58. Taffe B.G., Larminat F., Laval J., Croteau D.L., Anson R.M., Bohr V.A.. Gene-specific nuclear and mitochondrial repair of formamidopyrimidine DNA glycosylase-sensitive sites in Chinese hamster ovary cells. Mutat. Res. 1996; 364:183–192. [DOI] [PubMed] [Google Scholar]
- 59. LeDoux S.P., Wilson G.L., Beecham E.J., Stevnsner T., Wassermann K., Bohr V.A.. Repair of mitochondrial DNA after various types of DNA damage in Chinese hamster ovary cells. Carcinogenesis. 1992; 13:1967–1973. [DOI] [PubMed] [Google Scholar]
- 60. Akbari M., Visnes T., Krokan H.E., Otterlei M.. Mitochondrial base excision repair of uracil and AP sites takes place by single-nucleotide insertion and long-patch DNA synthesis. DNA Repair (Amst.). 2008; 7:605–616. [DOI] [PubMed] [Google Scholar]
- 61. Liu P., Qian L., Sung J.S., de Souza-Pinto N.C., Zheng L., Bogenhagen D.F., Bohr V.A., Wilson D.M. 3rd, Shen B., Demple B.. Removal of oxidative DNA damage via FEN1-dependent long-patch base excision repair in human cell mitochondria. Mol. Cell. Biol. 2008; 28:4975–4987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Sykora P., Wilson D.M. 3rd, Bohr V.A.. Repair of persistent strand breaks in the mitochondrial genome. Mech. Ageing Dev. 2012; 133:169–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Boveris A. Determination of the production of superoxide radicals and hydrogen peroxide in mitochondria. Methods Enzymol. 1984; 105:429–435. [DOI] [PubMed] [Google Scholar]
- 64. Han D., Antunes F., Canali R., Rettori D., Cadenas E.. Voltage-dependent anion channels control the release of the superoxide anion from mitochondria to cytosol. J. Biol. Chem. 2003; 278:5557–5563. [DOI] [PubMed] [Google Scholar]
- 65. Shokolenko I., Venediktova N., Bochkareva A., Wilson G.L., Alexeyev M.F.. Oxidative stress induces degradation of mitochondrial DNA. Nucleic Acids Res. 2009; 37:2539–2548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Chan S.W., Dedon P.C.. The biological and metabolic fates of endogenous DNA damage products. J. Nucleic Acids. 2010; 2010:929047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Dedon P.C. The chemical toxicology of 2-deoxyribose oxidation in DNA. Chem. Res. Toxicol. 2008; 21:206–219. [DOI] [PubMed] [Google Scholar]
- 68. Deng Z., Morse J.H., Slager S.L., Cuervo N., Moore K.J., Venetos G., Kalachikov S., Cayanis E., Fischer S.G., Barst R.J. et al. Familial primary pulmonary hypertension (gene PPH1) is caused by mutations in the bone morphogenetic protein receptor-II gene. Am. J. Hum. Genet. 2000; 67:737–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. International P.P.H.C., Lane K.B., Machado R.D., Pauciulo M.W., Thomson J.R., Phillips J.A. 3rd, Loyd J.E., Nichols W.C., Trembath R.C.. Heterozygous germline mutations in BMPR2, encoding a TGF-beta receptor, cause familial primary pulmonary hypertension. Nat. Genet. 2000; 26:81–84. [DOI] [PubMed] [Google Scholar]
- 70. Cheng Y., Ren X., Gowda A.S., Shan Y., Zhang L., Yuan Y.S., Patel R., Wu H., Huber-Keener K., Yang J.W. et al. Interaction of Sirt3 with OGG1 contributes to repair of mitochondrial DNA and protects from apoptotic cell death under oxidative stress. Cell Death Dis. 2013; 4:e731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Sun X., Nair J., Bartsch H.. A modified immuno-enriched 32P-postlabeling method for analyzing the malondialdehyde-deoxyguanosine adduct, 3-(2-deoxy-beta-D-erythro-pentofuranosyl)- pyrimido[1,2-alpha]purin-10(3H)one in human tissue samples. Chem. Res. Toxicol. 2004; 17:268–272. [DOI] [PubMed] [Google Scholar]
- 72. Sram R.J., Farmer P., Singh R., Garte S., Kalina I., Popov T.A., Binkova B., Ragin C., Taioli E.. Effect of vitamin levels on biomarkers of exposure and oxidative damage-the EXPAH study. Mutat. Res. 2009; 672:129–134. [DOI] [PubMed] [Google Scholar]
- 73. Blanchard J.L., Schmidt G.W.. Mitochondrial DNA migration events in yeast and humans: integration by a common end-joining mechanism and alternative perspectives on nucleotide substitution patterns. Mol. Biol. Evol. 1996; 13:537–548. [DOI] [PubMed] [Google Scholar]
- 74. Grivell L.A. Mitochondrial DNA. Sci. Am. 1983; 248:78–89. [DOI] [PubMed] [Google Scholar]
- 75. Cline S.D., Lodeiro M.F., Marnett L.J., Cameron C.E., Arnold J.J.. Arrest of human mitochondrial RNA polymerase transcription by the biological aldehyde adduct of DNA, M1dG. Nucleic Acids Res. 2010; 38:7546–7557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Richter C., Park J.W., Ames B.N.. Normal oxidative damage to mitochondrial and nuclear DNA is extensive. Proc. Natl. Acad. Sci. U.S.A. 1988; 85:6465–6467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Wang J., Xiong S., Xie C., Markesbery W.R., Lovell M.A.. Increased oxidative damage in nuclear and mitochondrial DNA in Alzheimer's disease. J. Neurochem. 2005; 93:953–962. [DOI] [PubMed] [Google Scholar]
- 78. Nakamoto H., Kaneko T., Tahara S., Hayashi E., Naito H., Radak Z., Goto S.. Regular exercise reduces 8-oxodG in the nuclear and mitochondrial DNA and modulates the DNA repair activity in the liver of old rats. Exp. Gerontol. 2007; 42:287–295. [DOI] [PubMed] [Google Scholar]
- 79. Nair J., Strand S., Frank N., Knauft J., Wesch H., Galle P.R., Bartsch H.. Apoptosis and age-dependant induction of nuclear and mitochondrial etheno-DNA adducts in Long-Evans Cinnamon (LEC) rats: enhanced DNA damage by dietary curcumin upon copper accumulation. Carcinogenesis. 2005; 26:1307–1315. [DOI] [PubMed] [Google Scholar]
- 80. Taylor R.W., Turnbull D.M.. Mitochondrial DNA mutations in human disease. Nat. Rev. Genet. 2005; 6:389–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.