Abstract
In plants, a key class of genes comprising most of disease resistance (R) genes encodes Nucleotide-binding leucine-rich repeat (NL) proteins. Access to common bean (Phaseolus vulgaris) genome sequence provides unparalleled insight into the organization and evolution of this large gene family (∼400 NL) in this important crop. As observed in other plant species, most common bean NL are organized in cluster of genes. However, a particularity of common bean is that these clusters are often located in subtelomeric regions close to terminal knobs containing the satellite DNA khipu. Phylogenetically related NL are spread between different chromosome ends, suggesting frequent exchanges between non-homologous chromosomes. NL peculiar location, in proximity to heterochromatic regions, led us to study their DNA methylation status using a whole-genome cytosine methylation map. In common bean, NL genes displayed an unusual body methylation pattern since half of them are methylated in the three contexts, reminiscent of the DNA methylation pattern of repeated sequences. Moreover, 90 NL were also abundantly targeted by 24 nt siRNA, with 90% corresponding to methylated NL genes. This suggests the existence of a transcriptional gene silencing mechanism of NL through the RdDM (RNA-directed DNA methylation) pathway in common bean that has not been described in other plant species.
Keywords: NB-LRR, DNA methylation, small RNAs, Phaseolus vulgaris, satellite DNA
1. Introduction
Plants have evolved a multi-layered defense system to detect and counter pathogen infection. As a first layer, plants can perceive relatively conserved small molecules, proteins and protein fragments, produced externally to the cell by microbes or host cell degradation products, and collectively referred to as Pathogen Associated Molecular Patterns (PAMPs). This recognition through plasma membrane pattern-recognition receptors (PRR) initiate a resistance response known as PAMP-triggered immunity (PTI).1 However, successful pathogens have developed proteins or small molecules termed effectors that can strike down PTI, resulting in Effector Triggered Susceptibility (ETS). As a second layer, plants have evolved the ability to recognize either directly or indirectly pathogen effectors using proteins encoded by resistance (R) genes. Genes encoding effectors that are recognized by R gene products, leading to effective plant resistance called Effector Triggered Immunity (ETI), are genetically defined as avirulence (Avr) genes. ETI drives a powerful immune response, epistatic to effector mediated immune suppression and sufficient to limit pathogen spread.2
R genes have been implicated in resistance against diverse and taxonomically unrelated pathogens including bacteria, viruses, nematodes, insects, filamentous fungi and oomycetes. Strikingly, regardless of the plant or the pathogen considered and despite the diversity of pathogen Avr proteins, the majority of cloned R genes encode intracellular proteins with a conserved central Nucleotide-Binding domain (NB), also known as NB-ARC (Nucleotide-Binding adaptor shared by Apaf1, certain R genes and CED4) and a more variable C-terminal Leucine-Rich Repeat (LRR) domain.3–5 With regard to their N-termini, two phylogenetically distinct major groups can be distinguished within the NB-LRR or NL proteins.6 The first group contains an N-terminal domain with homology to the Drosophila Toll and human Interleukin-1 Receptor (TIR), referred to as TIR-NB-LRR or TNL. The second group, corresponding to non-TNL group is usually known as CNL (for CC-NB-LRR), since many members of this group are predicted to form a coiled-coil (CC) structure in the N-terminus. Despite an overall lack of sequence similarity, most of characterized CC domains possess a small ‘EDVID’ motif.7,8 However, a less abundant subclass of CC domain has been described in which EDVID motif is not present and which presents closest sequence similarity to RPW8, a non-NL R protein from Arabidopsis thaliana that confers broad-spectrum resistance against powdery mildew (Erysiphe spp.).9,10
NL encoding R genes are present in all sequenced plant genomes in variable number, from less than 100 genes in papaya, cucumber, watermelon and melon11,12 to more than one thousand in apple.13 Variation in NL number is not necessarily proportional to the genome size and if NL-encoding genes often exhibit spectacular lineage-specific expansions, dramatic contractions have also been reported.14 This suggests that a limitation in the expansion of NL encoding gene number exists in a plant genome. This limitation possibly reflects fitness costs associated with resistance expression.15,16 Genome-wide investigation of NL gene family in various plant species reveal that NL encoding genes tend to be clustered in genomes. For instance, 66% of Arabidopsis thaliana NL genes, 76% of the rice NL genes, 73% of potato NL genes, 64% of cucumber NL genes, 81% of melon NL genes, 69% of watermelon NL genes, 81% of apple NL genes, and 70% of cassava NL genes are organized in clusters.12,13,17–20 These clusters vary in size and complexity and fall into two types based on the phylogenetic relationship of their NL sequences: homogeneous clusters containing phylogenetically related sequences that may have undergone frequent sequence exchanges and heterogeneous clusters containing phylogenetically distantly related NL and even both TNL and non-TNL encoding sequences.14 Coexistence of these two types of clusters suggests that different mechanisms are responsible of NL encoding genes evolution.
Cytosine methylation is an epigenetic modification that affects chromatin packaging and transcription and has been shown to be involved in various biological processes including biotic stress responses.21 Whereas in animals, methylation at CG dinucleotides predominates, in plant, cytosine methylation occurs in three sequence contexts controlled by distinct pathways: the symmetric CG and CHG contexts, as well as the asymmetric CHH context (where H= A, C, or T).22 DNA methylation is established in all three contexts through RNA-directed DNA methylation (RdDM) pathway, whereby 24-nucleotide (nt) small interfering RNAs (siRNAs) guide the DNA methyltransferase to the corresponding genomic DNA.22–24 Whereas methylation in symmetric CG and CHG contexts can be maintained through DNA replication by context-specific DNA methyltransferases, maintenance of asymmetric CHH methylation requires either CMT2 (at long Transposable Element -TE-) or RdDM (at short TE or genes), depending on the loci that are targeted.22,25,26 Different Arabidopsis hypomethylated mutants defective in the RdDM pathway showed a modified response to disease after pathogen infection,27–29 providing evidence that this pathway modulates immune responses against pathogen. The pattern and the role of DNA methylation differ depending on the targeted genomic features. For instance, methylation of repetitive sequences, such as transposable elements (TEs), generally occurs in all three contexts and acts as a genomic immune system, by suppressing transcription and proliferation of invading DNA elements, thereby maintaining genome integrity.30,31 In contrast, cytosines within the transcribed-region of protein coding genes are typically methylated at CG sites, referred to as CG gene-body methylation. Although the function of gene-body methylation remains poorly understood, it seems to modulate gene expression and is generally positively correlated with moderate and/or constitutive expression.32–35
Common bean (Phaseolus vulgaris) is the most important grain legume for direct human consumption in the world.36 As a major source of dietary protein, minerals and certain vitamins, common bean plays a significant role in human nutrition particularly in developing countries.37 Several diseases caused by diverse pathogens including, fungi, bacteria and viruses, threaten common bean productivity. Many R genes effective against these diseases have been genetically described in common bean.38,39 The use of resistant genotypes is the most economic and ecologically safe management strategy.40 As observed in other plant species, these resistance specificities effective against various types of pathogens are often organized in large gene clusters that co-locate with NL cluster. However, the specificity of common bean genome is that these large clusters are often located at the ends (rather than in the center) of linkage groups (LG).38,39 Molecular analysis of BAC clone sequences corresponding to two of these R clusters, B4 and Co-2 clusters, located at the end of linkage groups B4 and B11, respectively, revealed that these two complex R clusters consist of a tandem array of more than 40 CNL sequences.41,42 Fluorescence in situ hybridization (FISH) analysis revealed that these clusters are located in subtelomeric regions of chromosomes 4 and 11, in the vicinity to terminal knobs.41 Moreover, phylogenetic and comparative genomics analysis strongly suggest that the CNL present at the B4 R gene cluster derived from CNL of Co-2 R gene cluster, through an ectopic recombination event. A 528-bp satellite, named khipu, has been identified in B4 cluster closely intertwined to NL sequences.41Khipu is specific to the Phaseolus genus, displays a subtelomeric distribution in 17 out of 22 chromosome ends and is a component of the terminal knobs.41 Recently, the study of khipu sequences at the scale of the common bean genome reveals the existence of frequent sequence exchanges between non-homologous chromosomes in subtelomeric regions suggesting that the case reported for B4 and Co-2 clusters is not isolated.43
The genome sequence of common bean, G19833 Andean landrace, has recently been assembled.44 Among the 27, 197 annotated protein coding genes, 376 NL genes were predicted. Moreover, recently, a genome-wide analysis of DNA methylation using MethylC-seq has been performed in P. vulgaris G19833 genome.45 CG-gene body methylation, which is found both in plants and animals, and hypermethylation of TEs in all the three sequence contexts were observed in P. vulgaris. Significantly, the satellite repeats khipu were found highly methylated throughout the genome. However, the detailed analysis of the methylation pattern of the NL genes has not yet been investigated.
In this study, we present the classification of NL encoding gene family in common bean, their phylogenetic and positional relationships, and their physical organization as well as khipu satellite locations on the 11 chromosomes of P. vulgaris. As NL encoding genes are often present in close proximity to the methylated khipu repeated sequence, we investigated the methylation status of NL encoding genes using the MethylC-seq data that were previously published,45 as well as the 24-nt small RNA targeting and the expression level of NL. Our results provide significant insight into the evolution of NL and suggest a new regulation mechanism of NL transcription that has not been described in other plant species.
2. Materials and methods
2.1. Common bean genome resources and NL and khipu sequence datasets
This study was performed using the whole v 1.0 genome sequence of G19833 genotype downloaded from Phytozome (http://www.phytozome.net/). The complete set of common bean NL sequences used in this study was the initial set of 376 NL sequences annotated in common bean genome in Schmutz et al.,44 plus one additional NL gene identified after manual inspection and named Phvul.004G015950 (Supplementary Table S1). Among these 377 NL, 364 were on pseudomolecules and 13 on scaffolds. The khipu dataset used in this study is composed of 2,766 khipu sequences annotated in Richard et al.43
2.2. Phylogenetic analysis of NL encoding genes
NL genes were subdivided into two subsets, TNL (106 sequences) and non-TNL (271 sequences) subsets and distinct phylogenetic analyzes were performed for each subset. The region spanning from the P-loop to the MHD conserved protein motifs was used to construct the phylogenetic trees. Nucleic sequences were initially aligned using ClustalW with default parameters46 and then optimized by manual editing using Seaview47 (Supplementary Figs S1 and S2). Twenty non-TNL sequences, lacking a P-loop or MHD domains or presenting a highly divergent sequence were discarded from the alignment (Supplementary Table S1). We then checked for evidence of recombination among NL using a suite of programs implemented in RDP version 4,48 using parameters described in the literaute.49 Sequences showing significant evidence of recombination were eliminated from further analysis (Supplementary Table S1). Submitting the full alignments to Modelgenerator50 allowed to identify GTR + I+G and GTR + G models as the bests phylogenetic models to use for TNL and non-TNL, respectively. Maximum-likelihood trees were generated with PhyML.51 Bootstrap values were computed with the consensus of 1,000 trees generated with PhyML from alignments obtained with PHYLIP's seqboot program.52 The resulting phylogenetic trees were displayed using MEGA version 5.53
2.3. Methylation analysis of NL encoding genes
NL methylation analysis was performed on the methylome data of G19833 leaves developed by Kim et al.45 For each gene, analyses were performed on the genomic region delimited by the coordinates presented in Supplementary Table S2. For genes not supported by RNA sequencing data (RNAseq), it corresponds to the region from Start to Stop, including introns, while for genes supported by RNAseq data, upstream and downstream untranslated region (UTR) were also included.44 Weighted methylation levels54 of the 364 NL, 498 PPR and 116 Homeobox genes present in Phaseolus vulgaris G19833 pseudomolecules were calculated for CG, CHG and CHH sequence contexts, where H refers to A, C and T. For each gene and cytosine context, probability (one-tailed P-value) for the methylation difference from genome-wide average was calculated using the binomial distribution as described by Takuno and Gaut.55 Genes were classified into three categories based on the P-value: C methylated gene (PCHG < 0.05 or PCHH < 0.05), CG methylated gene (PCG < 0.05 and not C methylated gene), unmethylated gene (PCG > 0.95 and not C methylated gene). Integrative Genomics Viewer (IGV) was used to visualize methylome data on G19833 genome.56
2.4. Small RNA library construction, sequencing and analysis
RNA samples enriched for small fractions of G19833 young leaves were obtained with miRNeasy Mini Kit (#217004, QIAGEN®) and checked for their integrity on RNANano chip, using Agilent 2100 bioanalyzer (Agilent Technologies, Waldbroon, Germany). Small RNA-seq library was performed according to NEBNext® Multiplex Small RNA Library Prep Set for Illumina instructions (#E7300S, New England Biolabs, Inc.). Small RNA-seq libraries were checked for their quality on DNA1000 chip using Agilent 2100 bioanalyzer (Waldbroon, Germany) before Illumina sequencing (Illumina® California, U.S.A.). The SmallRNA-seq samples have been sequenced in Single-End (SE) with a read length of 100 bases on one lane of Hiseq2000 machine to obtained around 25 million reads. After adaptor trimming and t- and r-RNA removing, using Cutadapt57 and SortMeRNA,58 respectively, small RNAs of 24-nt length were selected using Prinseq v0.20.4.59 Redundant and non-redundant (NR) data sets of 24-nt sRNAs were then mapped on G19833 genome sequence including khipu, NL, PPR and homeobox gene family genomic sequences using Bowtie2 (v2.2.6) with ‘-l 24’ and ‘-n 0’ parameters60 and mapped reads were counted on each locus (as described for DNA methylation) and expressed in RPKM. A threshold of RPKM >5 in both redundant and non-redundant data sets was used to consider a sequence as mapped by small 24-nt RNA.
2.5. Analysis of NB-LRR genes expression
The publicly available leaves RNA-seq data of common bean G19833 was used to analyze the expression level of NL genes.44 FPKM (Fragments per kilobase of exon model per million fragments mapped) values corresponding to NL genes were extracted from Phytozome to perform a semi-quantitative categorization. Based on the FPKM scale defined in Hansey et al.61 expressed genes were divided into three classes: genes with a FPKM value below 5 are low expressed, genes with a FPKM value greater or equal to 5 and less than or equal to 200 are medium expressed, and genes with a FPKM value greater than 200 are highly expressed.
3. Results
3.1. Genome-wide annotation of NL in G19833
The present analysis was carried out on 377 NL genes identified in Schmutz et al.,44 including 364 NL genes located on the eleven pseudomolecules and 13 NL genes located on unanchored scaffolds. In brief, common bean NB-encoding genes were identified based on HMM search of the predicted protein sequences of P. vulgaris (G19833; JGI, version 1.0) to identify sequences containing NB-ARC domain, completed by a tBLASTn of the NB-encoding sequences identified in the first step, on the entire genome of G19833.44 HMM search led to the identification of 398 predicted proteins corresponding to 342 annotated genes and tBLAStn procedure identified 35 additional NL genes. On these 35 additional NL genes, 20 were missing in the JGI annotation and consequently a new identifier was created (the last digits are 50). After manual inspection of the 377 NB-encoding R genes candidates 268 are predicted as full length genes and 109 as pseudogenes (i.e. genes with a frameshift or a stop codon before the beginning of LRR domain) (Table 1 and Supplementary Table S1).
Table 1.
Number and classification of predicted NB-encoding R genes in Phaseolus vulgaris genome
| Predicted domains | #Full length genes | #Pseudogenes | #Total | % |
|---|---|---|---|---|
| TNL type | 82 | 24 | 106 | 28.1 |
| TIR-NB-LRR | 73 | 20 | 93 | |
| TIR-NB | 8 | 4 | 12 | |
| TIR-NB-TIR | 1 | 0 | 1 | |
| non-TNL type | 186 | 85 | 271 | 71.9 |
| CC-NB-LRR | 85 | 18 | 103 | |
| CC-NB | 3 | 1 | 4 | |
| NB-LRR | 86 | 64 | 150 | |
| NB | 5 | 2 | 7 | |
| CCRPW8-NB-LRR | 7 | 0 | 7 | |
| #Total | 268 | 109 | 377 |
Analysis of each NL candidate allowed us to classify them into TNL or non-TNL families (Table 1). From the complete set of 377 NL identified in common bean genome, 106 proteins belong to the TNL group including 82 full length and 24 pseudo TNL genes. The 82 full length genes were distributed as follows: 73 TNL, 8 TN, and one TNT gene (Phvul.008G195300). The remaining 271 NB-encoding genes were collectively grouped into the non-TNL type and were distributed as follows: 103 genes encode CNL, including 85 full length and 18 pseudo genes, four genes encode CN, including three full length and one pseudo genes, 150 genes encode NL, including 86 full length and 64 pseudo genes, and finally seven genes encode NB including five full length and two pseudo genes (Table 1). In addition, within the non-TNL type, seven genes have been identified to possess a variant of the CC domain, the CCRPW8 domain which share close sequence similarity with RPW8, a non-NL R protein from Arabidopsis thaliana conferring broad-spectrum resistance against powdery mildew (Erysiphe spp.).10 Overall more than two-third of the predicted NL-encoding genes are non-TNL genes and less than one third TNL genes.
3.2. NL physical organization
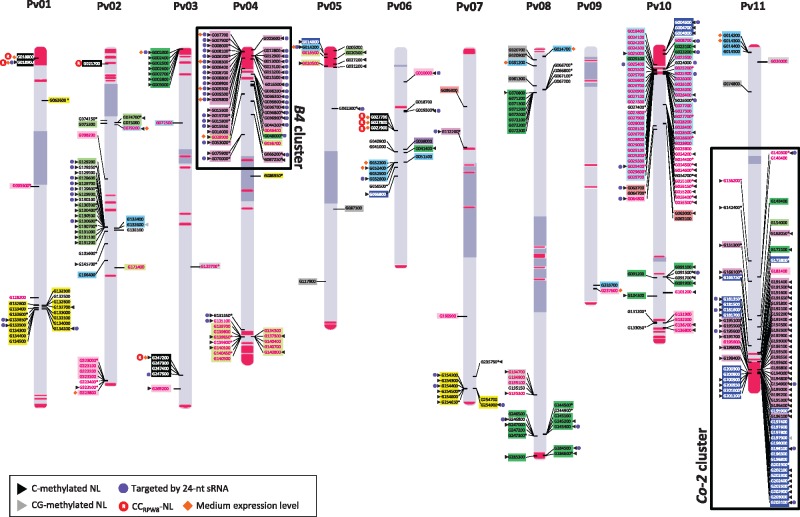
Physical map positions on the eleven pseudomolecules of G19833 genome were established for 364 (96.5%) of the annotated NL genes (Fig. 1). Both TNL and non-TNL are present on each pseudomolecule, in variable amounts. It is clear that the distribution of NL genes is not random among the chromosomes and that they tend to be organized in clusters. To identify NL clusters, we used a previous definition which specify that NL are grouped in clusters when they are not interrupted by more than eight non-NL genes.18 Using this criterion, we identified 43 clusters, containing 294 genes in total, on common bean pseudomolecules (Supplementary Table S1). Thus, on G19833 pseudomolecules, 80.8% of predicted NL encoding genes are organized in clusters while the remaining 19.2% (70 genes) are singleton genes. The size of the clusters varied across the genome from 2 to 40 NL. Remarkably, NL clusters in common bean tend to be at the end of chromosomes rather than in the center (Fig. 1). Particularly, three ‘super’ clusters of NL, consisting of several clusters, were located at the end of Pv04S (Pseudomolecule 4, short arm), Pv10S and Pv11L (Pseudomolecule 11, long arm) (Fig. 1 and Supplementary Table S1). The super cluster on Pv10S mainly consists of TNL sequences, while the two super clusters on Pv04S (41 NL) and Pv11L (71 NL) comprise mainly non-TNL sequences and correspond to the previously identified resistance cluster ‘B4’ and ‘Co-2’, respectively.39,41,62–64
Figure 1.
Chromosomal distribution of common bean NL encoding genes and khipu satellites. The relative map position of 364 NL encoding genes is shown on the individual pseudomolecules depicting the chromosomes Pv01 to Pv11 as identified in the literature.44 Each NL gene has a unique label representing the seven last informative digits from the annotation. For example, G132300 located on pseudomolecule one corresponds to the gene Phvul.001G132300. Genes encoded by the positive and negative DNA strands are depicted on the right and left sides of the chromosome, respectively. TNL sequences are presented in pink font and CNL sequences are presented in black or white font. Colours and background of the genes are identical to the phylogenetic trees (Fig. 2 and Supplementary Fig. S3). NLs corresponding to pseudogenes are denoted by an asterisk (*) after their name. Centromeric positions on individual pseudomolecules were identified using centromere satellite repeats identified in the literarure103 and are represented by dark blue bars on the pseudomolecules. The arm above centromeric region is designated as ‘short’ arm while the arm below is designated as the ‘long’ arm. Khipu satellite sequences are indicated with red bars on pseudomolecule structures according to Richard et al.,43 with block sizes proportional to number of khipu units. CG methylated genes and genes methylated in the three contexts are depicted by grey and black arrowheads, respectively.
3.3. NL phylogenetical and physical relationships
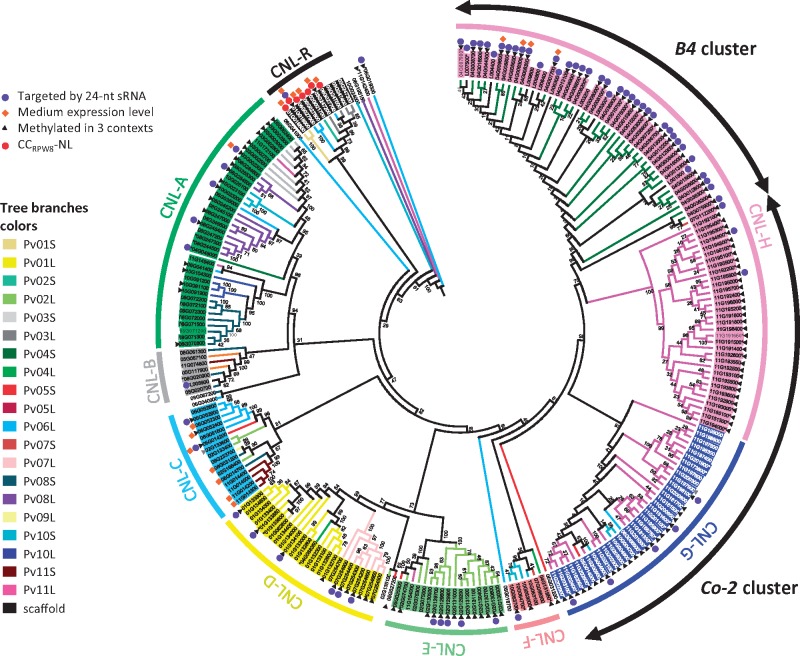
To study the evolutionary relationships among the predicted NL genes, two phylogenetic trees were estimated from the nucleic alignment of NB-ARC domains of non-TNL and TNL genes. The resulting two phylogenetic trees are composed of 243 and 100 non-TNL and TNL sequences, respectively (Fig. 2 and Supplementary Fig. S3). Non-TNL and TNL phylogenetic trees can be divided into nine and four major clades, respectively (Fig. 2 and Supplementary Fig. S3). In both trees, these clades are composed of NL sequences coming from different physical locations (Figs 1 and 2 and Supplementary Fig. S3). For example, the clade CNL-H is composed of 80 NL sequences that map, to two different chromosome ends Pv04S or the end of Pv11L and corresponding to resistance clusters B4 and Co-2, respectively. Consequently, the CNL present at the B4 cluster (B4-CNL) are more similar to the Co-2-CNL than to any other CNL in the bean genome. More heterogeneous clades are also observed. Clade CNL-C is one such example as it is composed of 16 NL sequences from seven different chromosome arms (Pv02S, Pv05S, Pv06L, Pv08S, Pv09L and Pv011S) (Figs 1 and 2). As expected the seven predicted CCRPW8-NL were gathered within the same clade (CNL-R). However, CCRPW8-NL reside on four different locations, end of Pv01S, end of Pv02S, Pv03L and Pv06L. The same observation holds true for many different CNL and TNL clades where phylogenetically close NL sequences can be spread on different chromosome arms, suggesting sequence exchange between non-homologous chromosomes. On the other hand, both non-TNL and TNL phylogenetic trees show that most subclades are composed of NL sequences from the same chromosome arm (Fig. 2 and Supplementary Fig. S3). For example, CNL-A clade is mostly composed of three subclades of NL from Pv03S, Pv08L and Pv08S, CNL-D clade is composed of two subclades of NL from Pv01L and Pv07L, CNL-E clade contains a huge subclade of Pv02L NL and TNL-A contains a subclade of Pv04L NL. If the previous examples clearly illustrate local duplication of NL sequences, clades CNL-G, CNL-H and TNL-D are even more striking (Fig. 2 and Supplementary Fig. S3). These impressive NL amplifications are clearly visible on the physical map, where blocks of NL sequences of the same colors are depicted (pink blocks at Pv04S and Pv11L, light blue block at Pv10S, and dark blue block at Pv11L, Fig. 1). In conclusion, phylogenic and positional relationships of common bean NL suggest both frequent sequence exchanges between non-homologous chromosomes and local amplification probably resulting from unequal crossing-over.
Figure 2.
Maximum Likelihood Phylogenetic analysis of the predicted P. vulgaris non-TNL genes. Each gene is labeled with its Phytozome identifiant without the ‘Phvul.’ prefix. One color was assigned to each chromosome arm named by a number (from 1 to 11), corresponding to the pseudomolecule number, followed by the letter L (long arm) or S (short arm) and consequently each branch in the tree is colored according to the location of the gene carried by this branch. Pseudogenes are indicated by an asterisk (*) in the beginning or at the end of their name. CG methylated genes and genes methylated in the three contexts are depicted by grey and black arrowheads, respectively.
3.4. NL colocalisation with the subtelomeric satellite DNA khipu
Khipu sequences annotated in Richard et al.,43 are depicted in red on the physical map of G19833 (Fig. 1). As previously shown, khipu sequences are present on all pseudomolecule ends except on Pv06S and Pv09S (Fig. 1). Khipu units are often in close proximity to NL sequences as noticeable at the end of Pv01S, Pv02S, Pv03S, Pv04S, Pv04L, Pv05S, Pv08L, Pv10S, Pv10L and Pv11L. Strikingly, there are a lot of khipu sequences in the three largest NL super clusters, on Pv04S, Pv10S and Pv11L (Fig. 1). However, several NL clusters do not co-locate with khipu blocks, such as NL clusters on chromosome 8 or 2 (Fig. 1).
3.5. NL DNA methylation
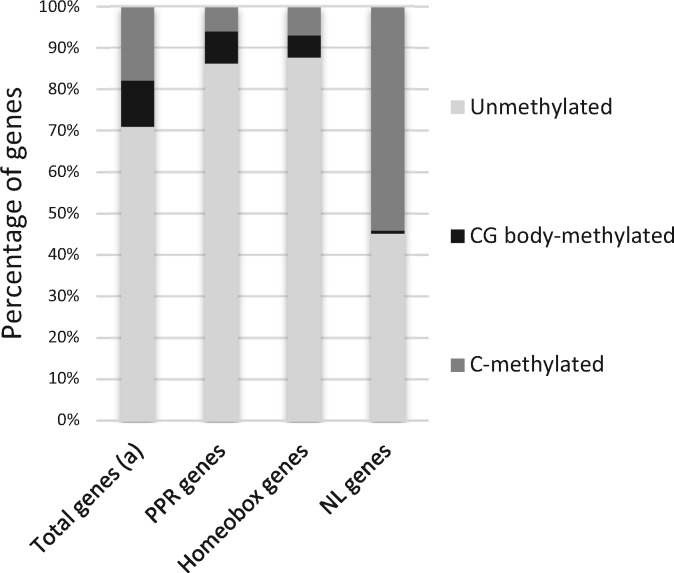
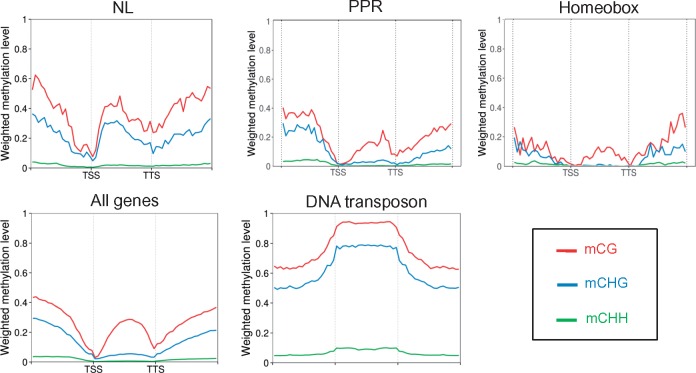
Due to their proximity to repeated and methylated khipu satellite sequences45 and, at least for B4 and Co-2 CNLs, to their proximity to terminal knobs,41 we decided to investigate the DNA methylation status of NL genes. Indeed, various examples of methylation spreading from repeated sequence to the neighboring genes have been described.65–67 DNA methylation analysis reveals that out of the 364 NL encoding genes present on the pseudomolecules, two (0.6%) are methylated in CG context, 197 (54.1%) are C-methylated genes (i.e. methylated in CHG and CHH contexts), and the remaining 165 genes (45.3%) are unmethylated (Fig. 3, Supplementary Table S1). Among the 104 TNL sequences, 31% are methylated while 64% of the 260 non-TNL sequences are methylated (Supplementary Fig. S4). Approximately, half of the full-length NL are methylated while almost 70% of the pseudogenes are methylated (Supplementary Fig. S4). This methylation mostly occurs on gene body (Fig. 4). Nearly all the C-methylated NL (89%) were also CG methylated (Supplementary Tables S1 and S3). Consequently, in common bean, ∼half of NL sequences are methylated in the three sequence contexts (CG, CHH, CHG), a pattern classically observed for repeated sequences. Manual inspection of each methylated NL genes revealed that out of 197 methylated NL, 34 correspond to NL methylated only in intron with most of these introns containing repeated sequences such as transposable elements (Supplementary Fig. S5). However, the remaining 163 C-methylated NL are methylated on their coding sequence with methylated area ranging from the complete ORF to half of the ORF (Supplementary Figs S5–S7). Inspection of nearby genes of NL genes revealed that they do not present this atypical DNA methylation profile even if located in the same genomic environment (Supplementary Fig. S7). In order to see if this high proportion of methylated genes within NL gene family is specific to this family or a common feature of gene families (that can be considered as a kind of repeated sequences), we looked at methylation pattern of two other gene families. We examined the methylation status of the 498 genes encoding PPR (pentatricopeptide repeat) protein family and the 116 genes encoding Homeobox transcription factor family in common bean genome. Three and two genes in the PPR and Homeobox family, respectively, present an intermediate level of CG methylation and were thus excluded from the following analysis. For the PPR and Homeobox gene family, only 13.6% (7.7% CG body and 5.9% C-methylated) and 13.3% (5.3% in CG body and 7% C-methylated) of the genes are methylated, respectively (Figs 3 and 4, Supplementary Table S2). Consequently, there is a much higher proportion of C-methylated genes in the NL family specifically. Moreover, NL genes present higher levels of DNA methylation when compared with Homeobox and PPR genes, whatever the sequence context (% CG: 53.43 versus 29.27 and 38.17, Wilcoxon test, all P > 0.05; % CHG: 39.44 versus 3.47 and 10.97, Wilcoxon test, all P > 0.05; %CHH: 2.72 versus 1.17 and 1.02, Wilcoxon test, all P > 0.05, for NL genes compared to Homeobox and PPR genes, respectively) (Table 2). In particular, level of CHG methylation is much higher for NL genes compared to other genes (‘all genes’, PPR and homeobox genes), although lower than what is observed for TEs (Fig. 4). In conclusion, compared to other gene families, NL gene family presents both a higher proportion of methylated genes and a higher methylation level.
Figure 3.
Distribution of the methylated status of the PPR, Homeobox and NL genes in P. vulgaris. aTotal genes analysis was described in Kim et al.45 and correspond to the 27,082 genes annotated in G19833 genome. Among the 498 studied PPR genes, three were found CG-methylated with an intermediate value and were then excluded from this analysis. Among the 116 studied Homeobox Transcription Factor genes, two were found CG-methylated with an intermediate value and were then excluded from this analysis. For the NL genes, the analysis was performed in the 364 NL genes mapped on the G19833 pseudomolecules.
Figure 4.
Average distribution of DNA methylation (for the three sequence contexts) of P. vulgaris NL, Homeobox and PPR gene families and their flanking regions. Flanking regions are 1 kb. TSS: Transcription Start Site. TTS: Transcription Termination Site. The metaplots for the 364 NL, 116 Homeobox and 498 PPR of G19833 pseudomolecules were performed in this study, using the methylome data of Kim et al.45 The metaplots corresponding to the DNA transposons and all genes were previously described in Kim et al.45
Table 2.
Average percentage of DNA methylation in the different sequence contexts in repeated elements and various gene families
| Genes | %CG | %CHG | %CHH |
|---|---|---|---|
| Total genesa | 14.65 | 6.48 | 0.58 |
| Transposable elementsa | 71.67 | 61.52 | 8.05 |
| Khipu satellite repeatsb | 93.29 | 72.06 | 5.89 |
| PPR genesc | 38.17 | 10.97 | 1.02 |
| Homeobox genesc | 29.27 | 3.47 | 1.17 |
| NL genesc | 53.43 | 39.44 | 2.72 |
aThe average percentage of methylation (see Methods) in the different sequence contexts were calculated for all genes and transposable elements annoted in Phaseolus vulgaris G19833 (in this study).
bThe average DNA methylation levels of khipu repeats were determined in Kim et al.45
cThe average percentage of methylation in NL, Homeobox and PPR genes were calculated only for the significantly methylated genes (C- and CG-methylated genes).
Phylogenetic and positional relationships of methylated NL genes have been studied and methylated NL are depicted by black arrowheads (Figs 1 and 2 and Supplementary Fig. S3). Concerning the three largest distal NL super clusters, contrasting patterns of methylation were observed since most NL sequences from the clusters on Pv04S (B4 cluster) and Pv11L (Co-2 cluster) are methylated, while few NL sequences of the cluster located on Pv10S are methylated (Figs 1 and 2 and Supplementary Fig. S3). The majority of methylated NL genes from Pv04S and Pv11L clusters belong to CNL-H clade (pink, Figs 1 and 2). Pv11L cluster is also composed of sequences located in clade CNL-G (dark blue, Figs 1 and 2) that are also methylated. Interestingly, this latter clade is also composed of the scarce methylated sequences present on the globally unmethylated Pv10S cluster. This suggests that methylation of NL encoding genes could be influenced by their sequence, as closely related sequences (e.g. belonging to the same clade) share the same methylation status. However, within the clade CNL-D (yellow, Figs 1 and 2), NL sequences located at the end of Pv07L are mostly methylated in contrast to NL sequences located on Pv01L (Figs 1 and 2). This is also the case for the clade CNL-A (green, Figs 1 and 2) where all the sequences of the end of Pv03S are methylated whereas only half of the NL of the end of Pv08L are methylated and only one sequence is methylated in the cluster of Pv08S. Interestingly, the methylated NL sequences of CNL-A clade that reside in Pv03S are organized in a cluster (cluster 03 A, Supplementary Table S1) tightly linked to khipu sequences, as the three methylated NL genes from the extremity of Pv08L in cluster 08 G (Phvul.008G284500, Phvul.008G284600, Phvul.008G285300) (Fig. 1 and Supplementary Table S1). These latest observations suggest that methylation on NL genes could also be influenced by physical location, and especially by the proximity of khipu repeats.
3.6. Common bean NL and khipu satellite DNA are massively targeted by 24-nt small RNAs
Because 24-nt sRNAs mediates RNA-directed DNA methylation (RdDM) and transcriptional gene silencing (TGS) of transposable elements, we then analyzed the relationship between khipu satellites, NL genes and 24-nt sRNA populations. 3, 087, 771 redundant and 1, 895, 583 non-redundant reads of 24-nt sRNAs were sequenced in G19833 young leaf library (Supplementary Tables S1 and S2). Khipu sequences were abundantly mapped by 24-nt sRNAs with 2596 out of 2677 khipu sequences mapped with the selected criteria (Supplementary Table S2). Ninety out of 377 (24%) NL genes are predicted to be targeted by 24-nt sRNAs. As for DNA methylation, NL gene family seems to have a different profile compared with other gene families since for PPR and Homeobox families only 7 out of 498 (1.4%) and 1 out of 116 (0.9%) are targeted by 24-nt sRNAs, respectively (Supplementary Table S2). The analysis of chromosomal repartition reveals that almost all C-methylated non-TNL sequences of the cluster located at Pv04S (B4 cluster) are also targeted by 24-nt sRNAs (Figs 1 and 2). Strikingly, a different situation is observed for the phylogenetically closely related NL sequences located at the end of Pv11L (Co-2 cluster; Clade G) that are also DNA methylated but are mostly not predicted to be targeted by 24-nt sRNAs (Figs 1 and 2).
3.7. NL expression
We used publicly available RNA-seq data of common bean G19833 to study NL expression level in young trifoliate leaf tissue. Globally, the level of expression of NL was either low (286 NL genes with RPKM < 5) or not detected (48 NL genes with RPKM = 0). Only 22 NL genes can be considered as moderately expressed (‘medium’, with RPKM comprised between 5 and 200) (Supplementary Table S1). Interestingly, with the exception of Phvul.002G021700, all the CCRPW8-NL genes belong to this ‘medium expression gene’ category. Another noteworthy element is that almost one third of the 22 moderately expressed NL genes, are from clade CNL-C (seven genes). Within the nine remaining moderately expressed NL genes, five reside on Pv04S, including one TNL and four CNL from clade CNL-H.
4. Discussion
Previously considered as an orphan crop, important genomic resources are now available in common bean thanks to the advent of next-generation sequencing (NGS) technologies.68 Based on multiple omics data from the sequenced common bean genotype G19833,44,45 our results indicate that gene movements play a crucial role in NL evolution in common bean genome and that NL present an atypical DNA methylation pattern.
4.1. NL evolution in common bean genome through both local amplification and sequence exchange between non-homologous chromosomes
We identified 377 NL-encoding genes in G19833 genome with approximately one third of TNL- and two thirds of non-TNL-encoding genes. As observed in other plant species, these NL sequences are mostly organized in clusters.18,19 In common bean most of the large NL clusters are located at the ends of the chromosomes and present a huge size compared to other species. This peculiar location has also been observed in several other plant species such as potato, tomato and cotton.19,69,70 However, this feature of NL clusters is not observed in all plant species and even in all Legume species since it has not been reported in Arabidopsis, Rice or Medicago.17,18,71 With regard to their huge size, it is particularly impressive for the NL clusters located at the end of Pv04S (B4 cluster), Pv11L (Co-2 cluster) and Pv10S, each containing more than 40 NL sequences. FISH analyses have revealed a subtelomeric location for B4 and Co-2 clusters.41 Distal regions of the chromosomes are highly recombinant compared to pericentromeric regions and are consequently favorable to promote NL amplification through unequal crossing-over.44,72,73 In agreement with that, we found that these three large NL clusters are composed of phylogenetically related NL, suggesting local amplifications of NL sequences. Similarly, in other plant species, extensive amplification of a few NL subfamilies have increased NL copy number considerably.74,75 On the other hand, some clusters are also composed of phylogenetically distant sequences, suggesting sequence exchange between non-homologous chromosomes. The present genome wide analysis shows that the B4 CNL are more similar to the Co-2 CNL that to any other CNL, confirming that the B4 cluster derives from the Co-2 cluster through an ectopic recombination between non-homologous chromosomes in subtelomeric regions.41 The same pattern has been observed in the phylogeny of the subtelomeric satellite DNA khipu, closely intermingled to NL sequences.43 These NL and khipu movement could have taken place in the context of sequence exchange between subtelomeres of non-homologous chromosomes as reported in human genome.76 Moreover, this proximity of NL with highly repeated sequences present on most of chromosome ends could promote unequal-crossing. Taking together these observations strongly suggest that in common bean, distal regions of the chromosomes are highly recombinant compared to pericentromeric regions and are consequently favorable to promote NL amplification through unequal crossing-over.
4.2. Common bean NL genes display a transposon-like methylation pattern in their coding region
MethylC-seq performed on G19833 allowed us to sensitively measure cytosine methylation within specific sequence contexts on NL genes. According to current views of DNA methylation patterns in plants, gene body methylation occurs mainly in a CG context whereas non-CG methylation is limited to repeated sequences such as transposable elements and non-protein-coding repeats.35,77–81 Contrasting with this view, we observed that more than half of common bean NL genes are methylated in their gene body not only in CG but also in CHG and CHH. Consequently, in common bean, most NL genes are methylated like transposons in their coding regions but with a lower level (Table 2 and Fig. 4). This non-CG methylation pattern seems specific to the NL gene family since it was not observed for genes belonging to other gene families such as PPR or Homeobox transcription, even if some members were also located in subtelomeric regions (Supplementary Fig. S8). Moreover, NL neighboring genes do not present this atypical DNA methylation profile even if located in the same genomic environment, strongly suggesting that this atypical methylation profile is specific to the NL gene family and is not simply due to their location (Supplementary Fig. S7). This unusual DNA methylation profile and targeting by 24-nt sRNAs of NL gene family have not been described yet in other plant species. Therefore, this analysis should be extended to other gene families in common bean as well as in other plant species to see if this phenomenon is specific of NL genes in common bean. In plants, other atypical genes displaying in their coding region a transposon-like methylation pattern have been reported including Asr1 gene in tomato82 and CRP (Cystein-Rich Peptide) gene family in Arabidopsis.83 Interestingly, a possible retrogene origin has been proposed for CRP genes.
4.3. What is responsible for DNA methylation on NL genes?
Phylogenetic and positional relationships of methylated NL genes in the G19833 pseudomolecules revealed that methylation status of NL genes could be influenced by their nucleotide sequence (as exemplified in clades CNL-G; -H, -R) or physical location. It is noteworthy that physically clustered genes often present the same methylation pattern. For example, genes of clade CNL-A are found mostly methylated in regions containing khipu repeats whereas they are mostly unmethylated in other chromosomal regions. What is responsible for DNA methylation of NL genes?
4.3.1. RdDM pathway mediated by 24-nt sRNAs
In plants, de novo cytosine methylation in the three sequence contexts and maintenance of methylation on asymmetric methylation sites (CHH) require the RdDM pathway, which involves numerous factors, including 24-nt sRNAs that guide methylation on homologous DNA sequences.22,84 While this pathway mainly targets repetitive elements, such as transposable elements, there are few reports of gene body methylation in the three contexts. One example is the CRP genes in Arabidopsis where cytosine methylation is partly dependent on the RdDM pathway83 and NL genes in common bean is an additional example. Indeed, in the present study, we showed that 90 NL genes (80/364 total NL genes mapped on G19833 pseudomolecules, 21.9%) are targeted by 24-nt sRNAs in their transcribed region. Strikingly, nearly all of them (76/80, 95%) are also highly gene-body methylated in the three sequence contexts as for example on Pv02L and Pv04S (B4 cluster) (Fig. 1 and Supplementary Fig. S9). Altogether, this suggests that in common bean, the RdDM pathway could target not only repeated sequences but also NL genes. This hypothesis is supported by the clear correlation between the NL gene body regions targeted by 24-nt sRNA and the methylated regions (Supplementary Figs S6 and S7). However, many C-methylated NL genes (61.4%; 121/197 mapped on G19833 pseudomolecules) are not targeted by 24-nt sRNAs, as for example most of the NL genes from Co-2 cluster (Pv11L), suggesting that another mechanism(s) (spreading of methylation see after), is responsible for cytosine methylation of these NL sequences. The B4 and Co-2 R clusters share many similarities: sequence content, subtelomeric physical location with proximity to khipu satellite sequences and high methylation status. However, they present a contrasting pattern regarding 24-nt sRNAs which target most of B4-NL but only few Co-2-NL (Fig. 1). Since the main difference between these two R clusters is the fact that B4 NL are young compare to Co-2 NL,41 it is tempting to speculate that this contrasting 24-nt pattern is due to their age difference. This is reminiscent of what has been described for retroelements, where RdDM, although targeted to transposons and repeats throughout the genome, is particularly notable at younger retroelements in Arabidopsis and in Gossypium raimondii.80,85–88
4.3.2. Spreading of DNA methylation from the satellite repeats, khipu
Methylated NL clustered on Pv03S, Pv04S, Pv08L and Pv11L present common features. They are all located at chromosome extremities which are enriched in the satellite sequence khipu, previously found to be highly methylated in the three sequence contexts (CG, CHG, CHH). Several studies in plants have detailed that DNA methylation can « spread » beyond repetitive elements (TEs or tandem repeats), over 200–1000 bp.89,90 Consequently, we propose that the methylation pattern observed on NL genes of these chromosomal regions could result from a methylation spreading originating from the khipu sequences, especially on Pv03S, Pv08L and Pv11L (Co-2 cluster) where methylated NL genes are not targeted by 24-nt sRNAs.
4.4. Biological role of cytosine methylation on NL genes: most of NL genes are expressed at a low level in common bean?
In plants, cytosine DNA methylation is involved in regulation of gene expression during normal development. DNA methylation in the promoter region of genes, whatever the cytosine context, is associated with a transcriptional silencing of the corresponding gene (for example FWA, SDC).90,91 Concerning DNA methylation in coding region, its influence on expression depends on the sequence context of methylated cytosine and on the methylation level. Indeed, genome-wide analyses of DNA methylation and gene expression revealed that CG-gene body methylation is positively correlated with gene expression34,35,92,93 while CHG or CHH gene body methylation can be negatively associated with gene expression levels.82,83,94 In that context, even if the biological role of the transposon-like methylation pattern of the NL gene bodies is not yet clear, we propose that it is likely related to gene silencing. Our results are consistent with these observations since 87.3% of C-methylated NL (172/197 C-methylated NL) are very poorly expressed or even not detected in leave RNA-seq data. Consequently, in addition to the well described post-transcriptional gene silencing mechanisms of NL involving microRNAs targeting NL mRNA,95–97 our results suggest the existence of an additional regulation mechanism of NL at the transcriptional level. Together, these mechanisms could be essential to down-regulate resistance expression in plant during normal growth condition, in absence of pathogen attack. Indeed, resistance genes may impose a fitness cost on host plants, and consequently their expression needs a high degree of control.16 Since increasing data suggest that DNA methylation dynamically responds to biotic stress,28,29,98 we propose that this methylation could be withdrawn in the presence of the pathogen allowing NL expression only when needed. Consequently, this peculiar subtelomeric genomic environment may favor the proliferation of large NL gene clusters due to, not only increased recombination but also to some form of silencing allowing a large amplification of NL sequences without fitness cost, as it has been proposed for F-box superfamily in Arabidopsis.99
5. Conclusion
In the present report, we have shown that NL sequences can move in the common bean genome and are methylated in the three sequence contexts like transposable elements. Another element that brings NL and transposable elements closer is their ability to damage DNA. Indeed, if this feature is well known for transposable elements that can integrate DNA after cleavage of the insertion site by a transposase or an integrase,100,101 a recent study has revealed the potential of NL proteins to bind genomic DNA in planta102 and to induce DNA damage through nicking in vitro. Altogether this suggests that NL genes could be considered as retrogenes.
Supplementary Material
Acknowledgements
We thank Christine Lelandais and Blake Meyers for fruitful discussions. This project was supported by grants from Institut National de la Recherche Agronomique, Centre National de la Recherche Scientifique, IDEEV, IFR87 and LABEX-SPS
Conflict of interest
None declared.
Supplementary data
Supplementary data are available at DNARES online.
References
- 1. Monaghan J., Zipfel C.. 2012, Plant pattern recognition receptor complexes at the plasma membrane, Curr. Opin. Plant Biol., 15, 349–57. [DOI] [PubMed] [Google Scholar]
- 2. Cui H.T., Tsuda K., Parker J. E.. 2015, Effector-Triggered Immunity: From pathogen perception to robust defense. In: Merchant, S. S. (ed), Annu. Rev. Plant Biol. , 66, 487–511. [DOI] [PubMed] [Google Scholar]
- 3. Meyers B.C., Dickerman A.W., Michelmore R.W., Sivaramakrishnan S., Sobral B.W., Young N.D.. 1999, Plant disease resistance genes encode members of an ancient and diverse protein family within the nucleotide-binding superfamily, Plant J., 20, 317–32. [DOI] [PubMed] [Google Scholar]
- 4. Dangl J.L., Jones J.D.G.. 2001, Plant pathogens and integrated defence responses to infection, Nature, 411, 826–33. [DOI] [PubMed] [Google Scholar]
- 5. McHale L., Tan X.P., Koehl P., Michelmore R.W.. 2006, Plant NBS-LRR proteins: adaptable guards, Genome Biol., 7, 212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Takken F.L.W., Goverse A.. 2012, How to build a pathogen detector: structural basis of NB-LRR function, Curr. Opin. Plant Biol., 15, 375–84. [DOI] [PubMed] [Google Scholar]
- 7. Rairdan G.J., Collier S.M., Sacco M.A., Baldwin T.T., Boettrich T., Moffett P.. 2008, The coiled-coil and nucleotide binding domains of the potato Rx disease resistance protein function in pathogen recognition and signaling, Plant Cell, 20, 739–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Collier S.M., Moffett P.. 2009, NB-LRRs work a “bait and switch” on pathogens, Trends Plant Sci., 14, 521–9. [DOI] [PubMed] [Google Scholar]
- 9. Collier S.M., Hamel L.P., Moffett P.. 2011, Cell death mediated by the N-terminal domains of a unique and highly conserved class of NB-LRR protein, Mol. Plant-Microbe Interact., 24, 918–31. [DOI] [PubMed] [Google Scholar]
- 10. Xiao S., Ellwood S., Calis O., et al. 2001, Broad-spectrum mildew resistance in Arabidopsis thaliana mediated by RPW8, Science, 291, 118–20. [DOI] [PubMed] [Google Scholar]
- 11. Porter B.W., Paidi M., Ming R., Alam M., Nishijima W.T., Zhu Y.J.. 2009, Genome-wide analysis of Carica papaya reveals a small NBS resistance gene family, Mol. Genet. Genomics , 281, 609–26. [DOI] [PubMed] [Google Scholar]
- 12. Lin X., Zhang Y., Kuang H.H., Chen J.J.. 2013, Frequent loss of lineages and deficient duplications accounted for low copy number of disease resistance genes in Cucurbitaceae, BMC Genomics, 14, 335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Arya P., Kumar G., Acharya V., Singh A.K., Ouzounis C.A.. 2014, Genome-wide identification and expression analysis of NBS-encoding genes in Malus x domestica and expansion of NBS genes family in Rosaceae, PLos One, 9, e107987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Michelmore R.W., Christopoulou M., Caldwell K.S.. 2013, Impacts of resistance gene genetics, function, and evolution on a durable future In: VanAlfen N. K. (ed), Annual Review of Phytopathology, pp. 291–319. Annual Reviews, Palo Alto. [DOI] [PubMed] [Google Scholar]
- 15. Tian D., Traw M.B., Chen J.Q., Kreitman M., Bergelson J.. 2003, Fitness costs of R-gene-mediated resistance in Arabidopsis thaliana, Nature, 423, 74–7. [DOI] [PubMed] [Google Scholar]
- 16. Purrington C.B. 2000, Costs of resistance, Curr. Opin. Plant Biol., 3, 305–8. [DOI] [PubMed] [Google Scholar]
- 17. Luo S., Zhang Y., Hu Q., et al. 2012, Dynamic Nucleotide-Binding site and Leucine-Rich Repeat-encoding genes in the grass family, Plant Physiol., 159, 197–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Meyers B.C., Kozik A., Griego A., Kuang H.H., Michelmore R.W.. 2003, Genome-wide analysis of NBS-LRR-encoding genes in Arabidopsis, Plant Cell, 15, 809–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jupe F., Pritchard L., Etherington G.J., et al. 2012, Identification and localisation of the NB-LRR gene family within the potato genome, BMC Genomics, 13, 75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lozano R., Hamblin M.T., Prochnik S., Jannink J.L.. 2015, Identification and distribution of the NBS-LRR gene family in the Cassava genome, BMC Genomics, 16, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Deleris A., Halter T., Navarro L.. 2016, DNA methylation and demethylation in plant immunity, Annu. Rev. Phytopathol., 54, 579–603. [DOI] [PubMed] [Google Scholar]
- 22. Law J.A., Jacobsen S.E.. 2010, Establishing, maintaining and modifying DNA methylation patterns in plants and animals, Nat. Rev. Genet., 11, 204–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. He X.J., Chen T., Zhu J.K.. 2011, Regulation and function of DNA methylation in plants and animals, Cell Res., 21, 442–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Matzke M. A., Mosher R. A.. 2014, RNA-directed DNA methylation: an epigenetic pathway of increasing complexity, Nat. Rev. Genet., 15, 394–408. [DOI] [PubMed] [Google Scholar]
- 25. Stroud H., Do T., Du J., et al. 2014, Non-CG methylation patterns shape the epigenetic landscape in Arabidopsis, Nat. Struct. Mol. Biol., 21, 64–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Stroud H., Greenberg M.V., Feng S., Bernatavichute Y.V., Jacobsen S.E.. 2013, Comprehensive analysis of silencing mutants reveals complex regulation of the Arabidopsis methylome, Cell, 152, 352–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Agorio A., Vera P.. 2007, ARGONAUTE4 is required for resistance to Pseudomonas syringae in Arabidopsis, Plant Cell, 19, 3778–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Dowen R.H., Pelizzola M., Schmitz R.J., et al. 2012, Widespread dynamic DNA methylation in response to biotic stress, Proc. Natl. Acad. Sci. U. S. A., 109, E2183–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lopez A., Ramirez V., Garcia-Andrade J., Flors V., Vera P.. 2011, The RNA silencing enzyme RNA polymerase V is required for plant immunity, PLoS Genet., 7, e1002434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Fultz D., Choudury S.G., Slotkin R.K.. 2015, Silencing of active transposable elements in plants, Curr. Opin. Plant Biol., 27, 67–76. [DOI] [PubMed] [Google Scholar]
- 31. Lisch D. 2009, Epigenetic regulation of transposable elements in plants, Annu. Rev. Plant Biol., 60, 43–66. [DOI] [PubMed] [Google Scholar]
- 32. Coleman-Derr D., Zilberman D.. 2012, DNA methylation, H2A.Z, and the regulation of constitutive expression, Cold Spring Harbor Symposia Quant Biol., 77, 147–54. [DOI] [PubMed] [Google Scholar]
- 33. To T.K., Saze H., Kakutani T.. 2015, DNA methylation within transcribed regions, Plant Physiol., 168, 1219–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zhang X., Yazaki J., Sundaresan A., et al. 2006, Genome-wide high-resolution mapping and functional analysis of DNA methylation in Arabidopsis, Cell, 126, 1189–201. [DOI] [PubMed] [Google Scholar]
- 35. Zilberman D., Gehring M., Tran R.K., Ballinger T., Henikoff S.. 2007, Genome-wide analysis of Arabidopsis thaliana DNA methylation uncovers an interdependence between methylation and transcription, Nat. Genet., 39, 61–9. [DOI] [PubMed] [Google Scholar]
- 36. Foyer C.H., Lam H.M., Nguyen H.T., et al. 2016, Neglecting legumes has compromised human health and sustainable food production, Nat. Plants, 2, 10. [DOI] [PubMed] [Google Scholar]
- 37. Broughton W.J., Hernandez G., Blair M., Beebe S., Gepts P., Vanderleyden J.. 2003, Beans (Phaseolus spp.)—model food legumes, Plant Soil, 252, 55–128. [Google Scholar]
- 38. Miklas P.N., Kelly J.D., Beebe S.E., Blair M.W.. 2006, Common bean breeding for resistance against biotic and abiotic stresses: from classical to MAS breeding, Euphytica, 147, 105–31. [Google Scholar]
- 39. Meziadi C., Richard M.M.S., Derquennes A., et al. 2016, Development of molecular markers linked to disease resistance genes in common bean based on whole genome sequence, Plant Sci., 242, 351–7. [DOI] [PubMed] [Google Scholar]
- 40. Bourguet D., Guillemaud T.. 2016, The Hidden and External Costs of Pesticide Use In: Lichtfouse E. (ed), Sustainable Agriculture Reviews, pp. 35–120. Springer International Publishing, Cham. [Google Scholar]
- 41. David P., Chen N.W.G., Pedrosa-Harand A., et al. 2009, A nomadic subtelomeric disease resistance gene cluster in common bean, Plant Physiol., 151, 1048–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Innes R.W., Ameline-Torregrosa C., Ashfield T., et al. 2008, Differential accumulation of retroelements and diversification of NB-LRR disease resistance genes in duplicated regions following polyploidy in the ancestor of soybean, Plant Physiol., 148, 1740–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Richard M.M.S., Chen N.W.G., Thareau V., et al. 2013, The subtelomeric khipu satellite repeat from Phaseolus vulgaris: lessons learned from the genome analysis of the Andean genotype G19833, Front. Plant Sci., 4, 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Schmutz J., McClean P.E., Mamidi S., et al. 2014, A reference genome for common bean and genome-wide analysis of dual domestications, Nat. Genet., 46, 707–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kim K.D., El Baidouri M., Abernathy B., et al. 2015, A comparative epigenomic analysis of polyploidy-derived genes in soybean and common Bean, Plant Physiol., 168, 1433–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Larkin M.A., Blackshields G., Brown N.P., et al. 2007, Clustal W and clustal X version 2.0, Bioinformatics, 23, 2947–8. [DOI] [PubMed] [Google Scholar]
- 47. Gouy M., Guindon S., Gascuel O.. 2010, SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building, Mol. Biol. Evol., 27, 221–4. [DOI] [PubMed] [Google Scholar]
- 48. Martin D.P., Lemey P., Lott M., Moulton V., Posada D., Lefeuvre P.. 2010, RDP3: a flexible and fast computer program for analyzing recombination, Bioinformatics, 26, 2462–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ashfield T., Egan A.N., Pfeil B.E., et al. 2012, Evolution of a complex disease resistance gene cluster in diploid Phaseolus and Tetraploid Glycine, Plant Physiol., 159, 336–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Keane T.M., Creevey C.J., Pentony M.M., Naughton T.J., McInerney J.O.. 2006, Assessment of methods for amino acid matrix selection and their use on empirical data shows that ad hoc assumptions for choice of matrix are not justified, BMC Evol. Biol., 6, 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Guindon S., Dufayard J.F., Lefort V., Anisimova M., Hordijk W., Gascuel O.. 2010, New algorithms and methods to estimate Maximum-Likelihood phylogenies: assessing the performance of PhyML 3.0, Syst. Biol., 59, 307–21. [DOI] [PubMed] [Google Scholar]
- 52. Felsenstein J. 1989, PHYLIP - Phylogeny Inference Package (Version 3.2), Cladistics, 5, 164–6. [Google Scholar]
- 53. Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S.. 2011, MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods, Mol. Biol. Evol., 28, 2731–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Schultz M.D., Schmitz R.J., Ecker J.R.. 2012, ′Leveling′ the playing field for analyses of single-base resolution DNA methylomes, Trends Genet., 28, 583–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Takuno S., Gaut B.S.. 2012, Body-methylated genes in Arabidopsis thaliana are functionally important and evolve slowly, Mol. Biol. Evol., 29, 219–27. [DOI] [PubMed] [Google Scholar]
- 56. Thorvaldsdottir H., Robinson J.T., Mesirov J. P.. 2013, Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration, Brief. Bioinf., 14, 178–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Martin M. 2011, Cutadapt removes adapter sequences from high-throughput sequencing reads, Embnet. J., 17, 10. [Google Scholar]
- 58. Kopylova E., Noe L., Touzet H.. 2012, SortMeRNA: fast and accurate filtering of ribosomal RNAs in metatranscriptomic data, Bioinformatics, 28, 3211–7. [DOI] [PubMed] [Google Scholar]
- 59. Schmieder R., Edwards R.. 2011, Quality control and preprocessing of metagenomic datasets, Bioinformatics, 27, 863–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Langmead B., Trapnell C., Pop M., Salzberg S.. 2009, Ultrafast and memory-efficient alignment of short DNA sequences to the human genome, Genome Biol., 10, R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Hansey C.N., Vaillancourt B., Sekhon R.S., et al. 2012, Maize (Zea mays L.) genome diversity as revealed by RNA-sequencing, Plos One, 7, e33071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Geffroy V., Sicard D., de Oliveira J.C.F., et al. 1999, Identification of an ancestral resistance gene cluster involved in the coevolution process between Phaseolus vulgaris and its fungal pathogen Colletotrichum lindemuthianum, Mol. Plant-Microbe Interact, 12, 774–84. [DOI] [PubMed] [Google Scholar]
- 63. Geffroy V., Macadre C., David P., et al. 2009, Molecular analysis of a large subtelomeric Nucleotide-Binding-Site-Leucine-Rich-Repeat family in two representative genotypes of the major gene pools of Phaseolus vulgaris, Genetics, 181, 405–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Creusot F., Macadre C., Cana E.F., et al. 1999, Cloning and molecular characterization of three members of the NBS-LRR subfamily located in the vicinity of the Co-2 locus for anthracnose resistance in Phaseolus vulgaris, Genome, 42, 254–64. [DOI] [PubMed] [Google Scholar]
- 65. Martin A., Troadec C., Boualem A., et al. 2009, A transposon-induced epigenetic change leads to sex determination in melon, Nature, 461, 1135–U1237. [DOI] [PubMed] [Google Scholar]
- 66. Talbert P.B., Henikoff S.. 2006, Spreading of silent chromatin: inaction at a distance, Nat. Rev. Genet., 7, 793–803. [DOI] [PubMed] [Google Scholar]
- 67. Vitte C., Fustier M.A., Alix K., Tenaillon M.I.. 2014, The bright side of transposons in crop evolution, Brief. Funct. Genomics., 13, 276–95. [DOI] [PubMed] [Google Scholar]
- 68. Varshney R.K., Ribaut J.M., Buckler E.S., Tuberosa R., Rafalski J.A., Langridge P.. 2012, Can genomics boost productivity of orphan crops?, Nat. Biotechnol., 30, 1172–6. [DOI] [PubMed] [Google Scholar]
- 69. Andolfo G., Jupe F., Witek K., Etherington G.J., Ercolano M.R., Jones J.D.G.. 2014, Defining the full tomato NB-LRR resistance gene repertoire using genomic and cDNA RenSeq, BMC Plant Biol., 14, 120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Wei H., Li W., Sun X., Zhu S., Zhu J., Wu R.. 2013, Systematic analysis and comparison of nucleotide-binding site disease resistance genes in a diploid cotton Gossypium raimondii, PLos One, 8, e68435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Young N.D., Debellé F., Oldroyd G.E.D., et al. 2011, The Medicago genome provides insight into the evolution of rhizobial symbioses, Nature, 480, 520–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Gore M.A., Chia J.M., Elshire R.J., et al. 2009, A first-generation haplotype map of Maize, Science , 326, 1115–7. [DOI] [PubMed] [Google Scholar]
- 73. Schmutz J., Cannon S.B., Schlueter J., et al. 2010, Genome sequence of the palaeopolyploid soybean, Nature, 463, 178–83. [DOI] [PubMed] [Google Scholar]
- 74. Seo E., Kim S., Yeom S.I., Choi D.. 2016, Genome-wide comparative analyses reveal the dynamic evolution of nucleotide-binding Leucine-Rich Repeat Gene Family among Solanaceae Plants, Front. Plant Sci., 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Wei C., Chen J., Kuang H., Wu K.. 2016, Dramatic number variation of R genes in Solanaceae species accounted for by a few R Gene Subfamilies, PLos One, 11, e0148708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Linardopoulou E.V., Williams E.M., Fan Y.X., Friedman C., Young J.M., Trask B.J.. 2005, Human subtelomeres are hot spots of interchromosomal recombination and segmental duplication, Nature, 437, 94–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Saze H., Kakutani T.. 2011, Differentiation of epigenetic modifications between transposons and genes, Curr. Opin. Plant Biol., 14, 81–7. [DOI] [PubMed] [Google Scholar]
- 78. Cokus S.J., Feng S., Zhang X.. 2008, Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning, Nature, 452, 215–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Feng S., Cokus S.J., Zhang X., et al. 2010, Conservation and divergence of methylation patterning in plants and animals, Proc. Natl. Acad. Sci. U.S.A, 107, 8689–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Lister R., O'Malley R.C., Tonti-Filippini J., et al. 2008, Highly integrated single-base resolution maps of the epigenome in Arabidopsis, Cell, 133, 523–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Zemach A., McDaniel I.E., Silva P., Zilberman D.. 2010, Genome-wide evolutionary analysis of eukaryotic DNA methylation, Science, 328, 916–9. [DOI] [PubMed] [Google Scholar]
- 82. Gonzalez R.M., Ricardi M.M., Iusem N.D.. 2011, Atypical epigenetic mark in an atypical location: cytosine methylation at asymmetric (CNN) sites within the body of a non-repetitive tomato gene, BMC Plant Biol., 11, 94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. You W., Tyczewska A., Spencer M., et al. 2012, Atypical DNA methylation of genes encoding cysteine-rich peptides in Arabidopsis thaliana, BMC Plant Biol., 12, 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Saze H., Tsugane K., Kanno T., Nishimura T.. 2012, DNA methylation in plants: relationship to small RNAs and histone modifications, and functions in transposon inactivation, Plant Cell Physiol., 53, 766–84. [DOI] [PubMed] [Google Scholar]
- 85. Matzke M. A., Kanno T., Matzke A. J. M.. 2015, RNA-directed DNA methylation: the evolution of a complex epigenetic pathway in flowering plants In: Merchant S. S. (ed), Annual Review of Plant Biology, pp. 243–267. Annual Reviews, Palo Alto. [DOI] [PubMed] [Google Scholar]
- 86. Zhong X.H., Hale C.J., Law J.A., et al. 2012, DDR complex facilitates global association of RNA polymerase V to promoters and evolutionarily young transposons, Nat. Struct. Mol. Biol., 19, 870–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Gong L., Masonbrink R.E., Grover C.E., Renny-Byfield S., Wendel J.F.. 2015, A cluster of recently inserted transposable elements associated with siRNAs in Gossypium raimondii, Plant Genome, 8, 8. [DOI] [PubMed] [Google Scholar]
- 88. Hollister J.D., Smith L.M., Guo Y.L., Ott F., Weigel D., Gaut B.S.. 2011, Transposable elements and small RNAs contribute to gene expression divergence between Arabidopsis thaliana and Arabidopsis lyrata, Proc. Natl. Acad. Sci. U. S. A, 108, 2322–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Ahmed I., Sarazin A., Bowler C., Colot V., Quesneville H.. 2011, Genome-wide evidence for local DNA methylation spreading from small RNA-targeted sequences in Arabidopsis, Nucl. Acids Res., 39, 6919–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Henderson I.R., Jacobsen S.E.. 2008, Tandem repeats upstream of the Arabidopsis endogene SDC recruit non-CG DNA methylation and initiate siRNA spreading, Genes Develop, 22, 1597–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Chan S.W., Zilberman D., Xie Z., Johansen L.K., Carrington J.C., Jacobsen S.E.. 2004, RNA silencing genes control de novo DNA methylation, Science , 303, 1336. [DOI] [PubMed] [Google Scholar]
- 92. Li X., Zhu J., Hu F., et al. 2012, Single-base resolution maps of cultivated and wild rice methylomes and regulatory roles of DNA methylation in plant gene expression, BMC Genomics, 13, 300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Schmitz R.J., Schultz M.D., Urich M.A., et al. 2013, Patterns of population epigenomic diversity, Nature, 495, 193–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Schmitz R.J., He Y., Valdes-Lopez O., et al. 2013, Epigenome-wide inheritance of cytosine methylation variants in a recombinant inbred population, Genome Res., 23, 1663–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Li F., Pignatta D., Bendix C., et al. 2012, MicroRNA regulation of plant innate immune receptors, Proc. Natl. Acad. Sci. U. S. A, 109, 1790–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Shivaprasad P.V., Chen H.M., Patel K., Bond D.M., Santos B.A., Baulcombe D.C.. 2012, A microRNA superfamily regulates nucleotide binding site-leucine-rich repeats and other mRNAs, Plant Cell, 24, 859–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Gonzalez V.M., Muller S., Baulcombe D., Puigdomenech P.. 2015, Evolution of NBS-LRR gene copies among dicot plants and its regulation by members of the miR482/2118 superfamily of miRNAs, Mol. Plant, 8, 329–31. [DOI] [PubMed] [Google Scholar]
- 98. Yu A., Lepere G., Jay F., et al. 2013, Dynamics and biological relevance of DNA demethylation in Arabidopsis antibacterial defense. Proc. Natl. Acad. Sci. U. S. A, 110, 2389–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Hua Z.H., Pool J.E., Schmitz R.J., et al. 2013, Epigenomic programming contributes to the genomic drift evolution of the F-Box protein superfamily in Arabidopsis, Proc. Natl. Acad. Sci. U. S. A, 110, 16927–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Wicker T., Sabot F., Hua-Van A., et al. 2007, A unified classification system for eukaryotic transposable elements, Nat. Rev. Genet., 8, 973–82. [DOI] [PubMed] [Google Scholar]
- 101. Sultana T., Zamborlini A., Cristofari G., Lesage P.. 2017, Integration site selection by retroviruses and transposable elements in eukaryotes, Nat. Rev. Genet., 18, 292–308. [DOI] [PubMed] [Google Scholar]
- 102. Fenyk S., Townsend P.D., Dixon C.H., et al. 2015, The potato Nucleotide-binding Leucine-rich Repeat (NLR) immune receptor Rx1 is a pathogen-dependent DNA-deforming protein, J. Biol. Chem., 290, 24945–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Iwata A., Tek A.L., Richard M.M.S., et al. 2013, Identification and characterization of functional centromeres of the common bean, Plant J., 76, 47–60. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.