Abstract
Dicer partner proteins Drosophila Loquacious-PB (Loqs-PB) and human TRBP tune the length of miRNAs produced by Dicer from a subset of pre-miRNAs and thereby alter their target repertoire, by an unknown mechanism. Here, we developed a novel high-throughput method that we named Dram-seq (Dice randomized pre-miRNA pool and seq) to study length distributions of miRNAs produced from thousands of different pre-miRNA variants. Using Dram-seq, we found that a base-mismatch in the pre-miRNA stem can alter the length of miRNAs compared with a base-pair at the same position in both Drosophila and human, and is important for the miRNA length tuning by Loqs-PB. Loqs-PB directly bound base-mismatched nucleotides in the pre-miRNA stem. We speculate that Loqs-PB tunes miRNA length by changing the conformation of base-mismatched nucleotides in the pre-miRNA stem to that of base-paired ones and thereby altering the distance of the pre-miRNA stem.
INTRODUCTION
MicroRNAs (miRNAs) regulate expression of target mRNAs in a sequence-specific manner and thus play important roles in various aspects of biological processes. miRNAs exist as several distinct isoforms called isomiRs, which exhibit heterogeneous ends and lengths (1,2). Cellular isomiR profiles change dynamically during animal development and cell differentiation (3,4). Dysregulated isomiR profiles are linked with cancers (5–9). Different isomiRs produced from the same precursor molecules regulate different mRNAs since they can have different seed sequence, which is located at nucleotide positions 2–8 counting from the 5′ end and determines the target repertoire of miRNAs (10,11). Also, alternative isomiR production can lead to alternative guide miRNA strand selection onto Argonaute for incorporation in a silencing complex, changing the target repertoire (12,13). Furthermore, miRNA length defines the reprogramming of the effector Argonaute complex; the non-canonical 22 nt length of miR-173 and miR-828 compared with the canonical 21 nt length of the majority of other miRNAs reprograms the Argonaute complex for production of secondary siRNAs called trans-acting siRNAs (tasiRNAs) in Arabidopsis (14–16). These studies underscore the importance of miRNA length and indicate that isomiR production is a regulated process leading to modulation of the regulatory potential of miRNAs.
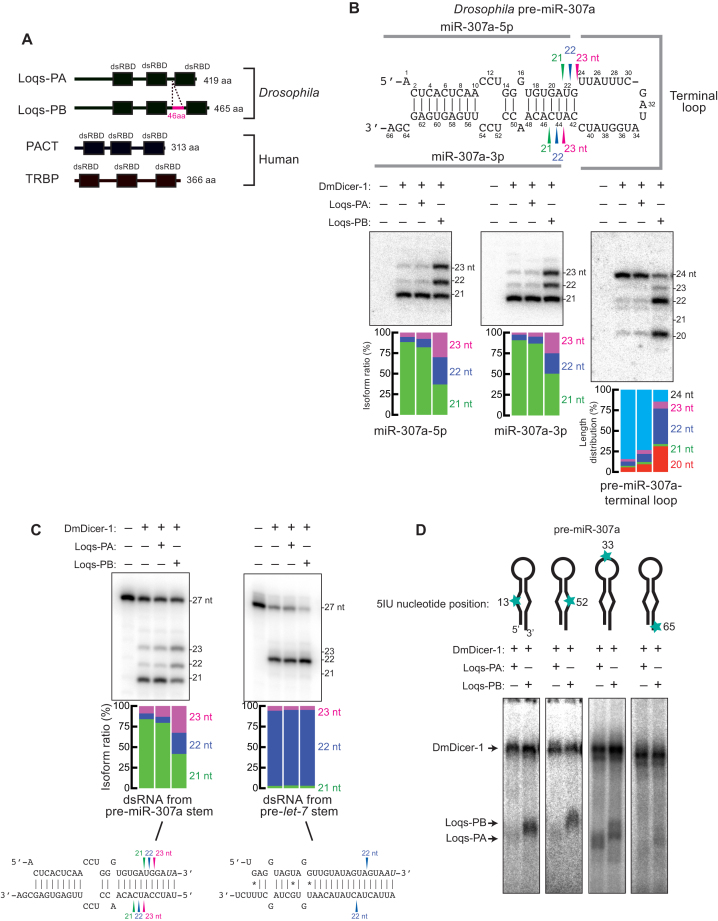
The Dicer enzymes produce miRNAs from their RNA hairpin precursors called pre-miRNAs. Both in Drosophila and human, Dicer associates with alternative partner proteins containing three dsRNA-binding domains (dsRBDs). Drosophila Dicer-1 (DmDicer-1) can bind Loquacious-PA (Loqs-PA) or Loquacious-PB (Loqs-PB), which are produced from the alternatively spliced isoforms of loqs mRNA (Figure 1A) (17–19). Compared with Loqs-PA, Loqs-PB has additional 46 aa residues in the linker between the second and third dsRBDs. Human Dicer (HsDicer) can bind PACT or TRBP (20–22). The first two dsRBDs of these Dicer partner proteins bind RNA substrates, and the third dsRBD binds the N-terminal helicase domain of Dicer (13,21,23–25).
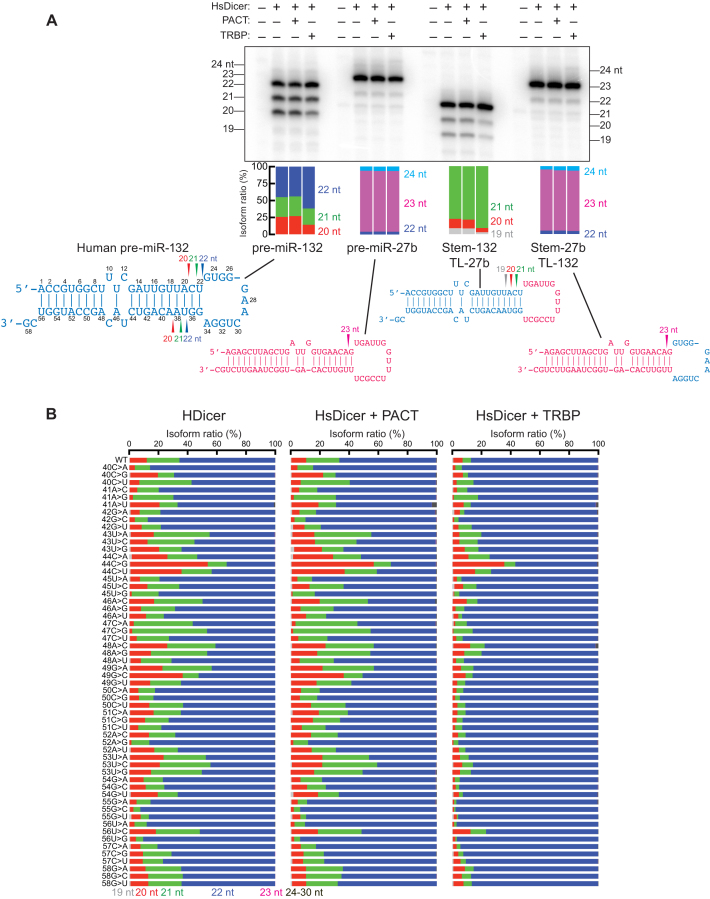
Figure 1.
Loqs-PB, but not Loqs-PA, binds the stem of pre-miR-307a and tunes the length of miR-307a. (A) Domain structures of Drosophila Loqs-PA, Drosophila Loqs-PB, human PACT, and human TRBP. Drosophila Loqs-PB has additional 46 aa residues in the linker between the second and third dsRBDs, compared with Loqs-PA. (B) In vitro dicing of 100 nM pre-miR-307a by 8 nM DmDicer-1 ± Loqs-PA or Loqs-PB for 120 min. 5′ arm-derived products (miR-307a-5p isomiRs), 3′ arm-derived products (miR-307a-3p isomiRs), and the terminal loop-derived products were detected by northern blot. DNA oligo probes 5′-CACACCCAGGTTGAGTGAGT-3′, 5′-TCGCTCACTCAAGGAGGTTG-3′, and 5′-TGGATACCATATCGAAATAA-3′ were used to detect miR-307–5p, miR-307–3p, and terminal loop, respectively. Representative gel images and length distributions revealed by quantification of gels (means of at least three independent trials) are shown. (C) In vitro dicing of 100 nM dsRNAs by 8 nM DmDicer-1 ± Loqs-PA or Loqs-PB for 120 min. The top strands are 5′ 32P-radiolabeled. The other end of the top strands was blocked by two deoxynucleotides (30). Representative gel images and length distributions revealed by quantification of gels (means of at least three independent trials) are shown. (D) Site-specific UV crosslinking of 5′ 32P-radiolabeled pre-miR-307a to 10 nM DmDicer-1 + Loqs-PA or Loqs-PB. Pre-miR-307a containing 5IU nucleotide modification at nucleotide positions 13, 52, 33 and 65 were used from left to right.
Previous studies by us and others showed that Drosophila Loqs-PB and human TRBP, but not Drosophila Loqs-PA or human PACT, regulate lengths of miRNAs produced from a subset of pre-miRNAs, producing isomiRs (10,12,13,26). For example, DmDicer-1 bound with Loqs-PB produces longer miR-307a isoforms (22 and 23 nt) than that produced by DmDicer-1 alone or DmDicer-1 bound with Loqs-PA (21 nt) in vitro and in vivo (10). Similarly, HsDicer bound with TRBP can produce a longer miR-132 isoform (22 nt) more than HsDicer alone or HsDicer bound with PACT does (10). Previous studies suggested biological significance and health relevance of isomiR production regulated by Dicer partner proteins. Loss of TRBP is associated with cancers, cardiac diseases, and spermatogenesis defect in mammals (27,28). Loss of Loqs-PB isoform causes female infertility and loss of germline stem cells in Drosophila (10,29). Better understanding of the regulatory mechanism in isomiR production by Dicer partner proteins will improve our understanding of the mechanisms by which cells modulate the regulatory potential of miRNAs. However, the molecular mechanism by which Loqs-PB and TRBP, but not Loqs-PA and PACT, tune lengths of miRNAs produced from a subset of pre-miRNAs is not understood.
In this study, we attempted to gain deeper insights into the molecular mechanism by which Loqs-PB and TRBP tune miRNA length. We developed a novel high-throughput method that we named Dram-seq (Dice randomized pre-miRNA pool and seq), which can determine length distributions of miRNA isoforms produced from thousands of different pre-miRNA variants. Using Dram-seq, we found that a base-mismatch in the pre-miRNA stem can alter miRNA length compared with a base-pair in both Drosophila and human. We also found that a base-mismatch can enable Loqs-PB to tune miRNA length. We speculate that Loqs-PB tunes miRNA length by changing the conformation of base-mismatched nucleotides in the pre-miRNA stem to that of base-matched ones.
MATERIALS AND METHODS
In vitro dicing assay
Recombinant proteins of Drosophila Dicer-1, Loqs-PA and Loqs-PB and human Dicer, PACT and TRBP were purified as described (10). In vitro dicing reaction was performed as described (10,30–32). All RNAs were gel-purified. Lengths of miRNA products were examined on urea-PAGE gels by using either 5′ 32P-radiolabeled pre-miRNA or non-radiolabeled pre-miRNA followed by northern blot, as described (10). At least three independent experiments were performed. Dried gels and membranes were exposed to image plates and analyzed with FLA-9500 and ImageQuant (GE Healthcare).
Site-specific crosslinking assay
Site-specific crosslinking assay was performed in a similar manner as previously described (31). 10 nM DmDicer-1 + Loqs-PA or Loqs-PB were incubated with a trace amount of 5′ 32P-radiolabeled pre-miR-307a containing 5-iodouridine (5IU) at nucleotide positions 13, 33 or 52, or 65 or pre-miR-999 variant containing mismatches at nucleotide positions 6 and 14 and 5IU at nucleotide positions 6, 14 or 29 in the buffer [30 mM Hepes–KOH (pH 7.4), 100 mM potassium acetate, 5 mM EDTA and 5 mM DTT] on ice for 30 min. After taking aliquots of the samples, the remaining mixtures were exposed to 302 nm UV light for 5 min, and were run on SDS-PAGE gels. The aliquots of the samples were run on a urea-PAGE gel, which confirmed that the 32P-radiolabeled pre-miRNAs were not cleaved during the incubation. At least three independent experiments were performed. Dried gels were exposed to image plates and analyzed with FLA-9500 and ImageQuant (GE Healthcare).
Fly strains
The pri-miR-307a transgene was generated by subcloning the 688 bp pri-miR-307a sequence comprised of the 66 bp pre-miR-307a sequence and 312 bp upstream and 310 bp downstream flanking regions, into a pUASPattB plasmid vector. The point mutations were introduced using PCR. The transgenes were integrated at the position attP2 on the third chromosome, using the BDSC fly strain 8622. A miR-307 null fly strain was created using the CRISPR system (33) and was used as trans-heterozygous with the previously published, independent miR-307 null strain (BDSC 58920) (34). We call the trans-heterozygous flies as miR-307anull. 3–5-day-old female flies fed with wet yeast paste were used for analysis.
Dram-seq
Partly randomized pre-miRNA pools containing randomized sequences at a defined ratio were prepared by chemical synthesis (Trilink). The 5′ arm-randomized pre-miR-307 pool comprised pre-miR-307 containing 85% wild-type sequence and 5% each of the three mutated sequences (‘85:5:5:5 ratio’) at nucleotide positions 1–20. For example, for position 1, whose wild-type sequence is A, 85% of the pre-miR-307 molecule in the pool has A at this position, 5% C, 5% G and 5% U. This ratio was chosen so that most of the pre-miRNA molecules in the pool have up to two point mutations compared with the wild-type sequence. The 3′ arm-randomized pre-miR-307 pool comprised pre-miR-307 containing the 85:5:5:5 ratio sequences at nucleotide positions 47–66. The 5′ arm-randomized pre-miR-132 pool comprised pre-miR-132 containing the 85:5:5:5 ratio sequences at nucleotide positions 1–17. The 3′ arm-randomized pre-miR-132 pool comprised pre-miR-132 containing the 85:5:5:5 ratio sequences at nucleotide positions 40–58. The 5′ arm-randomized pre-miR-282 pool comprised pre-miR-282 containing the 85:5:5:5 ratio sequences at nucleotide positions 1–19. The 3′ arm-randomized pre-miR-999 pool comprised pre-miR-999 containing the 85:5:5:5 ratio sequences at nucleotide positions 40–60.
Pre-miRNA pool was incubated with the recombinant Dicer and Dicer partner proteins as described (10). The miRNA products were gel-purified by urea-PAGE.
In this study, ‘nucleotide position’ refers to the position within a pre-miRNA primary sequence counting from the 5′ end, while ‘stem position’ refers to the position within the pre-miRNA stem, which is comprised of two nucleotides, one from the 5′ arm and the other from the 3′ arm of pre-miRNA. For example, stem position 13 of pre-miR-307a is comprised of the nucleotides at nucleotide positions 13 and 52.
Small RNA sequencing
Small RNA libraries were prepared and sequenced on Hiseq4000 (Illumina), as previously described (10,31,35–38). SRA accession number for the small RNA libraries reported in this paper is SRP102235.
RESULTS
The stem of pre-miR-307a is enough and the terminal loop is dispensable for Loqs-PB to tune miR-307a length
Recombinant DmDicer-1 protein bound with Loqs-PB (DmDicer-1 + Loqs-PB) produced longer miR-307a isoforms from pre-miR-307a compared with DmDicer-1 alone and DmDicer-1 bound with Loqs-PA (DmDicer-1 ± Loqs-PA) (Figure 1B, miR-307a-5p and miR-307a-3p) (10). From 66 nt long pre-miR-307a, DmDicer-1 ± Loqs-PA predominantly produced a 21 nt isoform of miR-307a, while DmDicer-1 + Loqs-PB produced longer (22 nt and 23 nt) isoforms of miR-307a. Consistent with the idea that this was caused by alternative DmDicer-1 cleavage positions within pre-miR-307a, the length of the terminal loop-derived product became shorter when DmDicer-1 was bound with Loqs-PB (Figure 1B, terminal loop). DmDicer-1 ± Loqs-PA predominantly produced a 24 nt long terminal loop-derived fragment, while DmDicer-1 + Loqs-PB produced more 20 and 22 nt long fragments.
DmDicer-1 produces the 22 nt miRNA isoform from pre-let-7 in the presence or absence of Loqs-PB in vitro (10). Using swap chimera pre-miRNAs between pre-miR-307a and pre-let-7, in which either the stem or the terminal loop of pre-miRNAs was swapped, we previously showed that the stem, but not the terminal loop, of pre-miR-307a enables Loqs-PB to tune miRNA length (10). To test this idea further, using dsRNA substrates lacking the terminal loop, we examined whether the stem of pre-miR-307a is enough for Loqs-PB to tune miRNA length. DmDicer-1 + Loqs-PB produced the longer miR-307a isoforms (22 and 23 nt) from the 27 bp dsRNA substrate derived from the pre-miR-307a stem more than DmDicer-1 ± Loqs-PA, which produced predominantly the 21 nt isoform (Figure 1C). In contrast, DmDicer-1 + Loqs-PB produced the same 22 nt isoform from the 27 bp dsRNA substrate derived from the pre-let-7 stem as that produced by DmDicer-1 ± Loqs-PA (Figure 1C). We concluded that the stem of pre-miR-307a is enough and the terminal loop is dispensable for Loqs-PB to tune the miRNA length.
Base-mismatched nucleotides in the pre-miR-307a stem are bound by Loqs-PB, but not Loqs-PA
Using site-specific UV-crosslinking, we identified the positions in pre-miR-307a to which Loqs-PA and Loqs-PB directly bind. Loqs-PB, but not Loqs-PA, was crosslinked to pre-miR-307a containing a UV-crosslinkable base modification (5-iodouridine [5IU]) at nucleotide position 13 or 52, which are base-mismatched and are located in the central region of the 5′ and 3′ arms, respectively (Figure 1D). In contrast, both Loqs-PA and Loqs-PB were crosslinked to pre-miR-307a containing the UV-crosslinkable base modification at nucleotide positions 33, which is located in the terminal loop (Figure 1D). Neither Loqs-PA nor Loqs-PB was efficiently crosslinked to pre-miR-307a containing the UV-crosslinkable base modification at nucleotide positions 65, which is located at the 3′ overhang (Figure 1D). DmDicer-1 was crosslinked efficiently to all four modified pre-miR-307a. These results indicated that Loqs-PB, but not Loqs-PA, directly binds base-mismatched nucleotides at the central region of the pre-miR-307a stem, while both bind the terminal loop.
Dram-seq: an unbiased and high-throughput method to study lengths of miRNAs produced from pre-miRNAs
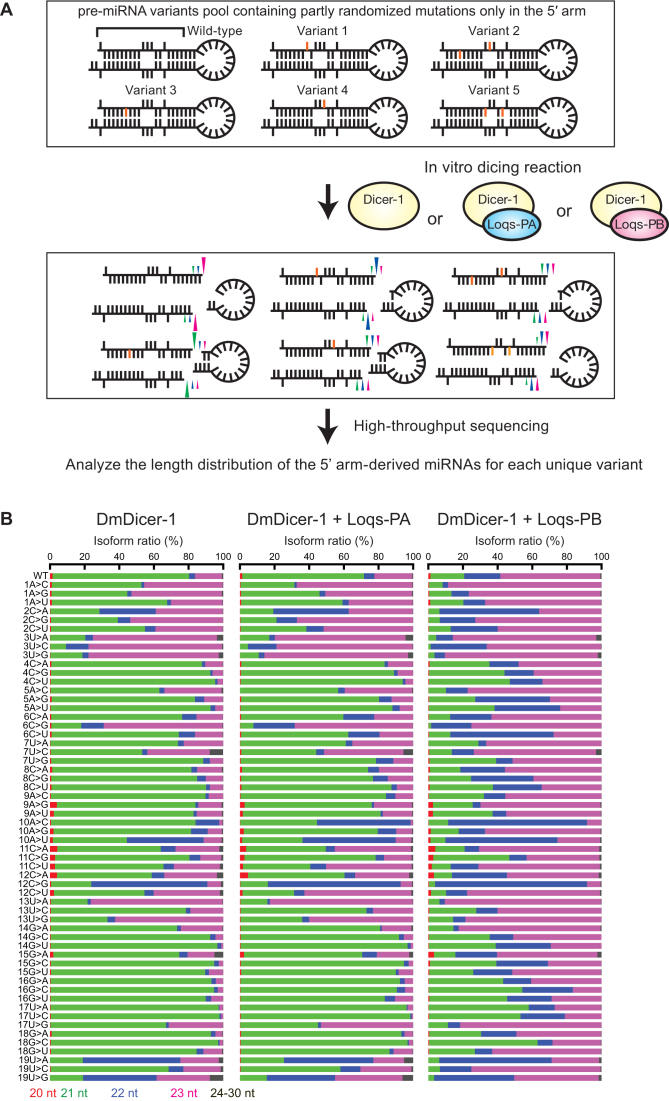
Our results suggested that Loqs-PB, but not Loqs-PA, tunes miR-307a length using a signature(s) present in the stem of pre-miR-307a. To identify the signature, we examined miRNA lengths produced from pre-miR-307a variants. We developed Dram-seq to study lengths of miRNA isoforms produced from thousands of different pre-miRNA variants in an unbiased and high-throughput manner (Figure 2A). Dram-seq uses a partially sequence randomized pre-miRNA variants pool as substrates for miRNA generation by Dicer. We used partially sequence-randomized pre-miR-307a variants pool as substrates for in vitro dicing reaction by recombinant DmDicer-1 ± Loqs-PA or Loqs-PB in Dram-seq. We prepared the 5′ arm-randomized pre-miR-307a variants pool in which each of the sequences at nucleotide positions 1–20 (5′ arm) of pre-miR-307a is randomized at a defined ratio, so that 85% of pre-miR-307a variant molecules in the pool have the wild-type sequence (A in the case of the position 1) and 5% each for mutated sequence (C, G, and U at the position 1) at each position. We performed in vitro dicing reaction using this pre-miR-307a variants pool and DmDicer-1 ± Loqs-PA or Loqs-PB, followed by gel-purification, cloning, and high-throughput sequencing of produced miRNAs. The randomization ratio (85% wild-type and 5% each for three mutated sequences) was determined based on our theoretical simulation to produce reasonably diverse variants and cover them with enough sequence reads (Supplementary Figure S1). The fraction for 3-point mutants was expected to become highest around this ratio (Supplementary Figure S1A). With this defined ratio, most of the unique pre-miRNA variant species in the pool that had significantly large enough fraction in the sequenced reads contained up to two point mutations (Supplementary Figure S1B). Too many point mutations per pre-miRNA molecule can vastly increase the diversity of the population and thus decrease the number of sequence reads per unique variant. Also, too many point mutations can result in an unstable or broken hairpin structure of pre-miRNA.
Figure 2.
Dram-seq. (A) Schematic representation of Dram-seq. An example using the 5′ arm-randomized pre-miRNA variants pool is shown. (B) Length distributions of miR-307a-5p isoforms produced from wild-type pre-miR-307a and its variants containing a single point mutation at nucleotide positions 1–19 by DmDicer-1 ± Loqs-PA or Loqs-PB, revealed by Dram-seq.
Since random mutations were introduced only at nucleotide positions 1–20 in this pool, only miRNAs derived from the 5′ arm of pre-miRNA (miR-307a-5p), but not miRNAs derived from the 3′ arm (miR-307a-3p) or the fragments derived from the terminal loop, contained mutations. Thus, for each miR-307a-5p sequence read, the entire sequence of the precursor pre-miR-307a variant can be determined. Then length distributions of miR-307a-5p isoforms produced from each of thousands of different pre-miR-307a variants can be determined. Using this approach, we could determine length distributions of miR-307a isoforms produced from wild-type pre-miR-307a, from 60 pre-miR-307a variants containing a single point mutation at nucleotide positions 1–20, and from 1710 pre-miR-307a variants containing double point mutations (Figure 2B and Supplementary Table S1). miRNA length distributions from many, but not all possible 30,780 variants containing three point mutations were also determined. Similarly, we performed Dram-seq using 3′ arm-randomized pre-miR-307 variants pool, in which nucleotide positions 47–66 were partially randomized at the defined ratio (85% wild-type and 5% each mutation), and determined length distributions of miRNA isoforms produced from wild-type, 60 single point mutant variants, 1710 double point mutant variants, and many (but not all possible 30 780) kinds of triple point mutant variants of pre-miR-307a (Supplementary Figure S2). Among the 10 626 miRNA length distributions determined from the 3541 pre-miRNA variants containing up to two point mutations by DmDicer-1 ± Loqs-PA or Loqs-PB, the mean and median number of reads for miRNAs were 4281 and 1512, respectively. 95.9% (10 188/10 626), 99.7% (10 592/10 626), and 100% (10 626/10 626) of them had the number of reads of at least 100, 20 and 2, respectively. Among the additional 184,680 miRNA length distributions potentially produced from the theoretically possible 61 560 pre-miRNA variants containing triple point mutations by DmDicer-1 ± Loqs-PA or Loqs-PB, 49.6% (91 662/184 680), 82.1% (151 694/184 680) and 98.2% (181 400/184 680) had the number of reads of at least 100, 20 and 2, respectively.
Importantly, the relative abundance of different variants in the initial pre-miRNA substrate pools or processed miRNAs does not influence length distributions of miRNAs produced from each unique pre-miRNA variant determined by Dram-seq. Therefore, even if there was any bias in a pre-miRNA substrate pool or if some mutations positively or negatively affected Dicer processing efficiency, it would not affect length distributions of miRNA isoforms determined by Dram-seq.
Dram-seq results are consistent with individual gel-based assay results
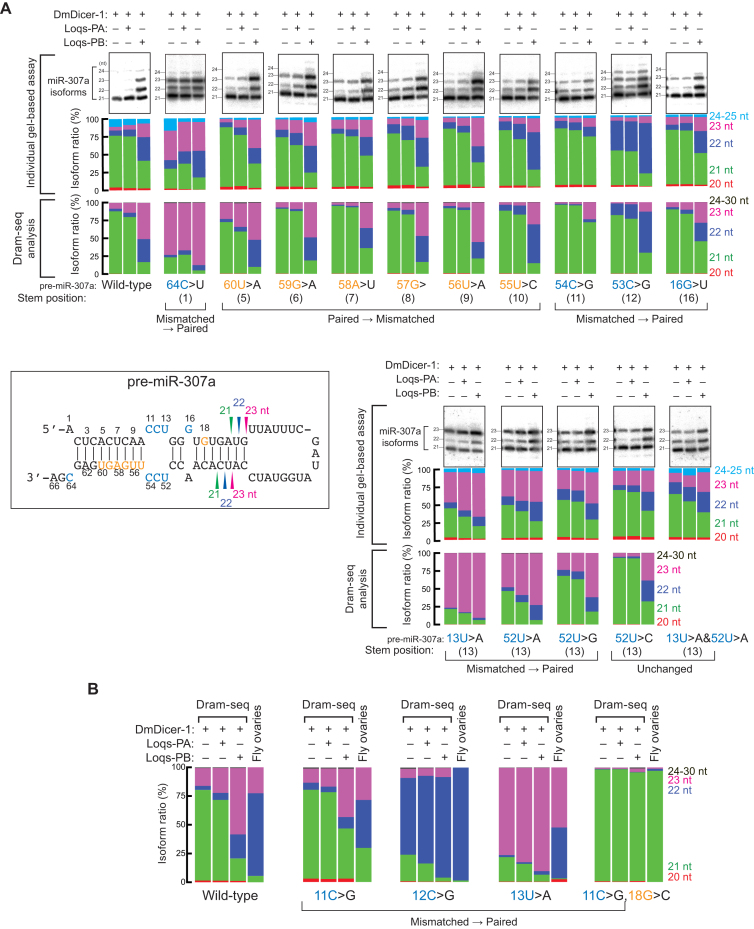
The Dram-seq analysis revealed that length distributions of miR-307a isoforms produced from many of pre-miR-307a variants were changed compared with wild-type pre-miR-307a in DmDicer-1 ± Loqs-PA or Loqs-PB (Figure 2B and Supplementary Figure S2A). Analysis also revealed that many pre-miR-307a variants resulted in altered length-tuning by Loqs-PB compared to wild-type pre-miR-307a. We first validated the Dram-seq results by comparing them with a low-throughput, gel-based length distribution analysis using several individually prepared pre-miR-307a variants. The Dram-seq results were comparable to the gel-based analysis (Figure 3A).
Figure 3.
miR-307a length distributions revealed by Dram-seq, individual gel-based in vitro assay, and sequencing from fly ovaries. (A) In vitro dicing of 100 nM pre-miR-307a variants by 8 nM DmDicer-1 ± Loqs-PA or Loqs-PB for 120 min. The miR-307a isoforms were detected by northern blot (top rows); length distributions revealed by quantification of gels (means of at least three independent trials) (middle rows); length distributions revealed by Dram-seq (bottom rows). (B) Length distributions of miR-307a isoforms produced from wild-type pre-miR-307a and its variants by recombinant DmDicer-1 ± Loqs-PA or Loqs-PB in vitro revealed by Dram-seq (left threes). Length distributions of miR-307a isoforms produced from the same pre-miR-307 variants in fly ovaries (right). Fly strains w1118; miR-307anull; UASP-miR-307a transgenes [wild-type and point mutants]/mat-15-Gal4.
The 64C>U mutation, which changed the base-mismatch at stem position 1 to a base-pair, resulted in production of longer miRNA isoforms by DmDicer-1 ± Loqs-PA or Loqs-PB compared with the isoforms produced form wild-type pre-miR-307a in both Dram-seq and gel-based assay (Figure 3A). Mutations in nucleotide positions 55–60, which changed base-pairs at stem positions 5–10 to base-mismatches, did not largely change miRNA length distributions in both Dram-seq and gel-based assay compared with wild-type pre-miR-307a. The 54C>G mutation, which changed the base-mismatch at stem position 11 to a base-pair, resulted in higher production of the 21 nt isoform by DmDicer-1 ± Loqs-PA or Loqs-PB compared with wild-type pre-miR-307a. The 53C>G mutation, which changed the base-mismatch at stem position 12 to a base-pair, resulted in higher production of the 22 nt isoform by DmDicer-1 ± Loqs-PA or Loqs-PB. The 16G>U mutation, which changed the base-mismatch at stem position 16 to a base-pair, did not largely change miRNA length. The 13U>A, 52U>A and 52U>G mutations, which changed the base-mismatch at stem position 13 to a base-pair, resulted in higher production of the 23 nt isoform by DmDicer-1 ± Loqs-PA or Loqs-PB. In this study, we considered a G-U wobble pair as base-matched. In contrast, the 52U>C mutation, which did not change the base-mismatch at stem position 13, did not largely change miRNA length compared with wild-type. For all these, Dram-seq results and gel-based assay results were largely consistent with each other, validating Dram-seq. The compensatory 13U>A&52U>A double mutations, which did not change the base-mismatch at stem position 13, did not largely change miRNA length in gel-based assay. Since 13U and 52U are located on different arms of pre-miRNA, the double mutant 13U>A&52U>A could not be examined by Dram-seq. These results together suggested that changes in miRNA length caused by the mutations in stem position 13 were largely due to the change in the base-pair/mismatch state rather than simply the change in sequence.
Dram-seq results are consistent with miRNA lengths produced in fly ovaries
We examined whether Dram-seq results have physiological relevance and are recapitulated in vivo. To test this, we created transgenic fly strains expressing transgenic wild-type or mutant pri-miR-307a in the background of the endogenous miR307anull. We high-throughput sequenced small RNAs prepared from ovaries (which express Loqs-PB (10,17)) of these flies and examined lengths of miR-307a isoforms (Figure 3B). The miRNA length profiles in the ovary samples would depend on both miRNA production and turnover, whereas those in the Dram-seq samples rely exclusively on miRNA production. Compared with the wild-type sequence, the 11C>G mutation, which changed the base-mismatch at stem position 11 to a base-pair, resulted in higher production of the 21 nt miR-307a isoform both in Dram-seq by DmDicer-1 + Loqs-PB and in fly ovaries. The 12C>G mutation, which changed the base-mismatch at stem position 12 to a base-pair, resulted in higher production of the 22 nt miR-307a isoform both in Dram-seq by DmDicer-1 ± Loqs-PA or Loqs-PB and in fly ovaries. The 13U>A mutation, which changed the base-mismatch at stem position 13 to a base-pair, resulted in higher production of the 23 nt isoform in Dram-seq by DmDicer-1 ± Loqs-PA or Loqs-PB and in fly ovaries. The 11C>G&18G>C mutation, which changed the base-mismatch at stem position 11 to a base-pair and the base-pair at stem position 18 to a base-mismatch, resulted in predominant production of the 21 nt isoform in Dram-seq by DmDicer-1 ± Loqs-PA or Loqs-PB and in fly ovaries. Thus, despite the additional mechanism (turnover) to determine the miRNA length profiles in vivo, Dram-seq results were largely consistent with in vivo results.
These in vitro and in vivo results together validated the Dram-seq results. We found that base-pair/mismatch state in pre-miRNA stem positions 11–13, especially at stem position 13, of pre-miRNA has a significant influence on miRNA lengths and on the ability of Loqs-PB to tune miRNA lengths.
Base-pairs at stem positions 11 and 13 lead to production of shorter and longer miRNA isoforms respectively
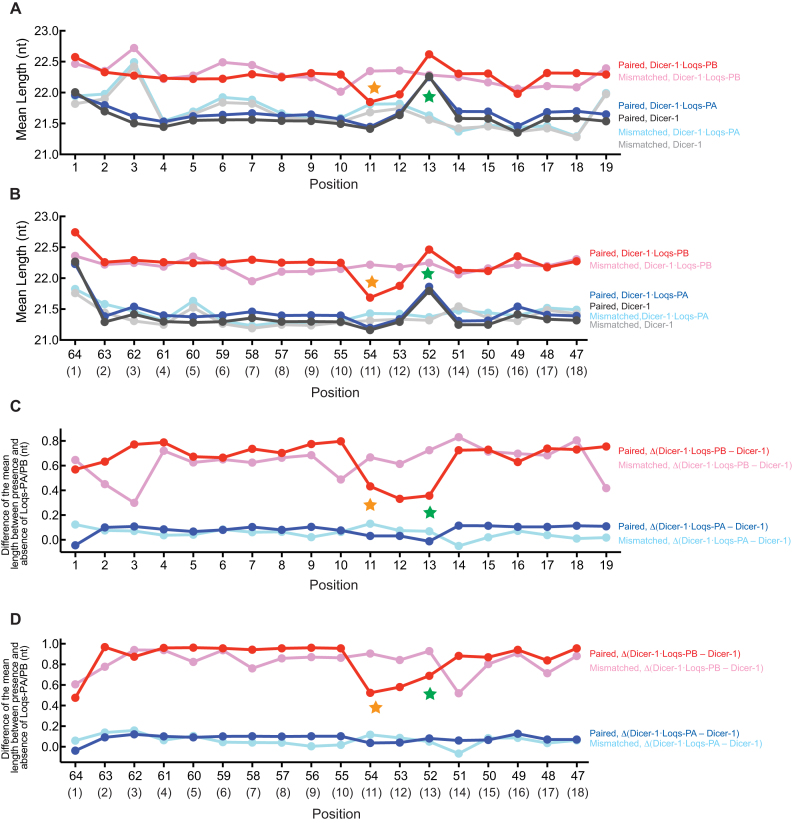
Next, we examined the effect of base-pair/mismatch state at each pre-miRNA stem position on miRNA lengths in a wider context. For this, we first calculated the mean length of miRNA isoforms produced from each unique pre-miR-307a variant. Then, we calculated the mean of the mean length of miRNA isoforms produced from pre-miR-307a variants containing a base-pair or base-mismatch at each stem position (Figure 4A and B, Supplementary Figure S3). We allowed up to one additional point mutation in nucleotide positions 1–19 (for the 5′ arm randomization) or 47–66 (for the 3′ arm randomization) other than the position of the interest. In Figure 4, each point shows the mean of mean miRNA length of 55–174 pre-miR-307 variants. This analysis allowed us to examine the effect of base-pair/mismatch at each position of the pre-miRNA stem on miRNA length in a wider context compared with examining single mutant results as we did above (Figure 3). We found that pre-miR-307a variants containing a base-pair at stem position 11 produced shorter miRNA isoforms by DmDicer-1 ± Loqs-PA or Loqs-PB compared with variants containing a base-mismatch at the same position in both the 5′ arm-randomized and 3′ arm-randomized Dram-seq (Figure 4A and B. Supplementary Figure S3). We also found that pre-miR-307a variants containing a base-pair at stem position 13 produced longer miRNA isoforms by DmDicer-1 ± Loqs-PA or Loqs-PB compared with variants containing a base-mismatch at the same position.
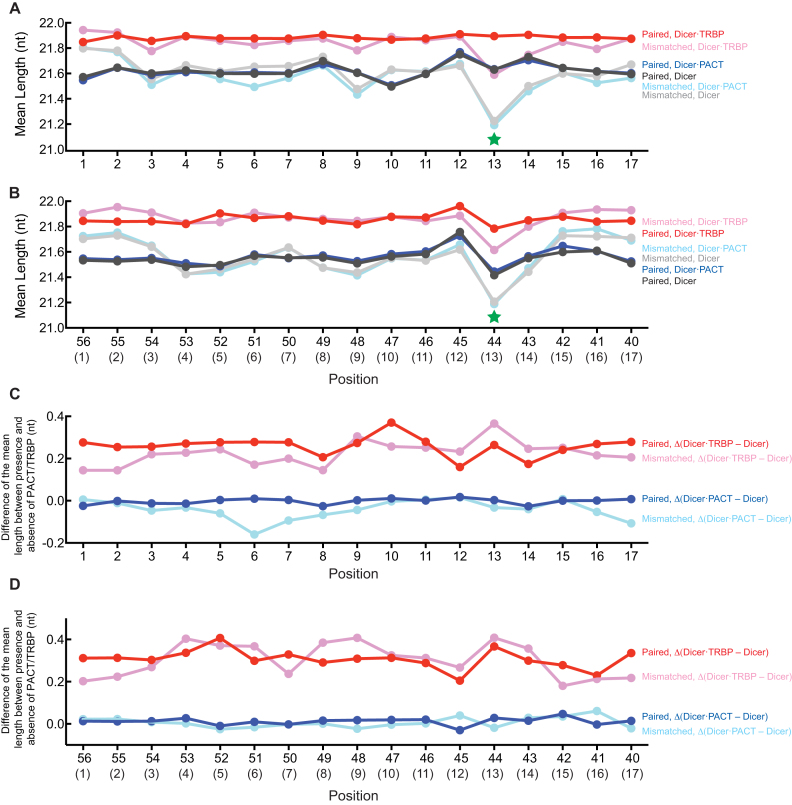
Figure 4.
Mean length of miR-307 isoforms produced from pre-miR-307a variants revealed by Dram-seq. (A, B) Mean of mean length of miRNAs produced by DmDicer-1 ± Loqs-PA or Loqs-PB from the (A) 5′ arm- and (B) 3′ arm- randomized pre-miR-307a variants containing a base-pair or base-mismatch at each position in the stem. The pre-miR-307a variants containing up to one mutation in addition to the mutation in the position of interest were analyzed. Orange and green stars at stem positions 11 and 13 indicate that the mean length was shorter and longer, respectively, when the positions were base-paired, compared with base-mismatched. (C, D) Mean of the difference of mean miRNA length produced from the (C) 5′ arm- and (D) 3′ arm- randomized pre-miR-307a variants containing a base-pair or base-mismatch at each position in the stem between DmDicer-1 + Loqs-PA and DmDicer-1 alone or between DmDicer-1 + Loqs-PB and DmDicer-1 alone. The difference in mean miRNA length was examined for each unique pre-miR-307a variant, and their means are shown. The stem position numbers are shown in parentheses in (B) and (D). Orange and green stars at stem positions 11 and 13 indicate that the Loqs-PB effect to make longer miRNAs was reduced when these positions were base-paired, compared with base-mismatched.
The effects of base-pair/mismatch at stem positions 11 and 13 on miRNA lengths seemed greater for DmDicer-1 ± Loqs-PA than for DmDicer-1 + Loqs-PB (Figure 4A and B. Supplementary Figure S3). To test this further, we examined the difference of mean miRNA length caused by the addition of Loqs-PA or Loqs-PB, compared with DmDicer-1 alone, for each pre-miR-307a variant. The effect of Loqs-PB to produce longer miRNAs than DmDicer-1 alone was reduced when stem position 11 or 13 had a base-pair compared with a base-mismatch (Figure 4C and D). Thus, we concluded that base-pairs at stem positions 11 and 13 lead to production of shorter and longer miR-307a isoforms, respectively, by DmDicer-1 ± Loqs-PA or Loqs-PB and weaken the effect of Loqs-PB to promote production of longer miR-307a isoforms, compared with base-mismatches. These findings were consistent with individual gel-based in vitro assay results and in vivo results from fly ovaries (Figure 3). Wild-type pre-miR-307a has base-mismatches at stem positions 11 and 13 (Figure 1B). Our results revealed that these base-mismatches are important for Loqs-PB to promote production of miR-307a isoforms that are longer than those produced by DmDicer-1 ± Loqs-PA.
Next, we examined length heterogeneity of miR-307a isoforms produced from each pre-miR-307a variant. We calculated length heterogeneity by using the equation H = 1 – Fmax, where H represents length heterogeneity and Fmax is the fraction of the most abundant miRNA isoform (2). We calculated the mean of length heterogeneity of miR-307a isoforms produced from pre-miR-307 variants containing a base-pair or base-mismatch at each stem position (Supplementary Figure S4A and B). Loqs-PB in general increased length heterogeneity compared with DmDicer-1 ± Loqs-PA. However, we found that when pre-miR-307a variants contained a base-pair at stem position 13, length heterogeneity of miRNA isoforms produced was higher for DmDicer-1 ± Loqs-PA and lower for DmDicer-1 + Loqs-PB compared with variants containing a base-mismatch at the same position. We calculated the mean of the difference of length heterogeneity caused by the addition of Loqs-PA or Loqs-PB, compared with DmDicer-1 alone, for each pre-miR-307 variant (Supplementary Figure S4C and D). The addition of Loqs-PB caused a decrease in length heterogeneity when stem position 13 had a base-pair, while it increased length heterogeneity when stem position 13 had a base-mismatch. These results revealed that the base-mismatch at stem position 13 is important for Loqs-PB to tune miRNA length.
Taken together, our Dram-seq and individual in vitro and in vivo analysis revealed that the base-mismatch at stem position 13 in pre-miR-307a promotes production shorter miR-307a isoforms by DmDicer-1 ± Loqs-PA compared with a base-pair and that the base-mismatch is important for Loqs-PB to promote production of longer miR-307a isoforms compared with DmDicer-1 ± Loqs-PA.
The stem of pre-miR-132, but not the terminal loop, contains a signature required for tuning of miRNA length by TRBP
Next, we studied human Dicer, PACT and TRBP in tuning miR-132 length. As we reported previously, HsDicer + TRBP produced longer miR-132 isoforms from pre-miR-132 more than HsDicer ± PACT, while the degree of the change caused by TRBP was much smaller than that by Loqs-PB (Figure 5A) (10). In contrast, the length of miR-27b produced from pre-miR-27b was not changed by TRBP. We created two swap chimera pre-miRNAs. The first chimera pre-miRNA has the stem derived from pre-miR-132 and the terminal loop derived from pre-miR-27b. HsDicer + TRBP produced the longer miRNA isoforms from this chimera pre-miRNA more than HsDicer ± PACT. The second chimera pre-miRNA has the stem derived from pre-miR-27b and the terminal loop derived from pre-miR-132. The length of miRNA produced from this chimera pre-miRNA was not changed by TRBP. Thus, we concluded that the stem of pre-miR-132, but not the terminal loop, contains a signature(s) required for TRBP to tune miR-132 length, consistent with what we found in the Drosophila Loqs-PB/pre-miR-307a system (Figure 1) (10).
Figure 5.
Dram-seq using human Dicer, PACT, TRBP, and pre-miR-132. (A) In vitro dicing of 100 nM 5′ 32P-radiolabeled pre-miR-132, pre-miR-27b, and their swap chimera pre-miRNAs by 6.5 nM HsDicer ± PACT or TRBP for 120 min. Representative gel image and length distributions revealed by quantification of gels (means of at least three independent trials) are shown. (B) Length distributions of miR-132a-3p isoforms produced from wild-type pre-miR-132 and its variants containing a single point mutation at nucleotide positions 40–58 by HsDicer ± PACT or TRBP, revealed by Dram-seq.
Dram-seq to reveal miR-132 isoform production by HsDicer ± PACT or TRBP
We performed Dram-seq using the 5′ arm- and 3′ arm- randomized pre-miR-132 variants pool and HsDicer ± PACT or TRBP (Figure 5B, Supplementary Figure S2B, and Supplementary Table S2). The sequences at nucleotide positions 1–17 and 40–58 were randomized at a defined ratio (85% wild-type 5% each for three mutations) in the 5′ arm- and 3′ arm- randomized pre-miR-132 variants pools, respectively. Among the 8,619 miRNA length distributions determined from the 2,872 pre-miRNA variants containing up to two point mutations by HsDicer ± PACT or TRBP, the mean and median number of reads for miRNAs were 2100 and 294, respectively. 86.4% (6340/8619), 98.7% (8506/8619) and 100% (8619/8619) of them had the number of reads of at least 50, 10 and 1, respectively. Among the additional 133,569 miRNA length distributions potentially produced from the theoretically possible 44,523 pre-miRNA variants containing triple point mutations by HsDicer ± PACT or TRBP, 28.6% (38 264/133 569), 53.5% (71 442/133 569) and 83.0% (110 917/133 569) had the number of reads of at least 50, 10 and 1, respectively. Length distributions of miR-132 isoforms produced from wild-type pre-miR-132 revealed by Dram-seq were consistent with that revealed by individual gel-based assay (Figure 5).
We analyzed the mean of the mean length of miR-132 isoforms produced from pre-miR-132 variants containing a base-pair or base-mismatch at each stem position. Interestingly, as in the Drosophila system, we found that pre-miR-132 variants containing a base-mismatch at stem position 13 produced shorter miRNA isoforms by HsDicer ± PACT or TRBP compared with variants containing a base-pair at the same position (Figure 6A and B and Supplementary Figure S5). In general, length heterogeneity of miRNA isoforms was decreased by TRBP (Supplementary Figure S6A and S6B). However, when pre-miR-132 variants contained a base-mismatch at stem position 13, length heterogeneity of miRNA isoforms produced by HsDicer + TRBP was higher compared with variants containing a base-pair at the same position.
Figure 6.
Mean length of miR-132 isoforms produced from pre-miR-132 variants revealed by Dram-seq. (A, B) Mean of mean length of miRNAs produced by HsDicer ± PACT or TRBP from (A) 5′ arm- and (B) 3′ arm- randomized pre-miR-132 variants containing a base-pair or base-mismatch at each position in the stem. The pre-miR-132 variants containing up to one mutation in addition to the mutation in the position of interest were analyzed. Green stars at the stem position 13 indicate that mean length was shorter, when the position was base-mismatched, compared with base-paired. (C, D) Mean of the difference of mean miRNA length produced from the (C) 5′ arm- and (D) 3′ arm- randomized pre-miR-132 variants containing a base-pair or base-mismatch at each position in the stem between HsDicer + PACT and HsDicer alone or between HsDicer + TRBP and HsDicer alone. The difference in mean miRNA length was examined for each unique pre-miR-132 variant, and their means are shown. Orange and green stars at stem positions 11 and 13 indicate that the Loqs-PB effect to make longer miRNAs was reduced when these positions were base-paired, compared with base-mismatched. The stem position numbers are shown in parentheses in (B) and (D).
We examined the difference of mean miRNA length and length heterogeneity caused by the addition of PACT or TRBP, compared with HsDicer alone, for each pre-miR-132 variant (Figure 6C and D and Supplementary Figure S6C and D). Unlike in the Drosophila system, we did not find any drastic change in the difference in mean length or length heterogeneity of miRNA isoforms caused by TRBP between variants containing a base-pair or base-mismatch at each stem position including stem position 13. These results suggest that although base-pair/mismatch status at stem position 13 in pre-miRNA is an important factor to determine miRNA length in human as in Drosophila, there may not be a single stem position that is crucially important for TRBP to tune miRNA length. In fact, wild-type pre-miR-132 has a base-pair at stem position 13.
Loqs-PB tunes miRNA length using base-mismatch in the pre-miRNA stem in a context-dependent manner
We tested whether the base-paired/mismatched shape of the pre-miR-307a stem alone is enough for Loqs-PB to tune miRNA length or the nucleotide sequence also contributes in a context-dependent manner. To test this, we examined whether we can engineer a pre-miRNA that is not responsive to Loqs-PB into Loqs-PB-responsive by introducing point mutations to mimic the base-paired/mismatched shape in the pre-miR-307 stem. We introduced several point mutations in pre-let-7 to mimic the shape of the pre-miR-307a stem and examined lengths of miRNAs produced. Loqs-PB did not tune the length of miRNAs produced from these pre-miRNAs (Supplementary Figure S7). Therefore, the base-paired/mismatched shape does not seem to be sufficient for tuning of miRNA length by Loqs-PB. The nucleotide sequence likely also plays a contextual role.
To test the idea of the context-dependent Loqs-PB effect further, we performed Dram-seq using the 3′ arm-randomized pre-miR-999 variants pool (Figure 7A, Supplementary Figure S8A, and Supplementary Table S3). Wild-type pre-miR-999 was not responsive to Loqs-PB, producing predominantly a 22 nt isoform of miR-999. As we found in the Drosophila pre-miR-307a and human pre-miR-132 systems, base-pairs/mismatches at a few of the stem positions in pre-miR-999 affected miRNA length (Figure 7B). As was observed in stem position 11 of Drosophila pre-miR-307a, pre-miR-999 variants containing a base-mismatch at stem position 5, 6 or 14, produced longer miR-999 isoforms compared with variants containing a base-pair at the same position. This effect seemed greater in DmDicer-1 ± Loqs-PA than in DmDicer-1 ± Loqs-PB. In fact, compared with DmDcer-1 alone, DmDicer-1 + Loqs-PB produced shorter miR-999 isoforms from pre-miR-999 variants containing a base-mismatch at stem position 5, 6 or 14 (Figure 7C). Thus, as was observed in stem position 13 in pre-miR-307a, a mismatch at stem position 5, 6 or 14 in pre-miR-999 variants enabled Loqs-PB to tune miRNA length, but this time to promote production of shorter miRNA isoforms. A base-mismatch at stem position 5, 6, or 14 of pre-miR-999 variants, caused an increase in length heterogeneity of miR-999 isoforms compared with a base-pair (Supplemental Figure S8B). Compared with Dicer-1 alone, Loqs-PB decreased length heterogeneity of miR-999 isoforms produced from pre-miR-999 variants containing a base-mismatch at stem position 5, 6 or 14, while it did not change length heterogeneity for variants containing a base-pair at these stem position (Supplemental Figure S8C).
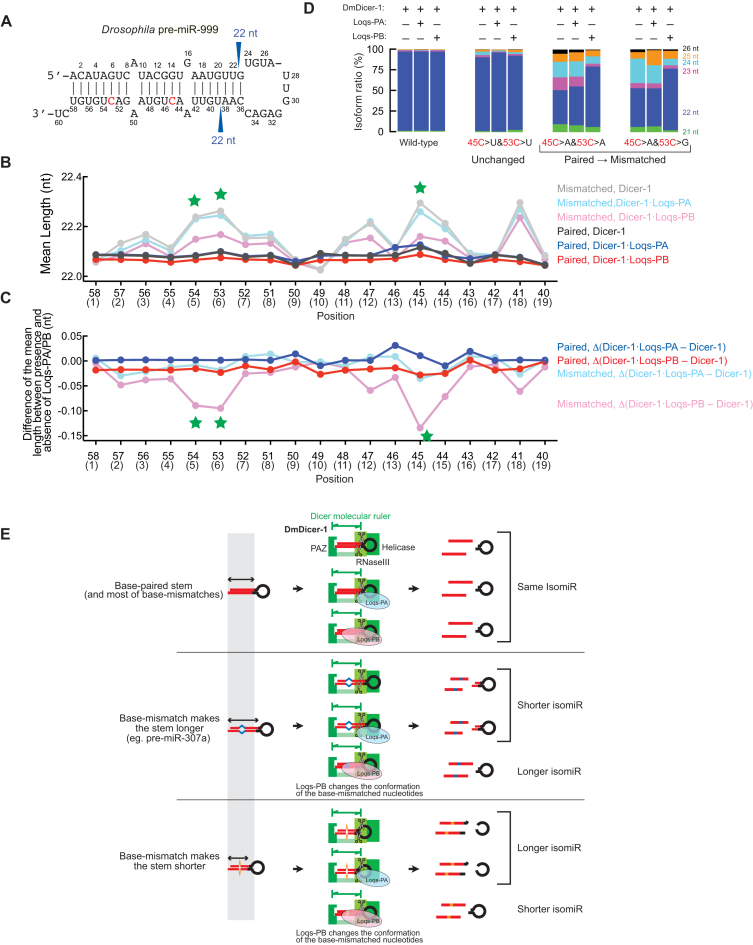
Figure 7.
Conversion of a Loqs-PB-non-responsive pre-miRNA into Loqs-PB-responsive by introducing base-mismatches in the pre-miRNA stem. (A) Drosophila pre-miR-999. Nucleotides C45 and C53, which were mutated in (D), are shown in red. (B) Mean of mean miRNA length produced by DmDicer-1 ± Loqs-PA or Loqs-PB from 3′ arm-randomized pre-miR-999 variants containing a base-pair or base-mismatch at each position in the stem determine by Dram-seq. The pre-miR-999 variants containing up to one mutation in addition to the mutation in the position of interest were analyzed. Green stars at stem positions 5, 6 and 14 indicate that the mean length was longer, when these positions were base-mismatched, compared with base-paired. (C) Mean of the difference of mean miRNA length produced from pre-miR-999 variants containing a base-pair or base-mismatch at each position in the stem between DmDicer-1 + Loqs-PA and DmDicer-1 alone or between DmDicer-1 + Loqs-PB and DmDicer-1 alone. The difference in mean miRNA length was examined for each unique pre-miR-999 variant, and their means are shown. Green stars at stem positions 5, 6, and 14 indicate that Loqs-PB promoted production of shorter miRNA isoforms than DmDicer-1 ± Loqs-PA when these positions were base-mismatched. The stem position numbers are shown in parentheses in (B) and (C). (D) Length distributions of miRNA isoforms produced from the wild-type and double point mutants of pre-miR-999 determined by Dram-seq. (E) The speculative model by which Loqs-PB tunes the length of miRNAs using base-mismatches in the pre-miRNA stem. The base-mismatched nucleotides at stem position 13 in pre-miR-307a take extended conformation and thus make the pre-miR-307a stem length longer compared with base-paired (Middle). Therefore, fewer pre-miR-307a stem nucleotides can fit in the Dicer molecular ruler, resulting in production of shorter miR-307a isoform (21 nt) by DmDicer-1 ± Loqs-PA compared with pre-miRNA variants containing a base-pair at the stem position 13 (Top). Loqs-PB binding changes the conformation of the base-mismatched nucleotides at stem position 13 of pre-miR-307a to that of base-paired ones (Middle). Therefore, upon binding of Loqs-PB, the length of the pre-miR-307a stem becomes shorter as in base-paired pre-miRNAa, resulting in production of longer miR-307a isoforms (22 and 23 nt) compared with DmDicer-1 ± Loqs-PA. In a certain context, base-mismatched nucleotides can take a more compact conformation, shorten the pre-miRNA stem, and results in production of longer miRNA isoforms. Upon Loqs-PB binding, their conformation is changed to that of base-paired ones, and the pre-miRNA stem becomes shorter, resulting in production of shorter miRNA isoforms compared with DmDicer-1 ± Loqs-PA. Most of mismatches in the pre-miRNA stem do not change the pre-miRNA stem length or miRNA length.
Wild-type pre-miR-999 has base-pairs at these three positions and was not responsive to Loqs-PB (Figure 7A and D). The 45C>A&53C>A and 45C>A&53C>G mutations, which changed base-pairs at stem positions 6 and 14 to base-mismatches, converted pre-miR-999 clearly responsive to Loqs-PB (Figure 7D). In contrast, the 45C>U&53C>U mutation, which did not change base-pairs at stem positions 6 and 14, did not convert pre-miR-999 responsive to Loqs-PB. Therefore, we succeeded in engineering a Loqs-PB-non-responsive pre-miRNA into Loqs-PB-responsive by introducing base-mismatches in the pre-miRNA stem. Unlike in tuning of miR-307 length, Loqs-PB promoted shorter miRNA production. Interestingly, we previously found that Loqs-PB promoted production of shorter miR-316 isoforms from pre-miR-316 in vitro and in vivo (10). Together, these results showed that Loqs-PB uses base-mismatches in the pre-miRNA stem to tune miRNA length and that whether Loqs-PB does so and whether it promotes production of longer or shorter miRNA isoforms are context-dependent.
We performed site-specific UV-crosslinking using pre-miR-999 variants containing base-mismatches at positions 6 and 14 and UV-crosslinkable 5IU base modification at nucleotide position 6, 14 or 29. Loqs-PB, but not Loqs-PA, was crosslinked to the pre-miR-999 variants containing a 5IU base modification at nucleotide position 6 or 14 (Supplemental Figure S8D). In contrast, both Loqs-PA and Loqs-PB were crosslinked to the pre-miR-999 variant containing a 5IU base modification at nucleotide positions 29, which is located in the terminal loop. DmDicer-1 was crosslinked efficiently to all these modified pre-miR-999 tested. These results indicated that Loqs-PB, but not Loqs-PA, directly binds base-mismatched nucleotides in the pre-miR-999 stem, while both Loqs-PA and Loqs-PB bind the terminal loop, as found in pre-miR-307a (Figure 1D).
Finally, in order to confirm that base-mismatches are important for Loqs-PB to change miRNA length, we tested processing of a pre-miR-307a variant (1A>G, 52U>A, 53C>G, 54C>G) in which four out of five base mismatches in the wild-type pre-miR-307a were replaced with canonical base-pairs in gel-based in vitro dicing assay (Supplementary Figure S9). The length distribution of the miRNA isoforms produced from this pre-miR-307a variant was not changed by the presence or absence of Loqs-PA or Loqs-PB. Therefore, we concluded that base-mismatches are important for Loqs-PB to change miRNA length.
DISCUSSION
We developed Dram-seq, a novel high-throughput method to examine miRNA isoform production from thousands of different pre-miRNA variants in a single reaction (Figure 2A). Dram-seq results were consistent with individually tested in vitro and in vivo results (Figure 3). Using Dram-seq, we showed that the base-mismatch at stem position 13 of pre-miR-307a leads to production of shorter miRNA isoforms by DmDicer-1 ± Loqs-PA compared with a base-pair at the same position (Figure 4). We also showed that the base-mismatch at this position is important for Loqs-PB to promote production of miR-307a isoforms that are longer than those produced by with DmDicer-1 ± Loqs-PA. These base-mismatched nucleotides at stem position 13 were directly bound by Loqs-PB, but not by Loqs-PA (Figure 1D). Furthermore, using Dram-seq, we were able to convert Loqs-PB-non-responsive pre-miR-999 into Loqs-PB-responsive by introducing base-mismatches in its stem (Figure 7D). Unlike in miR-307a, Loqs-PB promoted production of shorter miR-999 isoforms compared with DmDicer-1 ± Loqs-PA.
Based on these findings, we propose the following speculative model by which Loqs-PB tunes lengths of miRNAs produced from pre-miRNAs by DmDicer-1 (Figure 7E). The fixed distance between the Dicer PAZ domain, which binds the end of pre-miRNA, and RNaseIII active sites, which cleave pre-miRNA, serves as a molecular ‘ruler’ to measure the substrate RNA length (39). If more nucleotides within a pre-miRNA stem fit in the Dicer ruler, then longer miRNA isoforms are produced. If fewer nucleotides within a pre-miRNA stem fit in the Dicer ruler, then shorter miRNA isoforms are produced. The base-mismatched nucleotides at stem position 13 of pre-miR-307a take a non-canonical, extended conformation, which is different from the canonical A-form dsRNA helix conformation (Figure 7E, middle). Therefore, the distance of the stem becomes longer when stem position 13 has a base-mismatch, compared with a base-pair (Figure 7E, top). Thus fewer pre-miR-307a stem nucleotides fit in the Dicer ruler, resulting in production of shorter miRNA isoforms compared with pre-miRNAs containing a base-pair at stem position 13. Loqs-PB binding to the base-mismatched nucleotides at stem position 13 changes their conformation to that observed in the canonical A-form dsRNA helix. Thus, upon Loqs-PB binding, the extended pre-miR-307a stem becomes shorter and therefore more stem nucleotides fit within the Dicer ruler, resulting in production of longer miR-307a isoforms.
In contrast, in a certain context, base-mismatched nucleotides within a pre-miRNA stem can take a more compact conformation compared with the canonical A-form conformation, and thus can make the pre-miRNA stem shorter. Then more stem nucleotides fit in the Dicer ruler, producing longer miRNA isoforms (Figure 7E, bottom). Loqs-PB binding to these base-mismatched nucleotides changes their conformation to that observed in the canonical A-form dsRNA helix, extends the length of the pre-miRNA stem similar to the canonical one, and thus results in production of shorter miRNA isoforms compared with those produced by DmDicer-1 ± Loqs-PA.
The conformations of base-mismatches and their effects on the pre-miRNA stem length are context-dependent, likely influenced by sequence and base-pair/mismatch state in the flanking regions. The majority of base-mismatches observed in pre-miRNAs do not change pre-miRNA stem length, explaining why Loqs-PB tunes lengths of only miRNAs that are produced from a subset of pre-miRNAs. This model explains the different effects of Loqs-PB on different pre-miRNAs, since each pre-miRNA has different base-pair/mismatch in different contexts. In contrast, the model that Loqs-PB binding to DmDicer-1 changes the conformation of DmDicer-1 cannot explain the different effects of Loqs-PB on different pre-miRNAs.
Our model aligns well with previous structural models of Dicer bound with Dicer partner proteins determined by electron microscopy (40–43). Furthermore, supporting our model, previous crystal structures showed that a base-mismatch in a dsRNA duplex can change the length of the duplex (rise distance per base pair) (44–46). Some base-mismatches make a dsRNA duplex longer, while some make it shorter and others don’t change dsRNA length. The effects of a base-mismatch likely depend on sequences of both the mismatched bases themselves and the flanking nucleotides. The stem position 13 of pre-miR-307a has a U-U mismatch. In the previous crystal structure of 7 bp RNA duplex containing a U-U mismatch, four duplexes were present in the asymmetric unit (46). Each of the four duplexes in the asymmetric unit exhibits significant structural differences from the others. Such structural variability shows that the U-U mismatch-containing RNA duplex structure is flexible and that the energy differences between the conformations are small. On average, the U-U mismatch-containing RNA duplex had increased rise distance per base pair compared with a canonical A-form RNA helix (19.44 Å versus 16.86 Å for 7 bp RNA duplex, meaning 3.24 Å versus 2.81 Å per base pair) (46), supporting our idea that the position 13 mismatch in pre-miR-307 increases the distance of the stem. Furthermore, other previous crystal structures showed that dsRBDs of Dicer partner proteins bind a perfectly base-paired A-form dsRNA substrate in a canonical manner as observed in other dsRBDs (25), supporting our model that Loqs-PB binding changes the conformation of base-mismatched nucleotides to base-paired one. Previous studies showed that Loqs-PB helps DmDicer-1 process pre-miRNAs containing mismatches at the dicing cleavage site (47). Such pre-miRNAs are not efficiently processed by DmDicer-1 alone, while pre-miRNAs containing base-pairs at the dicing cleavage site are efficiently processed by DmDcer-1 in the presence or absence of Loqs-PB. This study also supports our model that Loqs-PB binding changes the conformation of base-mismatched nucleotides to base-paired one. Other studies suggested that asymmetric bulges within stems of pre-miRNAs are responsible for production of longer miRNAs because unpaired bases within bulges are not measured by the Dicer molecular ruler (2,14–16). We propose that a base-mismatch (meaning, unpaired bases within a symmetric bulge) can have similar effects but in a sequence context-dependent manner. Structural studies will be required to test this speculative model.
miR-307a seems to be a unique miRNA, which is by far the most responsive to Loqs-PB compared with any other miRNA. This may underscore its important biological and molecular roles. In fact, previous large-scale miRNA knockout studies in Drosophila revealed that miR-307a knockout female flies have shorter life spans and a larger number of primordial germ cells (34). However, the underling molecular mechanism, including the functions of each miR-307a isoform remains unknown. Using Dram-seq, we were able to identify pre-miR-307a variants that predominantly produced a specific isoform of miR-307a in vitro and in fly ovaries (Figure 3B). The identified pre-miR-307a variants and the transgenic fly strains created will be a useful resource for future study to examine biological and molecular roles of each miR-307a isoform.
Dram-seq is a useful tool to study production and function of isomiRs. The location, number, and ratio of sequence randomization can be customized for each different purpose. Random sequence insertion and deletion can also be introduced and examined. Dram-seq can also be performed using pre-miRNA pool in which the sequences in the terminal loop are randomized. Some pre-miRNAs contain a sequence motif that is recognized by a specific RNA-binding protein. For example, 5′ -GGAG-3′ motif in the terminal loop of the pre-let-7 family is bound by Lin28, which modulates production of let-7 (48). There might be sequence motif within the terminal loop of some pre-miRNAs that modulates isomiR production. Dram-seq can be used to identify such motifs.
Many human single nucleotide polymorphisms (SNPs) are found in pre-miRNA coding regions, some of which may affect isomiR production by Dicer from pre-miRNAs and may have physiological influence. Dram-seq can be used to conduct high-throughput analysis to identify SNPs that affect isomiR production.
DATA AVAILABILITY
SRA accession number for the small RNA libraries reported in this paper is SRP102235.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to Dr Sarah Wheelan (Johns Hopkins University) for help in the bioinformatics analysis of Dram-seq results. We are grateful to Dr Phillip Zamore (University of Massachusetts Medical School) for helpful discussion. We are grateful to Susan E. Liao, a member of Fukunaga lab, for her comments on the manuscript.
Author Contributions: Conceptualization, R.F.; Methodology, L.Z., S.K.K. and R.F.; Investigation, L.Z., S.K.K. and R.F. Writing–Original Draft, L.Z., S.K.K. and R.F.; Writing–Review & Editing, L.Z., S.K.K. and R.F. Funding Acquisition, R.F.; Supervision, R.F.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR online.
FUNDING
American Heart Association [15SDG23220028]; National Institutes of Health (NIH) [R01GM116841 to R.F.]. Funding for open access charge: NIH [R01GM116841].
Conflict of interest statement. None declared.
REFERENCES
- 1. Morin R.D., O’Connor M.D., Griffith M., Kuchenbauer F., Delaney A., Prabhu A.L., Zhao Y., McDonald H., Zeng T., Hirst M. et al. . Application of massively parallel sequencing to microRNA profiling and discovery in human embryonic stem cells. Genome Res. 2008; 18:610–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Starega-Roslan J., Krol J., Koscianska E., Kozlowski P., Szlachcic W.J., Sobczak K., Krzyzosiak W.J.. Structural basis of microRNA length variety. Nucleic Acids Res. 2011; 39:257–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Yuan Z., Ding S., Yan M., Zhu X., Liu L., Tan S., Jin Y., Sun Y., Li Y., Huang T.. Variability of miRNA expression during the differentiation of human embryonic stem cells into retinal pigment epithelial cells. Gene. 2015; 569:239–249. [DOI] [PubMed] [Google Scholar]
- 4. Fernandez-Valverde S.L., Taft R.J., Mattick J.S.. Dynamic isomiR regulation in Drosophila development. RNA. 2010; 16:1881–1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Li S.C., Liao Y.L., Ho M.R., Tsai K.W., Lai C.H., Lin W.C.. miRNA arm selection and isomiR distribution in gastric cancer. BMC Genomics. 2012; 13(Suppl. 1):S13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Babapoor S., Fleming E., Wu R., Dadras S.S.. A novel miR-451a isomiR, associated with amelanotypic phenotype, acts as a tumor suppressor in melanoma by retarding cell migration and invasion. PLoS One. 2014; 9:e107502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bajan S., Hutvagner G.. Regulation of miRNA processing and miRNA mediated gene repression in cancer. MicroRNA. 2014; 3:10–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cheng W.C., Chung I.F., Tsai C.F., Huang T.S., Chen C.Y., Wang S.C., Chang T.Y., Sun H.J., Chao J.Y., Cheng C.C. et al. . YM500v2: a small RNA sequencing (smRNA-seq) database for human cancer miRNome research. Nucleic Acids Res. 2014; 43:D862–D867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wu X., Zeng R., Wu S., Zhong J., Yang L., Xu J.. Comprehensive expression analysis of miRNA in breast cancer at the miRNA and isomiR levels. Gene. 2015; 557:195–200. [DOI] [PubMed] [Google Scholar]
- 10. Fukunaga R., Han B.W., Hung J.H., Xu J., Weng Z., Zamore P.D.. Dicer partner proteins tune the length of mature miRNAs in flies and mammals. Cell. 2012; 151:533–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tan G.C., Chan E., Molnar A., Sarkar R., Alexieva D., Isa I.M., Robinson S., Zhang S., Ellis P., Langford C.F. et al. . 5′ isomiR variation is of functional and evolutionary importance. Nucleic Acids Res. 2014; 42:9424–9435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lee H.Y., Doudna J.A.. TRBP alters human precursor microRNA processing in vitro. RNA. 2012; 18:2012–2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wilson R.C., Tambe A., Kidwell M.A., Noland C.L., Schneider C.P., Doudna J.A.. Dicer-TRBP complex formation ensures accurate mammalian microRNA biogenesis. Mol. Cell. 2015; 57:397–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chen H.M., Chen L.T., Patel K., Li Y.H., Baulcombe D.C., Wu S.H.. 22-Nucleotide RNAs trigger secondary siRNA biogenesis in plants. PNAS. 2010; 107:15269–15274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cuperus J.T., Carbonell A., Fahlgren N., Garcia-Ruiz H., Burke R.T., Takeda A., Sullivan C.M., Gilbert S.D., Montgomery T.A., Carrington J.C.. Unique functionality of 22-nt miRNAs in triggering RDR6-dependent siRNA biogenesis from target transcripts in Arabidopsis. Nat. Struct. Mol. Biol. 2010; 17:997–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Manavella P.A., Koenig D., Weigel D.. Plant secondary siRNA production determined by microRNA-duplex structure. PNAS. 2012; 109:2461–2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Forstemann K., Tomari Y., Du T., Vagin V.V., Denli A.M., Bratu D.P., Klattenhoff C., Theurkauf W.E., Zamore P.D.. Normal microRNA maturation and germ-line stem cell maintenance requires Loquacious, a double-stranded RNA-binding domain protein. PLoS Biol. 2005; 3:e236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jiang F., Ye X., Liu X., Fincher L., McKearin D., Liu Q.. Dicer-1 and R3D1-L catalyze microRNA maturation in Drosophila. Genes Dev. 2005; 19:1674–1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Saito K., Ishizuka A., Siomi H., Siomi M.C.. Processing of pre-microRNAs by the Dicer-1-Loquacious complex in Drosophila cells. PLoS Biol. 2005; 3:e235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chendrimada T.P., Gregory R.I., Kumaraswamy E., Norman J., Cooch N., Nishikura K., Shiekhattar R.. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature. 2005; 436:740–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Haase A.D., Jaskiewicz L., Zhang H., Laine S., Sack R., Gatignol A., Filipowicz W.. TRBP, a regulator of cellular PKR and HIV-1 virus expression, interacts with Dicer and functions in RNA silencing. EMBO Rep. 2005; 6:961–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lee Y., Hur I., Park S.Y., Kim Y.K., Suh M.R., Kim V.N.. The role of PACT in the RNA silencing pathway. EMBO J. 2006; 25:522–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Daniels S.M., Melendez-Pena C.E., Scarborough R.J., Daher A., Christensen H.S., El Far M., Purcell D.F., Laine S., Gatignol A.. Characterization of the TRBP domain required for dicer interaction and function in RNA interference. BMC Mol. Biol. 2009; 10:38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ye X., Paroo Z., Liu Q.. Functional anatomy of the Drosophila microRNA-generating enzyme. J. Biol. Chem. 2007; 282:28373–28378. [DOI] [PubMed] [Google Scholar]
- 25. Yang S.W., Chen H.Y., Yang J., Machida S., Chua N.H., Yuan Y.A.. Structure of Arabidopsis HYPONASTIC LEAVES1 and its molecular implications for miRNA processing. Structure. 2010; 18:594–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kim Y., Yeo J., Lee J.H., Cho J., Seo D., Kim J.S., Kim V.N.. Deletion of human tarbp2 reveals cellular microRNA targets and cell-cycle function of TRBP. Cell Rep. 2014; 9:1061–1074. [DOI] [PubMed] [Google Scholar]
- 27. Eulalio A., Mano M., Dal Ferro M., Zentilin L., Sinagra G., Zacchigna S., Giacca M.. Functional screening identifies miRNAs inducing cardiac regeneration. Nature. 2012; 492:376–381. [DOI] [PubMed] [Google Scholar]
- 28. Ding J., Chen J., Wang Y., Kataoka M., Ma L., Zhou P., Hu X., Lin Z., Nie M., Deng Z.L. et al. . Trbp regulates heart function through microRNA-mediated Sox6 repression. Nat. Genet. 2015; 47:776–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Park J.K., Liu X., Strauss T.J., McKearin D.M., Liu Q.. The miRNA pathway intrinsically controls self-renewal of Drosophila germline stem cells. Curr. Biol.: CB. 2007; 17:533–538. [DOI] [PubMed] [Google Scholar]
- 30. Cenik E.S., Fukunaga R., Lu G., Dutcher R., Wang Y., Tanaka Hall T.M., Zamore P.D.. Phosphate and R2D2 restrict the substrate specificity of Dicer-2, an ATP-driven ribonuclease. Mol. Cell. 2011; 42:172–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Fukunaga R., Colpan C., Han B.W., Zamore P.D.. Inorganic phosphate blocks binding of pre-miRNA to Dicer-2 via its PAZ domain. EMBO J. 2014; 33:371–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lin X., Steinberg S., Kandasamy S.K., Afzal J., Mbiyangandu B., Liao S.E., Guan Y., Corona-Villalobos C.P., Matkovich S.J., Epstein N. et al. . Common miR-590 variant rs6971711 present Only in African Americans reduces miR-590 biogenesis. PLoS One. 2016; 11:e0156065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kondo S., Ueda R.. Highly improved gene targeting by germline-specific Cas9 expression in Drosophila. Genetics. 2013; 195:715–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chen Y.W., Song S., Weng R., Verma P., Kugler J.M., Buescher M., Rouam S., Cohen S.M.. Systematic study of Drosophila microRNA functions using a collection of targeted knockout mutations. Dev. Cell. 2014; 31:784–800. [DOI] [PubMed] [Google Scholar]
- 35. Han B.W., Wang W., Li C., Weng Z., Zamore P.D.. Noncoding RNA. piRNA-guided transposon cleavage initiates Zucchini-dependent, phased piRNA production. Science. 2015; 348:817–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Han B.W., Wang W., Zamore P.D., Weng Z.. piPipes: a set of pipelines for piRNA and transposon analysis via small RNA-seq, RNA-seq, degradome- and CAGE-seq, ChIP-seq and genomic DNA sequencing. Bioinformatics. 2015; 31:593–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kandasamy S.K., Fukunaga R.. Phosphate-binding pocket in Dicer-2 PAZ domain for high-fidelity siRNA production. PNAS. 2016; 113:14031–14036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kandasamy S.K., Zhu L., Fukunaga R.. The C-terminal dsRNA-binding domain of Drosophila Dicer-2 is crucial for efficient and high-fidelity production of siRNA and loading of siRNA to Argonaute2. RNA. 2017; 23:1139–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Macrae I.J., Zhou K., Li F., Repic A., Brooks A.N., Cande W.Z., Adams P.D., Doudna J.A.. Structural basis for double-stranded RNA processing by Dicer. Science. 2006; 311:195–198. [DOI] [PubMed] [Google Scholar]
- 40. Lau P.W., Potter C.S., Carragher B., MacRae I.J.. Structure of the human Dicer-TRBP complex by electron microscopy. Structure. 2009; 17:1326–1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wang H.W., Noland C., Siridechadilok B., Taylor D.W., Ma E., Felderer K., Doudna J.A., Nogales E.. Structural insights into RNA processing by the human RISC-loading complex. Nat. Struct. Mol. Biol. 2009; 16:1148–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lau P.W., Guiley K.Z., De N., Potter C.S., Carragher B., MacRae I.J.. The molecular architecture of human Dicer. Nat. Struct. Mol. Biol. 2012; 19:436–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Taylor D.W., Ma E., Shigematsu H., Cianfrocco M.A., Noland C.L., Nagayama K., Nogales E., Doudna J.A., Wang H.W.. Substrate-specific structural rearrangements of human Dicer. Nat. Struct. Mol. Biol. 2013; 20:662–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Baeyens K.J., De Bondt H.L., Holbrook S.R.. Structure of an RNA double helix including uracil-uracil base pairs in an internal loop. Nat. Struct. Biol. 1995; 2:56–62. [DOI] [PubMed] [Google Scholar]
- 45. Holbrook S.R., Cheong C., Tinoco I. Jr, Kim S.H.. Crystal structure of an RNA double helix incorporating a track of non-Watson-Crick base pairs. Nature. 1991; 353:579–581. [DOI] [PubMed] [Google Scholar]
- 46. Sheng J., Larsen A., Heuberger B.D., Blain J.C., Szostak J.W.. Crystal structure studies of RNA duplexes containing s(2)U:A and s(2)U:U base pairs. J. Am. Chem. Soc. 2014; 136:13916–13924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Lim M.Y., Ng A.W., Chou Y., Lim T.P., Simcox A., Tucker-Kellogg G., Okamura K.. The Drosophila Dicer-1 partner loquacious enhances miRNA processing from hairpins with unstable structures at the dicing site. Cell Rep. 2016; 15:1795–1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Heo I., Joo C., Kim Y.K., Ha M., Yoon M.J., Cho J., Yeom K.H., Han J., Kim V.N.. TUT4 in concert with Lin28 suppresses microRNA biogenesis through pre-microRNA uridylation. Cell. 2009; 138:696–708. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
SRA accession number for the small RNA libraries reported in this paper is SRP102235.