Abstract
Chemically modified antisense oligonucleotides (ASOs) with phosphorothioate (PS) linkages have been extensively studied as research and therapeutic agents. PS-ASOs can enter the cell and trigger cleavage of complementary RNA by RNase H1 even in the absence of transfection reagent. A number of cell surface proteins have been identified that bind PS-ASOs and mediate their cellular uptake; however, the mechanisms that lead to productive internalization of PS-ASOs are not well understood. Here, we characterized the interaction between PS-ASOs and epidermal growth factor receptor (EGFR). We found that PS-ASOs trafficked together with EGF and EGFR into clathrin-coated pit structures. Their co-localization was also observed at early endosomes and inside enlarged late endosomes. Reduction of EGFR decreased PS-ASO activity without affecting EGF-mediated signaling pathways and overexpression of EGFR increased PS-ASO activity in cells. Furthermore, reduction of EGFR delays PS-ASO trafficking from early to late endosomes. Thus, EGFR binds to PS-ASOs at the cell surface and mediates essential steps for active (productive) cellular uptake of PS-ASOs through its cargo-dependent trafficking processes which migrate PS-ASOs from early to late endosomes. This EGFR-mediated process can also serve as an additional model to better understand the mechanism of intracellular uptake and endosomal release of PS-ASOs.
INTRODUCTION
Antisense oligonucleotides (ASOs) can induce sequence-specific cleavage of complementary RNAs by endonuclease RNase H1 (1). ASOs are widely used as both research tools and therapeutic agents (2). Most ASOs in clinical use and development have phosphorothioate (PS) backbones (3,4); in these oligonucleotides, sulfur replaces one of the non-bridging oxygen atoms of the phosphodiester linkage (5). The PS backbone is known to enhance protein binding in general (6,7). Typically, RNase H1-dependent PS-ASO gapmers are modified at 5 nucleotides with 2′-O-methoxyethyl (MOE) or 3 nucleotides with 2′-constrained ethyl (cEt) of 5′-end and 3′-end wing to enhance potency and other pharmacological properties (8,9).
Although pharmacokinetic properties of PS-ASOs have been well-studied in animals and humans (10), how these molecules are taken into cells is not fully understood (11). In the absence of transfection reagents or uptake-enhancing modifications such as N-acetyl galactosamine (12), efficiency of internalization depends on cell type and is thought to be a two-step process (13). The first step, adsorption, involves binding of PS-ASOs to extracellular proteins, membrane-associated proteins, or extracellular domains of transmembrane proteins. Internalization then occurs through endocytic pathways (14). Uptake pathways resulting in pharmacological effects are considered ‘productive’ (11). It is critical to identify which specific cell-surface proteins mediate PS-ASO uptake in productive and non-productive manners to make these agents more effective therapeutics.
Previous studies showed that incorporation of epidermal growth factor (EGF) into polyethylenimine (PEI) polymers resulted in a 10- to 100-fold increase in gene delivery (15–18). EGF is a 53-residue polypeptide that binds specifically with high affinity to the EGF receptor (EGFR) through hydrophobic interactions (19,20). The binding of EGF triggers the dimerization and internalization of the receptor at coated pits through a clathrin-mediated mechanism (21–24). These previous findings raised the possibility that an EGF-dependent or EGF-independent, EGFR-specific pathway might facilitate productive cellular uptake of PS-ASOs.
EGFR is a receptor tyrosine kinase with a large extracellular region, a single transmembrane (TM) domain, an intracellular juxta membrane (JM) region, and a cytoplasmic domain (23). The extracellular region of EGFR contains two homologous ligand binding domains, and the cytoplasmic region contains the tyrosine kinase domain and a C-terminal regulatory domain. Binding of EGF to the extracellular region triggers tyrosine phosphorylation of the cytoplasmic domain, which initiates EGFR endocytosis and degradation (25). EGFR is highly expressed in carcinomas and selected cancer cell lines such as the epidermoid A431 cells (21). In these carcinoma cells, EGFR is constitutively internalized and mediates a series of signaling cascades that are required for the survival of carcinoma cells (26). Thus, we sought to determine whether EGFR interacts with PS-ASOs and mediates their productive cellular uptake.
In this article, we first show that EGFR interacts with PS-ASOs. We then focus on the details of PS-ASO trafficking along EGFR-associated endocytic pathways. We find that PS-ASOs appear to travel together with EGF and EGFR from clathrin-coated pit structures, through early endosomes (EEs) to late endosomes (LEs), where EGFR may contribute to the release of PS-ASOs from LEs. We also show that EGFR mediated uptake is ‘productive’, i.e. observed decreased or increased PS-ASO mediated target reduction by reducing or overexpressing EGFR, respectively in in vitro cell systems. Thus, we conclude that one productive PS-ASO uptake pathway is mediated by EGFR.
MATERIALS AND METHODS
Reagents
Antibodies, siRNAs, and quantitative real-time PCR (qRT-PCR) primer probe sets are described in Supplementary Data. ASO sequences and chemical modifications are also listed in Supplementary Data.
Cell culture, transfection, PS-ASO free uptake and activity assay
A431 and HEK cells were grown according to the protocols provided by the American Type Culture Collection. Cells were seeded at 70% confluency one day before transfection or drug treatment. siRNAs were transfected at 3 nM final concentration using RNAiMAX (ThermoFisher Scientific) according to the manufacturer's protocol. A431 cells were transfected with plasmid expressing Rab5(Q79L) at 2 μg/million cells using Avalanche Transfection Reagent (EZ Biosystems) according to the manufacturer's protocol. At 48 h after transfection, cells were re-seeded in either 96-well plates or MatTek collagen-coated dishes at 50% confluency. Cells were incubated with PS-ASOs for 16 h, and then the PS-ASO activity assay or immunofluorescence analysis was performed. HEK cells were transfected with 2 μg plasmid for expression of EGFR using Lipofectamine 3000 (ThermoFisher Scientific) at 2 μg/million cells according to the manufacturer's protocol. These HEK cells were grown in G418, Geneticin, for 2 weeks to select cell clones with overexpression of EGFR for further use.
Analyses of protein binding with PS-ASOs
Analyses of protein binding to PS-ASOs were performed as described previously (7). Briefly, agarose neutravidin beads (ThermoFisher Scientific) were incubated with biotinylated PS-ASO (IONIS ID 451104) at 4°C for 1 h in buffer containing 50 mM Tris–HCl (pH 7.5), 100 mM KCl, 5 mM EDTA, and 0.1% NP-40. Beads were then incubated for 30 min in the same buffer supplemented with 10 mg/ml BSA and 0.2 mg/ml yeast tRNA. After washing with this buffer, PS-ASO-coated neutravidin beads were added to 1 mg cell lysate of A431 prepared in RIPA buffer (50 mM Tris–HCl, pH 7.4, 1% Triton X-100, 150 mM NaCl, 0.5% sodium deoxycholate, and 0.5 mM EDTA) supplemented with protease inhibitor cocktail (ThermoFisher Scientific). After incubation at 4°C for 3 h, the beads were washed with buffer containing 50 mM Tris–HCl (pH 7.5), 300 mM KCl, 5 mM EDTA, 0.1% NP-40 and 0.05% SDS. Bound proteins were eluted from 30 μl aliquots of the beads using 50 μl of 50 μM PS-ASO with various modifications by incubating for 10 min at room temperature. The eluted proteins were separated on 4–12% SDS-PAGE and were visualized by silver staining or were analyzed by western blot.
BRET assay for binding affinity
EGFR NanoLuc (Nluc) fusion protein construction, expression, and purification were performed essentially as described (27). Briefly, the human EGFR (NM_201282) extracellular and transmembrane domain was amplified by PCR from the full-length cDNA clone (Origene RC217223). The forward PCR primer (5′-GCTAGCAGCCACCATGCGACCCTCCGGGACG-3′) was comprised of sequence complimentary to the EGFR sequence, including the AUG start codon and preceded by a Kozak sequence and an NheI site (GCTAGCAGCCACC), whereas the reverse primer (5′-GCGCCACATCGT TCGGAAGGACTCGAG) was complementary to the EGFR sequence up to amino acid 676, then followed with an XhoI site. The PCR amplified product was ligated into NheI and XhoI sites of the NanoLuc expression vector pFC32K Nluc CMV-Neo (Promega).
The EGFR NLuc fusion protein was expressed by transfecting the plasmids into 6 × 105 HEK 293 cells using Effectene transfection reagent according to the manufacturer's protocol (Qiagen). Following a 24-h incubation, cells were removed from the plate by trypsinization, washed with PBS, then resuspended in 250 μl Pierce IP Lysis Buffer (Thermo Scientific). Lysates were incubated 1 h at 4°C while rotating, then debris pelleted by centrifugation at 15 000 rpm for 5 min. The fusion protein was immunoprecipitated using EGFR antibody overnight at 4°C followed by incubation with Pierce Protein G Magnetic Beads (Thermo Scientific) for 2 h. Beads were then washed 4 times with IP Lysis Buffer and finally suspended in 250 μl 2× binding buffer (0.2 M NaCl, 40 mM Tris, pH 7.5, 2 mM EDTA, 0.02% NP-40) before the binding assay.
BRET binding affinity assays were performed using 106 RLU/well of immune-precipitated EGFR NLuc fusion protein as described (27).
RNA preparation and qRT-PCR
Total RNA was prepared from cells grown in 96-well plates (around 104 cells per well) using a Qiagen RNeasy mini kit. qRT-PCR was performed using TaqMan primer probe sets essentially as described previously (7). Briefly, ∼50 ng total RNA in 5 μl water was briefly mixed with 0.3 μl solution containing 10 μM of each of forward and reverse primers and 3 μM fluorescently labeled probe, 0.3 μl RT enzyme mix (Qiagen), 4.4 μl RNase-free water, and 10 μl of 2 × PCR reaction buffer. Reverse transcription was performed at 48°C for 10 min; 40 cycles of PCR were conducted at 94°C for 20 s and 60°C for 20 s using the StepOne Plus RT-PCR system (Applied Biosystems). The mRNA levels were normalized to the amount of total RNA present in each reaction as determined for duplicate RNA samples by Ribogreen assay (ThermoFisher Scientific).
Immunofluorescence staining
Cells were fixed with 4% paraformaldehyde for 20 min at room temperature and were permeabilized with 0.05% saponin (Sigma) in PBS for 5 min. Cells were treated with blocking buffer (1 mg/ml BSA in PBS) for 30 min and then incubated with primary antibodies (1:100–1:200 in blocking buffer) at room temperature for 2–4 h or at 4°C overnight. After three washes with PBS, cells were incubated with fluorescently labeled secondary antibodies (1:200 in blocking buffer) at room temperature for 1–2 h. After washing, slides were mounted with Prolong Gold anti-fade reagent with DAPI (ThermoFisher Scientific) and imaged using a confocal microscope (Olympus FV-1000). Co-localization between PS-ASOs and different organelles was analyzed using FV10-ASW 3.0 viewer software.
Protein isolation and western blotting
Cells were lysed, and samples were incubated at 4°C for 30 min in RIPA buffer. Proteins were separated on 4–12% NuPAGE Bis-Tris gradient gels containing SDS (ThermoFisher Scientific) and electroblotted onto PVDF membranes using the iBLOT transfer system (ThermoFisher Scientific). The membranes were blocked with 5% non-fat dry milk in PBS at 4°C for 30 min. Membranes were then incubated with primary antibodies at room temperature for 1 h or at 4°C overnight. After three washes with PBS, the membranes were incubated with appropriate HRP-conjugated secondary antibodies (1:2000) at room temperature for 1 h, followed by image development using ECL reagents (Abcam) and quantification of protein levels using ImageLab (Bio-Rad).
Membrane binding assay for EGFR and EGF to PS-ASOs
Purified recombinant EGF (PHG0311L, ThermoFisher Scientific) or EGFR protein (PV3872, ThermoFisher Scientific) were incubated with FITC-labeled 2′-MOE gapmer PS-ASOs (IONIS ID 256903) or phosphodiester ASO (synthesized by Integrated DNA Technologies) in 100 μl binding buffer (20 mM Tris–HCl, pH 7.5, 150 mM NaCl, 1 mM DTT, 10% glycerol) as previously described (28). Reactions contained purified EGF at concentrations ranging from 3 nM to 3 μM or purified recombinant EGFR protein at concentrations ranging from 5 to 150 nM. After 1 h incubation at 37°C, the samples were loaded on a Hybond ECL nitrocellulose membrane (GE Healthcare) and soaked in wash buffer containing 20 mM Tris–HCl, pH 7.5 and 250 mM NaCl. The protein-bound ASOs were transferred to the membrane by applying a vacuum in a 96-well Bio-Rad Bio-Dot apparatus. After washing, the membranes were then air-dried and scanned using a PhosphorImager (GE Healthcare). The signal intensity was quantified using ImageJ, fitted as Hill Slope and the Kds were calculated using Prism.
Metabolic labeling of EGFR in A431 cells
Cells were starved 20 min in Met- and Cys-free media followed by 1 h chase with [35S]-labeled Met and Cys. After the chase, cell samples were collected in RIPA buffer at indicated times, and cell lysates were immunoprecipitated with EGFR antibody. Labeled EGFR immunocomplexes were resolved by SDS-PAGE and visualized and quantified by autoradiography using PhosphoImager.
Flow cytometry
Cy3-labeled PS-ASO was added to A431 cell culture. After 3 h, cells were washed with PBS, trypsinized, and re-suspended in PBS supplemented with 3% fetal bovine serum for analysis by flow cytometry using an Attune NxT Flow Cytometer (ThermoFisher Scientific).
In vivo study
Seven-week-old male BALB/c mice (Charles River Laboratories) were treated with PBS (Groups 1–4), control 3–10-3 cEt PS-ASO (IONIS ID 636727) (Groups 5–8), or PS-ASO targeting mouse EGFR (IONIS ID 542794) (Groups 9–12), twice, three days apart, at 50 mg/kg body weight to deplete liver EGFR. Four days after treatment of mouse EGFR ASO, mice were further injected with a 5–10–5 MOE PS-ASO targeting Factor XI (IONIS ID 404071) at different dose (0, 20, 40, 60 mg/kg). Animals were sacrificed 48 h after Factor XI ASO treatment for liver mRNA and protein expression analyses.
RESULTS
EGFR directly interacts with PS-ASOs
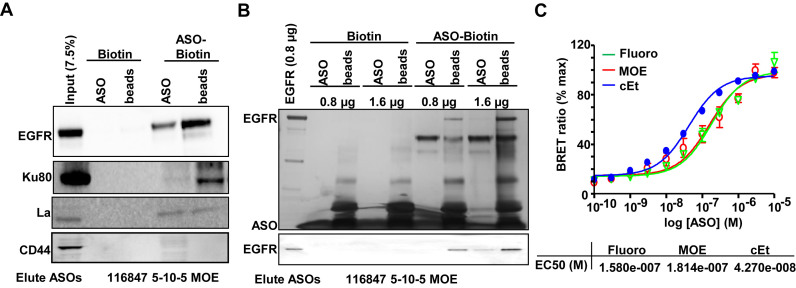
To determine whether EGFR interacts with PS-ASOs, we performed protein binding analyses using biotinylated 2′-MOE gapmer PS-ASOs (IONIS ID 451104) bound to neutravidin agarose beads. Beads were incubated with total A431 cell lysates, and bound proteins were eluted from the beads by competition using non-biotinylated 2′-MOE gapmer PS-ASO (IONIS ID 116847) with the same sequence. The eluted proteins and the residual bead-bound proteins were both dissolved in SDS-sample buffer and separated using SDS-PAGE and analyzed by western blot. EGFR was detected by western analyses in both eluted and bead-bound fractions as were Ku80 and La, which are known PS-ASO-interacting proteins (Figure 1A) (7). Another cell-surface glycoprotein CD44 was not detected in either fraction (29). EGFR could be eluted by 2′-cEt or 2′- Fluoro gapmer PS-ASOs as well (Supplementary Figure S1A). In addition, EGFR could also be eluted by three other competing PS-ASOs with different sequences (Supplementary Figure S1B). EGFR was similarly detected in eluted and bead-bound fractions using another biotinylated 2′-MOE gapmer PS-ASO with different sequence (Supplementary Figure S1C). These observations indicate that EGFR can interact with different PS-ASOs containing different 2′-modifications or sequences.
Figure 1.
EGFR interacts with PS-ASOs. (A) Western analyses of proteins that were eluted with PS-ASOs (ASO, IONIS ID 116847) or that remained on the biotinylated PS-ASO coated beads (beads). Neutravidin agarose beads pre-coated with either biotinylated 2′-MOE PS-ASOs (ASO-Biotin, IONIS ID 451104) or control biotin (Biotin) were incubated with cell lysates of A431, washed, and proteins were eluted using non-biotinylated PS-ASOs (IONIS ID 116847), which has the same modification and sequences (CTGCTAGCCTCTGGATTTGA) as the biotinylated PS-ASO (IONIS ID 451104). The experiments were repeated at least three times and representative results are shown. (B) Proteins eluted with PS-ASOs (ASO, IONIS ID 116847) or that remained on the beads (beads) were visualized by silver staining (Upper panel) in a 4–12% gradient SDS-PAGE. Blot of aliquoted proteins from the above experiment was probed for EGFR by western (lower panel). Affinity selection was performed using the biotinylated 2′-MOE PS-ASO (ASO-Biotin, IONIS ID 451104) or control biotin (Biotin), as (A), but with either 0.8 μg or 1.6 μg purified recombinant EGFR. The experiments were repeated at least three times and representative results are shown. (C) Relative affinities for 2′-Fluoro, 2′-cEt and 2′-MOE PS-ASOs were determined by competitive ASO binding to the NLuc/EGFR fusion protein containing extracellular domain and transmembrane domain in a BRET assay. 10 nM 5′- Alexa Fluor 594 conjugated cEt PS-ASO (Ionis ID: 766636) was competed with unconjugated 2′-Fluoro, 2′-MOE, or 2′-cEt PS-ASO at concentrations from 0.1 to 10,000 nM. Relative EC50 values are shown.
To determine whether the interaction between EGFR and PS-ASOs is direct, we conducted protein binding analyses using purified recombinant EGFR protein. Purified EGFR was detected in both eluted and bead-bound fractions as observed in silver-stained gels and by western blot (Figure 1B). This observation indicates that EGFR can directly interact with PS-ASOs.
To evaluate whether the interaction occurs between the PS-ASOs and the extracellular domain of EGFR and to understand the characteristics of the interaction, we performed a bioluminescence resonance energy transfer (BRET) assay utilizing the fusion protein of Nanoluc luciferase (Nluc) and the extracellular domain and transmembrane domain of EGFR as the BRET donor and PS-ASO tagged at the 3′ end with Alexa Fluor 594 as the BRET acceptor. The BRET signal can be used to construct a binding curve in which the mid-point is equivalent to the dissociation constant (EC50 or Kd) (30). The Nluc/EGFR fusion protein was expressed in HEK cells and identified at the expected molecular weight around 90 kDa (Supplementary Figure S2A). BRET signals were detected (Figure 1C) between Nluc/EGFR and 2′-MOE, 2′-cEt or 2′-Fluoro gapmer PS-ASO, indicating an interaction between PS-ASO and the transmembrane and extracellular domain of EGFR (Figure 1C).
BRET assays were also performed between Nluc/EGFR and other PS-ASOs to quantitively examine the effect of sequences of PS-ASOs on their binding affinity. Additional 10 sequences for each modification, including cEt, MOE or 2′-Fluoro, were tested. As expected, sequences can significantly affect the binding affinity to EGFR (EC50) (Supplementary Figure S2B–D). We also evaluated PS homopolymers to determine the overall range of affinities for different sequences. Except for polyG that can form intramolecular structures (31), all other polymers were tested and found to bind EGFR with different EC50s (Kd) (Supplementary Figure S2E). Thus, all PS-ASOs tested could bind to EGFR with affinities that are consistent with a biologically relevant interaction (27). Given this binding commonality between PS-ASOs and EGFR, we used 2′-MOE gapmer PS-ASOs in subsequent studies.
PS-ASOs traffic together with EGF and EGFR
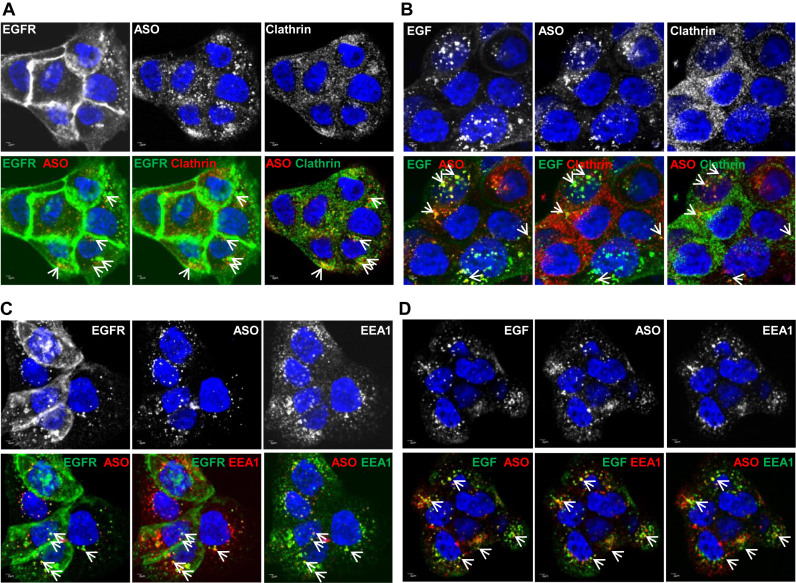
We next monitored internalization and trafficking of PS-ASOs in the presence of EGF. Upon EGF binding, EGFR is internalized through coated pits in a clathrin-mediated mechanism and then it traffics along endocytic pathways (24). A431 cells were incubated with Cy3-labeled PS-ASOs (IONIS ID 446654) and unlabeled-EGF (Figure 2A and C) or FITC-EGF (Figure 2B and D) for 30 mins and stained with antibodies to clathrin, EGFR, or EEA1, an EE marker. Co-localization was observed between PS-ASOs and EGFR and between PS-ASOs and EGF in both clathrin-stained (Figure 2A and B) and EEA1-stained structures (Figure 2C and D). Co-localization was also confirmed in Z-stack experiments (Supplementary Figures S3 and S4). These observations indicate that PS-ASOs can be internalized as cargos together with EGF and EGFR in clathrin-containing vesicles. It is likely that EGFR mediates this cellular uptake pathway for PS-ASOs in A431 cells.
Figure 2.
PS-ASOs traffic together with EGF and EGFR through the endocytic pathway. (A) Representative images of immunofluorescent staining for clathrin and EGFR in A431 cells incubated with non-labeled EGF and Cy3-labeled PS-ASOs for 30 min. The nuclei were stained with Hoechst 33342 (blue). The arrow indicates co-localization between EGFR (green) and PS-ASOs (red); between EGFR (green) and clathrin (red); between PS-ASOs (red) and clathrin (green). Scale bars, 2 μm. (B) Representative images of immunofluorescent staining for clathrin in A431 cells incubated with FITC-EGF and Cy3-labeled PS-ASOs for 30 min. The nuclei were stained with Hoechst 33342 (blue). The arrow indicates co-localization between EGF (green) and PS-ASOs (red); between EGF (green) and clathrin (red); between PS-ASOs (red) and clathrin (green). Scale bars, 2 μm. (C) Representative images of immunofluorescent staining for EEA1 and EGFR in A431 cells incubated with non-labeled EGF and Cy3-labeled PS-ASOs for 30 min. The nuclei were stained with Hoechst 33342 (blue). The arrow indicates co-localization between EGFR (green) and PS-ASOs (red); between EGFR (green) and EEA1 (red); and between PS-ASOs (red) and EEA1 (green). Scale bars, 2 μm. (D) Representative images of immunofluorescent staining for EEA1 in A431 cells incubated with FITC-EGF and Cy3-labeled PS-ASOs for 30 min. The nuclei were stained with Hoechst 33342 (blue). The arrow indicates co-localization between EGF (green) and PS-ASOs (red); between EGF (green) and EEA1 (red); between PS-ASOs (red) and EEA1 (green); Scale bars, 2 μm.
PS-ASOs interact with EGFR more tightly than with EGF
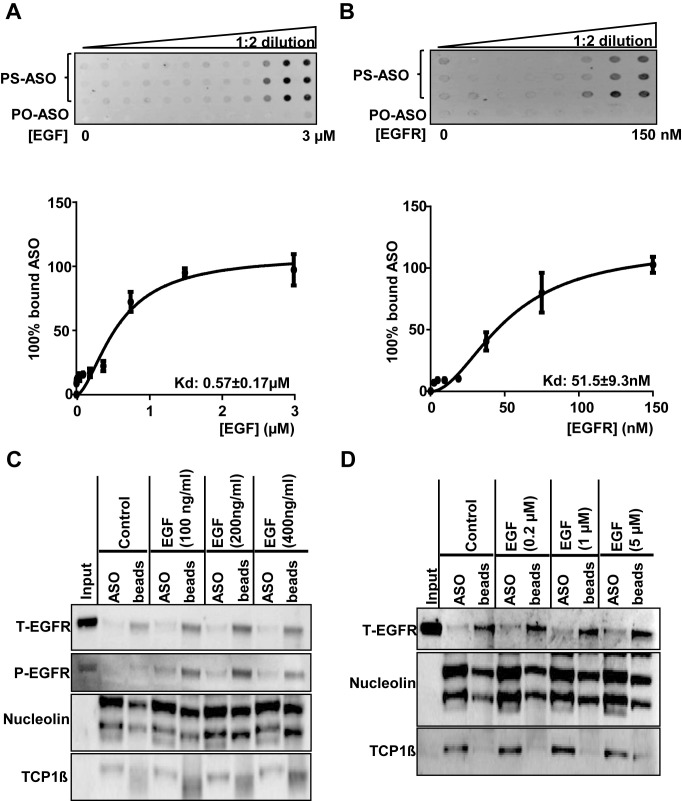
As PS-ASOs co-localized with EGF bound to EGFR, we examined the possibility that PS-ASOs interact with EGF. We determined the binding constants of EGF or EGFR for PS-ASOs in a membrane binding assay with purified proteins. FITC-labeled PS-ASO (IONIS ID 256903) was incubated with purified EGF or EGFR. The protein-bound PS-ASOs were transferred to a nitrocellulose membrane, and the signal intensity was determined. The binding constant of PS-ASOs to EGF was ∼570 nM and that to EGFR was ∼51 nM (Figure 3A and B). These results indicate that PS-ASOs bind EGFR far more tightly than EGF, suggesting that their co-trafficking is mainly mediated through the interaction between PS-ASOs and EGFR.
Figure 3.
PS-ASOs interact with EGFR more tightly than with EGF. (A and B) The membrane binding assay for EGF or EGFR and PS-ASOs. EGF at concentrations ranging from 3 nM to 3 μM or purified recombinant EGFR protein at concentrations ranging from 5 nM to 150 nM were incubated with PS-ASOs for 1 h at 37°C and the samples were loaded on a Hybond ECL nitrocellulose membrane. The signal intensities retained in nitrocellulose membrane for the protein bound form of the FITC-labeled 2′-MOE gapmer PS-ASO (PS-ASO, IONIS ID 256903) or phosphodiester ASO (PO-ASO) were quantified and the binding curves for EGF (A) and EGFR (B) were plotted using Prism. The error bars represent standard deviations from three experiments; (C) EGFR (total EGFR, T-EGFR) was blotted after PS-ASO affinity selection as shown in Figure 1, except that cell lysates were from A431 cells treated with indicated concentrations of EGF. The same blot was probed sequentially with antibodies to phosphorylated EGFR (P-EGFR), Nucleolin, and TCP1β. The experiments were repeated at least three times and representative results are shown; (D) EGFR (total EGFR, T-EGFR) was blotted after affinity selection as shown in Figure 1, except that the bound proteins were eluted by PS-ASOs in the presence of EGF at indicated concentrations. The same blot was probed sequentially with different antibodies against other ASO-binding proteins (Nucleolin and TCP1β). The experiments were repeated at least three times and representative results are shown.
We then examined whether EGF could interfere with the binding between PS-ASOs and EGFR. We compared levels of EGFR recovered from beads bound with biotinylated PS-ASOs (IONIS ID 451104) between lysates of cells treated with EGF or untreated (Figure 3C). The recovery rate of EGFR was comparable with and without EGF. Nucleolin and TCP1β, known PS-ASO-interacting proteins, were detected and served as assay controls (Figure 3C). The blot of phosphorylated EGFR (P-EGFR) indicates a response of cells to EGF treatment (Figure 3C). In addition, we included EGF with PS-ASOs at the elution step to see whether EGF altered the ASO elution of EGFR from beads bound with PS-ASOs. EGF did not significantly change the ASO elution of EGFR, nucleolin or TCP1β from the beads (Figure 3D). Thus, these results suggest that EGF does not interfere with the binding between PS-ASOs and EGFR, and that PS-ASOs and EGF likely bind to different sites on EGFR.
A BRET assay was also performed between Nluc/EGFR and an Alexa-594-tagged PS-ASO in the presence of EGF. The BRET signal between the Nluc/EGFR and PS-ASOs was not significantly different in the presence and absence of EGF (Supplementary Figure S5). Together, these results exclude the possibility that binding of PS-ASOs to EGF mediates the trafficking of PS-ASOs in EGFR-containing vesicles. These observations also demonstrate that the binding of PS-ASOs is not competitive with the binding of EGF to EGFR, suggesting that the PS-ASOs and EGF have distinct binding sites.
PS-ASOs do not affect EGFR synthesis, degradation, recycling or response to EGF
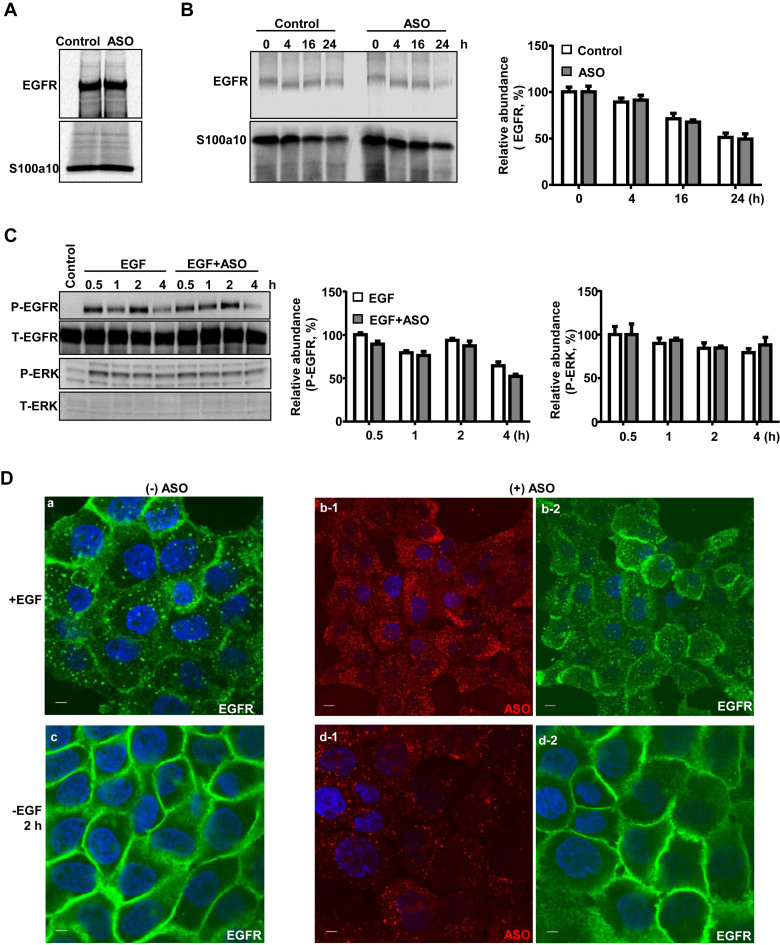
As PS-ASOs interact with EGFR, it is important to know whether PS-ASOs alter EGFR function. We first performed pulse-chase analyses in A431 cells labeled with [35S]-Met and [35S]-Cys to examine how PS-ASOs affect EGFR synthesis and degradation. Newly synthesized EGFR was immunoprecipitated from lysates of cells treated with or without PS-ASOs and separated by SDS-PAGE and visualized using autoradiography. Quantification of the gel images showed that EGFR synthesis (Figure 4A) and degradation rates (Figure 4B) were comparable in the presence and absence of PS-ASOs. In addition, more than 10 sequences of PS-ASOs were tested for potential effects on downstream signaling of EGFR. None of them significantly induced the phosphorylation of EGFR and ERK (Figure 4C, Supplementary Figure S6A and data not shown). These observations show that PS-ASOs, in general, do not induce EGFR signaling.
Figure 4.
PS-ASOs do not affect EGFR synthesis, degradation and recycling. (A) A431 cells were pulse labeled with [35S]-Met/Cys for 50 min. EGFR and S100a10 were immunoprecipitated using their specific antibodies, and analyzed by SDS-PAGE. (B) Intracellular levels of nascent EGFR were chased and analyzed by SDS-PAGE at indicated times after the labeling and were visualized and quantified by autoradiography, as in (A). S100a10 was detected and used as a loading control. (C) Protein samples were analyzed by western analyses from A431 cells treated with EGF or EGF and PS-ASOs. The blot was probed sequentially with antibodies specific to the various proteins and the images were quantified by ImageLab (Bio-Rad). (D) Representative images of immunofluorescent staining for EGFR (green) in A431 cells incubated with EGF (a) or EGF and Cy3-labeled PS-ASOs (red) (b-1 and b-2) for 16 h, and then 2 h after the removal of EGF (c) or EGF and Cy3-labeled PS-ASOs (d-1 and d-2). The nuclei were stained with Hoechst 33342 (blue). Scale bars, 5 μm.
The EGF-induced internalization and recycling of EGFR were also evaluated in the presence and absence of Cy3-labeled PS-ASOs (IONIS ID 446654). When cells were treated with EGF, EGFR staining was observed in plasma membrane and intracellularly, indicating that EGFR was internalized (Figure 4D, a). This pattern was not changed in cells co-treated with EGF and PS-ASOs (Figure 4D, b). Upon EGF removal, EGFR staining was mainly observed in plasma membrane, indicative of a recycling process (Figure 4D, c and d). This redistribution of EGFR to plasma membrane was not affected by PS-ASOs (Figure 4D, c and d). The effect of five other sequences of PS-ASOs on EGFR internalization and recycling were also evaluated using a PS-ASO antibody. None of them appeared to affect EGFR internalization and trafficking (Supplementary Figure S6B). These observations suggest that the normal function of EGFR is not compromised by the presence of PS-ASOs, and reiterate that EGF and PS-ASOs bind to different regions of EGFR.
EGF treatment increases PS-ASO activity in A431 cells
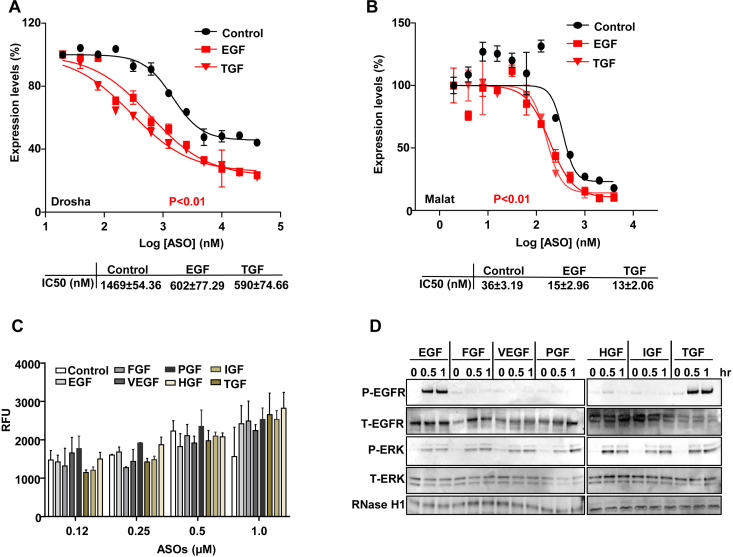
Since PS-ASOs can be internalized together with EGFR, we asked whether promoting this internalization process could increase PS-ASO activity. EGF drives EGFR internalization and activates its downstream signaling pathways, so we examined whether EGF treatment could increase PS-ASO activity in A431 cells. Cells were pre-treated with EGF, insulin growth factor (IGF), transforming growth factor (TGF), vesicular endothelial growth factor (VEGF), hepatocyte growth factor (HGF), platelet growth factor (PGF), or fibroblast growth factor (FGF) for 4 h before they were incubated with either Drosha- or Malat1-specific PS-ASOs. Only EGF and TGF treatment significantly increased activities of both PS-ASOs with the decrease in IC50 values ∼2- to 3-fold (Figure 5A, B and Supplementary Figure S4).
Figure 5.
EGF increases the activity but not uptake of PS-ASOs in A431 cells. (A and B) Reduction of Drosha (A) and Malat1 (B) RNA was determined by qRT-PCR analysis of A431 cells treated with EGF or TGF for 4 h before they were incubated with PS-ASOs. Data are relative to untreated cells. The error bars represent standard deviations from 3 independent experiments. Activity curves were fitted from data plotted in panels and IC50 was calculated based on a non-linear regression model. P (in red) <0.01, EGF or TGF treatment versus control; P values were computed by one-way ANOVA as the concentrations of PS-ASOs were set as random effect. (C) Intracellular fluorescence of Cy3-PS-ASOs was quantified by flow cytometry to determine PS-ASO uptake (RFU) in A431 cells treated with different growth factors for 4 h before they were incubated with Cy3-PS-ASOs for 2 h. The error bars represent standard deviations from 3 independent experiments. (D) Western analyses of proteins from A431 cells treated with different growth factors for indicated times. The blot was probed sequentially with different antibodies: phosphorylated EGFR (P-EGFR), total EGFR (T-EGFR), phosphorylated ERK (P-ERK), total ERK (T-ERK) and RNase H1.
Cells pre-treated with the same set of growth factors were also incubated with Cy3-labeled PS-ASOs at different concentrations for 4 h, and then PS-ASO uptake was analyzed by FACS. The levels of total internalized PS-ASOs were not significantly different among cells treated with different growth factors (Figure 5C). Lysates from those cells were further subjected to western analyses. The results showed that EGF and TGF triggered phosphorylation of both EGFR and ERK but other growth factors only activated ERK pathways (Figure 5D). In addition, none of these growth factors significantly changed protein levels of RNase H1. These results indicate that both EGF and TGF increased PS-ASO activity, potentially by promoting EGFR mediated PS-ASO internalization. As EGF or TGF treatment did not markedly affect total PS-ASO uptake, levels of PS-ASO internalization mediated by EGFR may not be substantial enough to be detected by analysis of total PS-ASO cellular levels.
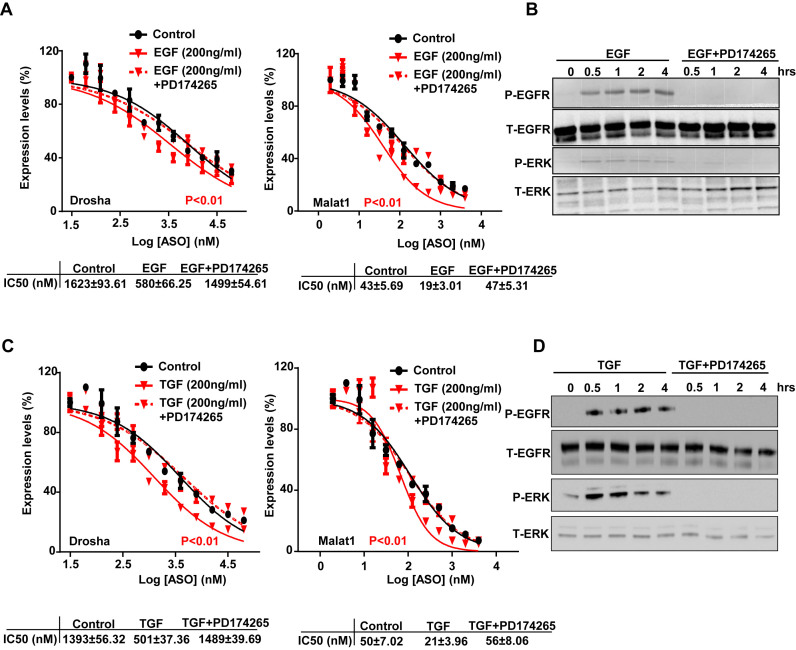
PD174265 reverses EGF and TGF driven increase in PS-ASO activity
To confirm that EGF and TGF increase PS-ASO activity through a mechanism involving EGFR, we co-treated cells with EGF and a specific EGFR tyrosine kinase inhibitor, PD174265 (32), to block EGFR internalization and signaling. PS-ASO activities were compared among cells that were pre-treated with EGF alone or EGF with PD174265 or cells without any treatment before they were incubated with either Drosha- or Malat1-specific PS-ASOs. In cells treated with EGF and PD174265, PS-ASO activities were similar to those in the absence of EGF with comparable IC50 values (Figure 6A). The results confirmed that EGF increased PS-ASO activities, but PD174265 abolished this effect. Western analyses of cell lysates showed that PD174265 completely blocked phosphorylation of EGFR and ERK (Figure 6B). Experiments were also conducted using TGF and PD174265. Similar to the experiment with EGF, PD174265 prevented TGF-induced phosphorylation of EGFR and ERK, and PS-ASO activities were similar to those in the absence of TGF with comparable IC50 values (Figure 6C and D). These results suggest that EGFR mediates the effects of EGF or TGF on PS-ASO activity.
Figure 6.
PD174265 reverses EGF or TGF driven increase in PS-ASO activity. (A) The levels of Drosha and Malat1 RNAs were quantified by qRT-PCR analysis of A431 cells treated with EGF or EGF and PD174265. Data are relative to untreated cells. The error bars represent standard deviations from 3 independent experiments. Activity curves were fitted from data plotted in panels based on a non-liner regression model and IC50 was calculated based on a non-linear regression model. P (in red) <0.01, EGF or TGF treatment versus control; P (in red) <0.01, EGF treatment versus control; P values were computed by one-way ANOVA as the concentrations of PS-ASOs were set as random effect. (B) Western analyses of proteins in A431 cells pre-treated with or without PD174265 followed by the treatment of EGF. The blot was probed sequentially with different antibodies for phosphorylated EGFR (P-EGFR), total EGFR (T-EGFR), phosphorylated ERK (P-ERK), total ERK (T-ERK). (C) Target reduction of Drosha and Malat1 RNAs was quantified by qRT-PCR analysis of A431 cells treated with TGF or TGF and PD174265. Data are relative to no PS-ASO control. The error bars represent standard deviations from three independent experiments. Activity curves were fitted from data plotted in panels and IC50 was calculated based on a non-liner regression model. P (in red) <0.01, TGF treatment versus control. P values were computed by One-way ANOVA as the concentrations of PS-ASOs were set as random effect. (D) Western analyses of proteins from A431 cells pre-treated with or without PD174265 followed by the treatment of TGF, as in (B).
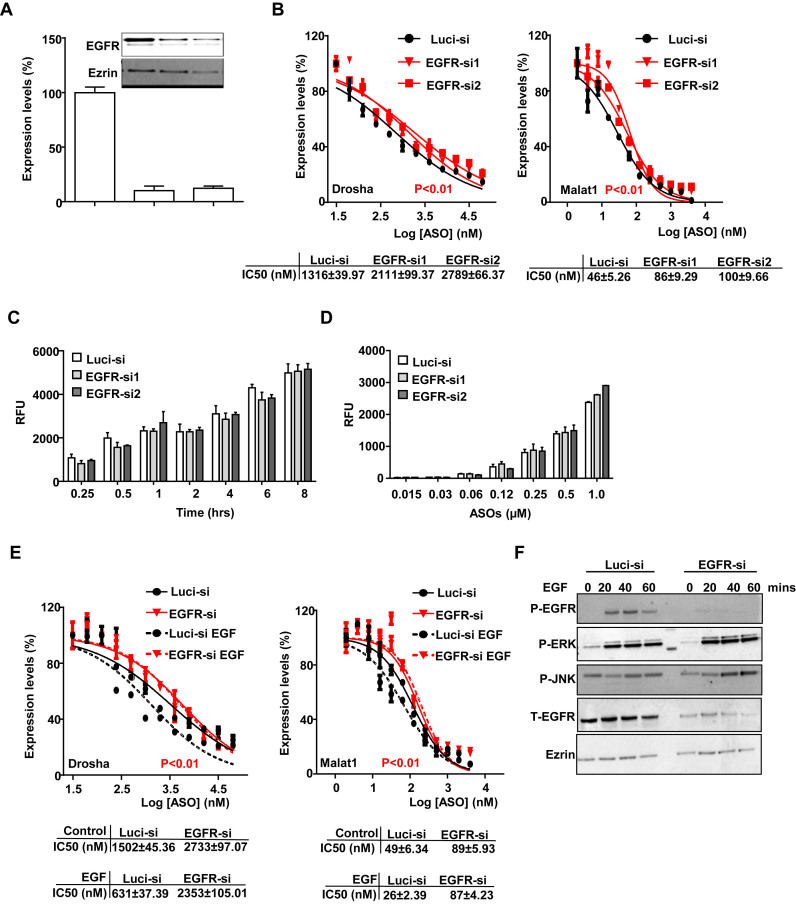
Reduction of EGFR decreases PS-ASO activity
We next reduced the levels of EGFR using siRNA and then evaluated PS-ASO activity. A431 cells were treated with two different siRNAs targeting EGFR and both decreased EGFR protein levels by more than 80% compared with control siRNA targeting luciferase (Figure 7A). Cells were then incubated with either Drosha- or Malat1-specific PS-ASOs. Reduction of EGFR decreased PS-ASO activities with the IC50 values elevated approximately 2-fold (Figure 7B). The levels of total PS-ASO uptake were also determined by flow cytometry using different concentrations of Cy3-labeled PS-ASOs in those cells. Uptake of Cy3-labeled PS-ASOs increased in a concentration-dependent manner as a function of time and did not depend significantly on the level of EGFR expression (Figure 7C and D). Thus, reduction of EGFR decreases PS-ASO activities without significantly changing total cellular uptake of PS-ASOs. Clearly, reduction of EGFR did not ablate activity, demonstrating that A431 cells have multiple pathways that can result in productive uptake of PS-ASOs.
Figure 7.
Reduction of EGFR decreases PS-ASO activity. (A) A431 cells were treated with control (Luci-si) or siRNAs targeting EGFR as indicated. RNA and protein levels of EGFR were determined by qRT-PCR and western analyses, respectively. RNA levels are relative to Luci-siRNA treated samples. Ezrin was served as a loading control. (B) Cells pre-treated with siRNAs for 48 h were incubated with PS-ASOs targeting either Drosha or Malat1 RNA for 16 h. RNA levels were quantified using qRT-PCR. The error bars represent standard deviations from three independent experiments. Activity curves were fitted from data plotted in panels and IC50 was calculated based on a non-linear regression model. P<0.01 (in red), EGFR-si1 or EGFR-si2 versus Luci-si; P values were computed by One-way ANOVA as the concentrations of PS-ASOs were set as random effect. (C and D) Intracellular fluorescence of Cy3-PS-ASOs was quantified by flow cytometry in siRNA-treated cells to determine uptake (RFU) as a function of time at 0.5 μM (C) or as a function of PS-ASO concentration at 2 h (D). The error bars represent standard deviations from three independent experiments. (E) qRT-PCR quantification of Drosha and Malat1 RNAs in A431 cells treated with control siRNA or EGFR siRNA, which were further incubated with EGF for 4 h before treatment with PS-ASOs. Data are relative to untreated cells. Activity curves were fitted from data plotted in panels and IC50 was calculated based on a non-liner regression model. The error bars represent standard deviations from 3 independent experiments. P< 0.01 (in red), EGFR-si versus Luci-si with or without EGF treatment; P values were computed by one-way ANOVA as the concentrations of PS-ASOs were set as random effect. (F) Western analyses for various proteins in A431 cells treated with control siRNA or siRNA targeting EGFR, which were incubated with EGF for indicated times.
To further confirm that the effect of EGF on PS-ASO activity is mediated through EGFR, we treated EGFR-depleted cells with EGF and examined whether the effect of EGF on PS-ASO activity was diminished. EGF increased activities of both Drosha- and Malat1-specific PS-ASOs in control cells with the decrease in IC50 values ∼2- to 3-fold, as in Figures 6 and 7, but that EGF effect was eliminated in EGFR-depleted cells (Figure 7E). Interestingly, results from western analyses of cell lysates showed that EGFR phosphorylation was significantly decreased but ERK phosphorylation was not changed in EGFR-reduced cells (Figure 7F). This observation suggests that it is the level of EGFR, and not downstream signaling mediated by EGFR, that regulates PS-ASO activity.
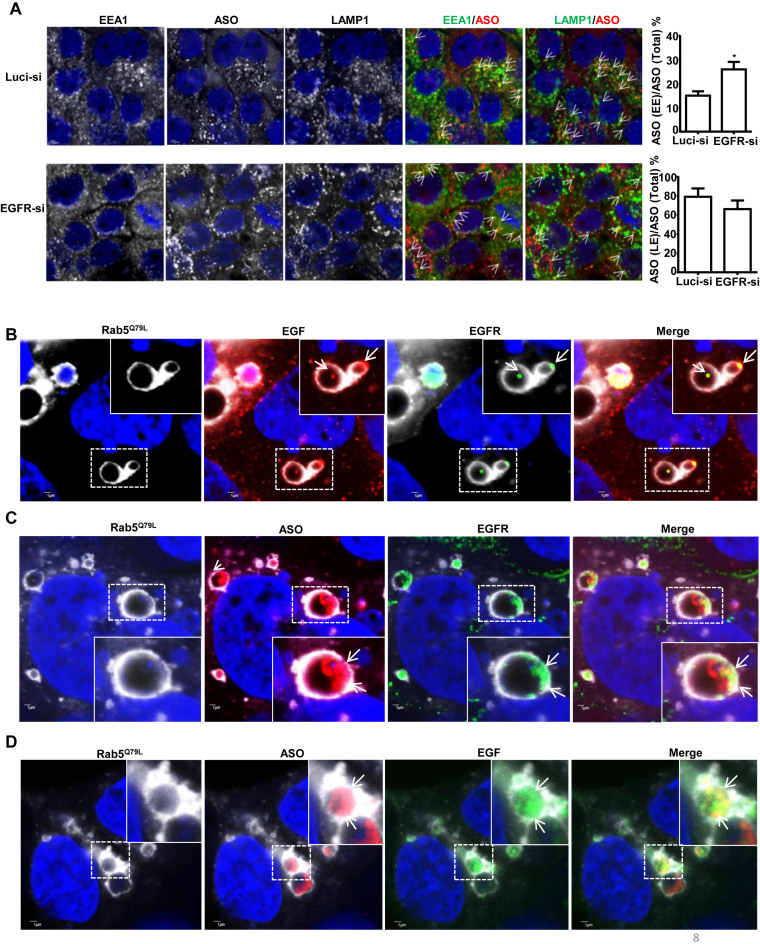
Reduction of EGFR delays PS-ASO trafficking from early to late endosomes
Previously, we reported that productive PS-ASOs are released primarily from the late endosomes, but not from early endosomes or during the fusion process from early to late endosomes (33). To evaluate the role of EGFR cargo in transporting ASO for late endosomal release, we examined the trafficking of PS-ASOs in control and EGFR-reduced cells. A431 cells were treated with EGFR siRNA for 48 h and were then incubated with 2 μM Cy3-labeled PS-ASO (IONIS ID 446654) for another 2 h. Cells were stained with EEA1 and LAMP1 to monitor the kinetics of PS-ASO trafficking from EEs to LEs. Quantification of PS-ASO-positive EEs (PS-ASO and EEA1 co-staining) and PS-ASO-positive LEs or lysosomes (PS-ASO and LAMP1 co-staining) showed that portions of EEs that contained PS-ASOs are increased in EGFR-reduced cells (Figure 8A). This result indicates that reduction of EGFR delays endocytic trafficking of PS-ASOs to LEs.
Figure 8.
Reduction of EGFR delays PS-ASO trafficking from early to late endosomes. (A) Representative images of immunofluorescent staining for EEA1 (green), and LAMP1 (green) in control (Luci-si) or EGFR reduced (EGFR-si) A431 cells incubated with Cy3-labeled PS-ASOs (red) for 2 h. Representative co-localization was indicated by arrows, between EEA1 or LAMP1 and PS-ASO; Scale bars, 2 μm. The PS-ASO-positive EEs or LEs were counted in 20 cells, and the percentage of the PS-ASO-positive EEs or LEs was calculated relative to the total numbers of the PS-ASO-positive foci; *P< 0.05, computed by Student's t-test. (B) Representative images of immunofluorescent staining for EGFR in RAB5(Q79L)-GFP overexpressing A431 cells treated with Alexa Fluor 647-EGF for 4 h. The nuclei were stained with Hoechst 33342 (blue). Enlarged images show co-localization, indicated by arrows, between EGFR (green) and EGF (red); Scale bars, 1 μm. (C) Representative images of immunofluorescent staining for EGFR in RAB5(Q79L)-GFP overexpressing A431 cells treated with Cy3-PS-ASOs and non-labeled EGF. The nuclei were stained with Hoechst 33342 (blue). Enlarged images show co-localization, indicated by arrows, between EGFR (green) and PS-ASOs (red); Scale bars, 1 μm. (D) Representative images of RAB5(Q79L)-GFP overexpressing A431 cells treated with Alexa Fluor 647-EGF and Cy3-PS-ASOs for 4 h. Enlarged images show co-localization, indicated by arrows, between PS-ASOs (red) and EGF (green); The nuclei were stained with Hoechst 33342 (blue). Scale bars, 1 μm.
To further evaluate the possibility that EGFR could participate in the EE to LE trafficking process, we examined the co-localization pattern between PS-ASOs and EGFR in cells with enlarged late endosomes. We overexpressed a constitutively active form of Rab5, Rab5(Q79L), which induces the formation of enlarged endosomes (34). A431 cells overexpressing RAB5(Q79L)-GFP were treated with Cy3-labeled PS-ASOs (IONIS ID 446654) and Alexa Fluor 647-EGF (Figure 8B and D) or unlabeled-EGF (Figure 8C) for 4 h and then stained for EGFR. EGF and EGFR were co-localized in dot-like structures, likely ILV, inside the lumen of enlarged endosomes marked by RAB5(Q79L)-GFP (Figure 8B). The localization pattern of PS-ASOs was very similar to that of EGF and EGFR, as we observed before, associated with ILV (35). Although the co-localization between PS-ASOs and EGFR or EGF was not as substantial as that between EGFR and EGF, it was abundant (Figure 8C and D). Their co-localization was also confirmed in Z-stack experiments (Supplementary Figure S8). In contrast, other ligands, such as dextran and LDL, were present in diffuse patterns inside enlarged endosomes without distinct co-localization with EGFR (Supplementary Figure S9). These observations suggest that the uptake of PS-ASOs is, at least in part, through EGFR at the cell surface, which mediates an efficient trafficking process for ASO migration from early to late endosomes to release.
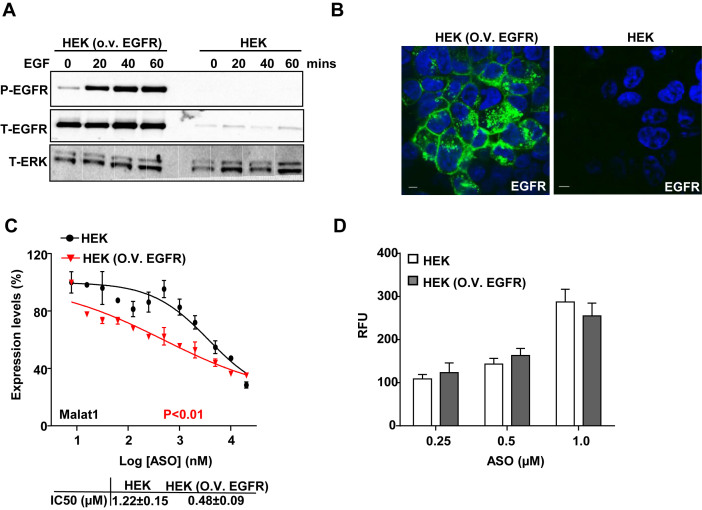
Overexpression of EGFR increases PS-ASO activity in HEK cells
Although EGFR is also expressed in liver hepatocytes, reduction of EGFR by a 3–10–3 cEt PS-ASO did not substantially affect 5–10–5 MOE PS-ASO activity in the liver (Supplementary Figure S10A–C). This is not surprising as the liver has multiple highly effective pathways for PS-ASO uptake (4) and EGFR levels in the liver are not as abundant as in A431 cells (36).
To further confirm that EGFR can mediate an uptake process that directs PS-ASOs into a productive release pathway, we generated a stable HEK cell line in which EGFR is overexpressed. HEK cells were used due to low basal expression levels of EGFR. In these engineered cells, levels of EGFR were at least 5-fold higher than that in control cells. Under conditions of EGFR overexpression, EGFR was responsive to EGF treatment as indicated by its autophosphorylation (Figure 9A) and it was mainly localized in the plasma membrane (Figure 9B). These results indicate that the overexpressed EGFR is functional. Overexpression of EGFR increased PS-ASO activity ∼3-fold (Figure 9C) but did not significantly change total levels of PS-ASO taken up by cells (Figure 9D). Thus, binding with EGFR facilitates PS-ASO trafficking through an endocytic pathway that results in productive intracellular release in two different cell lines.
Figure 9.
Overexpression of EGFR increases PS-ASO activity in HEK cells. (A) Western analyses for proteins in control or EGFR overexpressing HEK cells treated with EGF for indicated time. The blot was probed sequentially with different antibodies detecting phosphorylated EGFR (P-EGFR), total EGFR (T-EGFR) and total ERK (T-ERK) as loading control. (B) Representative images of immunofluorescent staining for EGFR (green) in HEK cells overexpressing EGFR. Nuclei were stained with Hoechst 33342 (blue). Scale bar, 5 μm. (C) Reduction of Malat1 RNA was quantified by qRT-PCR in HEK cells with or without overexpression of EGFR. Data are relative to untreated cells. Activity curves were fitted from data plotted in panels based on a non-linear regression model. The error bars represent standard deviations from 3 independent experiments. P< 0.01 (in red), HEK versus HEK (O.V. EGFR); P value was computed by One-way ANOVA as the concentrations of PS-ASOs were set as random effect; IC50 was calculated based on a non-liner regression model. (D) Intracellular fluorescence of Cy3-PS-ASO was quantified by flow cytometry to determine PS-ASO uptake (RFU) in HEK cells with or without overexpression of EGFR.
DISCUSSION
Receptor proteins have been implicated in binding and facilitating internalization of PS-ASOs into mammalian cells (11). Here we characterized an interaction between PS-ASOs and EGFR that appears to result in productive release of PS-ASOs as assessed by pharmacological activity. PS-ASOs were observed in clathrin-coated pits and EEs, co-localized with EGF and EGFR immediately after internalization. Reduction of EGFR delays PS-ASO trafficking from early to late endosomes and PS-ASOs and EGFR were co-localized inside enlarged late endosomes. These observations support our hypothesis that PS-ASO binding with EGFR initiates internalization and trafficking along endocytic pathways. We also demonstrated that EGFR is important for PS-ASO activities in A431 cells and that overexpression of EGFR increased PS-ASO activities in HEK cells. Thus, upon binding with PS-ASOs, EGFR can direct them to an appropriate trafficking process designated to LEs for their intracellular release.
Compared with ASOs with phosphodiester backbones, PS-ASOs are more hydrophobic and more promiscuously bind proteins with higher affinity (37,38). PS-ASOs bind with EGFR regardless 2′-modifications and sequences, but affinity does vary as a function of sequence. In addition, PS-ASOs do not interact with EGF as tightly as with EGFR or compete with EGF for its binding site at EGFR. Neither do they affect cell responses to EGF. Thus, PS-ASOs likely bind to regions of the extracellular domain of EGFR that differ from the site to which EGF binds. Upon binding, PS-ASOs can be loaded onto EGF-EGFR cargos trafficking along endocytic pathways.
Stabilin receptors were previously shown to bind and internalize PS-ASOs through a pathway that led to productive release (39). However, EGFR appears to bind and mediate PS-ASO release through a different mechanism. Although overexpression of either protein increased PS-ASOs activity, overexpression of EGFR did not increase total PS-ASO cellular uptake, whereas overexpression of Stabilin receptors did. Thus, EGFR does not affect total cellular uptake of PS-ASOs as much as Stabilin receptors, but directs PS-ASOs to a trafficking pathway that results in pharmacologically productive intracellular release.
The amount of internalized PS-ASO is not directly correlated with activity in different cell types (35). Differentially expressed cell surface proteins may route PS-ASOs into productive or nonproductive pathways depending on the type of cell (40). Such proteins constitute multiple pathways by which PS-ASOs enter cells and these pathways are independent, at least at the cell surface (11). If one pathway is inhibited, other pathways could compensate to maintain the total amount of cellular uptake. However, PS-ASOs taken up by other pathway(s) may not necessarily result in productive release of PS-ASOs. This is what we observed, as reduction of EGFR did not decrease total cellular uptake, but reduced activity of PS-ASOs. Thus, EGFR-mediated uptake could contribute to the productive pathway for PS-ASO intracellular release, but it is not the only productive pathway in A431 cells. Our observations reinforce the notion that the rate limiting step of PS-ASO activity is productive PS-ASO intracellular release but not cellular uptake (35).
It is believed that binding of PS-ASOs to different surface proteins results in internalization through different routes, some of which result in desired pharmacological activity and others that do not (41). Given the varied protein binding profile for PS-ASOs and the relative abundance of each protein, the same protein may not have the same contribution to productive uptake in different cells or tissues. This is exactly what was observed in our in vivo and in vitro studies, as the reduction of EGFR caused a decrease in PS-ASO activity in A431 cells, but not substantially in mouse livers. The relative abundance of PS-ASO binding proteins could ultimately affect the roles of those proteins in productive uptake in individual cell types. EGFR, as an example, is significantly more abundant in A431 cells than in the liver (36); therefore, it can proportionally mediate more uptake of PS-ASOs, which makes its role more prominent in productive release in A431 cells than in the liver. On the other hand, the asialoglycoprotein receptor (ASGR) was found to act as a PS-ASO binding protein and mediate productive uptake of PS-ASOs in the liver (42). That could be due to its high abundance in the liver. Thus, our present work provides a practical explanation that varied levels of protein expression at the cell surface could contribute to levels of productive uptake in different cells or organs (13,41).
Uptake of PS-ASOs through EGFR results in increased PS-ASO activities in A431 cells, suggesting that EGFR facilitates PS-ASO sorting into compartments with more efficient release than other receptors/pathways. We observed that PS-ASOs are co-localized with EGFR inside enlarged late endosomes and reduction of EGFR delays PS-ASO trafficking from EEs to LEs, suggesting that EGFR can initiate an appropriate trafficking process for ASO migration from early to late endosomes, which are the key steps for ASO release (33). Since the punctate structures of co-localization between EGFR and PS-ASOs appear to be associated with ILVs (35), the possibility of EGFR-mediated release mechanism of PS-ASOs through ILVs by back fusion cannot be excluded (35).
Internalized ligands either dissociate from receptors, such as LDL, or stay associated with receptors, such as EGF, inside LEs (43). Although co-localization between EGFR and PS-ASOs was observed, not all PS-ASOs were at EGFR-containing loci. One explanation is that some PS-ASOs dissociate from EGFR after they are internalized. Another explanation is that receptors other than EGFR could also mediate PS-ASO internalization and trafficking to LEs. It was also shown that different PS-ASO sequences vary in potency in a manner that is not entirely consistent with affinities for targeted RNAs (11). Affinity to EGFR, as an example, does vary as a function of sequence. It is likely that EGFR mediates considerably better uptake of PS-ASO with some sequences than the others. One of our goals is to develop a catalog of cell surface proteins and pathways that lead to productive PS-ASO uptake and to understand the key steps in the internalization pathways as well as the key proteins responsible for PS-ASO trafficking.
As binding to some receptors and other proteins facilitates cellular uptake of PS-ASOs, medicinal chemistry approaches could be employed to enhance productive interactions. It has been shown that relatively modest changes in 2′-modification of PS-ASOs can significantly impact interactions with cell surface proteins (44). Conjugation of ligands to PS-ASOs has been shown to improve delivery to desired tissues. For example, N-acetyl galactosamine, the ligand for the asialoglycoprotein receptor, conjugated to PS-ASOs (or to siRNAs) results in efficient delivery into hepatocytes (12). In addition, conjugation with ligands for G-protein-coupled receptors was also shown to increase PS-ASO productive uptake (45,46). As described here, the uptake pathway for PS-ASO through EGFR mediates a release mechanism, which can be considered to further optimize PS-ASO pharmacological activities.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr. Frank Bennett and Dr. Wen Shen for stimulating discussions.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Ionis Pharmaceuticals. Funding for open access charge: Ionis Pharmaceuticals.
Conflict of interest statement. None declared.
REFERENCES
- 1. Wu H., Lima W.F., Crooke S.T.. Properties of cloned and expressed human RNase H1. J. Biol. Chem. 1999; 274:28270–28278. [DOI] [PubMed] [Google Scholar]
- 2. Bennett C.F., Swayze E.E.. RNA targeting therapeutics: molecular mechanisms of antisense oligonucleotides as a therapeutic platform. Annu. Rev. Pharmacol. Toxicol. 2010; 50:259–293. [DOI] [PubMed] [Google Scholar]
- 3. Crooke S.T. Antisense strategies. Curr. Mol. Med. 2004; 4:465–487. [DOI] [PubMed] [Google Scholar]
- 4. Yu R.Z., Grundy J.S., Geary R.S.. Clinical pharmacokinetics of second generation antisense oligonucleotides. Expert Opin. Drug Metab. Toxicol. 2013; 9:169–182. [DOI] [PubMed] [Google Scholar]
- 5. Crooke S.T. Progress in antisense technology. Annu. Rev. Med. 2004; 55:61–95. [DOI] [PubMed] [Google Scholar]
- 6. Brown D.A., Kang S.H., Gryaznov S.M., DeDionisio L., Heidenreich O., Sullivan S., Xu X., Nerenberg M.I.. Effect of phosphorothioate modification of oligodeoxynucleotides on specific protein binding. J. Biol. Chem. 1994; 269:26801–26805. [PubMed] [Google Scholar]
- 7. Liang X.H., Sun H., Shen W., Crooke S.T.. Identification and characterization of intracellular proteins that bind oligonucleotides with phosphorothioate linkages. Nucleic Acids Res. 2015; 43:2927–2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Allerson C.R., Sioufi N., Jarres R., Prakash T.P., Naik N., Berdeja A., Wanders L., Griffey R.H., Swayze E.E., Bhat B.. Fully 2′-modified oligonucleotide duplexes with improved in vitro potency and stability compared to unmodified small interfering RNA. J. Med. Chem. 2005; 48:901–904. [DOI] [PubMed] [Google Scholar]
- 9. Prakash T.P., Allerson C.R., Dande P., Vickers T.A., Sioufi N., Jarres R., Baker B.F., Swayze E.E., Griffey R.H., Bhat B.. Positional effect of chemical modifications on short interference RNA activity in mammalian cells. J. Med. Chem. 2005; 48:4247–4253. [DOI] [PubMed] [Google Scholar]
- 10. Geary R.S., Norris D., Yu R., Bennett C.F.. Pharmacokinetics, biodistribution and cell uptake of antisense oligonucleotides. Adv. Drug Deliv. Rev. 2015; 87:46–51. [DOI] [PubMed] [Google Scholar]
- 11. Crooke S.T., Wang S., Vickers T.A., Shen W., Liang X.H.. Cellular uptake and trafficking of antisense oligonucleotides. Nat. Biotechnol. 2017; 35:230–237. [DOI] [PubMed] [Google Scholar]
- 12. Prakash T.P., Graham M.J., Yu J., Carty R., Low A., Chappell A., Schmidt K., Zhao C., Aghajan M., Murray H.F. et al. Targeted delivery of antisense oligonucleotides to hepatocytes using triantennary N-acetyl galactosamine improves potency 10-fold in mice. Nucleic Acids Res. 2014; 42:8796–8807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Juliano R.L., Ming X., Carver K., Laing B.. Cellular uptake and intracellular trafficking of oligonucleotides: implications for oligonucleotide pharmacology. Nucleic Acid Therap. 2014; 24:101–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Juliano R.L., Carver K., Cao C., Ming X.. Receptors, endocytosis, and trafficking: the biological basis of targeted delivery of antisense and siRNA oligonucleotides. J. Drug Target. 2013; 21:27–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Blessing T., Kursa M., Holzhauser R., Kircheis R., Wagner E.. Different strategies for formation of pegylated EGF-conjugated PEI/DNA complexes for targeted gene delivery. Bioconjugate Chem. 2001; 12:529–537. [DOI] [PubMed] [Google Scholar]
- 16. Yuan Q., Lee E., Yeudall W.A., Yang H.. Dendrimer-triglycine-EGF nanoparticles for tumor imaging and targeted nucleic acid and drug delivery. Oral Oncol. 2010; 46:698–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Supaprutsakul S., Chotigeat W., Wanichpakorn S., Kedjarune-Leggat U.. Transfection efficiency of depolymerized chitosan and epidermal growth factor conjugated to chitosan-DNA polyplexes. J. Mater. Sci. Mater. Med. 2010; 21:1553–1561. [DOI] [PubMed] [Google Scholar]
- 18. Bohl Kullberg E., Bergstrand N., Carlsson J., Edwards K., Johnsson M., Sjoberg S., Gedda L.. Development of EGF-conjugated liposomes for targeted delivery of boronated DNA-binding agents. Bioconjugate Chem. 2002; 13:737–743. [DOI] [PubMed] [Google Scholar]
- 19. Miller K., Beardmore J., Kanety H., Schlessinger J., Hopkins C.R.. Localization of the epidermal growth factor (EGF) receptor within the endosome of EGF-stimulated epidermoid carcinoma (A431) cells. J. Cell Biol. 1986; 102:500–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ogiso H., Ishitani R., Nureki O., Fukai S., Yamanaka M., Kim J.H., Saito K., Sakamoto A., Inoue M., Shirouzu M. et al. Crystal structure of the complex of human epidermal growth factor and receptor extracellular domains. Cell. 2002; 110:775–787. [DOI] [PubMed] [Google Scholar]
- 21. Zidovetzki R., Johnson D.A., Arndt-Jovin D.J., Jovin T.M.. Rotational mobility of high-affinity epidermal growth factor receptors on the surface of living A431 cells. Biochemistry. 1991; 30:6162–6166. [DOI] [PubMed] [Google Scholar]
- 22. Barbieri M.A., Roberts R.L., Gumusboga A., Highfield H., Alvarez-Dominguez C., Wells A., Stahl P.D.. Epidermal growth factor and membrane trafficking. EGF receptor activation of endocytosis requires Rab5a. J. Cell Biol. 2000; 151:539–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Alexander A. Endocytosis and intracellular sorting of receptor tyrosine kinases. Front. Biosci. 1998; 3:d729–d738. [DOI] [PubMed] [Google Scholar]
- 24. Sorkin A., Carpenter G.. Interaction of activated EGF receptors with coated pit adaptins. Science. 1993; 261:612–615. [DOI] [PubMed] [Google Scholar]
- 25. Chen P., Gupta K., Wells A.. Cell movement elicited by epidermal growth factor receptor requires kinase and autophosphorylation but is separable from mitogenesis. J. Cell Biol. 1994; 124:547–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sunaga N., Tomizawa Y., Yanagitani N., Iijima H., Kaira K., Shimizu K., Tanaka S., Suga T., Hisada T., Ishizuka T. et al. Phase II prospective study of the efficacy of gefitinib for the treatment of stage III/IV non-small cell lung cancer with EGFR mutations, irrespective of previous chemotherapy. Lung Cancer. 2007; 56:383–389. [DOI] [PubMed] [Google Scholar]
- 27. Vickers T.A., Crooke S.T.. Development of a quantitative BRET affinity assay for nucleic acid-protein interactions. PLoS One. 2016; 11:e0161930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Rio D.C. Filter-binding assay for analysis of RNA-protein interactions. Cold Spring Harbor Protoc. 2012; 2012:1078–1081. [DOI] [PubMed] [Google Scholar]
- 29. Fiore M.M. Cellular specificity for the activation of fibroblast growth factor-2 by heparan sulfate proteoglycan. Biochem. Biophys. Res. Commun. 2001; 284:384–388. [DOI] [PubMed] [Google Scholar]
- 30. Machleidt T., Woodroofe C.C., Schwinn M.K., Mendez J., Robers M.B., Zimmerman K., Otto P., Daniels D.L., Kirkland T.A., Wood K.V.. NanoBRET—a novel BRET platform for the analysis of protein-protein interactions. ACS Chem. Biol. 2015; 10:1797–1804. [DOI] [PubMed] [Google Scholar]
- 31. Phan A.T., Kuryavyi V., Luu K.N., Patel D.J.. Structure of two intramolecular G-quadruplexes formed by natural human telomere sequences in K+ solution. Nucleic Acids Res. 2007; 35:6517–6525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tong-Ochoa N., Kopra K., Syrjanpaa M., Legrand N., Harma H.. Homogeneous single-label tyrosine kinase activity assay for high throughput screening. Anal. Chim. Acta. 2015; 897:96–101. [DOI] [PubMed] [Google Scholar]
- 33. Wang S., Sun H., Tanowitz M., Liang X.H., Crooke S.T.. Annexin A2 facilitates endocytic trafficking of antisense oligonucleotides. Nucleic Acids Res. 2016; 44:7314–7330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Li G., Stahl P.D.. Structure-function relationship of the small GTPase rab5. J. Biol. Chem. 1993; 268:24475–24480. [PubMed] [Google Scholar]
- 35. Wang S., Sun H., Tanowitz M., Liang X.H., Crooke S.T.. Intra-endosomal trafficking mediated by lysobisphosphatidic acid contributes to intracellular release of phosphorothioate-modified antisense oligonucleotides. Nucleic Acids Res. 2017; 45:5309–5322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Thul P.J., Akesson L., Wiking M., Mahdessian D., Geladaki A., Ait Blal H., Alm T., Asplund A., Bjork L., Breckels L.M. et al. A subcellular map of the human proteome. Science. 2017; 356:815–820. [DOI] [PubMed] [Google Scholar]
- 37. Shen W., Liang X.H., Crooke S.T.. Phosphorothioate oligonucleotides can displace NEAT1 RNA and form nuclear paraspeckle-like structures. Nucleic Acids Res. 2014; 42:8648–8662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Liang X.H., Shen W., Sun H., Kinberger G.A., Prakash T.P., Nichols J.G., Crooke S.T.. Hsp90 protein interacts with phosphorothioate oligonucleotides containing hydrophobic 2′-modifications and enhances antisense activity. Nucleic Acids Res. 2016; 44:3892–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Miller C.M., Donner A.J., Blank E.E., Egger A.W., Kellar B.M., Ostergaard M.E., Seth P.P., Harris E.N.. Stabilin-1 and Stabilin-2 are specific receptors for the cellular internalization of phosphorothioate-modified antisense oligonucleotides (ASOs) in the liver. Nucleic Acids Res. 2016; 44:2782–2794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bausch-Fluck D., Hofmann A., Bock T., Frei A.P., Cerciello F., Jacobs A., Moest H., Omasits U., Gundry R.L., Yoon C. et al. A mass spectrometric-derived cell surface protein atlas. PLoS One. 2015; 10:e0121314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Juliano R.L., Carver K.. Cellular uptake and intracellular trafficking of oligonucleotides. Adv. Drug Deliv. Rev. 2015; 87:35–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Tanowitz M., Hettrick L., Revenko A., Kinberger G.A., Prakash T.P., Seth P.P.. Asialoglycoprotein receptor 1 mediates productive uptake of N-acetylgalactosamine-conjugated and unconjugated phosphorothioate antisense oligonucleotides into liver hepatocytes. Nucleic Acids Res. 2017; 45:12388–12400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Back N., Kanerva K., Kurutihalli V., Yanik A., Ikonen E., Mains R.E., Eipper B.A.. The endocytic pathways of a secretory granule membrane protein in HEK293 cells: PAM and EGF traverse a dynamic multivesicular body network together. Eur. J. Cell Biol. 2017; 96:407–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Mou T.C., Gray D.M.. The high binding affinity of phosphorothioate-modified oligomers for Ff gene 5 protein is moderated by the addition of C-5 propyne or 2′-O-methyl modifications. Nucleic Acids Res. 2002; 30:749–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ming X., Alam M.R., Fisher M., Yan Y., Chen X., Juliano R.L.. Intracellular delivery of an antisense oligonucleotide via endocytosis of a G protein-coupled receptor. Nucleic Acids Res. 2010; 38:6567–6576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Alam M.R., Dixit V., Kang H., Li Z.B., Chen X., Trejo J., Fisher M., Juliano R.L.. Intracellular delivery of an anionic antisense oligonucleotide via receptor-mediated endocytosis. Nucleic Acids Res. 2008; 36:2764–2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.