Abstract
Leishmania parasites are unicellular pathogens that are transmitted to humans through the bite of infected sandflies. Most of the regulation of their gene expression occurs post-transcriptionally, and the different patterns of gene expression required throughout the parasites’ life cycle are regulated at the level of translation. Here, we report the X-ray crystal structure of the Leishmania cap-binding isoform 1, LeishIF4E-1, bound to a protein fragment of previously unknown function, Leish4E-IP1, that binds tightly to LeishIF4E-1. The molecular structure, coupled to NMR spectroscopy experiments and in vitro cap-binding assays, reveal that Leish4E-IP1 allosterically destabilizes the binding of LeishIF4E-1 to the 5′ mRNA cap. We propose mechanisms through which Leish4E-IP1-mediated LeishIF4E-1 inhibition could regulate translation initiation in the human parasite.
INTRODUCTION
Leishmania are protozoan parasites that have a digenetic life cycle (1–3). They reside as flagellated promastigotes in sandflies. They are then transmitted to humans during blood meals and taken up by macrophages and dermal dendritic cells. Within these cells, the higher temperature and lower pH trigger a gene expression program essential to their differentiation from promastigotes into amastigotes (4–6). Amastigotes are obligatory intracellular and non-motile forms of the parasite (7). In the absence of conserved mechanisms for transcriptional activation, post-transcriptional mechanisms drive the developmental program of gene expression (8) and translation regulation plays a central role.
In higher eukaryotes, the translational initiation complex assembles at the m7GTP cap of messenger RNAs (mRNAs) (9) through the eukaryotic initiation factor 4F complex (eIF4F) (10). eIF4F comprises the cap-binding protein eIF4E, the DEAD-box RNA helicase eIF4A, and the scaffold protein eIF4G (11,12). eIF4G recruits the small ribosomal subunit indirectly by binding eIF3. eIF4G also interacts with eIF4E, partly through a consensus binding motif, Y(X)4LΦ (where X is any amino acid, and Φ is a hydrophobic residue) (13). The 4E binding proteins (4E-BPs), which also contain a Y(X)4LΦ motif, regulate the eIF4E/eIF4G interaction by competing with eIF4G for binding to eIF4E. 4E-BPs are phosphorylation dependent regulators of translation initiation, and only non-phosphorylated or minimally phosphorylated 4E-BPs bind to eIF4E. Phosphorylation of 4E-BP1 by the serine/threonine kinase mammalian/mechanistic target of rapamycin (mTOR) complex 1 (mTORC1) induces its dissociation from eIF4E, allowing eIF4E to interact with eIF4G and enabling translation initiation (14–18).
Leishmania parasites are ancient eukaryotes, and their protein-coding genes are organized as large chromosomal units that are mostly transcribed polycistronically. Transcripts are further trans-spliced and polyadenylated, yielding mature monocistronic mRNAs (19). During trans-splicing, a conserved spliced leader RNA (SL RNA) is added to the 5′ end of all mRNAs, providing a unique cap structure that is hypermethylated over the first four nucleotides (cap-4: m7Gpppm36,2,2’Apm2’Apm2’ Cpm23,2’U) (20,21). In promastigotes, translation initiation requires a canonical ‘Leishmania IF4F’ complex (LeishIF4F) comprising LeishIF4E-4, LeishIF4G-3 and LeishIF4A-1 (22–25). In amastigotes, the human infective stage, the components of an equivalent LeishIF4F complex remain elusive. Trypanosomatids encode several isoforms of the same initiation factor; there are six highly diverged paralogs of the human eIF4E cap-binding protein (LeishIF4E-1 to LeishIF4E-6). These have a small degree of conservation among themselves (40–60%) and also differ significantly from their counterparts in higher eukaryotes (30–40% sequence identity) (Supplementary Figure S1A) (24,26,27). Of the six paralogs, LeishIF4E-1 is highly expressed in axenic amastigotes and binds the m7GTP at 37°C, suggesting its involvement in translation initiation in amastigotes (23). LeishIF4E-1 binds the m7GTP cap and cap-4 with comparable affinities and pulls down other Leishmania translation initiation factors when purified over a m7GTP cap-binding affinity column (24). However, a ‘LeishIF4G’-like scaffold protein has yet to be identified as a LeishIF4E-1 binding partner. In our search for such a factor, we previously identified an 83 kDa LeishIF4E-1 interacting protein (Leish4E-IP1) that contains a Y(X)4LΦ near its N terminus (23). Secondary structure prediction algorithms [PSIPRED, (28)] suggest that Leish4E-IP1 is mostly unstructured, except for a segment near its N terminus (Supplementary Figure S1B). Leish4E-IP1 does not have any homologs in higher eukaryotes and does not contain the typical HEAT/MIF domains usually found in mammalian IF4Gs.
We determined the crystal structure of LeishIF4E-1 (residues 13–210) bound to a 40-residue N-terminal fragment of Leish4E-IP1 (residues 4–43). This fragment contains the α-helical Y(X)4LΦ consensus-binding motif, followed by a flexible linker and a second α-helix. Analysis of the crystal structure coupled with NMR spectroscopy experiments reveal that the binding of Leish4E-IP1 to LeishIF4E-1 alters the conformation of the tryptophan residues within its cap-binding pocket. Cap-binding affinity purification experiments, using parasite lysates, show that Leish4E-IP1 causes LeishIF4E-1 to dissociate from the cap. Taken together, our results suggest that Leish4E-IP1 is a repressor of the cap-binding protein LeishIF4E-1 in amastigotes, the human infective stage of Leishmania parasites, and uncover a fundamental mechanism of cap-binding regulation that is likely also used in other organisms.
MATERIALS AND METHODS
Constructs and protein purification
We obtained the plasmids encoding full-length LeishIF4E-1 (Accession number: LmjF27.1620) and Leish4E-IP1 (Accession number: LmjF35.3980) (23). We expressed LeishIF4E-1 as a fusion protein with a hexahistidine (H6) tag followed by a Tobacco Etch Virus (TEV) protease cleavage site at its N terminus (pHis-TEV-LeishIF4E-1 construct). We expressed fragments of Leish4E-IP1 (residues 1–100, 1–52) as fusion proteins with a H6 tag, followed by the domain B1 of a protein G (GB1) solubility enhancement tag (29) and a TEV cleavage site at their N terminus, generating pHis-GB1-TEV-Leish4E-IP1 constructs. We also subcloned the above-described constructs of Leish4E-IP1 into a pcDNA3.1-eGFP (eGFP, enhanced green fluorescent protein) mammalian expression vector using HindIII-XhoI restriction sites (Addgene plasmid 13031). These constructs allow the expression of the Leish4E-IP1 fragments as fusion proteins with eGFP at their C terminus (pCDNA3.1-eGFP/Leish4E-IP1 constructs).
We transformed Escherichia coli (BL21) cells with the appropriate plasmids and grew the cells in Luria-Bertani Broth (LB). We used 1 mM of isopropyl β-d-1-thiogalactopyranoside (IPTG) to induce His-TEV-LeishIF4E-1 or His-GB1-TEV-Leish4E-IP1 protein expression and allowed the cells to grow at 20°C for 16 h. We harvested and re-suspended the cells in a Leishmania purification binding buffer (LPBB) containing 50 mM NaH2PO4/Na2HPO4, pH 7.4, 500 mM NaCl, 10 mM imidazole, 5 mM 2-β-mercaptoethanol, benzonase and EDTA-free protease inhibitor cocktail tablet (Roche). We lysed the cells by sonication at 4°C and centrifuged the cells at 38 000 × g for 40 min. We loaded the supernatant on a nickel-affinity column (Ni-NTA; Qiagen) previously equilibrated with a protease inhibitor-free LPBB. We rinsed the Ni-NTA resin bound to proteins with LPBB supplemented with 20 mM imidazole. We eluted the proteins from the resin with LPBB supplemented with 100 mM imidazole and 100 mM EDTA. When required, we digested overnight the eluted proteins with TEV protease to cleave the His or the His-GB1 tags. We concentrated the eluted proteins by ultrafiltration using a 10 kDa-cutoff centrifuge filter. We further purified the proteins using a Superdex 75 HiLoad 16/60 preparative size exclusion column (GE Healthcare Bio-Sciences) equilibrated with a buffer containing 50 mM NaH2PO4/Na2HPO4, pH 7.4, 100 mM NaCl, 2 mM DTT (FPLC buffer). For Leish(IF4E-1/4E-IP1) complex preparation, we grew cells expressing the individual proteins of interest and harvested cells as described above. However, before sonication, we mixed both lysates and purified His-TEV-LeishIF4E-1/His-GB1-TEV-Leish4E-IP1 complex using a nickel-affinity column followed by a Superdex 75 HiLoad 16/60 preparative size exclusion column.
We expressed mouse IF4E (mIF4E) (residues 27–217) or full-length human 4E-BP1 (h4E-BP1) in E. coli as fusion proteins, containing respectively, a GB1-TEV tag and a His-GB1-TEV at their N terminus. We harvested the cells and purified GB1-TEV-mIF4E over an adipic-agarose-m7GDP column (30), and we purified His-GB1-TEV-h4E-BP1 over a nickel-affinity column as previously described (31). We further purified un-cleaved or cleaved proteins by TEV protease using a Superdex 75 HiLoad 16/60 preparative size exclusion column.
We performed isothermal calorimetry measurements at 25°C using a MicroCal ITC200 (Malvern, Center for Macromolecular Interactions, Harvard Medical School). The protein samples were dissolved in buffer containing 20 mM HEPES pH 7.8, 150 mM NaCl, 5 mM TCEP. At intervals of 5 min, 5 μl of approximately 80 μM Leish4E-IP1 fragments were injected into the 1.4 ml sample cell containing approximately 5 μM IF4E-1. After the baseline correction, the data were analyzed using software provided by the manufacturer (MicroCal). The first data point was excluded from the analysis. For each interaction, two independent titration experiments were performed.
Structural studies
NMR experiments
We grew E. coli strain BL21 (DE3) cells expressing either LeishIF4E-1 or Leish4E-IP1 fragments in M9 minimal media containing 95% 2H2O supplemented with 15NH4Cl and 13C6-d-glucose, when required, as the sole nitrogen and carbon sources, respectively. We purified individual proteins or Leish(IF4E-1/4E-IP1) complexes as described in the Constructs and Protein Purification methods section. After size exclusion purification, we exchanged the FPLC buffer to an NMR buffer (50 mM NaH2PO4/Na2HPO4, pH 6.5, 100 mM NaCl, 2 mM DTT, 5% D2O, concentrated to 0.1–0.4 mM) using a desalting PD-10 column (GE Healthcare). NMR spectroscopy experiments were recorded at 298 K either on Bruker or Varian spectrometers, operating at high field strengths of 600, 750 and 900 MHz, all equipped with a cryogenically cooled probe.
The backbone assignments of Leish4E-IP11–52 in the complex Leish(IF4E-1/4E-IP11–52), where Leish4E-IP11–52 is 15N and 13C labeled, and LeishIF4E-1 is unlabelled, were done using TROSY version of the traditional backbone triple resonance experiments, namely HNCA, HNCOCA, HNCO, HNCACO, HNCACB and HNCOCACB. Non-Uniform Sampling (NUS) in the two indirect dimensions was used to collect triple resonance data, in which 12–15% of the indirect grid was sampled using Poisson Gap Sampling (32). The data were reconstructed and processed using the hmsIST program (33).
Relaxation times T1, T2 and heteronuclear NOE were measured using a LeishIF4E-1/13C-15N-Leish4E-IP11-52 sample. We used TROSY based experiments to acquire T1, T2 and Heteronuclear NOE measurements. We used the following inversion recovery delays for T1 experiments: 10, 50, 75, 100, 150, 300, 600, 900, 1000 and 1800 ms. For T2 experiments, the relaxation delays were 16.32, 32.64, 48.96, 65.28, 81.6, 97.92, 114.24, 130.56 ms. A compensating Carr-Purcell-Meiboom-Gill (CPMG) block was applied to all the T2 experiments to ensure that the heating of all the individual measurements was equivalent to that of the 130.56 ms experiment. We performed heteronuclear NOE experiments with a saturation time of 2.5 s while the off-saturated time was 0. After data processing using NMRPipe (34), peak intensities were extracted using Sparky (35), fitted to exponential decays, and transverse relaxation rates (R1, R2 and Net NOE) were plotted in function of the residue number of Leish4E-IP11–52.
Crystallization, data collection, and structure determination
We expressed and purified His-TEV-LeishIF4E-1/His-GB1-TEV-Leish4E-IP11–52 complex as described in the Constructs and Protein Purification section. We cleaved the tags overnight using TEV protease and separated the Leish(IF4E-1/4E-IP11–52) complex from cleavage products by loading the protein preparation over a Superdex 75 HiLoad 16/60 preparative column previously equilibrated with a buffer containing 5 mM Tris–HCl (pH 7.4), 50 mM NaCl and 2 mM DTT (Crystallization Buffer, CB). We screened crystallization conditions with the Leishmania protein complex (10 mg/ml) using the hanging drop method. Crystals grew at room temperature in reservoir buffer containing 0.1 M Tris–HCl, pH 7.4, 3% polyethylene glycol 400 and 2.125–2.175 M ammonium sulfate. Crystals were flash frozen in reservoir buffer containing 15% (v/v) glycerol for data collection. We collected X-ray diffraction data at wavelength of 0.999 and temperature of 100 K at BCSB beamline ID-8.2.2 at the Advance Light Source (Lawrence Berkeley National Laboratory). The Leish(IF4E-1/4E-IP11–52) complex crystallized in the P6222 space group with unit cell dimensions of a = 68.22 Å, b = 68.22 Å, c = 220.20 Å (Supplementary Table S1). We processed diffraction data using XDS (36,37). Crystals diffracted up to 2.2 Å, but ice rings present between 2.7 and 2.2 Å compromised data quality. The data were truncated to 2.7 Å using XDS. We determined the structure by molecular replacement with PHASER (38) using as a search model a modified eIF4E/4E-BP1 complex (PDB ID: 1WKW). We observed one copy of the complex within the asymmetric unit, with a solvent content of 54.22%. We performed iterative model building with COOT (39), and refined with PHENIX (40). The final model converged to a Rwork of 23.96% and to a Rfree of 28.16%. 97.97% of all residues were in the most favored region of the Ramachandran space, with the additional 2.03% falling in the allowed regions as defined by MolProbity. We used PyMol to generate figures (41). We deposited the coordinates of the Leish(IF4E-1/4E-IP11–52) complex in the Protein Data Bank (PDB ID: 5WB5).
Biological assays
Leishmania amazonensis cells (∼2 × 108 cells) episomally expressing an SBP-tagged version of LeishIF4E-1 were harvested and lysed in 4 ml of Leishmania binding buffer (LBB: 20 mM HEPES pH 7.4, 50 mM NaCl, 2 mM EDTA, 1 mM DTT, 1% Triton X100). After clarifying the lysate by centrifugation, we added 1 ml of the lysate to 30 μl of packed m7GTP agarose resin (Jena Bioscience, Germany) pre-equilibrated in LBB. We purified the recombinant His-GB1-TEV-Leish4E-IP11–52, as described in the Constructs and Protein Purification section, and added this protein separately to the SBP-LeishIF4E-1 lysate bound to the m7GTP agarose resin (from 0 to 3 μg). After an incubation of 2 h at 4°C on a rocking platform, we washed the resin three times with 1 ml of LPBB for 2 min. We then resuspended the resin in SDS loading buffer and boiled it for 5 min. We determined the amount of SBP-LeishIF4E-1 bound to the m7GTP agarose resin in the absence or presence of His-GB1-TEV-Leish4E-IP1 by western blot using an anti-SBP mouse monoclonal antibody. We performed a densitometry analysis of the blot using ImageJ software (42). This allowed us to calculate the ratio of the total SBP-LeishIF4E-1 protein initially loaded on the resin versus the total of SBP-LeishIF4E-1 protein bound to the resin after being incubated with His-GB1-TEV-Leish4E-IP11–52 and three washes.
RESULTS
Crystallization of a Leish(IF4E-1/4E-IP11-52) complex
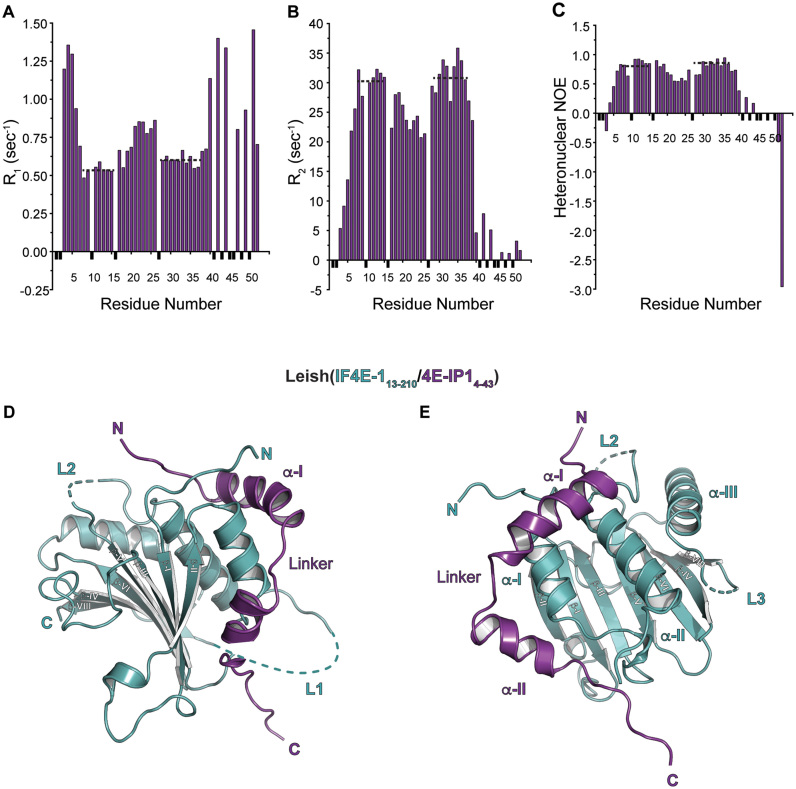
We expressed and purified LeishIF4E-1 from E. coli as a soluble protein (Supplementary Figure S1C). The transverse relaxation-optimized spectroscopy (TROSY)-heteronuclear single quantum correlation (HSQC) spectrum of LeishIF4E-1 is well-dispersed in the 1H dimension, confirming that the recombinant protein is folded (Supplementary Figure S2A). LeishIF4E-1 interacts with Leish4E-IP1, an 83 kDa protein of unknown function that has orthologs in various trypanosomatids (23). We could express and purify from E. coli constructs of Leish4E-IP1 that comprised the first 100 or 52 residues (Leish4E-IP11–100 and Leish4E-IP11–52) (Supplementary Figure S1D). Isothermal titration calorimetry (ITC) experiments indicated that Leish4E-IP11-100 and Leish4E-IP11–52 bind to LeishIF4E-1 with similar affinity (kD of about 90 and 10 nM, respectively, Table 1), suggesting that the minimal binding site for LeishIF4E-1 is retained in the shorter construct. The addition of unlabeled Leish4E-IP11-52 to a 15N labeled sample of LeishIF4E-1 resulted in a number of chemical shift perturbations in the 15N TROSY-HSQC spectrum of LeishIF4E-1, confirming their interaction (Supplementary Figure S2B). We also used NMR-based relaxation experiments to assess the dynamic properties of Leish4E-IP1 when bound to LeishIF4E-1 (Figure 1A–C). Within the Leish4E-IP11-52 construct, we observed that the N-terminal residues 1–5 and the C-terminal residues 39–52 are flexible when bound to LeishIF4E-1, with T2 values in the 100 ms range, suggesting that these regions are not part of the binding interface. We detected two stable stretches of residues (residues 5–20 and 27–38), with T2 values in the 30 ms range, indicating that these residues are stabilized in the Leish(IF4E-1/4E-IP1) complex. Indeed, the first segment of Leish4E-IP1 (residues 5–20) contains the consensus LeishIF4E-binding motif (8Y(X)4LΦ14), which is expected to interact with LeishIF4E-1. For structural studies, we thus prepared an Leish(IF4E-1/4E-IP11–52) complex purified initially by nickel-affinity followed by size exclusion chromatography (Supplementary Figure S1E). As we originally sought to crystallize a cap-bound complex, we added five molar equivalents of m7GTP (the authentic cap-4 is not available commercially and difficult to synthesize). The Leish(IF4E-1/4E-IP11–52) complex crystallized in the P6222 space group.
Table 1. Thermodynamic analysis of Leish4E-IP1 binding to LeishIF4E-1 as determined by isothermal titration calorimetry.
| Titrants | Leish4E-IP11–100 | Leish4E-IP11–52 |
|---|---|---|
| K (cal/mol) | 1.148 × 107 ± 6.645 × 106 | 1.03 × 108 ± 4.54 × 107 |
| ΔH (cal/mole) | –3108 ± 80.36 | –1.55 × 104 ± 281.8 |
| ΔS (cal/mol/°) | –21.9 | –15.3 |
| ΔG (kcal/mole) | –2.56 | –15.2 |
| K D (nM) | 87 | 9.7 |
Enthalpy changes (ΔH), entropy changes (ΔS), free energy changes (ΔG) and dissociation constants (KD) derived from ITC measurements at 25°C.
Figure 1.
NMR spectroscopy and crystal structure of LeishIF4E-1 bound to a fragment of Leish4E-IP1. (A) R1, (B) R2, (C) Heteronuclear NOE data are shown for a sample of ∼0.2 mM 15N13C-Leish4E-IP11-52 bound to LeishIF4E-1 in 95:5 H2O/D2O at pH 6.5. Data points corresponding to unassigned residues or to residues for which measurements could not be obtained are shown in black and were given an arbitrary negative value. Dashed lines indicate two-stable stretches of residues in solution compared to other regions in the Leish4E-IP11–52 fragment (See also Supplementary Figures S1 and S2). (D) Cartoon representation of the Leish(IF4E-1/4E-IP11–52) complex (PDB ID: 5WB5), where residues 13 to 210 of LeishIF4E-1 and residues 4–43 of Leish4E-IP11–52 are shown (See also Supplementary Figures S1 and S2). LeishIF4E-1 and Leish4E-IP1 are shown in cyan and purple, respectively. The N and C termini of each protein and secondary structure elements are indicated. Disordered segments in loops (L1 and L2) from LeishIF4E-1 are shown in dashed lines. LeishIF4E-1 UniProt accession number is E9ADE1 and Leish4E-IP1 accession number is E9AFM3. (E) Dorsal view of the complex described in (D). Disordered segments in loops (L2 and L3) from LeishIF4E-1 are shown in dashed lines.
Structure of the Leish(IF4E-1/4E-IP1) complex
We determined the structure of the Leish(IF4E-1/4E-IP11–52) complex by molecular replacement using as a search model a truncated version of an eIF4E/m7GTP/4E-BP1 complex [PDB ID: 1WKW, (43)] in which we removed the atoms corresponding to eIF4E loops and kept only the 4E-BP1 residues corresponding to the consensus binding motif. We then performed iterative model building and refinement (Supplementary Table S1). The final model contains one copy of the Leish(IF4E-1/4E-IP11–52) complex (PDB ID: 5WB5) in the asymmetric unit, with residues 13 to 210 of LeishIF4E-1 and residues 4 to 43 of Leish4E-IP11–52 (Figure 1D and E).
LeishIF4E-1 adopts a similar fold to the IF4E proteins from higher eukaryotes (a ‘cupped hand’ structure), including that of Drosophila, yeast, and of the human orthologs (44–47). The fold comprises a curved β-sheet (with strands β-I through β-VII) with three α–helices (helices α-I through α-III) on its dorsal side. LeishIF4E-1 lacks the final C-terminal β-strand found in human eIF4E. LeishIF4E-1 has a 17 residue insertion in Loop 1 (L1, 104–120) and a 7 residue insertion in Loop 2 (L2, 151–157) (Supplementary Figure S1A), which are disordered in our crystal structure (residues 101–121, residues 149–155). Moreover, we did not observe any densities for residues 198–201 (Loop 3). The N-terminal segment of Leish4E-IP1 binds the dorsal side of LeishIF4E-1 and comprises two α-helices (α-I and α-II) separated by a short linker.
Comparison of Leish(IF4E-1/4E-IP1) complex to the structures of other IF4E complexes
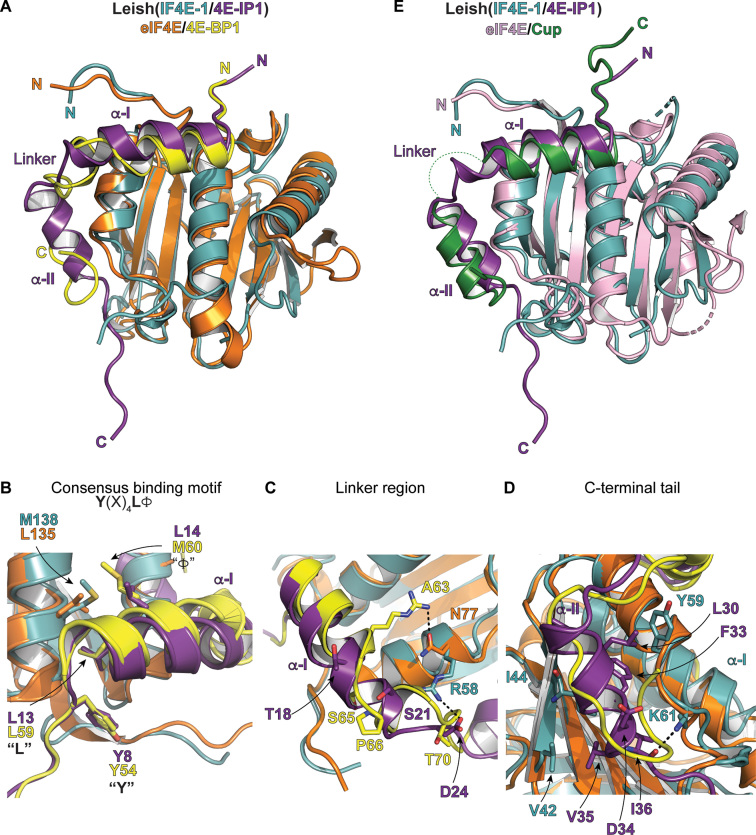
There are no known homologs of Leish4E-IP1. To obtain insight into its function, we compared the structure of Leish(IF4E-1/4E-IP11–52) to complexes of eIF4E bound to associated proteins known to modulate the function of eIF4E. Comparison of the Leish(IF4E-1/4E-IP11–52) structure to a complex of eIF4E bound to 4E-BP1 (eIF4E/4E-BP1) (31,48) (Figure 2A) reveals similarities and significant differences. Leish4E-IP1 binds to LeishIF4E-1 using the consensus binding motif 8Y(X)4LΦ14 (Figure 2B), where Y8 and L13 are crucial for the interaction (Supplementary Figure S3A). Leish4E-IP1 residues Y8 and L13 overlap perfectly with the corresponding residues in 4E-BP1 (Y54 and L59). However, covariation evolution is evident over the Φ position of the motif: in Leishmania major, a leucine found in Leish4E-IP1 (L14) interacts with a methionine in LeishIF4E-1 (M138), while in the human proteins, 4E-BP1 contributes a methionine (M60) and eIF4E provides a leucine (L135). These sequence differences could explain why Leish4E-IP1 has a 10-fold lower binding affinity for human eIF4E (data not shown). Indeed, consistent with a weakened interaction, using a bicistronic translation reporter system in human cells (HEK293T), we found that Leish4E-IP11–52 can only modestly repress translation initiation (15%), when compared to 4E-BP1 in the same assay (75% repression) (Supplementary Figure S3B).
Figure 2.
Comparison of key interaction motifs between Leish(IF4E-1/4E-IP1) complex and human eIF4E/4E-BP1 or Drosophila eIF4E/Cup complexes. (A) Cartoon representation of the structural alignment of the Leish(IF4E-1/4E-IP11-52) and the mammalian eIF4E/m7GTP/4E-BP1 complex [PDB ID: 5BXV, (31)]. The Leish(IF4E-1/4E-IP11–52) complex is represented and colored as described in Figure 1. eIF4E and 4E-BP1 are shown in orange and yellow, respectively. The m7GTP cap in the eIF4E/4E-BP1 complex is omitted for clarity. (B) As described in (A) but showing key interactions centered on the consensus binding motifs [Y(X)4LΦ] of Leish4E-IP1. Key residues are indicated. (C) As described in (A) but showing interactions centered on the linker region of Leish4E-IP1 (See also Figure 1A–C). (D) As described in (A) but showing interactions over the C-terminal tail of Leish4E-IP1. (E) Structural alignment of Leish(IF4E-1/4E-IP11–52) and the eIF4E/Cup complex from Drosophila melanogaster [PDB ID: 4AXG, (46)]. eIF4E and Cup are shown in pink and green, respectively. The linker segment of Cup is disordered in the complex and is shown as dashed lines (See also Supplementary Figure S1B and S4).
There are also several differences in the Leish4E-IP1 (residues 22–27) and 4E-BP1 (residues 64–73) linkers (Figure 2C). In the structure of eIF4E/4E-BP1, the α-I helix, which contains the consensus binding motif in 4E-BP1, ends with a salt bridge formed between 4E-BP1 R63 and eIF4E N77. This salt bridge in 4E-BP1 is followed by a small 310 segment induced by a proline-turn (P66), which exposes two critical phosphorylation sites (S65 and T70) on each side of the short helical segment (31,48). In the Leishmania complex, the Leish4E-IP1 α-I helix is extended by an additional turn. The shorter linker in Leish4E-IP1 does not contain a proline turn or any potential phosphorylation site. While several interactions anchor the 4E-BP-1 linker to eIF4E, the main interaction holding the linker in place in the Leish(IF4E-1/4E-IP11–52) structure is a salt bridge between LeishIF4E-1 R58 and Leish4E-IP1 D24. As expected, unlike the linker region in 4E-BP1, the linker portion of Leish4E-IP1 is more flexible than its neighbouring segments (the α-I and α-II helices), which is supported by the NMR relaxation experiments shown in Figure 1. Finally, there is a network of hydrophobic interactions between LeishIF4E-1 and Leish4E-IP1 over the α-II segment (Figure 2D), where residues from Leish4E-IP1 (V29, L30, F33, V35 and I36) are in close vicinity from either the β-II strand of LeishIF4E-1 (M45, I44, and V42) or the α-I of LeishIF4E-1 (Y59).
Among the several available structures of cap-binding proteins in complex with their regulators, the Leish4E-IP1 α-II helix resembles the most to the α-II helix of the Drosophila Melanogaster Cup protein bound to its corresponding eIF4E (46) (Figure 2E, See also Supplementary Figure S1B). In both Drosophila and Leishmania complexes, the residues within this region form an α-helical structure as opposed to the unstructured C-terminal tail observed in the eIF4E/4E-BP1 complex. Cup represses translation initiation of developmentally essential mRNAs in Drosophila (49,50). It is recruited to the 3′ UTR of mRNAs through adaptor proteins that bind specifically to sequence/structure elements in the mRNA. Cup then competes with eIF4G to bind with eIF4E, impairing the formation of the eIF4F complex.
The cap-binding pocket of LeishIF4E-1
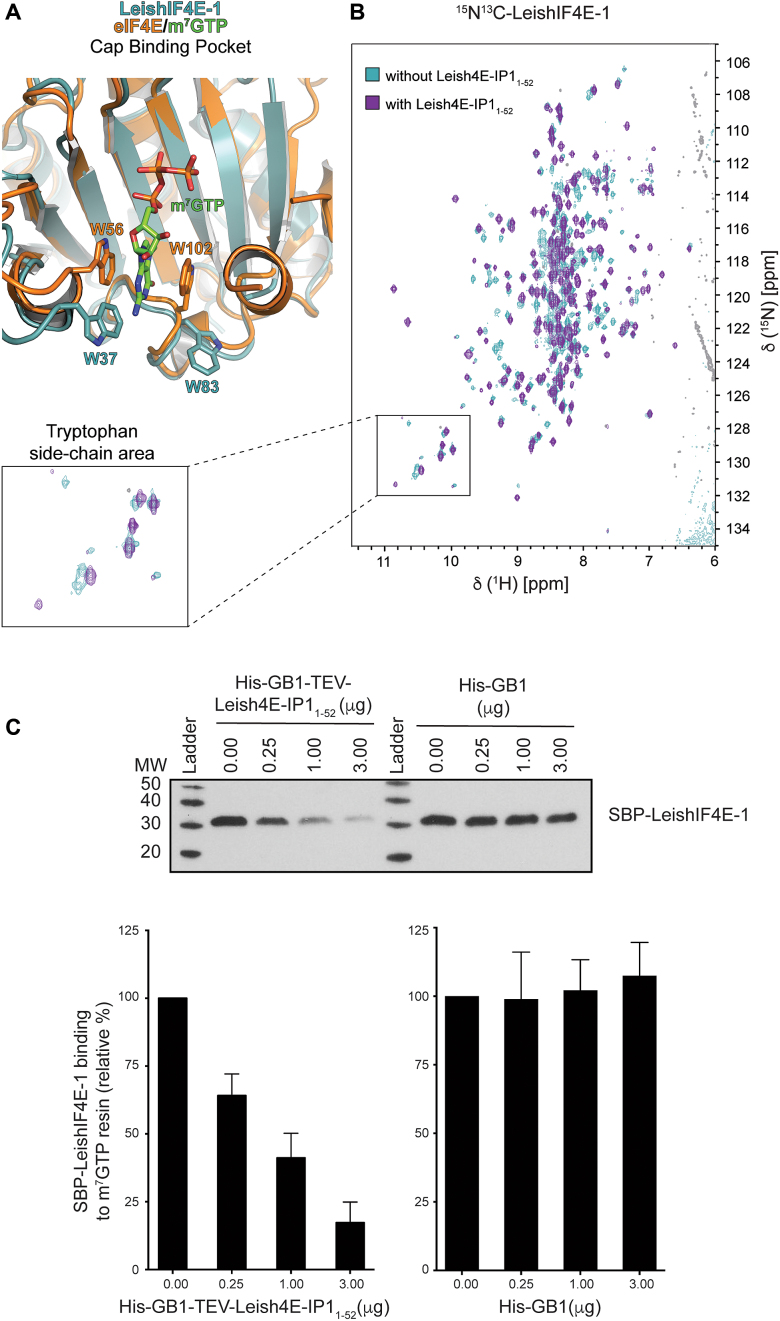
Cup binding increases the affinity of eIF4E to its m7GTP cap-ligand (46). The structural similarities between Leish4E-IP1 to Cup and the observation that Cup modifies eIF4E’s cap binding activity led us to closely examine the cap-binding residues in our complex. We did not observe density within the cap-binding pocket of LeishIF4E-1 for m7GTP despite the addition of a molar excess of m7GTP in the crystallization drops. However, in the Leish(IF4E-1/4E-IP11–52) complex we crystallized, two LeishIF4E-1 tryptophans (W37 and W83) expected to sandwich the m7GTP cap, are in a flipped out conformation (Figure 3A). This conformation has been observed in other crystallographic structures in which the m7GTP cap was not present in the complex [apo-eIF4E (47), eIF4E/4EB-P1 complex (51)]. It is possible that the m7GTP did not co-crystallize with the complex because the affinity of the LeishIF4E-1 for m7GTP is weak (Kas 0.16 μM-1) (24). It is also possible that the binding of Leish4E-IP11–52 binding to LeishIF4E-1 induces conformational changes in the LeishIF4E-1 cap-binding pocket that interfere with its ability to bind the cap. To look for evidence of conformational changes induced by Leish4E-IP11-52 binding to LeishIF4E-1, we recorded TROSY-HSQC spectra of LeishIF4E-1 unbound or bound to unlabeled Leish4E-IP11–52. In the presence of Leish4E-IP11–52, we observed chemical shift perturbations of five tryptophan side chains (LeishIF4E-1 has a total of seven tryptophan residues). Based on the crystal structure of Leish(IF4E-1/4E-IP11-52) complex (Figure 1), four tryptophans are located within the cap-binding pocket, and none are expected to interact directly with Leish4E-IP1. Although the backbone assignments for these tryptophans residues are unknown, the chemical shift perturbations of the five tryptophan residue side chains in LeishIF4E-1 support a conformational change within the cap-binding pocket upon the addition of Leish4E-IP11–52 (Figure 3B, see also Supplementary Figure S2).
Figure 3.
Effect of Leish4E-IP1 on the m7GTP cap-binding affinity of LeishIF4E-1. (A) Structural alignment of the Leish(IF4E-1/4E-IP11-52) and the mammalian eIF4E/m7GTP/4E-BP1 complex [PDB ID: 5BXV, (31)] centered on the cap-binding pocket. LeishIF4E-1 and eIF4E are colored in cyan and orange, respectively. For clarity, Leish4E-IP11–52 and 4E-BP1 are omitted. The side chains of W56 and W102 from eIF4E, when bound to 4E-BP1, are in a ‘flipped-in’ conformation, while the side chains of W37 and W83 from LeishIF4E-1, when bound to Leish4E-IP1, are in a ‘flipped-out’ conformation. (B) 15N-1H-TROSY-HSQC spectra overlay between 15N13C-LeishIF4E-1 (cyan) acquired on 600 MHz spectrometer and 15N13C-LeishIF4E-1 bound to unlabeled Leish4E-IP11-52 in an equimolar ratio (purple). An enlarged portion of the spectra shows the tryptophan side chain area (See also Supplementary Figure S2). (C) Upper panel: Pull-down experiments with Streptavidin-binding peptide (SBP)-tagged LeishIF4E-1 (SBP-LeishIF4E-1) from L. amazonensis. Cell extracts were affinity purified over an adipic-agarose m7GDP resin in the absence or presence of an increasing amount of recombinant His-GB1-TEV-Leish4E-IP11–52 or His-GB1 protein. Proteins that remained bound to the resin after several washes were separated on SDS-PAGE and analyzed by Western blot using an anti-SBP antibody, detecting LeishIF4E-1. Lower panel: Densitometry analysis of the blot shown in the upper panel. The effect of His-GB1-Tev-Leish4E-IP11–52 or His-GB1 on the binding of LeishIF4E-1 to the m7GDP resin is presented as a percentage value when compared to SBP-LeishIF4E-1 in the absence of Leish4E-IP11–52 (SBP-LeishIF4E-1, 0 μg, set as 100% signal). The data are represented as the mean of at least three independent experiments. Error bars indicate the standard error of the mean.
To further study the relevance of the conformational changes observed within the cap-binding pocket of LeishIF4E-1 in the presence of Leish4E-IP11–52, we assessed LeishIF4E-1’s ability to bind the cap in the presence or absence of Leish4E-IP11–52 (Figure 3C). We purified a Streptavidin-Binding Peptide (SBP)-tagged version of LeishIF4E-1 (SBP-LeishIF4E-1) from L. amazonensis parasites. After loading the parasite lysate on an adipic-agarose-m7GTP affinity column, we added increasing amount of Leish4E-IP11-52. The addition of Leish4E-IP11–52 to LeishIF4E-1 leads to its dissociation from the m7GTP cap (densitometry analysis showed a reduction of 75%). We observed no effect upon the addition of a GB1 polypeptide (negative control). Our results show that Leish4E-IP11–52 is a functional repressor of the cap-binding activity of LeishIF4E-1.
DISCUSSION
Despite the availability of high resolution structures of the Leishmania large ribosomal subunit (52) and of the Trypanosoma ribosome (53), which highlight irregular rRNA modifications in Leishmania, there are no structures yet available for factors involved in the critical step of translation initiation. The uniqueness of translation initiation in Leishmania parasites affords unexplored opportunity to identify specific drug targets (54,55), but this goal first entails broadening our understanding of the translation pathways in these organisms. Towards this aim, we determined the crystal structure of the LeishIF4E-1 isoform from Leishmania major in complex with an N-terminal fragment of a 4E-interacting protein, Leish4E-IP1 (Figure 1). Although the sequence of Leish4E-IP1 could not be used to deduce its function, the structure allowed us to come to a number of conclusions. First, Leish4E-IP1’s interface extends beyond the consensus motif, involving a short linker and a second alpha-helical segment that engages the dorsal surface of LeishIF4E-1. In 4E-BP1, two important phosphorylation sites (S65 and T70) are found within the linker, and phosphorylation of these sites is critical to promote the dissociation of 4E-BP1 from eIF4E. Leish4E-IP1 does not seem to contain phosphorylation sites in its linker portion (Figure 2C). Hence, it is unclear how the interaction between LeishIF4E-1 and Leish4E-IP1 is regulated. We observed a nine-fold difference between the binding affinity of Leish4E-IP11–100 (90 nM) and Leish4E-IP11–52 (10 nM) for LeishIF4E-1 (Table 1). The extra Leish4E-IP1 residues (amino acids 53–100), while not included in the structure, could be involved in modulating the interaction with LeishIF4E-1 (either directly or allosterically). Additionally, binding of Leish4E-IP1 could be regulated by phosphorylation on sites located outside the N-terminal fragment used in this study, or LeishIF4E-1 itself could get phosphorylated to modulate its interaction with Leish4E-IP1. The Leish(IF4E-1/4E-IP1) interaction could also be regulated through post-translational modifications other than phosphorylation. Another possibility is that there is no need for active regulation of the interaction between the two proteins and that the dynamics of the interaction are controlled through ligand availability and concentration. Second, we observed that the binding of Leish4E-IP1 to LeishIF4E-1 leads to conformational changes in the side chains of tryptophans in LeishIF4E-1, which are most likely located in the cap-binding pocket. This is consistent with our observation that the addition of Leish4E-IP11-52 to LeishIF4E-1 isolated from Leishmania parasites leads to the dissociation of LeishIF4E-1 from the m7GTP cap (Figure 3).
Our results suggest that Leish4E-IP1 is a cap-binding repressor of LeishIF4E-1. The molecular effect of Leish4E-IP1 on LeishIF4E-1 is thus different from the effect of 4E-BPs/eIF4Gs/Cup on eIF4E, which all have been shown to increase the affinity of eIF4E for the cap (56,57) and share a similar footprint on eIF4E (Supplementary Figure S4). Leish4E-IP1 may be more similar to Really Interesting New Gene (RING) domain containing proteins, including the tumor suppressor promyelocytic leukemia protein (PML) and the arenavirus Z protein, which both reduce the affinity of eIF4E for m7GTP (58,59). Residues of eIF4E involved with Z interaction have been described (60); however, no structure of an eIF4E/Z or eIF4E/PML complex is available for direct comparison. Therefore, the co-crystal structure of the Leishmania complex presented here, in combination with the NMR data, most clearly define a mechanism for cap-binding dysregulation. The identification of such a mode of regulation in human cells (whether it be by PML or by the viral Z protein), and now in a lower eukaryote (as presented here), further suggests that this mechanism is found in highly divergent life forms.
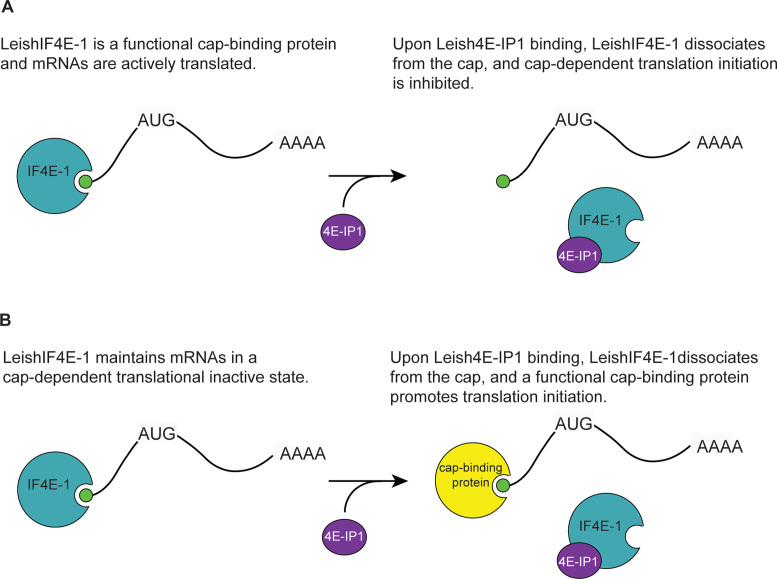
Despite LeishIF4E-1’s cap binding activity during prolonged exposure of parasites at increased temperatures and in acidic pH, an environment that would be encountered in the human host during infection, it is not known whether LeishIF4E-1 promotes translation initiation or keeps mRNAs in an inactive state by binding to their 5′ end of mRNAs and preventing cap-dependent translation. In light of our data, we propose two models to explain the role of Leish(IF4E-1/4E-IP1) complex formation during protein synthesis (Figure 4). In one model, LeishIF4E-1 is a functional cap-binding translation initiation factor. Upon binding of Leish4E-IP1, LeishIF4E-1 dissociates from the cap, which inhibits cap-dependent translation initiation mediated by LeishIF4E-1 (Figure 4A). An alternative model is that binding of LeishIF4E-1 to the cap maintains mRNAs in a cap-dependent translation inactive state (Figure 4B). Upon binding of Leish4E-IP1 to LeishIF4E-1, LeishIF4E-1 dissociates from the cap allowing a functional, yet unidentified, cap-binding protein to initiate translation. Additional studies would be required to support either model.
Figure 4.
Alternative models of Leish(IF4E-1/4E-IP1) function. (A) LeishIF4E-1 (IF4E-1, cyan) is a functional cap-binding protein and activates translation initiation. The binding of Leish4E-IP1 (4E-IP1, purple) inhibits translation initiation by preventing the binding of LeishIF4E-1 to the cap. (B) LeishIF4E-1 maintains mRNAs in a cap-dependent translational inactive state. Upon binding of Leish4E-IP1, LeishIF4E-1 dissociates from the cap, which frees the mRNAs and allows them to interact with a functional cap-binding protein (yellow) and to be actively translated. For clarity of the figure, proteins are not represented to scale.
DATA AVAILABILITY
Atomic coordinates and structure factors for the LeishIF4E-1 bound to Leish4E-IP11–52 crystal structure have been deposited with the Protein Data bank under accession number 5WB5.
Supplementary Material
ACKNOWLEDGEMENTS
We thank S.A. Robson and S.G. Hyberts for their help with NMR experiments. We thank S. Jenni, D.D. Raymond and D. Olal for their help with X-ray crystallography experiments. We thank the staff at BCSB (Advanced Light Source, Lawrence Berkeley National Laboratory) for assistance with X-ray data collection.
Author contributions: Conceptualization, G.W., M.S. and M.L.-A.; Methodology, H.A. and M.L.-A.; Investigation, S.M., H.A., A.Z., A.B. and M.L.-A; Resources, G.W., and M.S.; Writing-Original Draft, M.L.-A.; Writing-Review & Editing, S.M., H.A., M.S., G.W. and M.L.-A.; Supervision, G.W., M.S. and M.L.-A.; Funding Acquisition, G.W., M.S. and M.L.-A.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
US National Institutes of Health [R01-AI108718, R01-CA200913 to G.W.]; Harvard Catalyst|The Harvard Clinical and Translational Science Center (National Center for Advancing Translational Sciences, National Institutes of Health Award [UL1 TR001102]; Funding for open access charge: National Institutes of Health [R01-AI108718].
Conflict of interest statement. None declared.
REFERENCES
- 1. Dostálová A., Volf P.. Leishmania development in sand flies: parasite-vector interactions overview. Parasit Vectors. 2012; 5:276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kaye P., Scott P.. Leishmaniasis: complexity at the host-pathogen interface. Nat. Rev. Microbiol. 2011; 9:604–615. [DOI] [PubMed] [Google Scholar]
- 3. Bates P.A. Transmission of Leishmania metacyclic promastigotes by phlebotomine sand flies. Int. J. Parasitol. 2007; 37:1097–1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Shapira M., McEwen J.G., Jaffe C.L.. Temperature effects on molecular processes which lead to stage differentiation in Leishmania. EMBO J. 1988; 7:2895–2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zilberstein D., Shapira M.. The role of pH and temperature in the development of Leishmania parasites. Annu. Rev. Microbiol. 1994; 48:449–470. [DOI] [PubMed] [Google Scholar]
- 6. Rosenzweig D., Smith D., Opperdoes F., Stern S., Olafson R.W., Zilberstein D.. Retooling Leishmania metabolism: from sand fly gut to human macrophage. FASEB J. 2008; 22:590–602. [DOI] [PubMed] [Google Scholar]
- 7. Sacks D., Perkins P.. Identification of an infective stage of Leishmania promastigotes. Science. 1984; 223:1417–1419. [DOI] [PubMed] [Google Scholar]
- 8. Clayton C.E. Gene expression in Kinetoplastids. Curr. Opin. Microbiol. 2016; 32:46–51. [DOI] [PubMed] [Google Scholar]
- 9. Shatkin A.J. mRNA cap binding proteins: essential factors for initiating translation. Cell. 1985; 40:223–224. [DOI] [PubMed] [Google Scholar]
- 10. Kumar P., Hellen C.U.T., Pestova T.V.. Toward the mechanism of eIF4F-mediated ribosomal attachment to mammalian capped mRNAs. Genes Dev. 2016; 30:1573–1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gingras A.C., Raught B., Sonenberg N.. eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu. Rev. Biochem. 1999; 68:913–963. [DOI] [PubMed] [Google Scholar]
- 12. Sonenberg N., Hinnebusch A.G.. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell. 2009; 136:731–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mader S., Lee H., Pause A., Sonenberg N.. The translation initiation factor eIF-4E binds to a common motif shared by the translation factor eIF-4 gamma and the translational repressors 4E-binding proteins. Mol. Cell. Biol. 1995; 15:4990–4997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Beretta L., Gingras A.C., Svitkin Y.V., Hall M.N., Sonenberg N.. Rapamycin blocks the phosphorylation of 4E-BP1 and inhibits cap-dependent initiation of translation. EMBO J. 1996; 15:658–664. [PMC free article] [PubMed] [Google Scholar]
- 15. Gingras A.C., Raught B., Gygi S.P., Niedzwiecka A., Miron M., Burley S.K., Polakiewicz R.D., Wyslouch-Cieszynska A., Aebersold R., Sonenberg N.. Hierarchical phosphorylation of the translation inhibitor 4E-BP1. Genes Dev. 2001; 15:2852–2864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Holz M.K., Ballif B.A., Gygi S.P., Blenis J.. mTOR and S6K1 mediate assembly of the translation preinitiation complex through dynamic protein interchange and ordered phosphorylation events. Cell. 2005; 123:569–580. [DOI] [PubMed] [Google Scholar]
- 17. Shin S., Wolgamott L., Roux P.P., Yoon S.-O.. Casein kinase 1ϵ promotes cell proliferation by regulating mRNA translation. Cancer Res. 2014; 74:201–211. [DOI] [PubMed] [Google Scholar]
- 18. Shin S., Wolgamott L., Tcherkezian J., Vallabhapurapu S., Yu Y., Roux P.P., Yoon S.-O.. Glycogen synthase kinase-3β positively regulates protein synthesis and cell proliferation through the regulation of translation initiation factor 4E-binding protein 1. Oncogene. 2014; 33:1690–1699. [DOI] [PubMed] [Google Scholar]
- 19. Agabian N. Trans splicing of nuclear pre-mRNAs. Cell. 1990; 61:1157–1160. [DOI] [PubMed] [Google Scholar]
- 20. Bangs J.D., Crain P.F., Hashizume T., McCloskey J.A., Boothroyd J.C.. Mass spectrometry of mRNA cap 4 from trypanosomatids reveals two novel nucleosides. J. Biol. Chem. 1992; 267:9805–9815. [PubMed] [Google Scholar]
- 21. Michaeli S. Trans-splicing in trypanosomes: machinery and its impact on the parasite transcriptome. Future Microbiol. 2011; 6:459–474. [DOI] [PubMed] [Google Scholar]
- 22. Yoffe Y., Léger M., Zinoviev A., Zuberek J., Darzynkiewicz E., Wagner G., Shapira M.. Evolutionary changes in the Leishmania eIF4F complex involve variations in the eIF4E-eIF4G interactions. Nucleic Acids Res. 2009; 37:3243–3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zinoviev A., Léger M., Wagner G., Shapira M.. A novel 4E-interacting protein in Leishmania is involved in stage-specific translation pathways. Nucleic Acids Res. 2011; 39:8404–8415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yoffe Y., Zuberek J., Lerer A., Lewdorowicz M., Stepinski J., Altmann M., Darzynkiewicz E., Shapira M.. Binding specificities and potential roles of isoforms of eukaryotic initiation factor 4E in Leishmania. Eukaryotic Cell. 2006; 5:1969–1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Dhalia R., Reis C.R.S., Freire E.R., Rocha P.O., Katz R., Muniz J.R.C., Standart N., de Melo Neto O.P.. Translation initiation in Leishmania major: characterisation of multiple eIF4F subunit homologues. Mol. Biochem. Parasitol. 2005; 140:23–41. [DOI] [PubMed] [Google Scholar]
- 26. Freire E.R., Vashisht A.A., Malvezzi A.M., Zuberek J., Langousis G., Saada E.A., Nascimento J.D.F., Stepinski J., Darzynkiewicz E., Hill K. et al. eIF4F-like complexes formed by cap-binding homolog TbEIF4E5 with TbEIF4G1 or TbEIF4G2 are implicated in post-transcriptional regulation in Trypanosoma brucei. RNA. 2014; 20:1272–1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Freire E.R., Malvezzi A.M., Vashisht A.A., Zuberek J., Saada E.A., Langousis G., Nascimento J.D.F., Moura D., Darzynkiewicz E., Hill K. et al. Trypanosoma brucei translation initiation factor homolog EIF4E6 forms a tripartite cytosolic complex with EIF4G5 and a capping enzyme homolog. Eukaryotic Cell. 2014; 13:896–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Jones D.T. Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol. 1999; 292:195–202. [DOI] [PubMed] [Google Scholar]
- 29. Bao W.-J., Gao Y.-G., Chang Y.-G., Zhang T.-Y., Lin X.-J., Yan X.-Z., Hu H.-Y.. Highly efficient expression and purification system of small-size protein domains in Escherichia coli for biochemical characterization. Protein Expr. Purif. 2006; 47:599–606. [DOI] [PubMed] [Google Scholar]
- 30. Edery I., Altmann M., Sonenberg N.. High-level synthesis in Escherichia coli of functional cap-binding eukaryotic initiation factor eIF-4E and affinity purification using a simplified cap-analog resin. Gene. 1988; 74:517–525. [DOI] [PubMed] [Google Scholar]
- 31. Sekiyama N., Arthanari H., Papadopoulos E., Rodriguez-Mias R.A., Wagner G., Léger-Abraham M.. Molecular mechanism of the dual activity of 4EGI-1: Dissociating eIF4G from eIF4E but stabilizing the binding of unphosphorylated 4E-BP1. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:E4036–E4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hyberts S.G., Takeuchi K., Wagner G.. Poisson-gap sampling and forward maximum entropy reconstruction for enhancing the resolution and sensitivity of protein NMR data. J. Am. Chem. Soc. 2010; 132:2145–2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hyberts S.G., Milbradt A.G., Wagner A.B., Arthanari H., Wagner G.. Application of iterative soft thresholding for fast reconstruction of NMR data non-uniformly sampled with multidimensional Poisson Gap scheduling. J. Biomol. NMR. 2012; 52:315–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Delaglio F., Grzesiek S., Vuister G.W., Zhu G., Pfeifer J., Bax A.. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995; 6:277–293. [DOI] [PubMed] [Google Scholar]
- 35. Goddard T.D., Huang C.C., Ferrin T.E.. Software extensions to UCSF chimera for interactive visualization of large molecular assemblies. Structure. 2005; 13:473–482. [DOI] [PubMed] [Google Scholar]
- 36. Kabsch W. Integration, scaling, space-group assignment and post-refinement. Acta Crystallogr. D Biol. Crystallogr. 2010; 66:133–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kabsch W. XDS. Acta Crystallogr. D Biol. Crystallogr. 2010; 66:125–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. McCoy A.J. Solving structures of protein complexes by molecular replacement with Phaser. Acta Crystallogr. D Biol. Crystallogr. 2007; 63:32–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Emsley P., Cowtan K.. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004; 60:2126–2132. [DOI] [PubMed] [Google Scholar]
- 40. Adams P.D., Afonine P.V., Bunkóczi G., Chen V.B., Echols N., Headd J.J., Hung L.W., Jain S., Kapral G.J., Grosse-Kunstleve R.W. et al. The Phenix software for automated determination of macromolecular structures. Methods. 2011; 55:94–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Schrodinger, LLC The PyMOL Molecular Graphics System. 2015; Version 1.8. [Google Scholar]
- 42. Schindelin J., Rueden C.T., Hiner M.C., Eliceiri K.W.. The ImageJ ecosystem: an open platform for biomedical image analysis. Mol. Reprod. Dev. 2015; 82:518–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Tomoo K., Matsushita Y., Fujisaki H., Abiko F., Shen X., Taniguchi T., Miyagawa H., Kitamura K., Miura K.-I., Ishida T.. Structural basis for mRNA Cap-Binding regulation of eukaryotic initiation factor 4E by 4E-binding protein, studied by spectroscopic, X-ray crystal structural, and molecular dynamics simulation methods. Biochim. Biophys. Acta. 2005; 1753:191–208. [DOI] [PubMed] [Google Scholar]
- 44. Marcotrigiano J., Gingras A.C., Sonenberg N., Burley S.K.. Cocrystal structure of the messenger RNA 5′ cap-binding protein (eIF4E) bound to 7-methyl-GDP. Cell. 1997; 89:951–961. [DOI] [PubMed] [Google Scholar]
- 45. Matsuo H., Li H., McGuire A.M., Fletcher C.M., Gingras A.C., Sonenberg N., Wagner G.. Structure of translation factor eIF4E bound to m7GDP and interaction with 4E-binding protein. Nat. Struct. Biol. 1997; 4:717–724. [DOI] [PubMed] [Google Scholar]
- 46. Kinkelin K., Veith K., Grünwald M., Bono F.. Crystal structure of a minimal eIF4E-Cup complex reveals a general mechanism of eIF4E regulation in translational repression. RNA. 2012; 18:1624–1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Volpon L., Osborne M.J., Topisirovic I., Siddiqui N., Borden K.L.B.. Cap-free structure of eIF4E suggests a basis for conformational regulation by its ligands. EMBO J. 2006; 25:5138–5149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Peter D., Igreja C., Weber R., Wohlbold L., Weiler C., Ebertsch L., Weichenrieder O., Izaurralde E.. Molecular architecture of 4E-BP translational inhibitors bound to eIF4E. Mol. Cell. 2015; 57:1074–1087. [DOI] [PubMed] [Google Scholar]
- 49. Clouse K.N., Ferguson S.B., Schüpbach T.. Squid, Cup, and PABP55B function together to regulate gurken translation in Drosophila. Dev. Biol. 2008; 313:713–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Macdonald P.M. Translational control: a cup half full. Curr. Biol. 2004; 14:R282–R283. [DOI] [PubMed] [Google Scholar]
- 51. Siddiqui N., Tempel W., Nedyalkova L., Volpon L., Wernimont A.K., Osborne M.J., Park H.-W., Borden K.L.B.. Structural insights into the allosteric effects of 4EBP1 on the eukaryotic translation initiation factor eIF4E. J. Mol. Biol. 2012; 415:781–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Shalev-Benami M., Zhang Y., Matzov D., Halfon Y., Zackay A., Rozenberg H., Zimmerman E., Bashan A., Jaffe C.L., Yonath A. et al. 2.8-Å Cryo-EM Structure of the Large Ribosomal Subunit from the Eukaryotic Parasite Leishmania. Cell Rep. 2016; 16:288–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Hashem Y., des Georges A., Fu J., Buss S.N., Jossinet F., Jobe A., Zhang Q., Liao H.Y., Grassucci R.A., Bajaj C. et al. High-resolution cryo-electron microscopy structure of the Trypanosoma brucei ribosome. Nature. 2013; 494:385–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Brito Querido J., Mancera-Martínez E., Vicens Q., Bochler A., Chicher J., Simonetti A., Hashem Y.. The cryo-EM Structure of a Novel 40S Kinetoplastid-Specific Ribosomal Protein. Structure. 2017; 25:1785–1794. [DOI] [PubMed] [Google Scholar]
- 55. Shalev-Benami M., Zhang Y., Rozenberg H., Nobe Y., Taoka M., Matzov D., Zimmerman E., Bashan A., Isobe T., Jaffe C.L. et al. Atomic resolution snapshot of Leishmania ribosome inhibition by the aminoglycoside paromomycin. Nat Commun. 2017; 8:1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. von Der Haar T., Ball P.D., McCarthy J.E.. Stabilization of eukaryotic initiation factor 4E binding to the mRNA 5′-Cap by domains of eIF4G. J. Biol. Chem. 2000; 275:30551–30555. [DOI] [PubMed] [Google Scholar]
- 57. Ptushkina M., von Der Haar T., Karim M.M., Hughes J.M., McCarthy J.E.. Repressor binding to a dorsal regulatory site traps human eIF4E in a high cap-affinity state. EMBO J. 1999; 18:4068–4075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Kentsis A., Dwyer E.C., Perez J.M., Sharma M., Chen A., Pan Z.Q., Borden K.L.. The RING domains of the promyelocytic leukemia protein PML and the arenaviral protein Z repress translation by directly inhibiting translation initiation factor eIF4E. J. Mol. Biol. 2001; 312:609–623. [DOI] [PubMed] [Google Scholar]
- 59. Cohen N., Sharma M., Kentsis A., Perez J.M., Strudwick S., Borden K.L.. PML RING suppresses oncogenic transformation by reducing the affinity of eIF4E for mRNA. EMBO J. 2001; 20:4547–4559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Volpon L., Osborne M.J., Capul A.A., de la Torre J.C., Borden K.L.B.. Structural characterization of the Z RING-eIF4E complex reveals a distinct mode of control for eIF4E. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:5441–5446. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Atomic coordinates and structure factors for the LeishIF4E-1 bound to Leish4E-IP11–52 crystal structure have been deposited with the Protein Data bank under accession number 5WB5.