Abstract
In eukaryotes, ribosome biogenesis requires folding and assembly of the precursor rRNA (pre-rRNA) with a large number of proteins and snoRNPs into huge RNA-protein complexes. In spite of intense genetic, biochemical and high-resolution cryo-EM studies in Saccharomyces cerevisiae, information about the structure of the 35S pre-rRNA is limited. To overcome this, we performed high-throughput SHAPE chemical probing on the 35S pre-rRNA within 90S pre-ribosomes. We focused our analyses on external (5′ETS) and internal (ITS1) transcribed spacers as well as the 18S rRNA region. We show that in the 35S pre-rRNA, the central pseudoknot is not formed and the central core of the 18S rRNA is in an open configuration but becomes more constrained in 20S pre-rRNA. The essential ribosome biogenesis protein Mrd1 influences the structure of the 18S rRNA region locally and is involved in organizing the central pseudoknot and surrounding structures. We demonstrate that U3 snoRNA dynamically interacts with the 35S pre-rRNA and that Mrd1 is required for disrupting U3 snoRNA base pairing interactions in the 5′ETS. We propose that the dynamic U3 snoRNA interactions and Mrd1 are essential for establishing the structure of the central core of 18S rRNA that is required for processing and 40S subunit function.
INTRODUCTION
Eukaryotic ribosome biogenesis requires more than 200 trans-acting factors, including proteins and small nucleolar RNAs (snoRNAs) that aid in the folding and maturation of precursor rRNA (pre-rRNA) (1,2). These trans-acting factors associate with the pre-rRNA at different stages, from synthesis of the pre-rRNA until translation competent ribosomes are formed in the cytoplasm (3). In Saccharomyces cerevisiae, the nascent RNA pol I transcript is folded and assembled co-transcriptionally into a 90S complex, also called small subunit (SSU) processome, containing U3 snoRNP (4,5). In exponentially growing cells, about 70% of the transcripts are cleaved during transcription at sites A0, A1 and A2 (A0–A2) to separate maturation of the SSU from the large subunit (LSU), and to remove internal (ITS) and external (ETS) transcribed spacers (6,7). The remaining 30% become full-length 7 kb long 35S transcripts that are substrates for post-transcriptional processing. More than 70 trans-acting factors are associated with the 90S pre-ribosome (2,4,5). The molecular function of most of the trans-acting factors is however not known. The 90S pre-ribosome is assembled in a hierarchical manner (8) and contains several subcomplexes (9). There are good reasons to assume that the 90S pre-ribosome is structurally highly dynamic. Cryo electron microscopy (cryo-EM) models have provided structural information for the 18S rRNA domains and many protein binding sites in the U3 processome containing partially processed rRNA (cleaved at site A0 but not at sites A1 and A2) (10–14). Furthermore, they position the U3 snoRNA, which is essential for A0–A2 cleavage (15), in a central position, base pairing to the pre-rRNA. However, knowledge about the folding of 35S pre-rRNA prior to A0–A2 cleavage, and the role of different trans-acting factors affecting such folding, is lacking so far.
Mrd1 is an essential biogenesis protein crucial for A0–A2 cleavage (16,17). Mrd1 contains five RNA Binding Domains (RBDs), can be cross-linked to nucleotides that form helix (H) 27 and H28 in the mature 18S rRNA (18) and is involved in compaction of the 90S pre-ribosome prior to cotranscriptional cleavage (19). Mrd1 is also required for release of U3 snoRNA from base pairing with 35S pre-rRNA (17). U3 snoRNA associates with the RNA pol I transcript early during transcription and is present in the pre-rRNA terminal balls observed by EM (4,6). Chemical probing data corroborate the influence of U3 snoRNA on folding the 18S rRNA sequence in the 35S pre-rRNA (20). Experimental data exist for base pairing between the hinge regions of U3 snoRNA and the 5′ETS (20–23). The 5′ETS base-pairing with the U3 5′-hinge was discovered and experimentally proven in yeast (21,22), whereas the 5′ETS base-pairing with the U3 3′-hinge was first described and proven in trypanosomes and Xenopus (24–27) where it was also suggested to occur in yeast. The latter was subsequently validated in yeast (20,23). In addition, U3 snoRNA has been suggested to base pair to several sites within the 18S rRNA sequence (12–14,28,29). One site, predicted based on evolutionary conserved sequence complementarity, corresponds to H2 of the central pseudoknot (30). The 18S rRNA central pseudoknot is a structural element including two stems, H1 and H2 and adjacent nucleotides. It is important for the overall 18S rRNA architecture and the decoding center. Two additional sites, suggested by cross-linking, ligation and sequencing (31), are located in sequences included in H26 and H28/H44, close to the central pseudoknot in the 40S subunit. Furthermore, cryo-EM based models of pre-ribosomal complexes containing partially processed pre-rRNA show that U3 base pairs to sequences of H27, also located close to the central pseudoknot (12–14). The U3 snoRNA–35S pre-rRNA base pairing interactions would likely contribute to bringing the beginning and central part of the 18S rRNA region close to each other in space. Furthermore, U3 snoRNA release is posited to be necessary for establishing the structure of the central parts of 18S rRNA, including the central pseudoknot. Collectively, this suggests that U3 snoRNA plays an important role when the 18S rRNA region of 35S pre-rRNA is compacted during processing, but how U3 snoRNA carries out this important function requires further analyses.
We recently developed a protocol to investigate the structure of pre-rRNA present in affinity purified 90S pre-ribosomes by combining SHAPE (selective 2′-hydroxyl acylation analyzed by primer extension) with high-throughput sequencing (ChemModSeq) (32,33). The SHAPE chemical 1M7 generically reacts with 2′-OH of all four nucleotides in flexible regions (34). Here, we used this protocol to analyse the pre-40S part of the 35S pre-rRNA, i.e. the 5′ETS, 18S rRNA sequence and ITS1. We show that in the 35S pre-rRNA, the central region of the 18S rRNA is present in a more open configuration than in 20S pre-rRNA and that the central pseudoknot is not formed in the 35S pre-rRNA. Our SHAPE analyses of proposed U3 interactions in the 35S pre-rRNA indicate that not all the interactions are present in 90S pre-ribosomes at the same time, arguing that U3 snoRNA dynamically interacts with the 35S pre-rRNA and that the interactions between U3 snoRNA and the pre-rRNA are remodeled during processing of the 90S pre-ribosome. We also investigated the structure of 35S pre-rRNA in cells expressing wild type or mutated Mrd1. We find that a deleterious deletion of the fifth RBD of Mrd1 (Δ5) (17) affects folding of the 35S pre-rRNA in very specific regions, while the majority of nucleotides are structurally unaltered. Our results demonstrate that binding of Mrd1 to central positions in the 18S rRNA portion of the pre-rRNA influences surrounding structures, including interactions of U3 snoRNA with the pre-rRNA and formation of the central pseudoknot. Mrd1 is therefore an important biogenesis factor that assists in establishing essential RNA configurations in the 90S pre-ribosome.
MATERIALS AND METHODS
Yeast strains and genetic manipulations
The following strains have been described previously: PLY094 (17), ASY055 (18), Nop58-TAP (35). The FLY012 and FLY015 strains were obtained by genomic integration of PCR generated cassettes as described in (36).
The strains have the following genotypes:
PLY094: MATa; ura3–52; leu2–3, 112; his3Δ200; lysΔ201
ASY055: MATa; ura3–52; leu2–3, 112; his3Δ200; lysΔ201; trp1Δ; Mrd1-ΔRBD5-TAP-TRP1; PGAL1–3HA-Mrd1::K.l.URA3
FLY012: MATα; ura3–52; his3Δ200; trp1Δ; lysΔ201; GAL2; Mrd1-TAP-TRP1; PGAL1–3HA-Erb1-K.l.URA3
FLY015: MATα; ura3–52; his3Δ200; trp1Δ; lysΔ201; GAL2; Utp4-TAP-TRP1; PGAL1–3HA-Mrd1-HIS3
Nop58-TAP: MATα; His3Δ1; leu2Δ0; met15Δ0; ura3Δ0; Nop58-TAP::K.l.URA3
Northern blot
Northern blot analysis was performed as previously described (19). The oligonucleotides rDNA-2 and U3_91 were used for hybridization.
Oligonucleotides
rDNA-2: 5′-GCTCTCATGCTCTTGCC-3′
U3_91: 5′-GGGGTACAAAGGTTATG-3′
18S_155: 5′-TACCACAGTTATACCATGTAGT-3′
18S_1191: 5′-AGTCAAATTAAGCCGCAG-3′
5′ETS_568: 5′-GCTTTTTCAGGTCTCTCTGCTG-3′
5′ETS_355: 5′-GCTATTCAACAAGGCATT-3′
5′ETS_393: 5′-AGGAGGTTACACTTGAAG-3′
Oligonucleotides were ordered from Eurofins Genomics.
Affinity purification of pre-ribosomes, 1M7 probing and isolation of 35S pre-rRNA
Overnight cultures of FLY012, ASY055 and FLY015 strains grown in YPG (1% yeast extract, 2% peptone and 2% galactose) were transferred to YPD (1% yeast extract, 2% peptone and 2% glucose) and grown exponentially for 8 h at 30°C, 250 rpm to an OD600 of 0.5. The cells were harvested, washed in PBS and frozen in liquid nitrogen. 500 OD600 units of cells were lysed in 4 ml TMN150 buffer (50 mM Tris–HCl pH 8.0, 1.5 mM MgCl2, 150 mM NaCl, 0.1% NP-40 and 5 mM β-mercaptoethanol), Complete Protease Inhibitor Cocktail, EDTA free (Roche) by addition of 6 ml 0.5 mm Zirconia-Silica beads (BioSpec) and vortexing at 4°C for five times one minute. 6 ml of TMN150 buffer was added and the extracts were clarified by centrifugation at 3200 × g for 15 min, followed by centrifugation at 20 000 × g for 20 min. The pH was adjusted to 7.5 and the extract was incubated with 400 μl IgG sepharose slurry (GE Healthcare) that had been pre-equilibrated in TMN150, at 4°C for 1 h. After transfer to a Poly-Prep Chromatography column (Bio-Rad) and washing five times with 10 ml TMN150, the beads were resuspended in 1 ml of 1M7 buffer (50 mM Tris–HCl pH 8.0, 1.5 mM MgCl2, 150 mM NaCl). After transfer to a microcentrifuge tube, the beads were suspended in 1M7 buffer in a total volume of 600 μl. 30 μl of 250 mM 1M7 (Prime Organics Inc., Woburn, MA, USA) in DMSO was added to the IgG beads (or only DMSO for unmodified controls) followed by incubation for 3 min at room temperature.
The RNA was subsequently extracted by addition of guanidine thiocyanate-phenol and incubation at 65°C for 10 min. The RNA was further purified by phenol–chloroform and chloroform extraction and precipitated by adding glycogen (50 μg/ml), 1/10 volume of NaOAc, pH 5.2 and 2.5 volumes of 99% ethanol. The RNA was washed in 70% ethanol, dried and resuspended in 10 μl H2O and 10 μl formamide loading buffer (formamide, 0,025% xylene cyanol) was added. The RNA was separated in SeaPlaque GTG low melting temperature agarose (Lonza, Rockland, Maine) containing 0.5 μg/ml ethidium bromide. Specific RNA species were visualized at 302 nm using a Mini-Transilluminator (Bio-Rad) in preparative mode setting. The 35S pre-rRNA band was purified from the agarose gel as previously described (37).
Purification of 12S and 90S complexes containing U3 snoRNA
Extracts of cells containing Nop58-TAP were centrifuged in 10–50% sucrose gradients as previously described (19). 12S and 90S fractions containing U3 snoRNA, according to localization of U3 snoRNA by northern hybridization and sedimentation of 40S, 60S and 80S ribosomal complexes, were used for affinity purification with IgG Sepharose and 1M7 treatment as described above.
Primer extension and data analysis
Before primer extension, the RNA was concentrated using RNA Clean & Concentrator™-5 kit (Zymo Research). 10 pmol of the desired primer was labeled with γ-32P-ATP or γ-33P-ATP using T4 polynucleotide kinase (Thermo Fisher Scientific). Primer extension was performed using SuperScript III reverse transcriptase (Life Technologies). RNA was denatured at 85°C for 3 min in a thermocycler, followed by primer annealing at 45°C for 2 min. Primer extension was performed at 45°C for 45 min or at 45°C for 10 min followed by a 30 min incubation at 49°C. Two units of ExoI and 1 unit of RNase I (Thermo Fisher Scientific) were added followed by incubation at 37°C for 30 min. An equal volume of formamide loading buffer was added to the primer extension reactions, heated at 99°C for 1 min, loaded onto 6% poly-acrylamide sequencing gel together with sequencing ladders generated using a plasmid containing an rDNA repeat and Sequenase™ Version 2.0 DNA sequencing kit (Thermo Fisher Scientific). Gels were dried and primer extension pattern was developed and quantified using the FLA-3000 phosphorimager system (Fujifilm).
The control (=DMSO) profile was scaled so that peak intensities within minimally reactive nucleotides in the 1M7 lane were equal to corresponding nucleotides in the control lane, as recommended (38). Signal decay correction was done using exponential function (b = 0.999 for DMSO and b = 0.995 for 1M7 treated samples). 1M7 reactivities were calculated by subtracting band intensity in the control lane from that of the corresponding nucleotide in the 1M7 lane. Occasionally obtained negative values were set to zero. 2–8% normalization was performed as previously described (39). 1M7/DMSO ratios were calculated and reactivities of nucleotides with 1M7/DMSO < 1.5 were also set to zero to avoid false-positives. In all figures, a threshold of 0.25 for the reactivities was chosen, based on reproducibility and to confidently distinguish from noise.
ChemModSeq and data analyses
ChemModSeq was performed as described (32). Data analysis was performed as previously described (33) using the ChemModSeq pipeline (https://bitbucket.org/sgrann/chemmodseqpipeline). The resulting data were then used to calculate SHAPE reactivities using the StructureFold (40) and BUM-HMM (41) algorithms. The SHAPE reactivities were subsequently scaled using a 2–8% normalization step (39). To compare SHAPE reactivities between Δ5 and ΔErb1 we used the raw (unscaled) SHAPE reactivities and ΔSHAPE algorithm as previously described (42). The ΔSHAPE results were subsequently compared to BUM-HMM posterior probabilities for nucleotide reactivity, calculated as described (41). Nucleotides with posterior probabilities equal to or higher than 0.95 were considered modified by 1M7. For a nucleotide to be considered as differently modified it had to be selected by the ΔSHAPE algorithm and be considered modified by BUM-HMM in at least one of the two samples. The 20S ChemModSeq data were previously described (32) and were generated using Enp1-HTP as bait.
RESULTS
Shape analysis of 90S pre-ribosomal particles
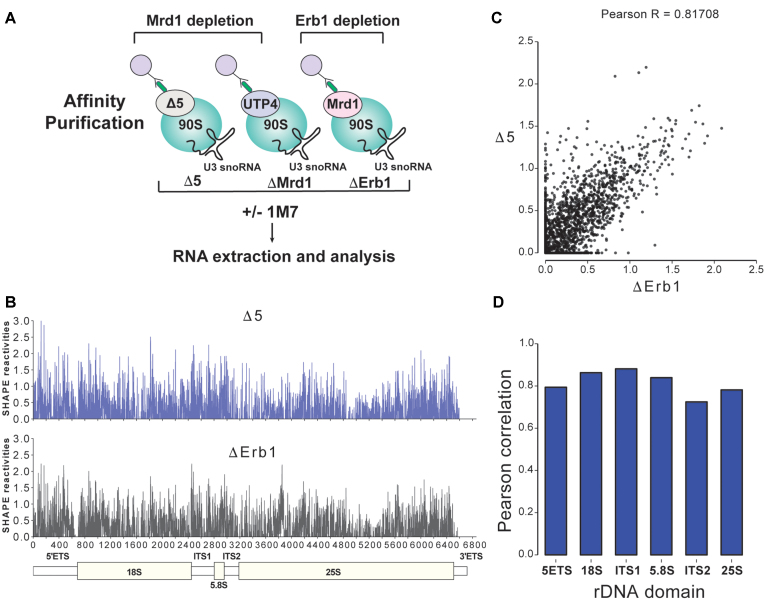
Analysis of the primary RNA pol I transcript, 35S pre-rRNA in yeast, is experimentally challenging as it is rapidly processed (7) and susceptible to degradation. We developed a protocol for isolation and structural analyses of 35S pre-rRNA present in the 90S pre-ribosome. This also allowed us to study if and how Mrd1 influences the folding of 35S pre-rRNA. To obtain sufficient amounts of 35S pre-rRNA, we used an Mrd1 mutant lacking the fifth RNA binding domain (17), and as a control a strain in which Erb1, a large subunit (LSU) processing factor, could be depleted. Depletion of pre-60S factors, such as Erb1, results in accumulation of 35S that can be processed into mature 18S rRNA (43), whereas depletion of Mrd1 (ΔMrd1) or mutations in Mrd1, such as Δ5, results in accumulation of 35S that can be processed into 25S and 5.8S rRNA, but not into mature 18S rRNA (16,17). We reasoned that the SSU part of the 35S pre-rRNA in ΔErb1 is likely to represent wild-type characteristics, whereas 35S pre-rRNA in Mrd1 mutants was expected to display structural differences reflecting essential functions of Mrd1 during maturation of the SSU processome. Depletion of Erb1 may induce structural changes in the LSU region of 35S pre-rRNA, but a detailed analysis of such alterations were beyond the scope of the present study. As Mrd1 and Erb1 are essential proteins we generated conditional mutants where the corresponding genes were placed under control of the Gal1-promoter (PGal1), which allows Mrd1 or Erb1 depletion in glucose containing medium. In these strains, appropriate proteins were TAP-tagged to allow affinity purification. We purified 90S pre-ribosomes containing 35S pre-rRNA from PGal1-ERB1/Mrd1-TAP, PGal1-MRD1/Mrd1Δ5-TAP and PGal1-MRD1/UTP4-TAP strains following depletion of Mrd1 or Erb1 in glucose containing medium. For simplicity, these three strains are referred to as ΔErb1, Δ5 and ΔMrd1, respectively (Figure 1A).
Figure 1.
1M7 Chemical probing of 90S pre-ribosomal complexes. (A) Experimental setup. After depletion of either WT Mrd1 or Erb1, 90S pre-ribosomal complexes (blue circles) were affinity purified using TAP (green rods) tagged Mrd1Δ5, UTP4 (for ΔMRD1) or Mrd1 (for ΔErb1). Purified complexes were treated with 1M7 or only the solvent (DMSO). 35S pre-rRNA was extracted from agarose gels prior to analysis by primer extension and ChemModSeq. (B) 35S pre-rRNA SHAPE reactivities from the Δ5 and ΔErb1 samples. A schematic representation of the various domains of 35S pre-rRNA is shown at the bottom. (C) Scatter plot comparing the 35S pre-rRNA SHAPE reactivities for Δ5 and ΔErb1. (D) Pearson correlation for the various domains of 35S pre-rRNA in Δ5 and ΔErb1.
Affinity purified 90S pre-ribosomal complexes were treated with the SHAPE reagent 1-methyl-7-nitroisatoic anhydride (1M7) (Figure 1A) that reacts with all four nucleotides in flexible (single-stranded) regions and acylates the 2′-OH of the ribose (34). To minimize contamination by co-precipitated mature rRNAs and processing intermediates, 35S pre-rRNA was extracted from agarose gels and purified after electrophoretic separation. (Supplementary Figure S1). Subsequently, cDNA libraries were generated by random priming and high-throughput sequenced using the ChemModSeq protocol. These data enabled us to generate a snapshot of SHAPE reactivity in the entire 35S pre-RNA (Supplementary Table S1 and Figure 1B). To validate our findings, we also performed primer extension with radioactively labeled primers. The 35S pre-rRNA SHAPE reactivity profiles of the Δ5 and ΔErb1 ChemModSeq samples were highly correlated (Pearson R = 0.81708; Figure 1B–D) arguing that the flexibility of the vast majority of nucleotides in Δ5 was not perturbed overall compared to ΔErb1. Specific clusters of nucleotides showing differential 1M7 reactivity were further analysed as described below.
The 18S rRNA region in 35S pre-rRNA is more flexible than in 20S pre-rRNA
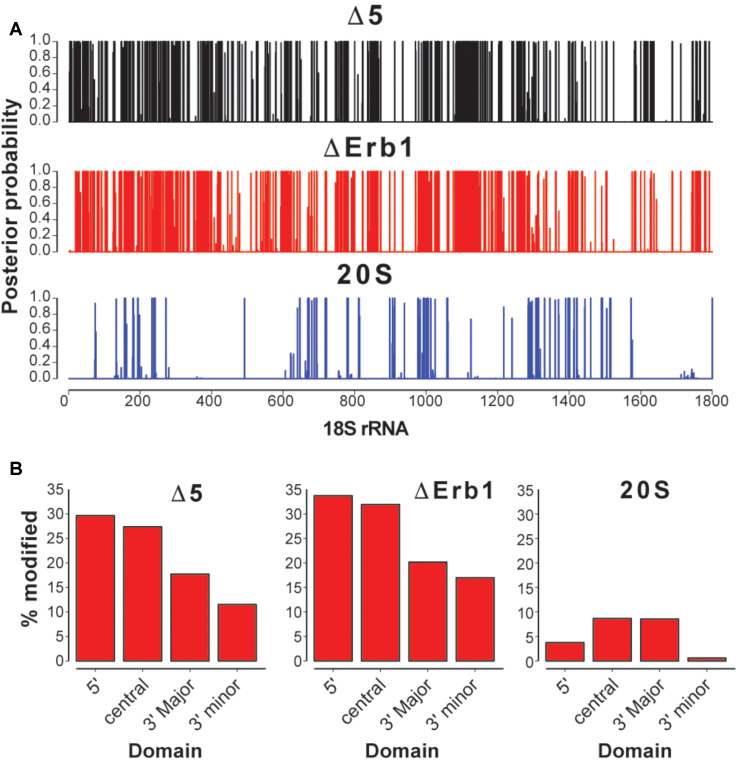
To identify sites that were very likely modified by 1M7, we utilized the Beta-Uniform Mixture Hidden Markov Model, BUM-HMM (41), a statistical approach for modeling reactivity scores derived from ChemModSeq count data. The BUM-HMM algorithm accounts for biological variability and biases in the data, such as coverage and sequence bias, to identify nucleotides that are significantly more modified compared to control samples. BUM-HMM calculates a posterior probability of modification for each nucleotide within 35S pre-rRNA, which indicates the likelihood that the observed degree of modification cannot be explained by random variability alone. When applied to the 18S rRNA region, we identified considerably more modified nucleotides in the 35S pre-rRNA (ΔErb1 and Δ5) compared to 20S pre-rRNA data (Figure 2A and B). This disparity was observed for all domains of the 18S rRNA region (Figure 2B), and the 5′ and central domains showed the highest number of modified nucleotides in the 35S pre-rRNA (27–34%). Within 20S pre-rRNA, most of the modified nucleotides are located at positions that in the mature 18S rRNA correspond to loops or single stranded regions (Supplementary Figure S2B). In contrast, many of the modified nucleotides in 35S pre-rRNA correspond to base paired nucleotides in the mature 18S rRNA (Supplementary Figure S2A). We conclude that the 18S rRNA region of the 35S pre-rRNA is a lot more open and flexible and therefore structurally distinct compared to the 20S pre-rRNA.
Figure 2.
BUM-HMM analyses of SHAPE reactivities for the 18S rRNA region. (A) The 18S rRNA region within 35S pre-rRNA in Mrd1Δ5 (black), within 35S pre-rRNA in ΔErb1 (red) and within 20S pre-rRNA (blue). Numbers show nucleotide positions from the 5′ to the 3′ end of 18S rRNA. (B) Percentage modified nucleotides (posterior probabilities ≥ 0.95) in each of the 18S rRNA domains in Δ5, ΔErb1 and 20S.
Based on analysis of the modification status at positions corresponding to helices in mature 18S rRNA (as shown for ΔErb1 in Supplementary Figure S2A), we propose that several 18S rRNA helices are already formed in the 35S pre-rRNA. Such examples can be found in all four major secondary structural domains of 18S rRNA, such as for example H11 (5′-domain), H23 (central domain), H41 (3′-major domain) and bottom part of H44 (3′-minor domain). The high number of modified nucleotides and their positions (Supplementary Figure S2A), suggest that many of the helices have not been formed in the 5′-domain. In the central region, where the different secondary structural domains of mature 18S rRNA converge, 35S but not 20S pre-rRNA contained a large number of modified nucleotides in the areas corresponding to H2, H3, H25, H26a H27, H28 and the top part of H44 in mature 18S rRNA (Supplementary Figure S2A). These data suggest that although some helices are likely formed already in the 35S pre-rRNA, the 90S pre-ribosomal particle is not fully compacted and that the central region is particularly flexible.
Mrd1 affects nucleotide reactivity at specific sites in the 5′ETS
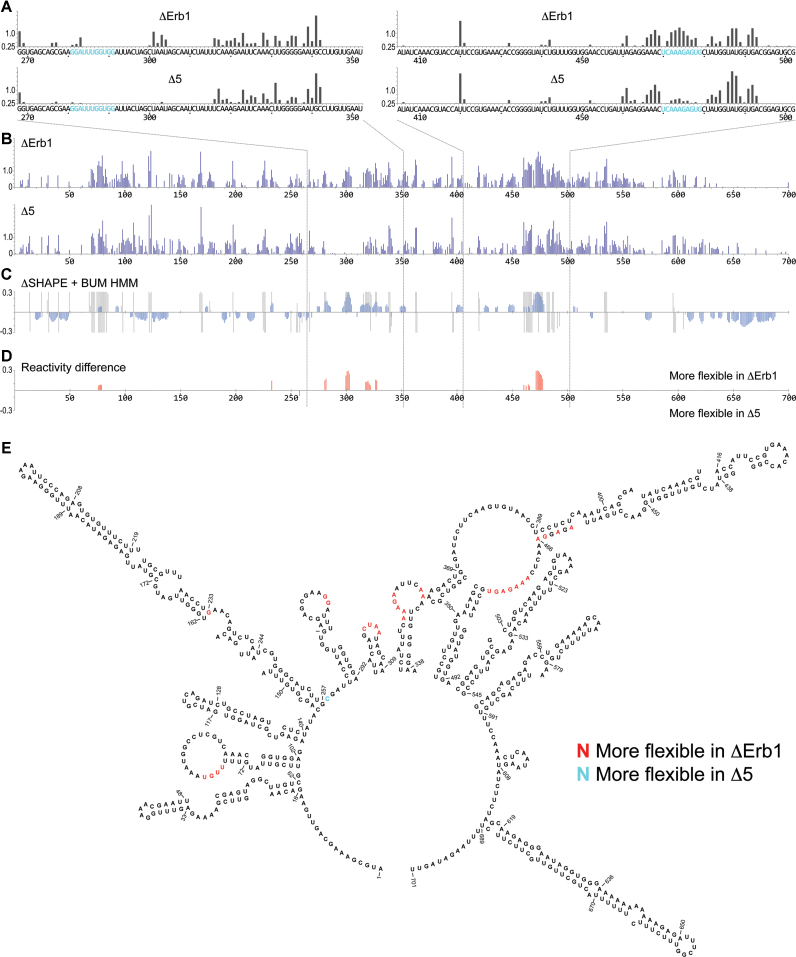
Although we found a strikingly overall similar pattern of 1M7 reactivity in ΔErb1 and Δ5, we identified distinct differences in specific regions (Figure 1B and C). Analyses of the 5′ETS region are shown in Figure 3A–E. ChemModSeq SHAPE reactivities for the 5′ETS region in ΔErb1 and Δ5 (Figure 3B) were compared using the ΔSHAPE algorithm (Figure 3C) (42). The ΔSHAPE algorithm identifies nucleotides that show differentential SHAPE reactivity in the ΔErb1 and Δ5 samples. This algorithm subtracts the raw SHAPE reactivities from the two samples and subsequently performs a statistical test to select only those nucleotides for which the difference in reactivity is statistically significant. We also analyzed our data using the BUM-HMM algorithm (41), to identify regions that were reproducibly modified (Figure 3C). In addition, to increase the stringency, only nucleotides that the BUM-HMM algorithm predicted to be modified in at least one of the two 35S samples were selected (Figure 3D and E). This showed that the ΔErb1 and Δ5 SHAPE reactivity patterns in the 5′ETS were significantly different in two regions which were further analysed by primer extension, nucleotides 270–350 and nucleotides 410–500 (Figure 3A). These regions contain two proposed U3 base pairing sites (281–291 and 470–479) (Figure 3A). According to cryo-EM data, the latter region can be extended to 464–479 (12–14). The nucleotides 281–291 were mostly unreactive in both samples, but nucleotides 282 and 283 were more reactive in ΔErb1 than in Δ5. More striking differences were observed for the nucleotides 470–479, which were considerably more reactive in ΔErb1 compared to Δ5 (Figure 3A). In both samples, nucleotides 466–469 were modified, suggesting that they were flexible and therefore likely not involved in base pairing interactions. The primer extension SHAPE reactivity pattern was very similar to that obtained by ChemModSeq (Figure 3A and B, Supplementary Table S1). Also, the ΔMrd1 and Δ5 reactivity patterns were almost identical (Supplementary Figure S4). Notably, the nucleotides 470–479 were also reactive in primer extension analysis of pre-rRNA purified using a wild type yeast strain where Nop58 was TAP-tagged (Supplementary Figure S3B, 90S/1M7). This supports the idea that the ΔErb1 strain represents wild-type characteristics regarding nucleotide flexibility of the SSU part of the 35S pre-rRNA. We conclude that in ΔErb1 35S pre-rRNA, the nucleotides 470–479 are largely reactive and thus unlikely to be base paired, whereas in the two Mrd1 mutants these nucleotides are more constrained. This suggests that the region is base paired, representing a striking structural alteration in the 90S pre-ribosome, induced by the Mrd1 mutations.
Figure 3.
SHAPE reactivity pattern for the 5′ETS region in 35S pre-rRNA. (A) Primer extension determined SHAPE reactivities of nucleotides 268–355 (left) and nucleotides 405–504 (right) in ΔErb1 and Δ5. U3 base pairing sites, proposed in (20–23), are shown in blue. (B) ChemModSeq analyses for the entire 5′ETS (nucleotides 1–700) of 35S pre-rRNA in ΔErb1 and Δ5. (C) Comparison of SHAPE reactivities in ΔErb1 and Δ5, using ΔSHAPE analyses and BUM-HMM probabilities. Blue bars show the ΔSHAPE values. Gray regions mark nucleotides with posterior probabilities of ≥0.95 according to BUM-HMM analyses for ΔErb1 (top) and Δ5 (bottom). (D) ΔSHAPE reactivity differences between ΔErb1 and Δ5 that overlap with the BUM-HMM probabilities. In red, more flexibility in ΔErb1, in blue, more flexibility in Δ5. The regions analysed by primer extension are indicated by dashed lines. (E) Reactivity differences marked in a secondary structure model of the 5′ETS (44). Red nucleotides were more reactive in ΔErb1, blue nucleotides were more reactive in Δ5.
Structural models for the 5′ETS have previously been proposed based on chemical and enzymatic probing, in silico predictions and cryo-EM data (11,12,14,44,45). These models are compact, structurally similar and include 9–10 helices. In agreement with such compact models, we identified relatively few modified nucleotides in the 5′ETS of ΔErb1 (10.5% in contrast to 17–34% for the 18S rRNA domains shown in Figure 2B). In the proposed models these nucleotides were generally drawn as part of single-stranded loops. As a consequence of the relatively small number of SHAPE reactive nucleotides, it was not possible to confidently predict a revised structural model of the 5′ETS. Furthermore, our SHAPE results do not more strongly support one model over the others as the models are structurally similar. Note that in Figure 3E, the U3 snoRNA is not included in the model. The key finding of our analyses was the apparent structural differences in the 5′ETS found in the ΔErb1 and Δ5 samples, which were located in and around sequences proposed to base pair to U3 snoRNA.
Analysis of the 18S rRNA region of the 35S pre-rRNA
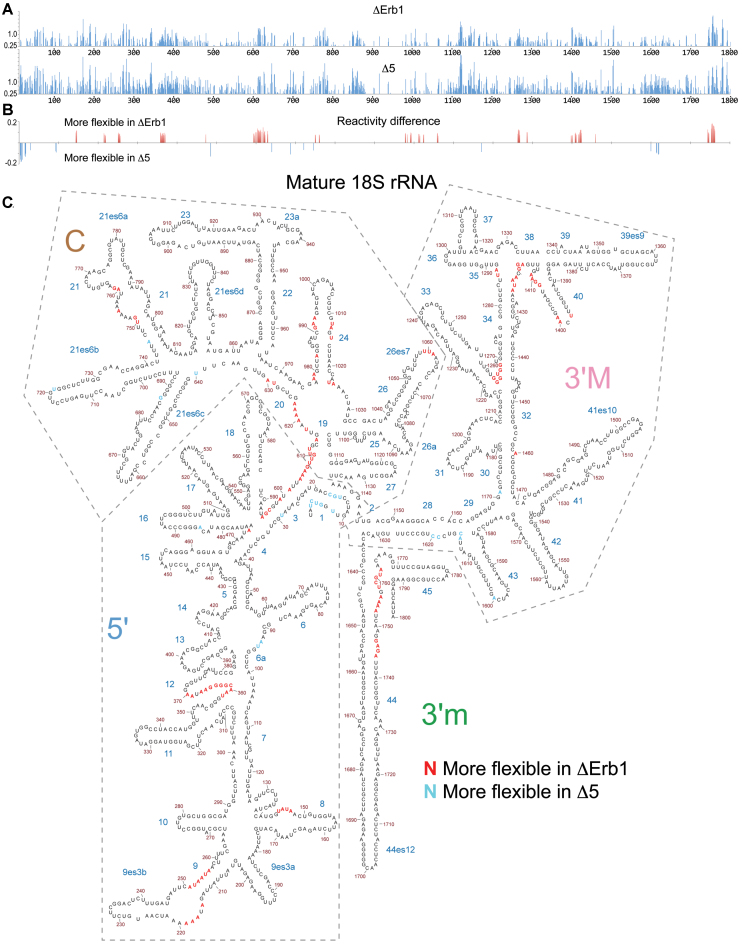
ΔSHAPE analyses together with BUM-HMM filtering revealed significant differences in nucleotide reactivity at specific sites within the 18S rRNA region of 35S pre-rRNA when comparing ΔErb1 and Δ5 (Figure 4A–C). Many of the differences cluster in the secondary structure of mature 18S rRNA (Figure 4C), suggesting that the Δ5 mutation affects the structure of specific regions. Such clusters are present at or close to H1, H3, H9, H12, H19, H24, H28, H34, H40 and top of H44. Some of the clusters are at the central region of the 18S rRNA portion of the pre-rRNA (close to H1, H3, H19, H28 and the upper part of H44) and may be a direct consequence of impaired Mrd1 function in the Δ5 mutant, as Mrd1 cross-links to H27 and H28 in vivo (18).
Figure 4.
SHAPE reactivity pattern for the 18S rRNA region in 35S pre-rRNA. (A) ChemModSeq analysis of SHAPE reactivities of the 18S rRNA region in 35S pre-rRNA in ΔErb1 and Δ5. (B) ΔSHAPE differences between ΔErb1 and Δ5 that overlap with the BUM-HMM probabilities ≥0.95. Data analysis as in Figure 3. Red bars represent nucleotides with more reactivity in ΔErb1 and blue bars nucleotides more reactive in Δ5. (C) Distribution of SHAPE reactivity differences between ΔErb1 and Δ5 in 18S rRNA within 35S pre-rRNA. Reactivity differences from Figure 4B are marked in the mature 18S rRNA secondary structure. Red marks nucleotides where ΔErb1 reactivity is higher and blue marks nucleotides where Δ5 is higher. Domains in the 18S rRNA are indicated by dashed lines.
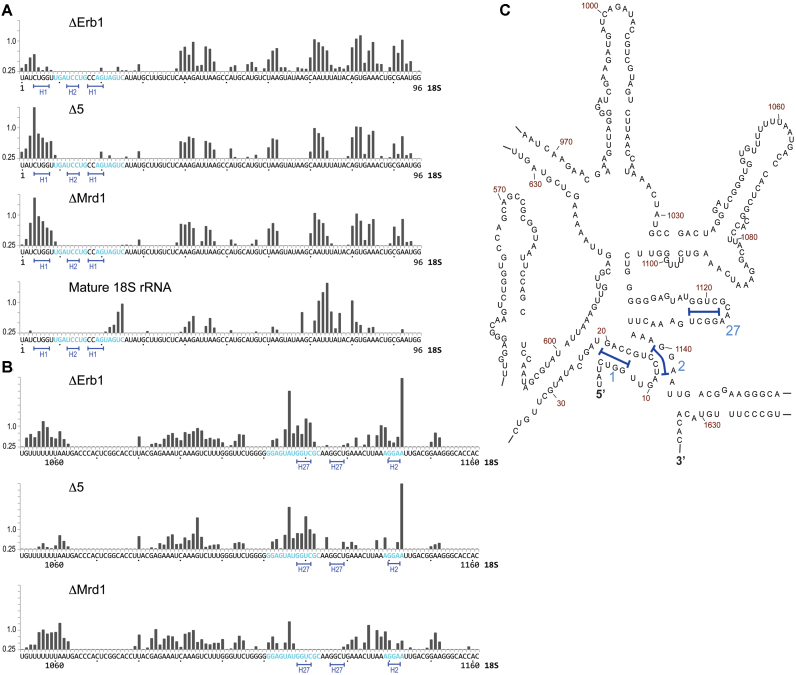
The biggest difference detected in the Δ5 strain was an increased reactivity in the 5′-end of the 18S rRNA portion corresponding to H1 (Figure 4). Primer extension analysis of the pre-rRNA confirmed that nucleotides 4–8 in the 5′-end of 18S rRNA were more reactive in Δ5 than in the ΔErb1 strain (Figure 5A) where only moderate reactivity was detected. No reactivity was observed for this segment in 18S rRNA (Figure 5A). The reactivity patterns of Δ5 and ΔMrd1 for this segment were very similar (Figure 5A).
Figure 5.
SHAPE reactivity pattern for the 5′ and central regions of the 18S rRNA part of 35S pre-rRNA from ΔErb1, Δ5 and ΔMrd1, as revealed by primer extension. (A) Nucleotides 1–100 and (B) nucleotides 1052–1161. Blue letters indicate nucleotides proposed to base pair to U3 snoRNA (28,30). Underlined nucleotides 4–8 and 16–19 are involved in H1, nucleotides 12–14 and 1140–1142 in H2. Nucleotides 1118–1121 and 1126–1129 are involved in H27. (C) Secondary structure of the central region in 18S rRNA. Helices are indicated by numbers. H1, H2 and H27 (as marked in A and B) are indicated by blue brackets.
Our primer extension analysis confirmed that many nucleotides in the central region, spanning nucleotides 1050–1160, were 1M7 modified (Figure 5B), including the nucleotides involved in formation of H2. The SHAPE reactivity profiles were very similar between Δ5 and ΔErb1, but we identified differences as compared to ΔMrd1. In mature 18S rRNA, nucleotides 1118–1121 base pairs to 1126–1129 to form H27. In ΔErb1 and Δ5 but not in ΔMrd1, nucleotides 1118–1122 were reactive (Figure 5B). Therefore, binding of Mrd1 to nucleotides 1126–1128 (18) could prevent formation of H27. Furthermore, we consistently found in our primer extension experiments that nucleotide A1143 was hyperreactive in ΔErb1 and Δ5, but not in ΔMrd1 (Figure 5B).
Our data argue that the central pseudoknot is not formed in the 35S pre-rRNA. The central pseudoknot consists of nucleotides 4–20, 1138–1144 and includes H1 and H2. Nucleotides involved in the central pseudoknot were reactive in all three 35S pre-rRNA samples (Figure 5A and B, Supplementary Table S1). The increased reactivity of nucleotides 4–8 (Figure 5A) in the Mrd1 mutants as compared to ΔErb1 indicates that Mrd1 is involved in formation of H1 of the central pseudoknot.
Different nucleotide flexibility of U3 snoRNA in 12S and 90S complexes
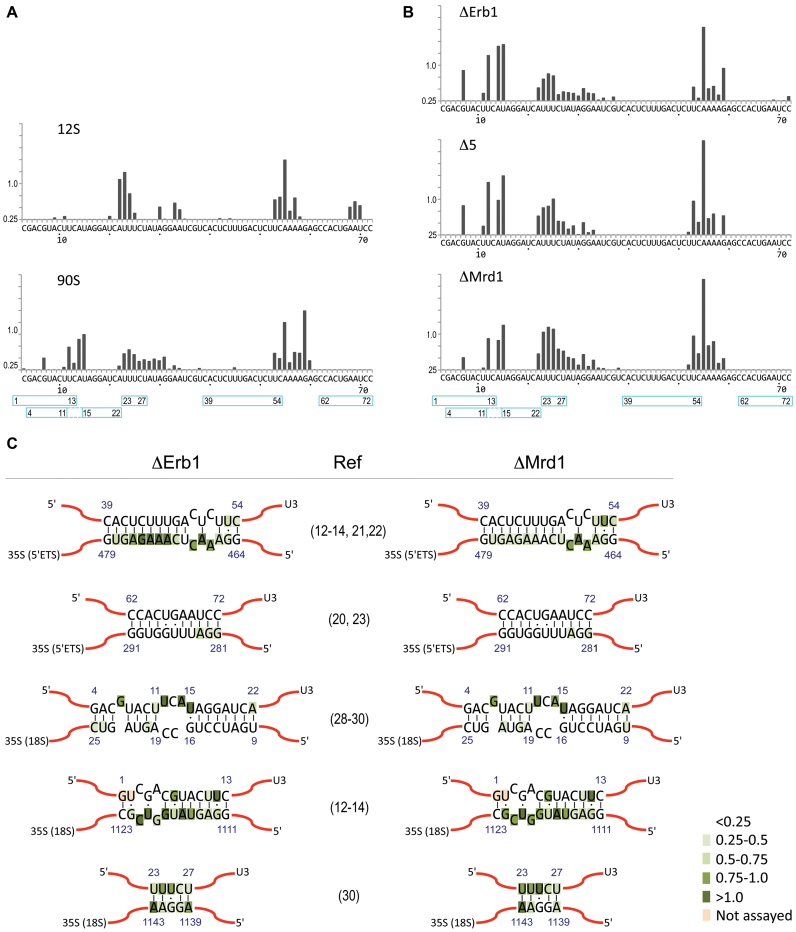
Some of the differences in reactivity pattern between 35S pre-RNA samples were observed at or near proposed U3 interaction sites. We therefore examined the SHAPE reactivity of the 5′-end of U3 snoRNA. The U3 snoRNP exists in the cell as a free 12S complex and as part of the 90S pre-ribosome. Sucrose gradient centrifugation was applied to separate 12S and 90S complexes with subsequent affinity purification using Nop58-TAP as bait in a WT strain. The U3 containing complexes, called U3 12S and U3 90S, were subjected to SHAPE analysis (Figure 6).
Figure 6.
SHAPE reactivity pattern of U3 snoRNA. Reactivity pattern of nucleotides 3 to 72 at the 5′ end of U3 snoRNA as assayed by primer extension in (A) for 12S and 90S complexes and (B) for 90S pre-ribosomes in ΔErb1, Δ5 and ΔMrd1. Blue boxes with numbers (beginning and end) mark nucleotides involved in proposed/demonstrated U3–35S interactions. (C) SHAPE reactivities at the proposed or experimentally demonstrated sites of base pairing between U3 snoRNA and 35S pre-rRNA. References are given. Reactivity of the nucleotides, based on our primer extension results, is shown in shades of green (light to dark green represents increasing SHAPE reactivity, as indicated).
Comparison of U3 12S and U3 90S identified both similarities and differences in reactivity patterns in U3 snoRNA present in these complexes (Figure 6A). Notably, a cluster of modified nucleotides (nucleotides G7 and 12–15) were present only in U3 90S and a cluster of three nucleotides (nucleotides 68–70) were present only in U3 12S. We also noted that in the region consisting of nucleotides 22–35, the distribution of reactive nucleotides differed between the two complexes, with a more continuous and extended region of reactivity in U3 90S. Our results show that U3 snoRNA has a different structure in the 90S pre-ribosome compared to in 12S complexes. Some of these differences may be due to base pairing interactions between U3 snoRNA and 35S pre-rRNA. The observed pattern of nucleotide reactivity of U3 snoRNA in the 90S processome in our data (Figure 6C) is compatible with several, but not all, of the previously proposed base pairing models (see Discussion).
Analysis of the U3 snoRNA nucleotide flexibility in 90S pre-ribosomes from ΔMrd1 and Δ5 cells showed very similar results as for ΔErb1 cells (Figure 6B). We noted that this flexibility pattern was similar to the one seen for U3 snoRNA in the 90S processome (Figure 6A), where Erb1 was present. We conclude that no clear influence of Mrd1 mutants could be observed on the reactivity of nucleotides in the 5′-part of U3 snoRNA.
DISCUSSION
Experimental protocol for structural probing of 35S pre-rRNA
Folding of pre-rRNA in the context of an RNA–protein complex is essential for ribosome synthesis in eukaryotes. We provide structural information about the early folding of the 35S pre-rRNA in S. cerevisiae that so far has been lacking. These data, together with recent cryo-EM studies (10–14) and with SHAPE data on the 25S rRNA component of the 35S pre-rRNA (33) provide a better understanding of the early events of ribosome maturation. We developed an experimental strategy that made it possible to specifically probe the flexibility of the nucleotides of the entire 35S pre-rRNA in 90S pre-ribosomes in SHAPE experiments. In exponentially growing yeast cells, ∼30% of the ribosomal gene transcripts become full-length 35S pre-rRNA (6,7) to be processed into mature ribosomes. In 70% of the cases, processing of the nascent transcripts take place during transcription, liberating pre-40S subunits. The complexes purified in this study contain full length 35S pre-rRNA. This, however, does not exclude the possibility that these complexes exhibit some compositional and/or structural heterogeneity.
The 18S rRNA is largely folded in connection with formation of the 43S pre-ribosome
We provide experimental data that demonstrate that the 18S rRNA part of 35S pre-rRNA is more flexible and structurally distinct compared to the 18S rRNA part of 20S pre-rRNA (Figure 2). Notably, the 5′-domain of 18S rRNA in 35S pre-rRNA is particularly devoid of formed helices (Supplementary Figure S2). This is in contrast to cryo-EM based models of SSU processomes containing partially processed pre-rRNA that has been cleaved at site A0 (10–14), suggesting that the 5′-domain is folded following or in connection to A0-cleavage. Our SHAPE data show that the mature 18S rRNA structure is largely established only within the 43S pre-ribosome or concomitant with the generation of this complex (Figure 2 and Supplementary Figure S2). The high proportion of flexible nucleotides in the 35S pre-rRNA likely reflects intermediate structures and extensive pre-rRNA-protein interactions. Our results in particular highlight a high proportion of flexible nucleotides in the 35S pre-rRNA that will become part of the structurally and functionally important central region of 18S rRNA. These data provide support for a model in which the coordinated formation of the central structures is crucial for the maturation and cleavage of the pre-ribosome.
Mrd1 is involved in structuring central regions of the 18S rRNA part in 35S pre-rRNA required for processing
A large number of trans-acting factors are associated with the 90S pre-ribosome and involved in A0–A2 processing of the pre-rRNA. It is likely, although not previously demonstrated that some of these factors function by inducing or stabilizing defined pre-rRNA structures. Our SHAPE analyses implicate Mrd1 in structuring the 35S pre-rRNA at specific sites (Supplementary Table S1, Figures 2–5, Supplementary Figure S4). This indicates that the function of Mrd1 includes establishing and/or maintaining interactions that directly and/or indirectly are required for structures that are consistent with 35S pre-rRNA processing. The most striking nucleotide reactivity differences observed between 35S pre-rRNA from Δ5 and ΔErb1 were found in regions involved in U3 snoRNA-5′ETS interactions and within the 18S rRNA sequences corresponding to helices H1, H3, H28 and H44 in mature 18S rRNA. Depletion of Mrd1 (ΔMrd1) also resulted in similar changes in reactivity in the 5′ETS and H1 (Supplementary Figure S4, Figure 5A), but in addition in sequences corresponding to H27 (Figure 5B). Our data show that H27, important for the function of the decoding center of the ribosome (46), is not formed in the presence of Mrd1 (Figure 5B), implying that Mrd1 needs to be removed to allow H27 formation. Reactivity differences observed in helices H1, H3, H27, H28 and top of H44 show a striking overlap with helices which have been proposed to constitute a distinct structural central core in 18S rRNA, important for 40S subunit structure and function (47). Formation of this central core is therefore likely to represent an important maturation step in 35S pre-rRNA. Based on our SHAPE data and considering that Mrd1 cross-links to sequences in pre-rRNA corresponding to H27 and H28 (18), we propose that Mrd1 is directly involved in structuring the central regions within 35S pre-rRNA to allow the compaction of pre-ribosomes observed in vivo prior to transcript cleavage (6,19). EM analysis of chromatin spreads demonstrated that the Δ5 mutation still allows formation of large knobs, although they are not compacted and will not be cleaved at A0–A2 (17). Together with our SHAPE results, this suggests that Δ5 mutation partially perturbs the structure of 90S pre-ribosomes by introducing local 35S pre-rRNA structural alterations. These alterations are coupled to essential conformational changes in the pre-ribosome that enable pre-rRNA processing. Therefore, our results suggest that conformation changes precede pre-rRNA cleavages.
Recently a structure for yeast ITS1 was proposed that was based on genetic and phylogenetic analyses (48). The BUM-HMM analysis of the ITS1 region in the Δ5 and ΔErb1 are consistent with the proposed structure (Supplementary Figure S5). Few SHAPE reactivity differences between the Δ5 and ΔErb1 samples were observed in ITS1, where the A2 cleavage site is located (Supplementary Figure S6). It is unlikely that these minor differences reflect structural changes that explain why the Mrd1 mutations impair A2 cleavage (17). It is more probable that structural changes induced by the Mrd1 mutations elsewhere directly or indirectly affect A2 cleavage. Erb1 is required for processing of pre-rRNA at site A3 (49). We note that several nucleotides in ITS1 just upstream of A3 showed significantly higher reactivity in the ΔErb1 mutant. Thus, our data indicate that the local structure near the A3 site is likely perturbed in the absence of Erb1.
The central pseudoknot is not formed in the 35S pre-rRNA
Our analysis reveals that the central pseudoknot is not formed in 90S pre-ribosomal complexes, while it is formed in 20S pre-rRNA. We base this conclusion on nucleotides involved in both H1, H2 and adjacent tertiary interactions that appear to be open in 35S pre-rRNA (Figures 4, 5A and B), but not in 20S pre-rRNA (Supplementary Figure S2). In accordance, our data suggest that U3 snoRNA base pairs with nucleotides 9–25 (Figure 6). These data support the likelihood that processing of pre-rRNA at sites A0-A2 occurs in connection with the formation of the central pseudoknot. In agreement, H1 has been implicated in cleavage of the pre-rRNA at site A1 (50), which generates the 5′-end of 18S rRNA. Interestingly, we found that nucleotides 4–8 involved in H1 were modified to a greater extent in the Mrd1 mutants compared to in ΔErb1 (Figures 4, 5A). These data implicate Mrd1 in structuring H1 and thus formation of the central pseudoknot. Possibly, the intermediate reactivity of nucleotides 4–8 observed in the ΔErb1 could indicate that H1 is formed in part of the population of 90S pre-ribosomes. An attractive model is that a protein fails to bind and stabilize nucleotides 4–8 to allow central pseudoknot formation and pre-rRNA processing in the Mrd1 mutants. Such a protein could be an assembly factor, possibly Mrd1, or Nop9, both of which bind close to the central pseudoknot (18,51). It could also be rpS2/uS5, which is the primary central pseudoknot binding r-protein that interacts with nucleotides 4–7 in the mature ribosome (52).
The U3-35S pre-rRNA interactions within the 90S pre-ribosome are dynamic
Multiple interactions between yeast U3 snoRNA and the pre-rRNA have been proposed or experimentally determined (12–14,20–23,29,31). However, the coordination and the kinetics of the U3 snoRNA interactions with the pre-rRNA are not fully characterized, including how they are promoted, stabilized and disrupted. It has been suggested that the interactions are formed in a specific order (20,53,54), but it is not clear if all interactions take place simultaneously or if some are short-lived.
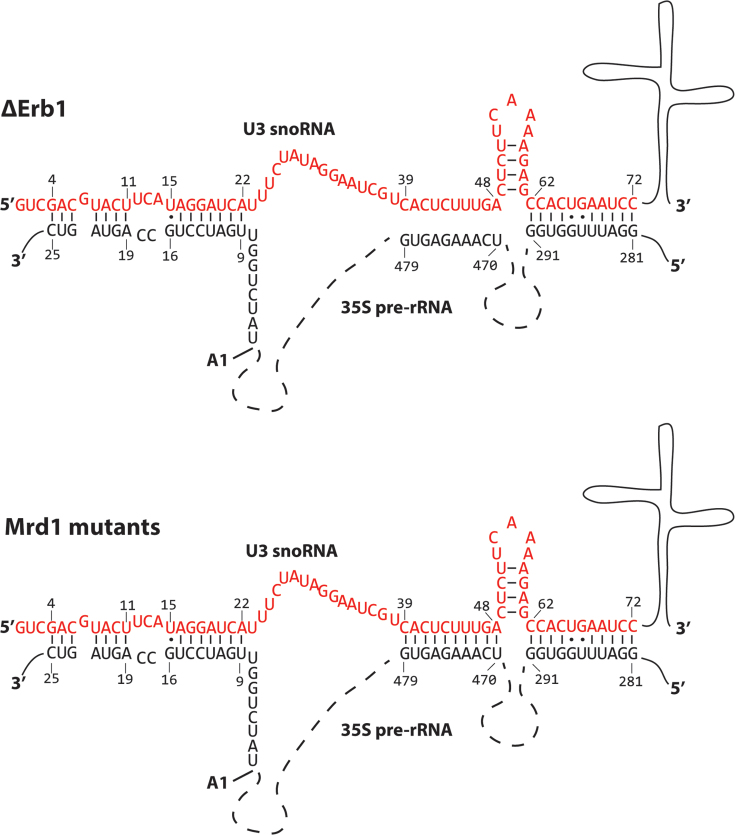
Suggested or experimentally proven base pair interactions between U3 snoRNA and 35S pre-rRNA are summarized in Figure 6C. In the presently analysed 90S pre-ribosomal complexes the following base pair interactions are compatible with our determined SHAPE reactivities: Nucleotides 281–291 of the 5′ETS and U3 62–72, 18S 9–15 and U3 16–22, 18S 19–25 and U3 4–11 (Figures 3–6). These interactions are schematically shown in Figure 7. We did not observe an interaction between U3 23–27 and nucleotides 1139–1143 of 18S rRNA that will be part of the central pseudoknot (Figure 6C) as has been previously proposed (30). Mutational analyses have also failed to provide experimental support for such an interaction (29) and the interaction has not been observed in recent cryo-EM models (10–14). Interactions proposed by CLASH analyses (31) were not included in our analyses because primer extension data was not obtained for the corresponding sequences. Our data show that nucleotides 470–479 of the 5′ETS in the ΔErb1 and Nop58-TAP strains are not base paired to U3 39–48, but that this interaction likely is present in the Mrd1 mutants (Figure 3, Supplementary Figure S4B). This is in agreement with Mrd1 being required to release base pairing interactions between U3 snoRNA and 35S pre-rRNA (17). The SHAPE reactivity patterns in the analysed 90S complexes from ΔErb1 or Δ5 were not compatible with the extended interactions involving nucleotides U3 49–54 and 35S 459–464, that were proposed by cryo-EM (12–14). Nucleotides 39–48 in U3 snoRNA were not modified in U3 12S complexes (Figure 6A), suggesting that these nucleotides adopt a similar conformation in U3 12S as in 90S pre-ribosomes purified from the ΔErb1 and Nop58-TAP strains.
Figure 7.
Schematic interpretation of U3 snoRNA-35S pre-rRNA interactions in ΔErb1 and Δ5 based on the SHAPE reactivity results presented in this study. The nucleotides of the 5′ end of U3 snoRNA are represented in red and the sequence of relevant parts of 35S pre-rRNA in black, other parts are indicated by broken lines. Regions involved in base pair interactions are delimited by numbers and base pairs are shown by connecting lines or dots. Numbering for the 5′ETS starts at the 5′ end of 35S pre-rRNA. The numbering of the beginning of the 18S rRNA part of 35S pre-rRNA starts at the A1 cleavage site (pointed out).
Based on our chemical probing data and previous studies showing that U3 snoRNA base pairs to multiple sites within the pre-rRNA, we conclude that not all U3–pre-rRNA interactions take place at the same time, suggesting that they occur sequentially. Two of our findings support this idea. Firstly, our chemical probing data are compatible with a subset, but not all, of the proposed interactions (Figure 6C). Secondly, we find that base pairing at the 5′ETS site, 470–479, occurs transiently. In the ΔErb1 and Nop58-TAP strains this interaction has been mostly resolved, but is retained in the Mrd1 mutants. Collectively, our results support a model in which U3 snoRNA base pairs at multiple pre-rRNA sites and that U3 occupancy at these sites shifts during maturation of the 90S pre-ribosome.
Recent cryo-EM models of the S. cerevisiae (12,14) and Chaetomium thermophilum (13) SSU processome containing partially processed pre-rRNA have demonstrated an interaction between U3 1–11 and nucleotides 1111–1124 of the 18S rRNA portion of the pre-rRNA, corresponding to 18S rRNA H27. This interaction does not take place in the 90S pre-ribosomal complexes analysed here, since we found nucleotides 1111–1124 to be modified (Figure 5B). Furthermore, our SHAPE reactivity data favor an interaction between U3 4–11 and 18S 19–25 (Figure 6C). Possibly the release of a U3-pre-rRNA base pairing interaction allows U3 snoRNA to switch its interaction partner from 18S 19–25 to 1111–1124. Considering that Mrd1 directly binds to H27 (18), we speculate that Mrd1 may be involved in regulating such a switch.
Mrd1 dependent remodeling of the 90S pre-ribosome
Cryo-EM studies of SSU processomes containing partially processed pre-rRNA display details of where different assembly factors bind to this complex (12–14). Notably, the endonuclease Utp24 implicated in A1 and A2 cleavage is not positioned in direct contact with the A1 site, implying that remodeling must occur to allow it to access its substrate (12–14). The structural differences of the 35S pre-rRNA between the ΔErb1 strain and the Mrd1 mutants may reflect such a remodeling event. Cryo-EM models of the S. cerevisiae and C. thermophilum SSU processome reveals that the U3–5′ETS duplex (nucleotides 470–479) is located in close proximity to the A1 cleavage site (12–14). The Mrd1-dependent release of U3 snoRNA from 470–479 (Figure 3) may lead to structural remodeling that positions Utp24 in close proximity to its substrate. We propose that the release of U3 snoRNA from 470–479 may result in structural remodeling near the A1 cleavage site via the assembly factor Utp7. In a cryo-EM model (14), Utp7 stabilizes the 470–479 U3–35S-pre-rRNA interaction and also binds to, or adjacent to, nucleotides 300–303, 318–322 that display differences in SHAPE reactivity between ΔErb1 and the Mrd1 mutants (Figure 3, Supplementary Table S1), suggesting that Utp7 binding is altered in the Mrd1 mutants. Notably, Utp7 has been described as an Mrd1 interaction partner (55). Utp7 interacts with Utp14 and Sof1, the latter which is interacting with sequences adjacent to the A1 cleavage site (14). We propose that the binding of Utp7 to the 5′ETS is altered upon Mrd1 dependent release of U3 snoRNA from the 470–479 interaction site, resulting in altered binding of Sof1 and reorganisation near the A1 binding site.
In addition to Mrd1, several other components are required for U3 release and pseudoknot formation. The action of the helicase Dhr1 (56) and the assembly factor Esf2 (57) that is an activator of the helicase Dbp8 (58) are required for release of U3 snoRNA. The methyltransferase Bud23 physically and functionally interacts with Dhr1 and methylates G1575 in 18S rRNA, in a helix which stacks coaxially with the central pseudoknot (52,59–61). Nop9 binds to one strand of H2 and H28 (51) and presumably aids in formation of the central pseudoknot. The absence of Mrd1, Dhr1, Esf2, Bud23 and Nop9 in cryo-EM models suggest that these factors enter the appropriate sites in the 90S pre-ribosome in connection with formation of the central pseudoknot and processing of the pre-rRNA. The precise localization of Mrd1, the conformation of its five RBDs and its contacts with other proteins and the pre-rRNA will be important to evaluate the critical role of Mrd1 in ribosome biogenesis.
DATA AVAILABILITY
The ChemModSeq next generation sequencing data and processed data (counts for RT stops and read coverage over 35S) is available from the NCBI Gene Expression Omnibus (GEO) under accession number GSE106868.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Petra Björk for technical support.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR online.
FUNDING
Swedish Research Council [621-2010-5236 to L.W.]; Swedish Cancer Society (to L.W.); Carl Tryggers Foundation [CTS 15:531 to L.W.]; EMBO short term fellowship [F.L., ASTF 568-2012] Wellcome Trust [091549 to S.G., 096997 to E.B.]; Wellcome Trust Centre for Cell Biology core grant [092076]. Next generation sequencing was carried out by Edinburgh Genomics, the University of Edinburgh. Edinburgh Genomics is partly supported through core grants from NERC [R8/H10/56, MRC (MR/K001744/1), BBSRC (BB/J004243/1)]. Funding for open access charge: Swedish Research Council [621-2010-5236].
Conflict of interest statement. None declared.
REFERENCES
- 1. Henras A.K., Soudet J., Gérus M., Lebaron S., Caizergues-Ferrer M., Mougin A., Henry Y.. The post-transcriptional steps of eukaryotic ribosome biogenesis. Cell. Mol. Life Sci. 2008; 65:2334–2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Woolford J.L., Baserga S.J.. Ribosome biogenesis in the yeast Saccharomyces cerevisiae. Genetics. 2013; 195:643–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Tschochner H., Hurt E.. Pre-ribosomes on the road from the nucleolus to the cytoplasm. Trends Cell Biol. 2003; 13:255–263. [DOI] [PubMed] [Google Scholar]
- 4. Dragon F., Gallagher J.E.G., Compagnone-Post P.A., Mitchell B.M., Porwancher K.A., Wehner K.A., Wormsley S., Settlage R.E., Shabanowitz J., Osheim Y. et al. . A large nucleolar U3 ribonucleoprotein required for 18S ribosomal RNA biogenesis. Nature. 2002; 417:967–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Grandi P., Rybin V., Baßler J., Petfalski E., Strauß D., Marzioch M., Schäfer T., Kuster B., Tschochner H., Tollervey D. et al. . 90S pre-ribosomes include the 35S pre-rRNA, the U3 snoRNP, and 40S subunit processing factors but predominantly lack 60S synthesis factors. Mol. Cell. 2002; 10:105–115. [DOI] [PubMed] [Google Scholar]
- 6. Osheim Y.N., French S.L., Keck K.M., Champion E.A., Spasov K., Dragon F., Baserga S.J., Beyer A.L.. Pre-18S ribosomal RNA is structurally compacted into the SSU processome prior to being cleaved from nascent transcripts in Saccharomyces cerevisiae. Mol. Cell. 2004; 16:943–954. [DOI] [PubMed] [Google Scholar]
- 7. Koš M., Tollervey D.. Yeast pre-rRNA processing and modification occur cotranscriptionally. Mol. Cell. 2010; 37:809–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Pérez-Fernández J., Román Á., De Las Rivas J., Bustelo X., Dosil M.. The 90S preribosome is a multimodular structure that Is assembled through a hierarchical mechanism. Mol. Cell. Biol. 2007; 27:5414–5429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Krogan N.J., Peng W.-T., Cagney G., Robinson M.D., Haw R., Zhong G., Guo X., Zhang X., Canadien V., Richards D.P. et al. . High-definition macromolecular composition of yeast RNA-processing complexes. Mol. Cell. 2004; 13:225–239. [DOI] [PubMed] [Google Scholar]
- 10. Kornprobst M., Turk M., Kellner N., Cheng J., Flemming D., Koš-Braun I., Koš M., Thoms M., Berninghausen O., Beckmann R. et al. . Architecture of the 90S pre-ribosome: a structural view on the birth of the eukaryotic ribosome. Cell. 2016; 166:380–393. [DOI] [PubMed] [Google Scholar]
- 11. Chaker-Margot M., Barandun J., Hunziker M., Klinge S.. Architecture of the yeast small subunit processome. Science. 2017; 355:eaal1880. [DOI] [PubMed] [Google Scholar]
- 12. Sun Q., Zhu X., Qi J., An W., Lan P., Tan D., Chen R., Wang B., Zheng S., Zhang C. et al. . Molecular architecture of the 90S small subunit pre-ribosome. Elife. 2017; 6:e22086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cheng J., Kellner N., Berninghausen O., Hurt E., Beckmann R.. 3.2-A-resolution structure of the 90S preribosome before A1 pre-rRNA cleavage. Nat. Struct. Mol. Biol. 2017; 24:954–964. [DOI] [PubMed] [Google Scholar]
- 14. Barandun J., Chaker-Margot M., Hunziker M., Molloy K.R., Chait B.T., Klinge S.. The complete structure of the small-subunit processome. Nat. Struct. Mol. Biol. 2017; 24:944–953. [DOI] [PubMed] [Google Scholar]
- 15. Hughes J.M., Ares M. Jr. Depletion of U3 small nucleolar RNA inhibits cleavage in the 5’ external transcribed spacer of yeast pre-ribosomal RNA and impairs formation of 18S ribosomal RNA. EMBO J. 1991; 10:4231–4239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jin S.B., Zhao J., Rk P.B., Schmekel K., Ljungdahl P.O., Wieslander L.. Mrd1p is required for processing of pre-rRNA and for maintenance of steady-state levels of 40 S ribosomal subunits in yeast. J. Biol. Chem. 2002; 277:18431–18439. [DOI] [PubMed] [Google Scholar]
- 17. Lundkvist P., Jupiter S., Segerstolpe A., Osheim Y.N., Beyer A.L., Wieslander L.. Mrd1p is required for release of base-paired U3 snoRNA within the preribosomal complex. Mol. Cell. Biol. 2009; 29:5763–5774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Segerstolpe Å., Granneman S., Björk P., de Lima Alves F., Rappsilber J., Andersson C., Högbom M., Tollervey D., Wieslander L.. Multiple RNA interactions position Mrd1 at the site of the small subunit pseudoknot within the 90S pre-ribosome. Nucleic Acids Res. 2013; 41:1178–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Segerstolpe Å., Lundkvist P., Osheim Y.N., Beyer A.L., Wieslander L.. Mrd1p binds to pre-rRNA early during transcription independent of U3 snoRNA and is required for compaction of the pre-rRNA into small subunit processomes. Nucleic Acids Res. 2008; 36:4364–4380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Dutca L.M., Gallagher J.E.G., Baserga S.J.. The initial U3 snoRNA:pre-rRNA base pairing interaction required for pre-18S rRNA folding revealed by in vivo chemical probing. Nucleic Acids Res. 2011; 39:5164–5180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Beltrame M., Tollervey D.. Identification and functional analysis of two U3 binding sites on yeast pre-ribosomal RNA. EMBO J. 1992; 11:1531–1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Beltrame M., Tollervey D.. Base pairing between U3 and the pre-ribosomal RNA is required for 18S rRNA synthesis. EMBO J. 1995; 14:4350–4356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Marmier-Gourrier N., Cléry A., Schlotter F., Senty-Ségault V., Branlant C.. A second base pair interaction between U3 small nucleolar RNA and the 5′-ETS region is required for early cleavage of the yeast pre-ribosomal RNA. Nucleic Acids Res. 2011; 39:9731–9745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hartshorne T. Distinct regions of U3 snoRNA interact at two sites within the 5’ external transcribed spacer of pre-rRNAs in Trypanosoma brucei cells. Nucleic Acids Res. 1998; 26:2541–2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hartshorne T., Toyofuku W.. Two 5’-ETS regions implicated in interactions with U3 snoRNA are required for small subunit rRNA maturation in Trypanosoma brucei. Nucleic Acids Res. 1999; 27:3300–3309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Borovjagin A.V., Gerbi S.A.. Xenopus U3 snoRNA docks on pre-rRNA through a novel base-pairing interaction. RNA. 2004; 10:942–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Borovjagin A.V., Gerbi S.A.. An evolutionary intra-molecular shift in the preferred U3 snoRNA binding site on pre-ribosomal RNA. Nucleic Acids Res. 2005; 33:4995–5005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Méreau A., Fournier R., Grégoire A., Mougin A., Fabrizio P., Lührmann R., Branlant C.. An in vivo and in vitro structure-function analysis of the Saccharomyces cerevisiae U3A snoRNP: protein-RNA contacts and base-pair interaction with the pre-ribosomal RNA. J. Mol. Biol. 1997; 273:552–571. [DOI] [PubMed] [Google Scholar]
- 29. Sharma K., Tollervey D.. Base pairing between U3 small nucleolar RNA and the 5’ end of 18S rRNA is required for pre-rRNA processing. Mol. Cell. Biol. 1999; 19:6012–6019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hughes J.M. Functional base-pairing interaction between highly conserved elements of U3 small nucleolar RNA and the small ribosomal subunit RNA. J. Mol. Biol. 1996; 259:645–654. [DOI] [PubMed] [Google Scholar]
- 31. Kudla G., Granneman S., Hahn D., Beggs J.D., Tollervey D.. Cross-linking, ligation, and sequencing of hybrids reveals RNA—RNA interactions in yeast. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:10010–10015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hector R.D., Burlacu E., Aitken S., Bihan T. Le, Tuijtel M., Zaplatina A., Cook A.G., Granneman S.. Snapshots of pre-rRNA structural flexibility reveal eukaryotic 40S assembly dynamics at nucleotide resolution. Nucleic Acids Res. 2014; 42:12138–12154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Burlacu E., Lackmann F., Aguilar L.-C., Belikov S., Van Nues R., Trahan C., Hector R.D., Dominelli-Whiteley N., Cockroft S.L., Wieslander L. et al. . High-throughput RNA structure probing reveals critical folding events during early 60S ribosome assembly in yeast. Nat. Commun. 2017; 8:714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mortimer S.A., Weeks K.M.. A fast-acting reagent for accurate analysis of RNA secondary and tertiary structure by SHAPE chemistry. J. Am. Chem. Soc. 2007; 129:4144–4145. [DOI] [PubMed] [Google Scholar]
- 35. Granneman S., Kudla G., Petfalski E., Tollervey D.. Identification of protein binding sites on U3 snoRNA and pre-rRNA by UV cross-linking and high-throughput analysis of cDNAs. Proc. Natl. Acad. Sci. U.S.A. 2009; 106:9613–9618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Longtine M.S., McKenzie A., Demarini D.J., Shah N.G., Wach A., Brachat A., Philippsen P., Pringle J.R.. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998; 14:953–961. [DOI] [PubMed] [Google Scholar]
- 37. Wieslander L. A simple method to recover intact high molecular weight RNA and DNA after electrophoretic separation in low gelling temperature agarose gels. Anal. Biochem. 1979; 98:305–309. [DOI] [PubMed] [Google Scholar]
- 38. Lusvarghi S., Sztuba-Solinska J., Purzycka K.J., Rausch J.W., Le Grice S.F.J.. RNA secondary structure prediction using high-throughput SHAPE. J. Vis. Exp. 2013; 75:e50243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Low J.T., Weeks K.M.. SHAPE-directed RNA secondary structure prediction. Methods. 2010; 52:150–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Tang Y., Bouvier E., Kwok C.K., Ding Y., Nekrutenko A., Bevilacqua P.C., Assmann S.M.. Structural bioinformatics StructureFold: genome-wide RNA secondary structure mapping and reconstruction in vivo. Biochemistry. 2015; 31:2668–2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Selega A., Sirocchi C., Iosub I., Granneman S., Sanguinetti G.. Robust statistical modeling improves sensitivity of high-throughput RNA structure probing experiments. Nat. Methods. 2016; 14:83–89. [DOI] [PubMed] [Google Scholar]
- 42. Smola M.J., Calabrese J.M., Weeks K.M.. Detection of RNA–protein interactions in living cells with SHAPE. Biochemistry. 2015; 54:6867–6875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Pestov D.G., Stockelman M.G., Strezoska Ž., Lau L.F.. ERB1, the yeast homolog of mammalian Bop1, is an essential gene required for maturation of the 25S and 5.8S ribosomal RNAs. Nucleic Acids Res. 2001; 29:3621–3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Yeh L.C., Lee J.C.. Structure analysis of the 5’ external transcribed spacer of the precursor ribosomal RNA from Saccharomyces cerevisiae. J. Mol. Biol. 1992; 228:827–839. [DOI] [PubMed] [Google Scholar]
- 45. Intine R.V.A., Good L., Nazar R.N.. Essential structural features in the Schizosaccharomyces pombe pre-rRNA 5′ external transcribed spacer11Edited by D. E. Draper. J. Mol. Biol. 1999; 286:695–708. [DOI] [PubMed] [Google Scholar]
- 46. Velichutina I. V, Dresios J., Hong J.Y., Li C., Mankin A., Synetos D., Liebman S.W.. Mutations in helix 27 of the yeast Saccharomyces cerevisiae 18S rRNA affect the function of the decoding center of the ribosome. RNA. 2000; 6:1174–1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Gulen B., Petrov A.S., Okafor C.D., Vander Wood D., O’Neill E.B., Hud N.V., Williams L.D.. Ribosomal small subunit domains radiate from a central core. Sci. Rep. 2016; 6:20885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Coleman A.W. Nuclear rRNA transcript processing versus internal transcribed spacer secondary structure. Trends Genet. 2015; 31:157–163. [DOI] [PubMed] [Google Scholar]
- 49. Sahasranaman A., Dembowski J., Strahler J., Andrews P., Woolford J.L.. Assembly of Saccharomyces cerevisiae 60S ribosomal subunits: role of factors required for 27S pre-rRNA processing. EMBO J. 2011; 30:4020–4032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Sharma K., Venema J., Tollervey D.. The 5’ end of the 18S rRNA can be positioned from within the mature rRNA. RNA. 1999; 5:678–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wang B., Ye K.. Nop9 binds the central pseudoknot region of 18S rRNA. Nucleic Acids Res. 2017; 45:3559–3567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ben-shem A., Garreau de Loubresse N., Melnikov S., Jenner L., Yusupova G., Yusupov M.. The structure of the eukaryotic ribosome at 3.0 Å resolution. Science. 2011; 334:1524–1529. [DOI] [PubMed] [Google Scholar]
- 53. Borovjagin A.V., Gerbi S.A.. Xenopus U3 snoRNA GAC-box A′ and box A sequences play distinct functional roles in rRNA processing. Mol. Cell. Biol. 2001; 21:6210–6221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Shah B.N., Liu X., Correll C.C.. Imp3 unfolds stem structures in pre-rRNA and U3 snoRNA to form a duplex essential for small subunit processing. RNA. 2013; 19:1372–1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Gavin A.-C., Bösche M., Krause R., Grandi P., Marzioch M., Bauer A., Schultz J., Rick J.M., Michon A.-M., Cruciat C.-M. et al. . Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature. 2002; 415:141–147. [DOI] [PubMed] [Google Scholar]
- 56. Sardana R., Liu X., Granneman S., Zhu J., Gill M., Papoulas O., Marcotte E.M., Tollervey D., Correll C.C., Johnson A.W.. The DEAH-box helicase Dhr1 dissociates U3 from the pre-rRNA to promote formation of the central pseudoknot. PLoS Biol. 2015; 13:1–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Hoang T., Peng W., Vanrobays E., Krogan N., Hiley S., Beyer A.L., Osheim Y.N., Greenblatt J., Hughes T.R., Lafontaine D.L.J.. Esf2p, a U3-associated factor required for small-subunit processome assembly and compaction. Mol. Cell. Biol. 2005; 25:5523–5534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Granneman S., Lin C., Champion E.A., Nandineni M.R., Zorca C., Baserga S.J.. The nucleolar protein Esf2 interacts directly with the DExD/H box RNA helicase, Dbp8, to stimulate ATP hydrolysis. Nucleic Acids Res. 2006; 34:3189–3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Sardana R., White J.P., Johnson A.W.. The rRNA methyltransferase Bud23 shows functional interaction with components of the SSU processome and RNase MRP. RNA. 2013; 19:828–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Sardana R., Zhu J., Gill M., Johnson A.W.. Physical and functional interaction between the methyltransferase Bud23 and the essential DEAH-box RNA helicase Ecm16. Mol. Cell. Biol. 2014; 34:2208–2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Létoquart J., Huvelle E., Wacheul L., Bourgeois G., Zorbas C., Graille M., Heurgué-Hamard V., Lafontaine D.L.J.. Structural and functional studies of Bud23–Trm112 reveal 18S rRNA N7-G1575 methylation occurs on late 40S precursor ribosomes. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:E5518–E5526. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The ChemModSeq next generation sequencing data and processed data (counts for RT stops and read coverage over 35S) is available from the NCBI Gene Expression Omnibus (GEO) under accession number GSE106868.