Abstract
The transcription factor PLZF (promyelocytic leukemia zinc finger protein) acts as an epigenetic regulator balancing self-renewal and differentiation of hematopoietic cells through binding to various chromatin-modifying factors. First described as a transcriptional repressor, PLZF is also associated with active transcription, although the molecular bases underlying the differences are unknown. Here, we reveal that in a hematopoietic cell line, PLZF is predominantly associated with transcribed genes. Additionally, we identify a new association between PLZF and the histone methyltransferase, EZH2 at the genomic level. We find that co-occupancy of PLZF and EZH2 on chromatin at PLZF target genes is not associated with SUZ12 or trimethylated lysine 27 of histone H3 (H3K27me3) but with the active histone mark H3K4me3 and active transcription. Removal of EZH2 leads to an increase of PLZF binding and increased gene expression. Our results suggest a new role of EZH2 in restricting PLZF positive transcriptional activity independently of its canonical PRC2 activity.
INTRODUCTION
Cell differentiation critically depends on combinations of transcriptional regulators controlling the development of individual lineages. Transcription factors (TFs) have long been recognized as major regulators of blood stem cell development and the subsequent differentiation into multiple mature hematopoietic lineages (1). PLZF, also known as Zbtb16, is a master transcriptional regulator with effects on growth, self-renewal and differentiation with a well-recognized activity on the regulation of hematopoietic cell differentiation (2). It was first identified in a patient with acute promyelocytic leukemia, where a reciprocal chromosomal translocation t(11;17)(q23;q21) results in a fusion with the RARA gene encoding retinoic acid receptor alpha (3). Initially described as a myeloid TF (4), PLZF is now known to play a role in spermatogonial, mesenchymal and neural progenitor cells by balancing self-renewal and differentiation (5). Within the hematopoietic tissue, PLZF is involved in hematopoietic stem cell maintenance, in the development of multiple hematopoietic cells and in regulating immune responses (6–8).
Most of the biological functions of PLZF can be explained by its transcriptional repressive activity. Indeed, PLZF was first described as a sequence-specific transcriptional repressor, binding regulatory elements of its target genes through its carboxyl seven zinc fingers (9). PLZF repressive activity was further documented by its ability to bind multi-protein complexes as the SMRT-Sin3-HDAC-NcoR complexes (10,11) or Polycomb group (PcG) complexes (12,13), leading to a local repressed chromatin state. If PLZF is well recognized as an epigenetic repressor, data also suggest that PLZF acts as a transcriptional activator, inducing its target gene expression. Initial evidence came with the identification of direct targets of PLZF in hematopoietic progenitors that were highly expressed in its presence but down regulated when its expression was decreased (14). The presence of PLZF on regulatory elements of active genes was confirmed by genome-wide approaches in different tissues (15,16). In the latter study, similar frequencies of genes were expressed or repressed in the presence of PLZF (16) challenging the idea of PLZF being principally a transcriptional repressor. Thus, as with many sequence-specific TFs, PLZF may function as a transcriptional repressor or activator depending on the molecular context, using mechanisms that are not fully understood. Cellular context, genomic architecture of the target loci or post-translational modifications may all influence PLZF transcriptional activity (17). Furthermore, through the N-terminal BTB/POZ domain, PLZF interacts with various chromatin-modifying factors (18) and thus switching from interaction with one molecule to another may radically change the transcriptional output of PLZF activity.
PcG proteins function as multi-subunit complexes through the concerted participation of at least two major complexes (PRC1/PRC2) allowing the initiation and maintenance of the H3K27me3 repressive epigenetic mark that is catalyzed by the histone methyltransferase (HMT) EZH2 that composes the PRC2 core complex (19). Besides its well-known function in promoting transcriptional repression, EZH2 acts as an oncogene or a tumor suppressor in hematopoietic neoplasms, depending on the cell context, suggesting a specificity of its activity that may be dictated by chromatin structure and/or cofactors (20,21). Partial explanation of EZH2 specificity may reside in variation of PRC2 composition and post-translational modifications that could influence EZH2 enzymatic activity (22). For example, it has been reported that PRC2 methylation of JARID2 promotes its catalytic activity (23). Methyltransferase activity of EZH2 requires at least SUZ12 and EED, two other PRC2 components (24) and increasing evidence suggests that EZH2 when not associated with canonical PRC2 components, such as SUZ12, occupies active genes independently of its H3K27me3 activity (25,26). This non-canonical function of EZH2 can be activated by JAK3–mediated phosphorylation in NK cells (27) and AKT signaling in memory T cells (28).
Our previous study pointed out a major role for PLZF in restricting mouse hematopoietic stem cell function by coordinating the expression of genes related to stemness, differentiation, cell cycle and aging (29). However, we were unable to determine whether the action of PLZF on gene expression was direct or not. To better understand the gene-regulated network controlled by PLZF in the hematopoietic system and to shed light on its molecular transcriptional effect we studied the association of PLZF, histone marks and EZH2 and their effects on gene activity. Here, we show that PLZF interacts with EZH2 in vivo in myeloid cells and that together, they regulate a group of common genes mostly associated with the active histone mark H3K4me3 and active transcription. Modulating EZH2 expression modifies PLZF chromatin binding, with according changes in histone marks and gene expression. These results bring new elements for the understanding of epigenetic regulation mechanisms involving the two multifaceted proteins PLZF and EZH2.
MATERIALS AND METHODS
Cell culture, T-rex system and knockdown
Human myeloblastic cell line KG1, 293T-rex cells (a generous gift from R. Margueron) and 293T cells were maintained at exponential growth in Roswell Park Memorial Institute (RPMI) or Dulbecco’s-modified Eagle’s medium respectively supplemented with 10% foetal calf serum, 1% penicillin-streptomycin and 1% sodium pyruvate. 293T-rex cells were stably transfected with PLZF mutants or Gal4 as control, selected with neomycin and induced with doxycycline (1 μg/ml) for 24 h. EZH2 and PLZF knockdown (KD) in KG1 cells were achieved using a doxycycline-induced short hairpin RNA (sh-RNA)-targeting EZH2 or PLZF (pTRIPZ-EZH2, Openbiosystem # V2THS_63066, pTRIPZ-PLZF, Openbiosystem # V2LHS_72096). A non-silencing sh-RNA (pTRIPZ-NS, openbiosystem # RHS4743) was used as control. Cells containing the pTRIPZ were selected on puromycine (1.75 μg/ml). KD of PLZF and EZH2 was obtained by the addition of doxycycline (2 μg/ml) to the cells during 7–10 days.
Nuclear extracts, immunoprecipitations and western blotting.
Nuclear protein extraction of KG1 and 293T cells was done using the dounce homogenizer with high salt concentration according to Dignam and Roeder. https://www.ncbi.nlm.nih.gov/pubmed/20150077. Cellular fractionation was carried out using the subcellular protein fractionation kit for cultured cells (Thermofisher). For co-immunoprecipitations (co-IPs) in KG1 cells, nuclear extracts were diluted in hypotonic buffer and incubated 3 h with 2 μg of anti-PLZF (Santa Cruz Biotechnology, H300) or 10 μl of anti-EZH2 (Active Motif, cat 39901 or AC22 antibody) antibody in the presence of Protein G beads (Dynabeads, Life Technologies) and resulting complexes were washed, denatured and eluted according to the manufacturer’s instructions.
Nuclear extracts of 293T cells were immunoprecipitated with the ProFound c-Myc tag IP/coIP kit (No 23622) following the manufacturer’s instructions. Immunoblotting was performed as previously described (13). Briefly, nuclear extracts were separated under denaturated conditions on polyacrylamide gels, blotted on nitrocellulose membrane and incubated with different primary antibodies listed in supplemental experimental procedure.
Chromatin immunoprecipitation
ChIP was performed as described (13) with minor modifications. Following chromatin extraction, DNA was sonicated to obtain 200–600 bp fragments (ultrasonic processor, Fischer Scientific, ref 75041, 40% amplitude, 30 s on and 30 s off for 20 min). 2 × 107 KG1 cells were used for PLZF, EZH2, SUZ12 and PolII ChIPs, and 5 × 106 cells for histone marks. After immunoprecipitation, DNA was purified with the I-Pure kit (Diagenode) for sequencing analyses or with chelex-100 (Bio-Rad Laboratories) for quantitative polymerase chain reaction (q-PCR) analyses.
RNA extraction and reverse transcription
RNA from KG1 cells were extracted using RNeasy mini Kit (Qiagen) according to manufacturer’s recommendations. RNA purity and concentration was determined using Nanodrop 2000 (Thermofisher Scientific). Reverse transcription was performed on 1 μg of total RNA using the Transcriptor High Fidelity cDNA synthesis kit (Roche Diagnosis).
Quantitative PCR analysis
Quantification of ChIPed DNA or cDNA was performed by real-time PCR. DNA target sequences were selected based on our ChIP-seq analyses in KG1 cells and primers were designed for amplifying the DNA location of interest with Primer-BLAST. Primer sequences are available in supplemental experimental procedure.
For normalization of ChIP-qPCR, Spike-in Drosophila Chromatin and Spike-in Antibody (Active motif cat# 53083) were added to the ChIP reaction as a minor fraction of the IP reaction. The immunoprecipitated Drosophila chromatin was quantified using Drosophila Positive Control Primer Set Pbgs (Active motif cat# 71037), allowing the attribution of a spike-in factor used for normalization of compared samples.
Real-time PCR was performed using Power SYBR Green PCR Master Mix (Applied Biosystems) according to the manufacturer’s instructions with the 7500 Fast Real Time PCR system (Life Technologies). For ChIPed DNA, IgG control ‘cycle over the threshold’ Ct values were subtracted to Input or IP Ct values and converted into bound value by 2(-(IP Ct or input Ct- IgG IP Ct)). Data are expressed as % of bound/input. For cDNA relative expression levels were determined by the 2∧ΔΔCT method using HPRT or PBG-D as housekeeping genes. Experiments were performed with two individual clones treated or not with the doxycycline. Technical triplicates are presented as mean values ± SD.
Sequencing and data processing
Libraries and sequencing were performed with a HiSeq 2000 next-generation sequencing platform (Illumina) using a 40 bp single-end protocol, either at the Montpellier GenomiX (MGX; Montpellier, France) or at the Massachusetts Institute of Technology BioMicro center (Cambridge, MA, USA). Sequencing was performed on libraries prepared from duplicates of ChIP. Computational analyses are developed in supplemental experimental procedure.
Genomic distribution of PLZF signal was visualized using cis-regulatory Element Annotation System (CEAS). For de novo TF motif discovery, PLZF, EZH2 or the common PLZF/EZH2 peaks were called with MACS2 (http://liulab.dfci.harvard.edu/MACS/) using a q-value cut-off set at 0.01, and changed into summit regions (−50/+50 pb from peak summit). The promoter and gene body peak summits were filtered and merged, and only the top 500 most significantly enriched regions (ranked by P-value) were analyzed using Regulatory Sequence Analysis Tools (RSAT) (30). The related binding motifs were found using JASPAR and HOMER databases.
Heatmap visualization of ChIP enrichments at EZH2 and PLZF-enriched non-redundant transcripts was performed using ngs.plot version 2.47 (31). Plotting of the enrichment profiles were done by ranking the genes using k-means clustering method (here set up at six clusters). The average profiles were calculated using the mean read coverage among the gene bodies in each resulted cluster. Regions that were differentially enriched for PLZF, H3K27me3 or H3K4me3 upon shPLZF or shEZH2 were identified using MACS2 bdgdiff module and diffReps with slight modifications (P-values re-adjusted in R using Bonferroni correction) (32) depending on their localization to distinguish promoter and gene body regions and converted into official gene symbols using biotools.fr. Differential enrichments were visualized using the Deeptools suite (bamCompare, computeMatrix, plotProfile) (33). Term enrichment analysis was performed for each group of genes using DAVID bioinformatics database (Only ‘Gene Ontology Biological Processes’ terms were retain for further analysis). Retained ontologies were clustered into specific thematic using custom bash script.
RESULTS
Characterization of PLZF function through its genomic localization
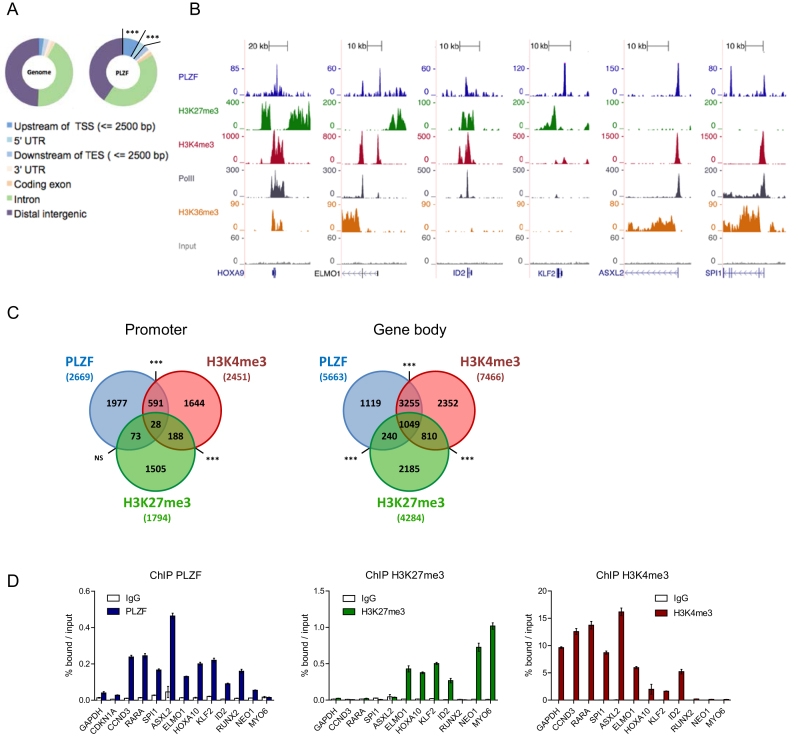
We profiled PLZF chromatin occupancy by chromatin immunoprecipitation coupled to high-throughput DNA sequencing (ChIP-seq) in KG1 cells, a myeloid cell line expressing the stem cell marker CD34+ (34). PLZF targets are listed in Supplementary Table S1. Analysis of PLZF peak location with respect to gene feature, showed an over-representation of PLZF binding at gene promoter regions defined as 2.5 kb upstream of TSS (3.6-fold enrichment compared to genome; P-value = 1.5e-322) (Figure 1A). As PLZF could be associated with active or repressive chromatin states, we profiled H3K27me3 and H3K4me3 histone marks that are associated with gene repression or activation, respectively. Examples of PLZF, H3K27me3 and H3K4me3 ChIP-seq coverage are shown in Figure 1B. PLZF was associated with H3K4me3, H3K27me3 or both marks (Figure 1B). When PLZF was associated with H3K4me3, the elongating mark H3K36me3 and RNA Polymerase II (Pol II) were also present indicating an active transcriptional state of PLZF-bound genes (Figure 1B). Independently of its localization at promoter or gene body regions, PLZF bound regions were mostly associated with H3K4me3, representing 23% of PLZF-bound promoters and 76% of PLZF bound gene bodies and to a much lesser extent with H3K27me3 (non-significant at promoter and 22% at gene bodies) (Figure 1C). Validation of PLZF bound regions was performed by ChIP-qPCR on a subset of selected genes involved in myeloid cell identity (Figure 1D). We validated that the developmental genes involved in hematopoietic stem cell self-renewal (HOXA10, KLF2, ID2) were enriched with the H3K27me3 (Figure 1D) whereas TFs involved in myeloid differentiation (RARA, SPI1) were enriched with H3K4me3 (Figure 1D). Gene ontology (GO) analysis using the DAVID database confirmed that regions bound by PLZF were enriched for genes associated with cell signaling, metabolic and immune system processes (Supplementary Figure S1). Interestingly, PLZF-target genes enriched in H3K27me3 were more associated with developmental processes (Supplementary Figure S1). Taken together our data indicate that PLZF correlates mainly with the H3K4me3 epigenetic mark in KG1 cells and suggest distinct target genes depending on its co-association with H3K4me3 or H3K27me3.
Figure 1.
Genomic localization of PLZF in CD34+ KG1 cell line. (A) Genomic distribution of PLZF obtained from PLZF ChIP-seq in KG1 cells compared to the whole-genome distribution (TSS: transcription start site; TES: transcription end site; UTR: untranslated region), ***fold change > 2 and P < 0.001 (hypergeometric test). (B) Representative UCSC Genome browser tracks of PLZF, H3K27me3, H3K4me3, PolII and H3K36me3 and Input ChIP-seq in KG1 cells. (C) Venn diagrams showing the association between PLZF, H3K27me3 and H3K4me3 bound regions at the Promoter (−2.5/+0.1 kb from TSS) or in the Gene body (+0.1 kb from TSS to TES). Association between two peaks was considered if the two peaks were localized on the same genomic feature of the same gene. NS not significant, ***P < 0.001 (hypergeometric test). (D) Validation of PLZF, H3K27me3 and H3K4me3 bound regions by ChIP-qPCR. GAPDH and CDKN1A served as negative controls for PLZF and positive controls for H3K4me3. NEO1 and MYO6 served as positive controls for H3K27me3. Percentages of bound DNA over input are shown as a mean ±SD of two independent experiments (n = 3–5).
PLZF interacts with EZH2 through its BTB-POZ domain
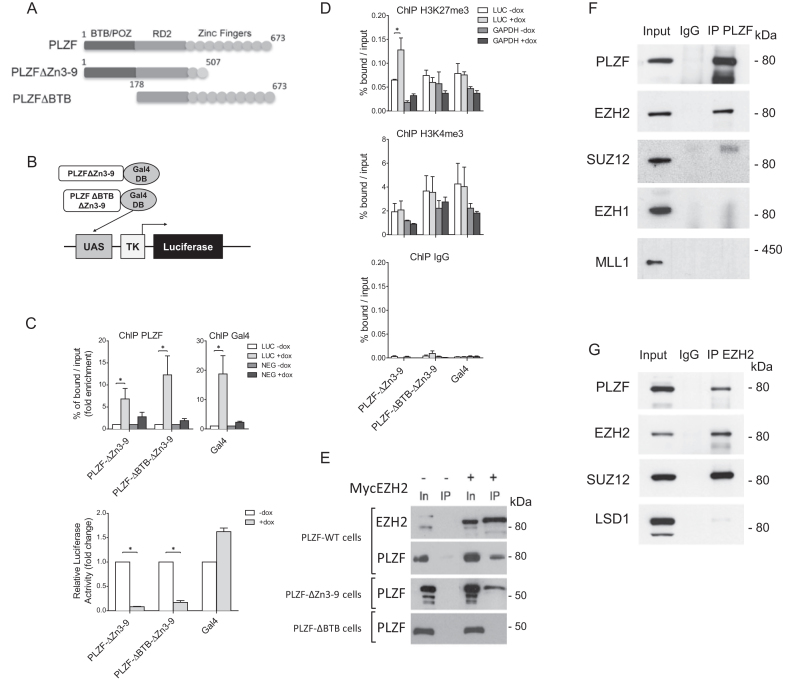
To assess the importance of PLZF on H3K27me3 or H3K4me3 levels we engineered cell lines stably integrated with a luciferase reporter gene controlled by a 5x Gal4 UAS sequence and expressing doxycycline-inducible wild-type or BTB-deleted PLZF (Figure 2A) in which the PLZF DNA binding domain (zinc fingers 3–9) has been swapped with the Gal4-DNA binding domain (Figure 2B). Upon expression, PLZF-Gal4 recruitment results in luciferase transcriptional repression (Figure 2C) suggesting that in this in vitro system, it is the repressive activity of PLZF that is prevailing which is in agreement with previous in vitro studies (35). Monitoring changes to the chromatin landscape by H3K27me3 and H3K4me3 ChIP, we found that only the H3K27me3 was significantly modified by PLZF recruitment and this modification is dependent on the presence of the BTB domain (Figure 2D). These results suggest that PLZF recruitment leads to BTB domain-dependent increase in H3K27me3 and transcriptional repression of the luciferase promoter.
Figure 2.
PLZF interacts with EZH2 but not SUZ12. (A) Schematic of PLZF mutant constructions used in this figure. (B) Schematic representation of the luciferase reporter under the control of TK promoter, with Gal4 DNA-binding domain fused to PLZF domain. (C) PLZF and Gal4 were immunoprecipitated and DNA enrichment was measured at the luciferase transgene using LUC primers that amplify the luciferase transgene and NEG primers used as negative control (upper panel). Luciferase activity was measured 24 h after doxycycline induction (+dox) on cellular clones expressing PLZFΔZn3–9Gal4 or PLZFΔBTBΔZn3–9Gal4; Gal4 alone was used as control (lower panel). (D) Measurement of H3K27me3 and H3K4me3 levels at the luciferase transgene (LUC primers), after PLZFΔZn3–9Gal4 or PLZFΔBTBΔZn3–9Gal4 induction. GAPDH was used as negative control for H3K27me3 enrichment and as positive control for H3K4me3. In panels C and D, results are expressed as the mean ±SD with *P < 0.05 (Anova and Kruskal Wallis) of independent experiments realized on three individual clones. (E) Nuclei of 293T cells co-transfected with PLZF (WT or mutants) and EZH2 were purified. Myc-EZH2 was immunoprecipitated with anti-Myc coupled-resin. IPs were immunoblotted with anti-EZH2 or anti-PLZF. Input lanes (In) represents 2% of the amount of samples processed in each immunoprecipitates (IP). (F) Nuclear extracts of KG1 cells were immunoprecipitated with anti-PLZF or anti-IgG antibodies. IPs were immunoblotted with anti-PLZF, anti-EZH2, anti-SUZ12, anti-EZH1 and anti-MLL1 antibodies. Inputs represent 5% of samples processed in each IP. (G) KG1 nuclear extracts were immunoprecipitated with anti-EZH2 or anti-IgG antibodies. IPs were immunoblotted with anti-PLZF, anti-EZH2, anti-SUZ12 and anti-LSD1 antibodies. Inputs represent 5% of samples processed in each IP.
As EZH2 is the HMT responsible for H3K27me3 deposition, we investigated a potential interaction between PLZF and EZH2 through co-IP in HEK293T cells over expressing a MYC-tagged EZH2 with either wild-type or truncated PLZF. We found that PLZF WT and PLZFΔZn3–9, co-precipitated EZH2, whereas PLZFΔBTB that lacks the BTB domain did not (Figure 2E), demonstrating that PLZF molecularly interacts with EZH2 through its BTB-POZ domain.
The potential interplay between PLZF and EZH2 was investigated by co-IPs in the KG1 cells in which PLZF and EZH2 are endogenously expressed. Western blot analysis on proteins co-precipitating with PLZF demonstrated that PLZF associated with EZH2 but not with SUZ12, a PRC2 component, EZH1 nor MLL1, two HMTs associated with H3K4me3 (Figure 2F). Reverse IP with an anti-EZH2 antibody confirmed the association of EZH2 with PLZF, which also confirmed that EZH2 additionally interacts with SUZ12 in these cells (Figure 2G). These results highlight the importance of the PLZF BTB-POZ domain for the interaction with and recruitment of EZH2 HMT activity characterized by an increase in H3K27me3, which is associated with transcriptional repression.
PLZF shares genomic targets with EZH2 in the absence of SUZ12
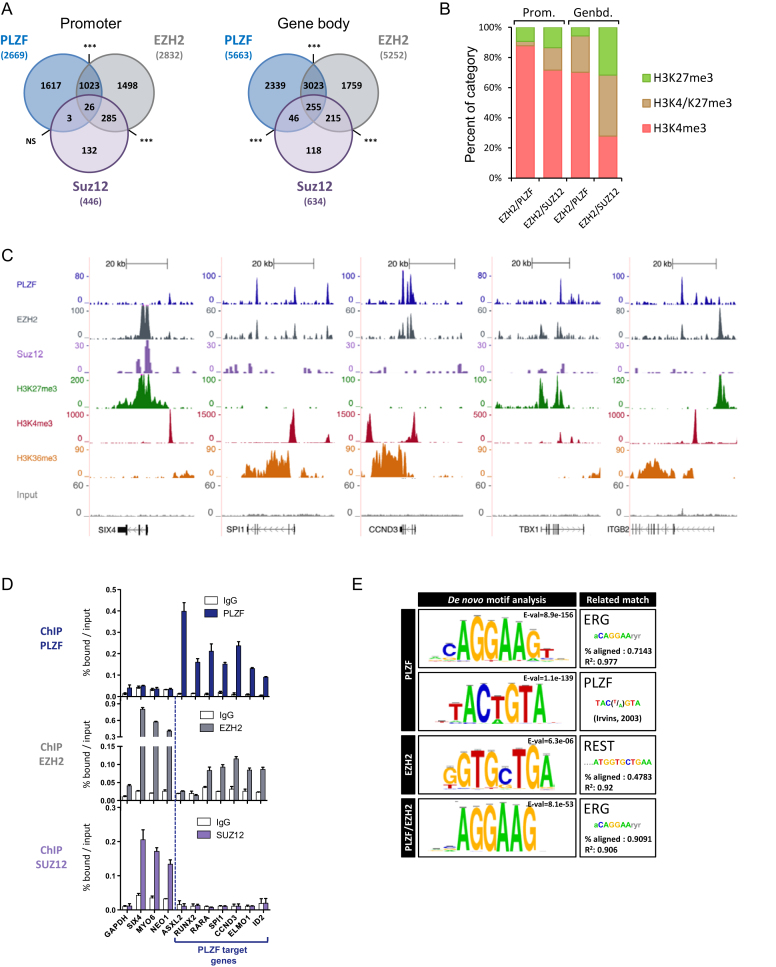
To ascertain whether PLZF associates with EZH2 on chromatin in vivo, we profiled EZH2 and SUZ12 chromatin occupancy in the KG1 cell line and processed the data as for PLZF-bound genomic regions. Data showed that PLZF and EZH2 co-occupied significantly numerous promoters (39% of PLZF bound) and gene bodies (58% of PLZF bound) (Figure 3A). However, PLZF, EZH2 and SUZ12 co-occupancy was low, 2.5% at PLZF/EZH2 promoter (NS) and 8% at PLZF/EZH2 gene bodies (Figure 3A). PLZF/EZH2 and EZH2/SUZ12 peaks were compared in terms of their enrichment in H3K4me3, H3K27me3 or both H3K27me3/H3K4me3 at gene body or promoter regions. EZH2 peaks lacking PLZF (EZH2-unique) were associated with a higher percentage of H3K27me3 or H3K27me3/H3K4me3 in comparison to EZH2/PLZF common bound regions (Figure 3B). Of note is that EZH2-unique bound regions associated with H3K4me3 showed less ChIP-seq signal as compared to those bound by H3K27me3 (see later, Figure 4A). Examples of PLZF, EZH2 and SUZ12 ChIP-seq coverage are shown in Figure 3C, where these typical examples illustrate the co-localization of EZH2 and PLZF at transcribed genes that do not contain SUZ12. We next validated some PLZF/EZH2-common or EZH2-unique bound loci by performing PLZF and EZH2 ChIP-qPCR (Figure 3D). These experiments revealed that although detectable by ChIP-qPCR, EZH2 binding was lower in the case of PLZF/EZH2 common genes (Figure 3D, middle panel) in comparison to EZH2-unique (SIX4, MYO6 and NEO1). This suggests that EZH2 recruitment occurs with different affinities at its unique PLZF-common sites. In addition, SUZ12 was present at unique EZH2 sites but not at EZH2/PLZF ones (Figure 3D, lower panel).
Figure 3.
PLZF and EZH2 share common target genes. (A) Venn diagrams showing the association between PLZF, EZH2 and SUZ12 bound regions at promoter (−2.5kb/+0.1 kb from TSS) and gene body (+0.1 kb from TSS to TES) regions. Association between two peaks was considered if they were localized on the same genomic feature of the same gene. NS not significant, ***P < 0.001 (hypergeometric test). (B) Percentages of H3K4me3 unique, H3K27me3 unique or H3K4me3/H3K27me3 marks in EZH2/PLZF (without SUZ12) or EZH2/SUZ12 (without PLZF) target genes, at promoter (Prom.) or gene body (Genbd.). (C) UCSC genome browser views of EZH2, PLZF and SUZ12 ChIP-seq and associated histone marks in KG1 cells. (D) ChIP-qPCR analysis of PLZF, EZH2 and SUZ12 on selected target genes. Percentages of bound DNA over input are shown as a mean ±SD of two independent experiments (n = 3–5). (E) Motif search at PLZF, EZH2 or PLZF/EZH2 summit regions.
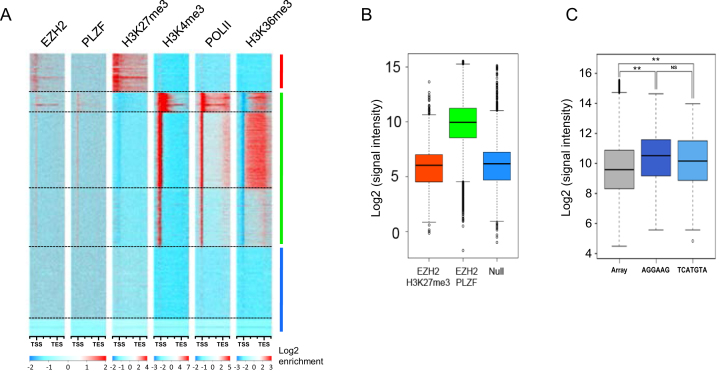
Figure 4.
PLZF/EZH2 common genes are active genes. (A) Heatmap of k-means clustered (n = 6) EZH2, PLZF, H3K27me3, H3K4me3, PolII and H3K36me3 ChIP-seq signals in KG1 cells on hg19 refGene (TSS to TES ±2 kb). All genes were scaled to have the same length. (B) Microrray generated-mRNA expression values are shown for the three groups defined in E: the EZH2/H3K27me3 target genes (upper cluster); the PLZF/EZH2 target genes (middle cluster) and null genes (lower cluster). (C) Box plot representing the microarray median expression values according to PLZF DNA recognition motif; NS: not significant **P < 0.01 (Anova and Kruskal Wallis test).
De novo DNA motif searching using RSAT (30) at the 500 highest-ranked (by q-value) PLZF ChIP-seq peaks identified enrichment of an ERG-like binding motif, present at 40% of PLZF-occupied sites and a motif with no known identity in JASPAR and HOMER databases, present at 15% of PLZF-occupied sites (Figure 3E). This novel motif is, however, matching perfectly to an experimentally-determined PLZF binding site (36). When motif discovery was done with the PLZF/EZH2 common peaks, only the ERG binding motif was overrepresented (Figure 3E), suggesting that PLZF can be associated with two binding site signatures that are distinguishable by the presence or not of EZH2. Motif search for EZH2 did not reveal a strong consensus-binding site confirming the current knowledge on the non-sequence specificity of EZH2 (37). All together, these results show that PLZF and EZH2 share common target genes that are characterized by the absence of SUZ12 and enriched with the H3K4me3 active mark.
PLZF/EZH2 target genes are active genes
To further explore the co-association of PLZF and EZH2 on chromatin, we performed enrichment analyses followed by k-means clustering of EZH2, PLZF, H3K27me3, H3K4me3, Pol II and H3K36me3 on refSeq genes (Figure 4A). This clustering confirmed that PLZF enriched genes were mostly associated with the active mark H3K4me3 and the elongation mark H3K36me3, a chromatin profile associated with gene activation. In addition, this clustering separated EZH2-targeted genes into two distinct groups, based on either the presence or absence of PLZF. The group of EZH2-targets lacking PLZF-binding was enriched in H3K27me3 and lacks Pol II, H3K4me3 and H3K36me3 enrichment (Figure 4A; upper cluster). The group of genes co-targeted by EZH2 and PLZF were highly enriched in Pol II, H3K4me3 and H3K36me3 but lacking H3K27me3 (Figure 4A; middle cluster). A third set of genes contained no enrichment for any of the tested factors (Figure 4A; lower cluster).
We next assessed the expression status of these three groups of genes using publically available expression profiles on KG1 cells (GSM1316691). This analysis showed that the genes occupied by EZH2 and H3K27me3-enriched (upper group) were poorly expressed and similar to genes containing no tested factors (lower group), whereas genes co-targeted by EZH2 and PLZF (middle group) were highly expressed (Figure 4B). Thus, PLZF or PLZF/EZH2 are mostly associated with gene activation in KG1 cells. To question whether the genes targeted by PLZF and highly expressed were preferentially associated with one of the two putative binding site motifs identified in Figure 3E, we investigated expression level of PLZF-targeted genes according to the presence or not of the putative binding sites. The results confirm that PLZF bound genes were significantly more expressed than the full transcriptome but independently of the nature of the identified PLZF binding motifs (Figure 4C).
To rule out the possibility of a result biased by the nature of our cell line (the KG1 cell line is a leukemic-derived cell line), we compared histone mark profiles from our defined PLZF-target genes in leukemic KG1 cells with ChIP-seq data available on primary CD34 positive hematopoietic stem cells (CD34+HSC) (GSM772951; GSM773042). H3K27me3, H3K4me3 and H3K27Ac CD34+ HSC ChIP-seq data were clustered according to EZH2 and PLZF enrichment in KG1 cells. The heatmap revealed highly comparable enrichment profiles for the histone marks in both KG1 and CD34+HSC cells (Supplementary Figure S2), demonstrating that PLZF target genes in KG1 cells are associated with a chromatin profile that is permissive to transcription in CD34+ HSC. Altogether the results show that the majority of PLZF or PLZF and EZH2 target genes are active genes.
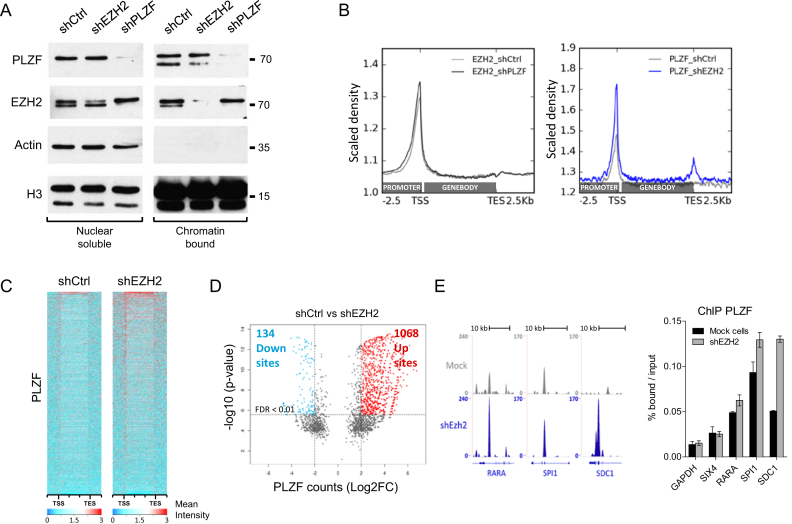
EZH2 influences PLZF chromatin activities
To further understand the interplay between PLZF and EZH2, we performed shRNA-directed KD experiments and analysed the effect of the KD on each other. KD of PLZF and EZH2 were obtained by infecting KG1 cells with lentivirus containing shRNA directed against either PLZF or EZH2. KG1 clones efficiently down regulating PLZF or EZH2 were selected (Supplementary Figure S3). Effect of the shEZH2 and shPLZF was studied on PLZF and EZH2 levels in nuclear soluble and chromatin bound fractions. Our results showed that both shRNA were efficient at the chromatin for knocking down their targets. As a consequence, PLZF KD diminished PLZF binding at target genes and EZH2 KD diminished EZH2 binding at its target genes (Supplementary Figure S4). EZH2 KD did not change PLZF nuclear soluble fractions but slightly changed the ratio between the two PLZF bands in chromatin bound fraction, increasing the upper band in comparison to the lower band (Figure 5A). PLZF KD however, did not modulate chromatin bound EZH2 but stabilized nuclear soluble EZH2 (Figure 5A). Visualization of ChIP-seq signals demonstrated that downregulation of PLZF did not globally change EZH2 signal, while downregulation of EZH2 resulted in an increase in PLZF signal (Figure 5B). This increase was mainly due to a gain in ChIP-seq intensity at PLZF bound–loci (Figure 5C) but also to the appearance of newly targeted loci (Supplementary Table S1). Quantification of differential PLZF-bound sites upon EZH2 KD showed a majority of sites with increased PLZF binding (Up sites) in comparison to sites showing reduced PLZF binding (down sites; Figure 5D and Supplementary Figure S5). Some of these loci containing increased PLZF upon shEZH2 were validated by spike-in-normalized ChIP-qPCR (Figure 5E). These results suggest that EZH2 negatively influences PLZF chromatin association.
Figure 5.
Effect of PLZF and EZH2 KDs on PLZF and EZH2 activities. (A) Immunoblot of PLZF and EZH2 in shCtrl, shPLZF or shEZH2 conditions in nuclear soluble or chromatin bound fractions; ACTIN was used as a control of fraction purity and H3 as a loading standard. (B) Density profile of EZH2 (right panel) and PLZF (left panel) signal tracks, after shPLZF or shEZH2 KD, respectively. (C) Heatmap of PLZF ChIP-seq signals from −2.5 kb upstream the TSS to +2.5 kb dowstream the TES in shCtrl and shEZH2 KG1 cells. (D) Volcano plot showing PLZF differentially regulated sites (1 kb sliding windows) in shCtrl versus shEZH2 conditions. Left rectangle highlights significantly downregulated sites, right rectangle highlights significantly upregulated sites (FDR < 0.01; log2 fold change > 2). FDR: false discovery rate (Bonferroni correction). (E) Representative UCSC Genome browser tracks (left panel) of PLZF in shCtrl and shEZH2 conditions and its associated spike in ChIP-qPCR validation (right panel).
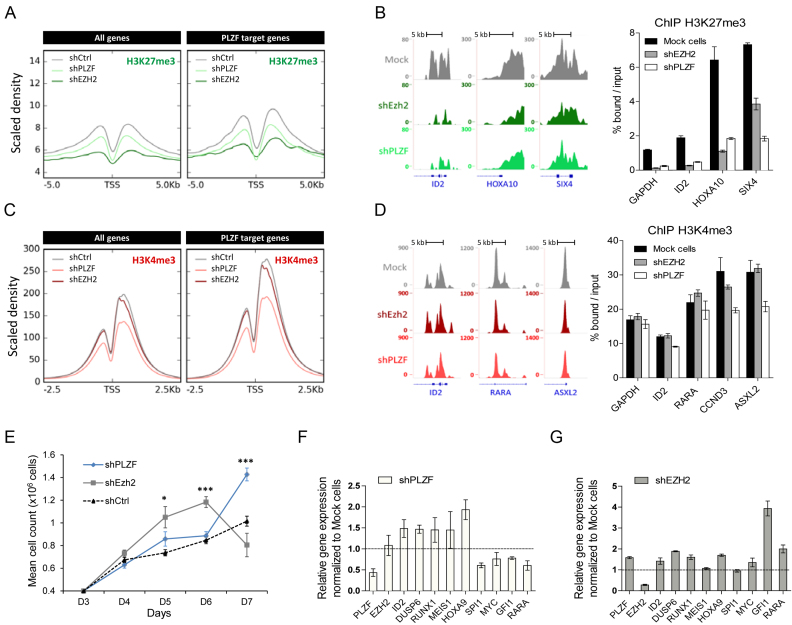
As EZH2 KD is increasing PLZF binding we next compared the effects of EZH2 KD to PLZF KD on the epigenetic landscape and gene expression. As expected, we found that EZH2 KD effectively erased the H3K27me3 signal (Figure 6A). Surprisingly, while not globally affecting EZH2 chromatin binding (Figure 5B), PLZF KD decreases H3K27me3 signal (Figure 6A). This decrease in signal was not specific to PLZF target genes as it was also observed on all genes (Figure 6A and B) and EZH2-only target genes (Supplementary Figure S6 and Figure 6B). GO analysis revealed that the H3K27me3-decreased genes upon PLZF KD were mainly associated with terms related to development processes (Supplementary Figure S7; Table S2A and C) suggesting that PLZF indirectly modifies the canonical activity of EZH2. Focusing on H3K4me3, we observed that PLZF KD decreases H3K4me3 signal at PLZF target genes, which is not observed with EZH2 KD (Figure 6C and D). By intersecting the H3K4me3-differentially enriched genes with PLZF-bound promoters, we observed that 60% of the PLZF-bound promoters were modified in their H3K4me3 level (Supplementary Figure S8). GO analysis revealed that H3K4me3-decreased genes were enriched in terms related to biosynthesis and genomic (transcription regulation and nucleic acid metabolism) suggesting a difference in PLZF activity depending on the gene identity (Supplementary Figure S8 and Table S2B-C). As these processes are critical in a wide variety of biological processes especially during cell cycle (38), we investigated the effect of PLZF KD on cell proliferation. Supportive of its role as a tumor suppressor (5), cells showed better growth upon PLZF KD than control cells (Figure 6E). In line with previous observation (Supplementary Figure S7), developmental genes were upregulated upon either PLZF or EZH2 KD (Figure 6F and G). However, PLZF and EZH2 KD displayed opposite effect on the expression of myeloid associated genes (decreased expression in PLZF KD and increased expression in EZH2 KD) which could reflect the modification of PLZF binding upon EZH2 observed in Figure 5.
Figure 6.
Effect of PLZF and EZH2 KDs on chromatin structure and gene expression. (A) Density profile of H3K27me3 signal tracks after PLZF or EZH2 KDs in KG1 cells. (B) Representative UCSC Genome browser tracks (left panel) of H3K27me3 after PLZF or EZH2 KDs and its associated spike-in ChIP-qPCR validation (right panel). (C) Density profile of H3K4me3 signal tracks after PLZF or EZH2 KDs in KG1 cells. (D) Representative UCSC Genome browser tracks (left panel) of H3K27me3 after PLZF or EZH2 KDs and its associated spike-in ChIP-qPCR (right panel) validation. (E) Proliferation assay of Trypan blue stained KG1 cells expressing shPLZF, shEZH2 or shCtrl. Data are expressed as mean cell count ±SEM of two individual clones (three replicates each) with *P < 0.05, ***P < 0.001 (t-test compared to shCtrl). (F) Gene expression analysis of PLZF target genes after PLZF KD. (G) Gene expression analysis on PLZF target genes after EZH2 KD. (F and G) Relative level of expression was evaluated using the 2∧ΔΔCT method with PBG-D as the reference gene. Data are presented as a mean ± SD of two independent clones (n = 3–6).
DISCUSSION
Gene regulation is a central cellular process governing cell fate in development and differentiation. It relies to a large extent on transcriptional regulation, mediated by TFs and chromatin regulators. However, the interplay between TFs and epigenetic chromatin modification is still poorly understood.
Like many TFs, PLZF acts as either a repressor or an activator of transcription. However, PLZF repressive activity has been the more recognized and extensively documented. Indeed, many structural studies have shown that the BTB/POZ domain of PLZF is a transcriptional repressive domain (39,35,40) that can achieve repression through sequence specific recruitment of corepressors such as N-CoR/HDAC or PcG complexes (18). In our study, we also highlight the repressive activity of PLZF when assayed using a Gal4/UAS reporter system. In this context, the Gal4-PLZF fusion is targeted to a gene reporter that is strongly repressed. Repression in that case was probably not uniquely due to EZH2 activity but was characterized by the accumulation of H3K27me3 when the BTB/POZ domain of PLZF was present. These data are consistent with our result showing that PLZF interacts with EZH2, the HMT of PRC2, via its BTB-POZ domain. Contrary to these data, our genome-wide approach in myeloid cells revealed that PLZF is mainly associated with active transcription. Indeed, PLZF has been shown to activate the transcription of some of its bound promoters in myeloid development (14) as well as in lymphoid cells where PLZF displays activator activity (16). While this transcriptional activation property of PLZF may be specific to cells of hematopoietic origin, it will be important to study PLZF activity globally in other tissues, such as spermatogenesis, where it plays an important role during differentiation (15). Nonetheless, our genome-wide study demonstrates that a minority of genes were directly repressed by PLZF, although these PLZF-repressed genes were enriched with developmental and differentiation genes strengthening the role of PLZF in controlling cellular development (12,41). The dual effect of PLZF on transcription could rely on many factors influencing its partner binding or its specific DNA motif recognitions. DNA motif search in our myeloid cells resulted in the emergence of two main recognition motifs. One of the discovered motifs is very similar to the consensus -A-T/G-G/C-T-A/C-A/C-A-G-G-T- found previously by in vitro analyses (42) and match perfectly with the consensus domain defined by Krumlauf and collaborators based on electrophoretic mobility shift assay and directed mutagenesis (37). The other discovered motif is the consensus of the ERG TF belonging to the ETS family. A canonical ETS consensus was also found as a PLZF binding motif in NKT cells (16) indicating a potential interplay between PLZF and ETS factors.
As we show that PLZF interacts with EZH2 and could ectopically recruit EZH2 activity to chromatin, one hypothesis is that EZH2 could influence PLZF activity and enhance its repressive activity in myeloid cells. However, our data do not support this hypothesis but instead revealed that common PLZF and EZH2 bound promoters were devoid of the PRC2 component, SUZ12, yet enriched with the active H3K4me3 histone mark. In addition, related coding regions of common PLZF/EZH2 targeted promoters were enriched for Pol II and H3K36me3, a histone mark associated with elongating Pol II, and were expressed. These data highlight a non-canonical EZH2 function when associated with PLZF. Indeed, previous studies have fostered a non-canonical activity of EZH2 associated with active gene expression: in hormone-refractory breast cancer and castration-resistant prostate cancer, EZH2 switches to a transcriptional activator and acts independently of the PRC2 complex (25,43); in glioblastoma, AKT signaling activation leads to phosphorylation of EZH2 and inhibits its H3K27me3 enzymatic activity (44). Thus, rather than EZH2 influencing PLZF repressive activity, could EZH2 influence PLZF positive transcriptional activity? Globally, the KD of EZH2 has an activating effect characterized by a decrease in H3K27me3 concomitant with an increase in gene expression. Interestingly, KD of EZH2 had a positive effect on chromatin-bound PLZF levels and influenced the transcriptional output of its target genes. This suggests that EZH2 could modulate PLZF transcriptional activity independently of its HMT activity. Previous studies have demonstrated that the transcriptional repression activity of PLZF is specifically dependent on the HAT activity of p300 or HAT1, which acetylates PLZF (45,46) and that SUMOylation in K242, K387 and K396, extensively modulates PLZF-mediated transcriptional repression (47). While preparing this manuscript an interesting study showed that in NKT cells EZH2 directly methylates PLZF, resulting in its degradation (48). Even though we did not observe changes in PLZF protein level upon shEZH2 in our system, the methylation of PLZF by EZH2 could afford an explanation of the positive transcriptional output resulting of the observed PLZF–EZH2 interaction.
Conversely, could PLZF be an influencing factor of EZH2 activity? Our KD experiments showed that downregulating PLZF had little influence on global EZH2 binding, suggesting that PLZF is not involved in EZH2 recruitment per se. However, our results showed that PLZF KD decreases H3K4me3 levels globally and to a lesser extend decreasing H3K27me3 levels of developmental genes, suggesting that loss of PLZF as an effect on EZH2 canonical activity. By analogy with what has been proposed for RBB4 (49) or lncRNA (50), where both factors have proposed to be sensors of EZH2 activity, PLZF could be also a modulator of EZH2 activity.
In conclusion, we provide evidence that PLZF and EZH2 co-associate at the chromatin level, but when located at the same loci the two proteins lose their respective repressive activity revealing a non-canonical EZH2 activity that regulates PLZF transcriptional output.
DATA AVAILABILITY
ChIP-seq data (FastQ and Bigwig files) are accessible in the NCBI Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/) under the accession number GSE109619.
In addition to our own ChIP-seq data, previously published datasets were retrieved from the Gene Expression Omnibus (GEO) database for re-analysis and comparisons. Sequencing datasets were thus retrieved for: human hematopoietic cells enriched within the immature cell surface marker CD34+ (HSC-CD34+) H3K27me3 (GSM772951), HSC-CD34+ H3K4me3 (GSM773041), (HSC-CD34+) H3K27ac. In addition, expression data from the HSC CD34+ cells of healthy donor (GSM308470) and expression data from human untreated KG1 cells (GSM1316691) were extracted.
Supplementary Material
ACKNOWLEDGEMENTS
Authors thank Dr Vincent Geli and Dr Ari Melnick for helpful discussions. Authors gratefully acknowledge the advice of Prof. Atsushi Iwama during revision of the manuscript. Authors gratefully acknowledge Montpellier MGX and MIT Biomicro Center Sequencing facilities for the ChIP-seq sequencing and the CRCM DISC and Cibi Platforms for the bioinformatics support.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Association pour la Recherche sur le Cancer [ARC2013-fixe]; Association Laurette Fugain [ALF2014/09]; Institut Thématique Multi-Organisme-cancer [C13104L to E.D., C.G.; P036560 to E.D., J.C.A.]; l’Institut National du Cancer [20141PLBIO06–1 to E.D., C.G., M.P.]; Ministère de l’Enseignement Supérieur (to M.K.); Ligue Contre le Cancer (to M.K.); cancéropôle PACA (to G.T., A.E.K.). Funding for open access charge: l’Institut National du Cancer.
Conflict of interest statement. None declared.
REFERENCES
- 1. Rosenbauer F., Tenen D.G.. Transcription factors in myeloid development: balancing differentiation with transformation. Nat. Rev. Immunol. 2007; 7:105–117. [DOI] [PubMed] [Google Scholar]
- 2. Dick J.E., Doulatov S.. The role of PLZF in human myeloid development. Ann. N Y Acad. Sci. 2009; 1176:150–153. [DOI] [PubMed] [Google Scholar]
- 3. Chen Z., Brand N.J., Chen A., Chen S.J., Tong J.H., Wang Z.Y., Waxman S., Zelent A.. Fusion between a novel Kruppel-like zinc finger gene and the retinoic acid receptor-alpha locus due to a variant t(11;17) translocation associated with acute promyelocytic leukaemia. EMBO J. 1993; 12:1161–1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Melnick A., Licht J.D.. Deconstructing a disease: RARalpha, its fusion partners, and their roles in the pathogenesis of acute promyelocytic leukemia. Blood. 1999; 93:3167–3215. [PubMed] [Google Scholar]
- 5. Liu T.M., Lee E.H., Lim B., Shyh-Chang N.. Concise review: balancing stem cell self-renewal and differentiation with PLZF. Stem Cells. 2016; 34:277–287. [DOI] [PubMed] [Google Scholar]
- 6. Savage A.K., Constantinides M.G., Han J., Picard D., Martin E., Li B., Lantz O., Bendelac A.. The transcription factor PLZF directs the effector program of the NKT cell lineage. Immunity. 2008; 29:391–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Labbaye C., Spinello I., Quaranta M.T., Pelosi E., Pasquini L., Petrucci E., Biffoni M., Nuzzolo E.R., Billi M., Foa R. et al. . A three-step pathway comprising PLZF/miR-146a/CXCR4 controls megakaryopoiesis. Nat. Cell Biol. 2008; 10:788–801. [DOI] [PubMed] [Google Scholar]
- 8. Kovalovsky D., Uche O.U., Eladad S., Hobbs R.M., Yi W., Alonzo E., Chua K., Eidson M., Kim H.J., Im J.S. et al. . The BTB-zinc finger transcriptional regulator PLZF controls the development of invariant natural killer T cell effector functions. Nat. Immunol. 2008; 9:1055–1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Li X., Lopez-Guisa J.M., Ninan N., Weiner E.J., Rauscher F.J. 3rd, Marmorstein R.. Overexpression, purification, characterization, and crystallization of the BTB/POZ domain from the PLZF oncoprotein. J. Biol. Chem. 1997; 272:27324–27329. [DOI] [PubMed] [Google Scholar]
- 10. David G., Alland L., Hong S.H., Wong C.W., DePinho R.A., Dejean A.. Histone deacetylase associated with mSin3A mediates repression by the acute promyelocytic leukemia-associated PLZF protein. Oncogene. 1998; 16:2549–2556. [DOI] [PubMed] [Google Scholar]
- 11. Hong S.H., David G., Wong C.W., Dejean A., Privalsky M.L.. SMRT corepressor interacts with PLZF and with the PML-retinoic acid receptor alpha (RARalpha) and PLZF-RARalpha oncoproteins associated with acute promyelocytic leukemia. Proc. Natl. Acad. Sci. U.S.A. 1997; 94:9028–9033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Barna M., Merghoub T., Costoya J.A., Ruggero D., Branford M., Bergia A., Samori B., Pandolfi P.P.. Plzf mediates transcriptional repression of HoxD gene expression through chromatin remodeling. Dev. Cell. 2002; 3:499–510. [DOI] [PubMed] [Google Scholar]
- 13. Boukarabila H., Saurin A.J., Batsche E., Mossadegh N., van Lohuizen M., Otte A.P., Pradel J., Muchardt C., Sieweke M., Duprez E.. The PRC1 Polycomb group complex interacts with PLZF/RARA to mediate leukemic transformation. Genes Dev. 2009; 23:1195–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Doulatov S., Notta F., Rice K.L., Howell L., Zelent A., Licht J.D., Dick J.E.. PLZF is a regulator of homeostatic and cytokine-induced myeloid development. Genes Dev. 2009; 23:2076–2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lovelace D.L., Gao Z., Mutoji K., Song Y.C., Ruan J., Hermann B.P.. The regulatory repertoire of PLZF and SALL4 in undifferentiated spermatogonia. Development. 2016; 143:1893–1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mao A.P., Constantinides M.G., Mathew R., Zuo Z., Chen X., Weirauch M.T., Bendelac A.. Multiple layers of transcriptional regulation by PLZF in NKT-cell development. Proc. Natl. Acad. Sci. U.S.A. 2016; 113:7602–7607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. McConnell M.J., Durand L., Langley E., Coste-Sarguet L., Zelent A., Chomienne C., Kouzarides T., Licht J.D., Guidez F.. Post transcriptional control of the epigenetic stem cell regulator PLZF by sirtuin and HDAC deacetylases. Epigenet. Chromatin. 2015; 8:38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Maeda T. Regulation of hematopoietic development by ZBTB transcription factors. Int. J. Hematol. 2016; 104:310–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Margueron R., Reinberg D.. The Polycomb complex PRC2 and its mark in life. Nature. 2011; 469:343–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Scelfo A., Piunti A., Pasini D.. The controversial role of the Polycomb group proteins in transcription and cancer: how much do we not understand Polycomb proteins. FEBS J. 2015; 282:1703–1722. [DOI] [PubMed] [Google Scholar]
- 21. Sashida G., Iwama A.. Multifaceted role of the polycomb-group gene EZH2 in hematological malignancies. Int. J. Hematol. 2017; 105:23–30. [DOI] [PubMed] [Google Scholar]
- 22. Holoch D., Margueron R.. Mechanisms regulating PRC2 recruitment and enzymatic activity. Trends Biochem. Sci. 2017; 42:531–542. [DOI] [PubMed] [Google Scholar]
- 23. Sanulli S., Justin N., Teissandier A., Ancelin K., Portoso M., Caron M., Michaud A., Lombard B., da Rocha S.T., Offer J. et al. . Jarid2 methylation via the PRC2 complex regulates H3K27me3 deposition during cell differentiation. Mol. Cell. 2015; 57:769–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Pasini D., Bracken A.P., Jensen M.R., Lazzerini Denchi E., Helin K.. Suz12 is essential for mouse development and for EZH2 histone methyltransferase activity. EMBO J. 2004; 23:4061–4071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lee S.T., Li Z., Wu Z., Aau M., Guan P., Karuturi R.K.M., Liou Y.C., Yu Q.. Context-specific regulation of NF-κB target gene expression by EZH2 in breast cancers. Mol. Cell. 2011; 43:798–810. [DOI] [PubMed] [Google Scholar]
- 26. Xu K., Wu Z.J., Groner A.C., He H.H., Cai C., Lis R.T., Wu X., Stack E.C., Loda M., Liu T. et al. . EZH2 oncogenic activity in castration-resistant prostate cancer cells is Polycomb-independent. Science. 2012; 338:1465–1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yan J., Li B., Lin B., Lee P.T., Chung T.H., Tan J., Bi C., Lee X.T., Selvarajan V., Ng S.B. et al. . EZH2 phosphorylation by JAK3 mediates a switch to noncanonical function in natural killer/T-cell lymphoma. Blood. 2016; 128:948–958. [DOI] [PubMed] [Google Scholar]
- 28. He S., Liu Y., Meng L., Sun H., Wang Y., Ji Y., Purushe J., Chen P., Li C., Madzo J. et al. . Ezh2 phosphorylation state determines its capacity to maintain CD8(+) T memory precursors for antitumor immunity. Nat. Commun. 2017; 8:2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Vincent-Fabert C., Platet N., Vandevelde A., Poplineau M., Koubi M., Finetti P., Tiberi G., Imbert A.M., Bertucci F., Duprez E.. PLZF mutation alters mouse hematopoietic stem cell function and cell cycle progression. Blood. 2016; 127:1881–1885. [DOI] [PubMed] [Google Scholar]
- 30. Thomas-Chollier M., Herrmann C., Defrance M., Sand O., Thieffry D., van Helden J.. RSAT peak-motifs: motif analysis in full-size ChIP-seq datasets. Nucleic Acids Res. 2012; 40:e31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Shen L., Shao N., Liu X., Nestler E.. ngs.plot: Quick mining and visualization of next-generation sequencing data by integrating genomic databases. BMC Genomics. 2014; 15:284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Shen L., Shao N.Y., Liu X., Maze I., Feng J., Nestler E.J.. diffReps: detecting differential chromatin modification sites from ChIP-seq data with biological replicates. PLoS One. 2013; 8:e65598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ramirez F., Ryan D.P., Gruning B., Bhardwaj V., Kilpert F., Richter A.S., Heyne S., Dundar F., Manke T.. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 2016; 44:W160–W165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Koeffler H.P., Golde D.W.. Acute myelogenous leukemia: a human cell line responsive to colony-stimulating activity. Science. 1978; 200:1153–1154. [DOI] [PubMed] [Google Scholar]
- 35. Melnick A., Ahmad K.F., Arai S., Polinger A., Ball H., Borden K.L., Carlile G.W., Prive G.G., Licht J.D.. In-depth mutational analysis of the promyelocytic leukemia zinc finger BTB/POZ domain reveals motifs and residues required for biological and transcriptional functions. Mol. Cell. Biol. 2000; 20:6550–6567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ivins S., Pemberton K., Guidez F., Howell L., Krumlauf R., Zelent A.. Regulation of Hoxb2 by APL-associated PLZF protein. Oncogene. 2003; 22:3685–3697. [DOI] [PubMed] [Google Scholar]
- 37. Benveniste D., Sonntag H.J., Sanguinetti G., Sproul D.. Transcription factor binding predicts histone modifications in human cell lines. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:13367–13372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lane A.N., Fan T.W.. Regulation of mammalian nucleotide metabolism and biosynthesis. Nucleic Acids Res. 2015; 43:2466–2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ahmad K.F., Engel C.K., Prive G.G.. Crystal structure of the BTB domain from PLZF. Proc. Natl. Acad. Sci. U.S.A. 1998; 95:12123–12128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Melnick A., Carlile G., Ahmad K.F., Kiang C.L., Corcoran C., Bardwell V., Prive G.G., Licht J.D.. Critical residues within the BTB domain of PLZF and Bcl-6 modulate interaction with corepressors. Mol. Cell. Biol. 2002; 22:1804–1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Barna M., Hawe N., Niswander L., Pandolfi P.P.. Plzf regulates limb and axial skeletal patterning. Nat. Genet. 2000; 25:166–172. [DOI] [PubMed] [Google Scholar]
- 42. Li J.Y., English M.A., Ball H.J., Yeyati P.L., Waxman S., Licht J.D.. Sequence-specific DNA binding and transcriptional regulation by the promyelocytic leukemia zinc finger protein. J. Biol. Chem. 1997; 272:22447–22455. [DOI] [PubMed] [Google Scholar]
- 43. Shi B., Liang J., Yang X., Wang Y., Zhao Y., Wu H., Sun L., Zhang Y., Chen Y., Li R. et al. . Integration of estrogen and Wnt signaling circuits by the polycomb group protein EZH2 in breast cancer cells. Mol. Cell. Biol. 2007; 27:5105–5119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kim E., Kim M., Woo D.H., Shin Y., Shin J., Chang N., Oh Y.T., Kim H., Rheey J., Nakano I. et al. . Phosphorylation of EZH2 activates STAT3 signaling via STAT3 methylation and promotes tumorigenicity of glioblastoma stem-like cells. Cancer Cell. 2013; 23:839–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Guidez F., Howell L., Isalan M., Cebrat M., Alani R.M., Ivins S., Hormaeche I., McConnell M.J., Pierce S., Cole P.A. et al. . Histone acetyltransferase activity of p300 is required for transcriptional repression by the promyelocytic leukemia zinc finger protein. Mol. Cell. Biol. 2005; 25:5552–5566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Sadler A.J., Suliman B.A., Yu L., Yuan X., Wang D., Irving A.T., Sarvestani S.T., Banerjee A., Mansell A.S., Liu J.P. et al. . The acetyltransferase HAT1 moderates the NF-kappaB response by regulating the transcription factor PLZF. Nat. Commun. 2015; 6:6795. [DOI] [PubMed] [Google Scholar]
- 47. Chao T.T., Chang C.C., Shih H.M.. SUMO modification modulates the transrepression activity of PLZF. Biochem. Biophys. Res. Commun. 2007; 358:475–482. [DOI] [PubMed] [Google Scholar]
- 48. Vasanthakumar A., Xu D., Lun A.T., Kueh A.J., van Gisbergen K.P., Iannarella N., Li X., Yu L., Wang D., Williams B.R. et al. . A non-canonical function of Ezh2 preserves immune homeostasis. EMBO Rep. 2017; 18:619–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Schmitges F.W., Prusty A.B., Faty M., Stutzer A., Lingaraju G.M., Aiwazian J., Sack R., Hess D., Li L., Zhou S. et al. . Histone methylation by PRC2 is inhibited by active chromatin marks. Mol. Cell. 2011; 42:330–341. [DOI] [PubMed] [Google Scholar]
- 50. Davidovich C., Zheng L., Goodrich K.J., Cech T.R.. Promiscuous RNA binding by Polycomb repressive complex 2. Nat. Struct. Mol. Biol. 2013; 20:1250–1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
ChIP-seq data (FastQ and Bigwig files) are accessible in the NCBI Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/) under the accession number GSE109619.
In addition to our own ChIP-seq data, previously published datasets were retrieved from the Gene Expression Omnibus (GEO) database for re-analysis and comparisons. Sequencing datasets were thus retrieved for: human hematopoietic cells enriched within the immature cell surface marker CD34+ (HSC-CD34+) H3K27me3 (GSM772951), HSC-CD34+ H3K4me3 (GSM773041), (HSC-CD34+) H3K27ac. In addition, expression data from the HSC CD34+ cells of healthy donor (GSM308470) and expression data from human untreated KG1 cells (GSM1316691) were extracted.