Abstract
Mitochondrial transcription factor A (TFAM) is essential for the maintenance, expression and transmission of mitochondrial DNA (mtDNA). However, mechanisms for the post-translational regulation of TFAM are poorly understood. Here, we show that TFAM is lysine acetylated within its high-mobility-group box 1, a domain that can also be serine phosphorylated. Using bulk and single-molecule methods, we demonstrate that site-specific phosphoserine and acetyl-lysine mimics of human TFAM regulate its interaction with non-specific DNA through distinct kinetic pathways. We show that higher protein concentrations of both TFAM mimics are required to compact DNA to a similar extent as the wild-type. Compaction is thought to be crucial for regulating mtDNA segregation and expression. Moreover, we reveal that the reduced DNA binding affinity of the acetyl-lysine mimic arises from a lower on-rate, whereas the phosphoserine mimic displays both a decreased on-rate and an increased off-rate. Strikingly, the increased off-rate of the phosphoserine mimic is coupled to a significantly faster diffusion of TFAM on DNA. These findings indicate that acetylation and phosphorylation of TFAM can fine-tune TFAM–DNA binding affinity, to permit the discrete regulation of mtDNA dynamics. Furthermore, our results suggest that phosphorylation could additionally regulate transcription by altering the ability of TFAM to locate promoter sites.
INTRODUCTION
The mitochondrial genome is compacted into nucleoprotein complexes known as mitochondrial nucleoids (1–3), of which mitochondrial transcription factor A (TFAM or mtTFA) is the most abundant protein (4,5). TFAM belongs to the high-mobility-group (HMG) box family of DNA binding proteins (6,7), and consists of two HMG box domains (HMG1 and HMG2) separated by a linker, coupled to a C-terminal tail via HMG2 (Figure 1). TFAM is essential for initiating transcription of mitochondrial DNA (mtDNA) by specifically recognizing the light-strand and heavy-strand promoters of the mitochondrial genome (8–12). TFAM tethers the N-terminal region of mitochondrial RNA polymerase (mtRNAP), resulting in the bending of promoter DNA around mtRNAP (13). The subsequent recruitment of TFB2M induces promoter melting required for transcription initiation (12–16). TFAM monomers have been shown to diffuse rapidly along non-specific DNA sequences via a 1D sliding mechanism (17). This represents one likely route by which TFAM locates the promoters of mtDNA to initiate transcription (17,18).
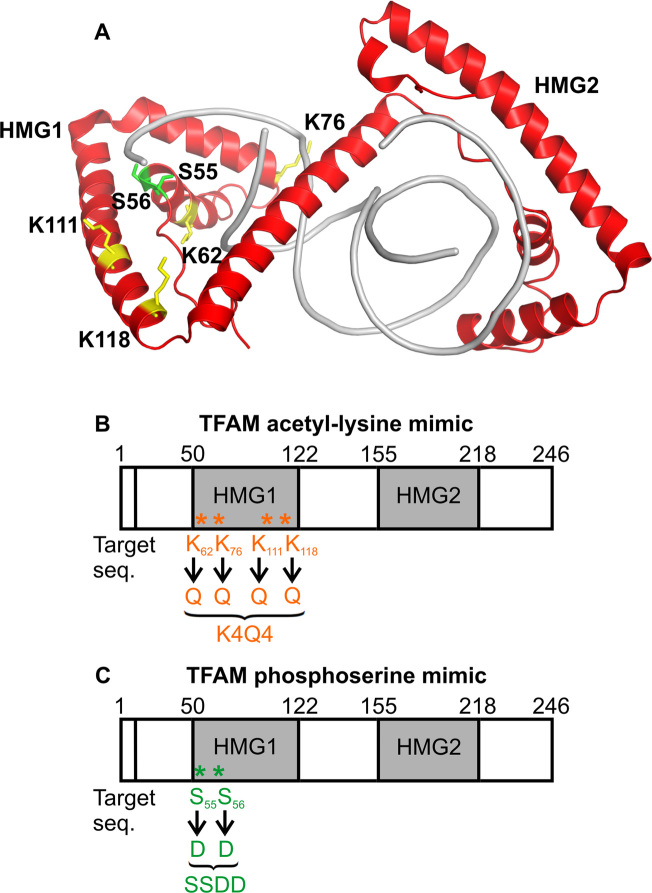
Figure 1.
Acetylation and phosphorylation of TFAM within HMG1. (A) Location of post-translationally modified lysine and serine residues in the X-ray structure of TFAM (PDB ID 4NNU) (31) bound to sequence non-specific DNA (gray). Phosphorylation of serine residues S55 and S56 (green) (35) and acetylation of lysine residues K62, K76, K111 and K118 (yellow) were identified by LC–MS/MS in TFAM purified from HEK293 cells (Supplementary Figure S1). (B) Schematic representation of TFAM acetyl-lysine mimic, TFAMK4Q4. (C) Schematic representation of TFAM phosphoserine mimic, TFAMSSDD.
In addition, studies have shown that TFAM can coat mtDNA extensively through non-specific DNA interactions (17,19), thereby facilitating the compaction of mtDNA into mitochondrial nucleoids (2,20–22), which is required for segregation in dividing cells (23). Such compaction has also been suggested to inhibit replication and transcription of the mitochondrial genome (24–26). The mechanisms by which TFAM mediates DNA compaction are still under debate (17,22,26–32). X-ray crystallographic studies demonstrate that human TFAM imposes a 180° U-turn on both sequence-specific and sequence non-specific DNA (27,29,31). Fluorescence resonance energy transfer data is further consistent with TFAM compacting non-specific DNA sequences through bending of the DNA backbone (28). However, other mechanisms have also been proposed. For instance, using combined optical tweezers and fluorescence microscopy, it has been shown that human TFAM can bind cooperatively to non-specific DNA sequences to form stable protein patches that increase the flexibility of the DNA. This enhanced flexibility is likely to arise through local base-pair melting of DNA induced by the binding of TFAM (the so-called flexible-hinge mechanism) and provides an effective means to compact mtDNA (17). Meanwhile, data from atomic force microscopy, super-resolution imaging and electron microscopy suggest that TFAM-mediated looping and cross-linking of DNA can additionally play a role in the compaction of non-specific sequences of mtDNA (22,32).
Little is known about post-translational mechanisms for regulating TFAM function. TFAM is modified by O-linked glycosylation (33), acetylation (34) and phosphorylation (35,36), but the specific sites of these modifications and their respective downstream consequences are only beginning to emerge. Our previous work demonstrated that human TFAM is phosphorylated at serines S55 and/or S56 within its HMG1 domain by cAMP-dependent protein kinase (PKA) in cultured cells (35). Phosphorylated TFAM, as well as a phosphoserine mimic at serines 55/56, impairs DNA binding, abrogates transcriptional activation and leads to rapid degradation by the Lon protease within mitochondria (35). TFAM has also been shown to be phosphorylated at serine S177 within HMG2 by ERK1/2 (36). A phosphoserine mimic of TFAM at serine S177 decreases DNA binding, down-regulating mitochondrial transcription and mitochondrial respiration in cells (36). However, the mechanisms underlying impaired DNA binding remain unclear; specifically, does phosphorylation decrease DNA-association, or increase DNA-release? Moreover, the influence of phosphorylation, as well as other post-translational modifications, on the ability of TFAM to both compact and slide on DNA is unknown.
Here we identify that human TFAM is acetylated at lysines 62, 76, 111 and 118 within HMG1 in cultured cells. Using a combination of bulk and single-molecule methods we then unravel how relevant acetyl-lysine and phosphoserine mimics of human TFAM influence its ability to bind, compact and diffuse on non-specific DNA sequences. We show that these mimics are able to compact non-specific DNA sequences via the flexible-hinge mechanism, similar to unmodified wild-type TFAM. However, in each case, a higher protein concentration is required to achieve this, reflecting a reduced binding affinity for double-stranded DNA (dsDNA). Furthermore, we demonstrate important distinctions between the two mimics; strikingly, the phosphoserine mimic slides on dsDNA faster, and dissociates more rapidly, than either the acetyl-lysine mimic or wild-type TFAM. Taken together, these findings indicate mechanistic pathways by which post-translational modifications could regulate both compaction and transcription of mtDNA in vivo.
MATERIALS AND METHODS
Identification of HMG1 acetylation sites within TFAM purified from HEK293 cells
TFAM carrying a carboxyl-terminal hexahistidine tag was transiently overexpressed in HEK293 cells and purified as described previously (35). Briefly, the isolated protein samples were separated by SDS-PAGE and the TFAM band was excised for in-gel trypsin digestion. Peptides were enriched by titanium dioxide tip (GL Sciences Inc.) and eluted with 5% NH4OH followed by C18 ZipTip desalting. The resulting peptides were analyzed by LC–MS/MS on an Ultimate 3000 LC system coupled with an Orbitrap Velos tandem MS instrument in the positive ion mode with a spray voltage of 2 kV and a capillary temperature of 275°C. Collision-induced dissociation was used for peptide fragmentation (Supplementary Figure S1).
Protein purification
PCR-based site-directed mutagenesis (QuikChange II, Stratagene) was employed to engineer the cDNAs encoding TFAMWT, TFAMSSDD and TFAMK4Q4, which all lacked their respective amino terminal mitochondrial targeting sequences and had a hexahistidine tag fused to their carboxyl-termini. TFAM variants were expressed from pET-22b (+) in the Escherichia coli strain Rosetta DE3 (EMD Millipore). Proteins were purified by column chromatography using Ni2+ agarose as well as ion exchange chromatography (35). Protein purity was analyzed by Coomassie Blue staining and immunoblotting with antibodies recognizing recombinant TFAM (Supplementary Figure S2).
Microscale thermophoresis (MST)
Experiments were performed using a Monolith NT0.115 instrument (NanoTemper Technologies) at 80% MST and LED power at 23°C. Thermophoresis was induced in a sample mixture containing Cy5-labeled DNA (20 nM) and varying concentrations of TFAM in a buffer of either (i) 25 mM HEPES pH 7.8, 100 mM NaCl and 0.2% pluronics (Figure 2), or (ii) 25 mM Tris–HCl pH 7.6, 20 mM NaCl and 0.1% pluronics (Supplementary Figure S3). The DNA consisted of a non-specific sequence forming an 18 bp duplex with a 13 nucleotide single-stranded overhang. Ratios of the fluorescence intensity associated with Cy5-DNA before and after thermophoresis (Fnorm) were calculated and averaged from 3 independent experiments as a function of TFAM concentration. The fraction of DNA–TFAM complexes, represented by the change in Fnorm, was fit to a quadratic equation to determine the equilibrium binding constant (Supplementary Methods, Eq. S1 and S2) (37).
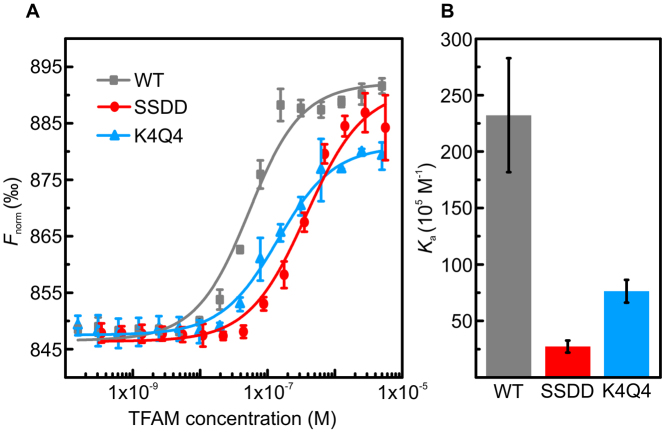
Figure 2.
TFAMK4Q4 and TFAMSSDD exhibit a reduced binding affinity for non-specific DNA compared with TFAMWT. (A) Data from MST experiments, showing the normalized fluorescence (Fnorm) resulting from titrating sequence non-specific Cy5-labeled DNA (20 nM) with TFAMWT, TFAMSSDD and TFAMK4Q4, respectively. In each case, three independent experiments were performed and the data were fitted using Eq. S2. (B) Equilibrium binding constant, Ka, determined from the fits in panel (A) for TFAMWT, TFAMSSDD and TFAMK4Q4. Measurements were performed using an 18 bp oligomer with a 13 nucleotide single-stranded overhang, at room temperature and in a buffer of 25 mM HEPES pH 7.8 with 100 mM NaCl and 0.2% pluronics. Errors are standard errors of the mean (S.E.M.).
Preparation of alexa-555-labeled TFAM
To obtain fluorescent TFAMWT, TFAMSSDD and TFAMK4Q4, cysteine residues of each protein were labeled with maleimide alexa-555 (Molecular Probes). To this end, purified samples of TFAMWT, TFAMSSDD and TFAMK4Q4 (in a buffer of 50 mM HEPES pH 7.5, 300 mM NaCl, 10 mM MgCl2 and 20% glycerol) were each incubated in excess dye (10-fold) for 2 h at 19°C. Unreacted dye was removed from the samples with size-exclusion spin-columns (Sephadex G-25, GE Healthcare). The labeling ratio (one fluorophore per TFAM monomer) was determined by analysis of the coefficient of absorption for tryptophan (associated with the protein) and that of the alexa dye in each sample.
Single-molecule assays
Single-molecule experiments were performed on a custom-made inverted microscope that combines wide-field fluorescence microscopy with dual-trap optical tweezers, described in detail previously (38,39). Biotinylated λ-DNA (∼48.5 kb, Roche) was tethered between two streptavidin-coated microspheres (diameter 4.5 μm, Spherotech Inc) in situ within a multichannel laminar flow-cell mounted on an automated XY-stage (39). Fluorescence from alexa-555-labeled TFAM was imaged on an EMCCD camera following excitation with a 532 nm excitation laser. All single-molecule experiments were performed at room temperature. Note that we employed conditions that were similar to those used in previous single-molecule studies of TFAMWT, namely with a buffer of 10 mM Tris–HCl pH 7.6 with 25 mM NaCl. In addition to aiding comparison with previous work (17), studies have shown that the concentration of sodium ions in mitochondria, [Na+]mito, can be significantly lower than 150 mM. For example, in MDCK cells, the baseline [Na+]mito is ∼50 mM as determined using the sodium-sensitive fluorescent probe CoroNa Red (40), which is preferentially taken up by polarized mitochondria. In astrocytes, the baseline [Na+]mito is ∼12–20 mM, as measured by CoroNa Red (41). Thus, our experimental conditions are within this range.
McGhee von Hippel analysis of DNA–TFAM binding
Force–extension curves of λ-DNA in the presence of TFAM were fit to the extensible worm-like chain (eWLC) model, for forces up to 30 pN, from which the persistence length (Lp) was extracted (42). The fractional protein coverage of the DNA (v) was then determined from the measured Lp, using the relation given by Farge et al. (17). By fitting the change in v as a function of TFAM concentration to the McGhee von Hippel model for cooperative binding (43,44) (Supplementary Methods, Eq. S3–S6), the binding affinity and cooperativity for TFAM–DNA interactions were extracted.
Determination of TFAM–DNA disassembly rates
Individual λ-DNA molecules were incubated in a relaxed configuration (<0.5 pN, with an end-to-end length ∼4 μm) in the presence of alexa-555-TFAM for 5 min. The concentration of fluorescently-labeled TFAMWT, TFAMSSDD and TFAMK4Q4, respectively, was sufficiently high so as to induce complete coating of the DNA (>80–100 nM). After incubation, the TFAM-bound dsDNA molecule was transferred rapidly to a protein-free buffer channel where it was held at a constant tension of 5 pN and visualized by stroboscopic illumination with a 532 nm excitation laser (Supplementary Methods, Eq. S7 and S8).
Determination of TFAM diffusion coefficient
λ-DNA molecules were incubated in a low concentration of alexa-555-labeled TFAM (<30 nM) such that only a few protein monomers were bound. The DNA molecules were then moved to a protein-free buffer channel where the displacement of each monomer on the DNA was measured over time by recording fluorescence movies with a short (0.5 s) frame rate. The corresponding kymographs were analyzed with a custom-written MATLAB-based program that tracked the position of each alexa-555-TFAM monomer as a function of time. Only traces spanning longer than 10 s and which did not cross another were considered. The diffusion constant (D) was determined from MSD plots (MSD = 2Dt + offset) for all trajectories measured (Supplementary Methods).
RESULTS
Acetyl-lysine and phosphoserine mimics have lower affinities than wild-type TFAM for non-specific DNA
Four acetylated lysine residues were identified by LC–MS/MS with high confidence interval, using human TFAM that was expressed recombinantly in HEK293 cells and purified as previously described (35) (Supplementary Figure S1). The acetylated residues were located at positions K62, K76, K111 and K118 within the HMG1 domain. Using the X-ray structure of TFAM bound to a sequence non-specific dsDNA oligonucleotide (31), our analysis shows that K62 and K76 directly contact DNA, whereas K111 and K118 do not (Figure 1A). K62 and K76 are in close proximity to serine residues S55 and S56 (also located within HMG1), which we have previously shown are phosphorylated by PKA in mitochondria (35). In the X-ray structure of TFAM, S55 interacts with DNA through a water molecule, whereas S56 contacts a deoxyribose in the DNA backbone (31). It is possible that acetylation of K62, K76, K111 and K118 occurs concomitantly in the cell. As the data were acquired using a bottom-up proteomic approach, in which the protein was trypsin-digested into peptides prior to LC–MS/MS analysis, we were not able to determine whether individual TFAM proteins contain all four acetylated sites, or a mixture of acetylation isoforms with single acetyl-lysine modifications.
To distinguish the effects of site-specific acetylation and phosphorylation of TFAM on sequence non-specific DNA binding, we produced recombinant TFAM variants with amino acid substitutions mimicking these PTMs (Materials and Methods). For the acetyl-lysine mimic (TFAMK4Q4), all four lysine residues K62, K76, K111 and K118 in HMG1 were replaced with glutamine, thus mimicking the effect of acetylation, which neutralizes positively charged side chains (Figure 1B and Materials and Methods). The phosphoserine mimic (TFAMSSDD) was engineered by replacing S55 and S56 with aspartic acid residues, as previously described (35), thereby mimicking the negative charge of phosphate modification (Figure 1C). In order to determine the relative DNA binding affinities of TFAMSSDD, TFAMK4Q4 and unmodified wild-type TFAM (TFAMWT), we first employed microscale thermophoresis (MST). In these experiments, we used a sequence non-specific DNA substrate (consisting of an 18 bp oligomer with a 13 nucleotide single-stranded overhang) in a buffer of 25 mM HEPES pH 7.8 with 100 mM NaCl. In this way, we determined the following trend in equilibrium binding constant, Ka: TFAMWT (Ka = 23.2 ± 5.1 × 106 M−1) > TFAMK4Q4 (Ka = 7.6 ± 1.0 × 106 M−1) > TFAMSSDD (Ka = 2.7 ± 0.5 × 106 M−1). This is shown in Figure 2 and Table 1. Note that the trend in binding affinity between the three TFAM species shows little dependence on ionic strength (Supplementary Figure S3 and Table S1).
Table 1. Summary of parameters describing the interaction of TFAMWT, TFAMSSDD and TFAMK4Q4 with non-specific dsDNA. All values were derived using single-molecule methods (in a buffer of 10 mM Tris–HCl pH 7.6 with 25 mM NaCl, as presented in Figures 3–5), except the final two columns, which report Kaand Kd obtained using MST (in a buffer of 25 mM HEPES pH 7.8 with 100 mM NaCl and 0.2% pluronics, as presented in Figure 2). All errors are S.E.M.
| TFAM variant | K a (105 M−1) | K d (10−7 M) | k off (10−3 s−1) | k on (102 M−1 s−1) | D (104 nm2s−1) | ω | K a (MST) (106 M−1) | K d (MST) (10−8 M) |
|---|---|---|---|---|---|---|---|---|
| TFAMWT | 16.3 ± 2.6 | 6.1 ± 1.0 | 3.0 ± 1.0 | 48.7 ± 17.8 | 10.1 ± 1.6 | 99 ± 9 | 23.2 ± 5.1 | 4.3 ± 0.9 |
| TFAMSSDD | 2.3 ± 0.2 | 43.7 ± 4.6 | 5.9 ± 1.4 | 13.6 ± 3.6 | 65.1 ± 15.5 | 76 ± 7 | 2.7 ± 0.5 | 36.5 ± 7.1 |
| TFAMK4Q4 | 1.7 ± 0.7 | 58.2 ± 24.1 | 2.9 ± 1.0 | 5.0 ± 2.6 | 15.1 ± 2.6 | 125 ± 46 | 7.6 ± 1.0 | 13.1 ± 1.7 |
TFAMWT, TFAMSSDD and TFAMK4Q4 can compact DNA comparably given sufficient protein concentration
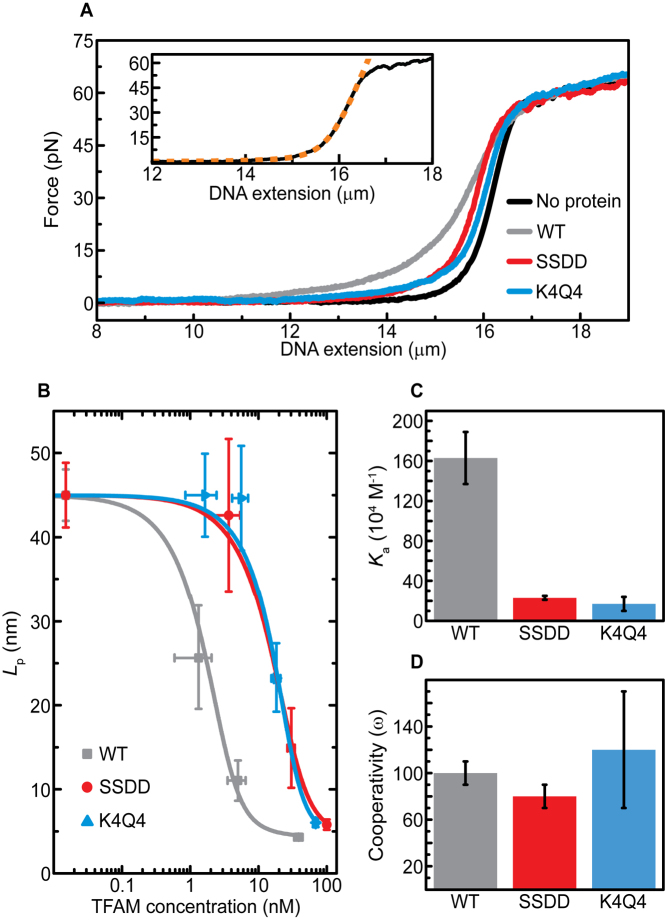
We have shown previously that TFAMWT binds to non-specific dsDNA sequences cooperatively and compacts the DNA molecule by increasing its flexibility (reflected in a decrease in the DNA’s persistence length, Lp, from ∼50 to ∼4 nm) (17). To investigate whether TFAMSSDD or TFAMK4Q4 can alter this mechanism of DNA organization compared to TFAMWT, we used optical tweezers to measure the force-extension behavior of individual dsDNA molecules (λ-phage; ∼48.5 kb) as a function of TFAM concentration. Sample force-distance (FD) curves, obtained under equilibrium conditions (Supplementary Figure S4), are highlighted in Figure 3A, with data recorded in a buffer of 10 mM Tris–HCl pH 7.6 with 25 mM NaCl. Note that the buffer conditions used here were chosen so as to provide a direct comparison with previous single-molecule studies of TFAMWT (17). Each measured FD curve was fit to the extensible worm-like chain (eWLC) model (Figure 3A inset), which describes the force-extension properties of dsDNA using three parameters: Lp, Lc (the contour length) and S (the stretch modulus). In this way, we extracted Lp as a function of protein concentration for TFAMWT, TFAMSSDD and TFAMK4Q4, respectively. The corresponding results are displayed in Figure 3B (solid points). From these data, a relatively steep decrease in Lp from ∼45 nm to ∼5 nm occurs as a function of both TFAMSSDD and TFAMK4Q4 concentration, analogous to that observed for TFAMWT here and reported previously (17). We thus conclude that, provided a sufficient protein concentration is present, TFAMWT, TFAMSSDD and TFAMK4Q4 can increase the flexibility of DNA to a comparable extent. Additionally, we resolve that TFAMSSDD and TFAMK4Q4 can each induce a similarly small increase in Lc as observed for TFAMWT, while the stretch modulus of DNA is unaffected by all three TFAM variants. Together, these results demonstrate that the phosphoserine and acetyl-lysine mimics can each compact sequence non-specific DNA through the flexible-hinge mechanism.
Figure 3.
TFAMWT, TFAMSSDD and TFAMK4Q4 can compact DNA comparably given sufficient protein concentration. (A) Sample FD curves for dsDNA in the absence of TFAM protein (black) and in the presence of 50 nM TFAMWT (gray), TFAMSSDD (red) and TFAMK4Q4 (blue), respectively. The inset shows an example FD curve (black, solid line) which is fitted (orange, dashed line) to the eWLC model. (B) Lp values (determined from eWLC fits to FD curves) for dsDNA as a function of increasing TFAM concentration. Gray, red and blue data points represent measured data for TFAMWT, TFAMSSDD and TFAMK4Q4, respectively. The solid lines correspond to McGhee von Hippel fits to the measured Lp values for each TFAM variant. (C) Equilibrium binding constant Ka for all three TFAM variants, determined from the fits in panel (B). (D) Cooperativity factor ω for all three TFAM variants, determined from the fits in panel (B). All data were obtained at room temperature and in a buffer of 10 mM Tris–HCl pH 7.6 with 25 mM NaCl. Errors are S.E.M.
Strikingly, however, a significantly higher concentration of either TFAMSSDD or TFAMK4Q4, relative to TFAMWT, is required to impose the observed maximum change in Lp (Figure 3B). Further, the sharp decrease in Lp as a function of protein concentration, in the case of all three TFAM variants, indicates that TFAM binds to dsDNA in a cooperative manner. Both the affinity and cooperativity of TFAM binding to DNA can be quantified by fitting the protein concentration dependence of Lp with the McGhee von Hippel model (Figure 3B, solid lines). In this model (Eq. S6), the binding of a protein to DNA is characterized by an equilibrium binding constant Ka (in M−1), a cooperativity factor ω and a footprint n. Using the footprint for TFAMWT on DNA as a fixed parameter (30 bp, as determined previously (17)), we freely fit Ka and ω for TFAMSSDD and TFAMK4Q4, as well as for TFAMWT. In the case of the phosphoserine and acetyl-lysine mimics, these fits indicate that Ka is ∼7–10 times lower than that of TFAMWT, while the binding cooperativity (ω) is similar to that of TFAMWT (∼100). These results are summarized in Figure 3C/D and Table 1. Note that the values of Ka and ω do not change significantly if a footprint of 22 bp, as has been suggested from crystallography studies (27,31), is used (Supplementary Table S2).
TFAMSSDD and TFAMK4Q4 show contrasting DNA on- and off-rates
The significant decrease in equilibrium DNA binding constant for both TFAMSSDD and TFAMK4Q4 compared with TFAMWT reflects changes in the on-rate (kon) and/or off-rate (koff) of the protein. To unravel the mechanisms underlying the reduced binding affinity of the phosphoserine and acetyl-lysine mimics, we quantified the off-rate for each TFAM species by means of combined optical tweezers, fluorescence microscopy and microfluidics. In these experiments, we incubated single dsDNA molecules in sufficient TFAM concentration so as to fully coat the DNA. Here, each TFAM variant was pre-labeled with an alexa-555 fluorescent dye in a 1:1 ratio. Once fully-coated with alexa-555-TFAM, we quickly transferred the DNA molecule into a microfluidic channel containing protein-free buffer and visualized the decrease in fluorescence intensity at a fixed (5 pN) force (Figure 4A). The decay in fluorescence intensity occurs due to unbinding of the fluorescently-labeled protein from the DNA and, to some extent, from bleaching of the dye. A sample fluorescence intensity decay curve for TFAMWT is shown in Figure 4B. Correcting for the bleaching (Supplementary Figure S6 and Eq. S8) yields the value of koff for TFAM–DNA complexes under the given buffer conditions.
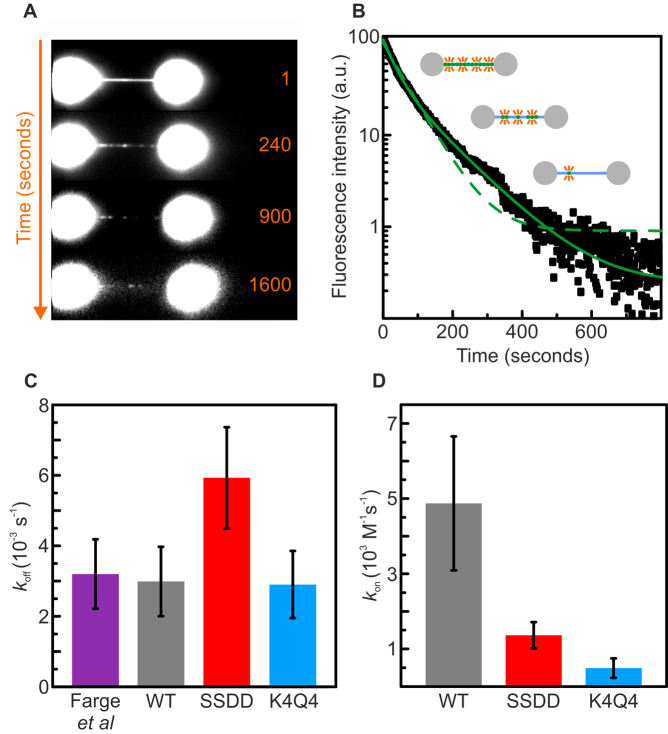
Figure 4.
TFAMSSDD dissociates from non-specific DNA faster than TFAMK4Q4 or TFAMWT. (A) Selected frames from a wide-field fluorescence movie recording the unbinding of alexa-555-labeled TFAMWT from dsDNA tethered between optically-trapped beads. The buffer contained no free TFAM. (B) Corresponding decay in fluorescence intensity for alexa-555-TFAMWT bound to a single dsDNA molecule in protein-free buffer. Fits to a bi-exponential and mono-exponential function (Supplementary Figure S5) are shown with solid and dashed green lines, respectively. (C) Histogram displaying the off-rates for TFAMWT (gray), TFAMSSDD (red) and TFAMK4Q4 (blue) extracted from mono-exponential fits to fluorescence intensity decay curves (Supplementary Methods). For reference, these data are compared with the value for TFAMWT reported previously by Farge et al. (17), also obtained using a mono-exponential fit (purple). (D) Histogram comparing the on-rates for TFAMWT (gray), TFAMSSDD (red) and TFAMK4Q4 (blue), calculated using the values for Ka and koff determined from Figures 3C and 4C, respectively. All data were obtained at room temperature and in a buffer of 10 mM Tris–HCl pH 7.6 with 25 mM NaCl. Errors are S.E.M.
It was reported previously that the unbinding of TFAMWT from DNA follows a mono-exponential decay, from which a decay time of ∼300 s can be extracted (17). This is recapitulated here in Figure 4C. Interestingly, our current analysis indicates that the unbinding of TFAM from DNA often deviates slightly from a pure mono-exponential decay and, in such cases, can be slightly better modeled as a bi-exponential decay (Figure 4B). This suggests that the DNA–TFAM disassembly process involves more than one unbinding mode (see Discussion for more details). More importantly in the current context, however, is the comparison of the overall off-rates for the three TFAM species. We found that the use of a bi-exponential function to quantify the fluorescence decay curves was highly sensitive to small variations in the experimental noise, making it difficult to provide a meaningful comparison between TFAMWT, TFAMSSDD and TFAMK4Q4 (Supplementary Methods). However, we note that a mono-exponential function remains a good approximation for the majority of the measured data, and also allows us to robustly compare the unbinding kinetics of TFAMWT, TFAMSSDD and TFAMK4Q4 to one another and directly to previous single-molecule analyses of TFAMWT (17). The results of this comparison, presented in Figure 4C, show that koff for TFAMSSDD is ∼2-fold higher than for both TFAMK4Q4 and TFAMWT (which exhibit an off-rate of ∼3 × 10−3 s−1). Since Ka = kon/koff, we can also calculate the on-rate for each TFAM variant, as highlighted in Figure 4D. In this way, we conclude that the decrease in Ka for TFAMK4Q4 (identified in Figure 3C) is primarily due to the reduction in kon, while in the case of TFAMSSDD, the decrease in Ka reflects both a ∼3.5-fold lower on-rate and a ∼2-fold higher off-rate, relative to TFAMWT. The values for Ka, kon and koff (obtained for λ-DNA in 10 mM Tris–HCl pH 7.6 with 25 mM NaCl) for the three TFAM variants are summarized in Table 1.
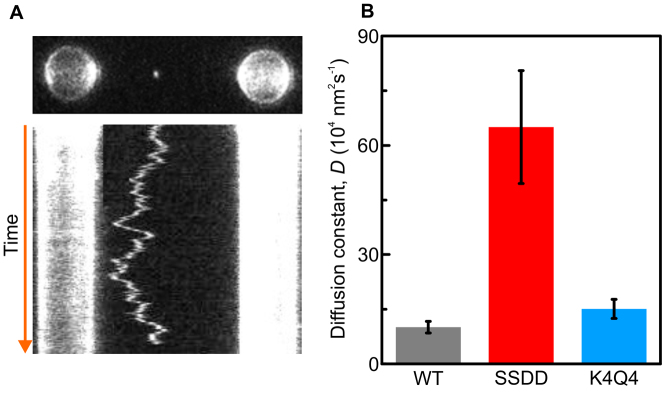
TFAMSSDD exhibits increased 1D diffusion
TFAMWT has been shown previously to exhibit relatively rapid 1D diffusion on dsDNA (17), whereby it moves randomly back and forth along the DNA with a measured diffusion constant of (8.6 ± 0.5) 104 nm2s−1. The diffusion of TFAMWT on DNA is salt-independent, indicating that it proceeds via a sliding, rather than a hoping or jumping mechanism (17). It has been suggested that this behavior facilitates cooperative binding of the protein and provides a mechanism to search for promoter sites on mtDNA (17,18). In light of this, we next set out to determine if the phosphoserine or acetyl-lysine mimics exhibit different 1D sliding dynamics on DNA relative to TFAMWT.
Using a combination of optical tweezers, fluorescence microscopy and microfluidics, monomers of alexa-555-labeled TFAM bound to dsDNA were visually tracked with nanometer precision in real time. Illustrative fluorescence images for TFAMWT on DNA are presented in Figure 5A. For all three TFAM variants, their motion on dsDNA is well described by a linear mean-squared displacement (MSD) as a function of time, consistent with free 1D diffusion (Supplementary Figure S7). A linear fit to such data provides the associated diffusion constant (D). As Figure 5B highlights, TFAMSSDD is found to diffuse ∼6.5-fold and ∼4.5-fold faster on dsDNA than TFAMWT and TFAMK4Q4, respectively. The faster diffusion on dsDNA, together with the increased unbinding rate (koff) measured for TFAMSSDD, relative to the other TFAM variants, suggests that phosphorylation results in a less stable binding of TFAM to DNA than in either wild-type or acetylated TFAM.
Figure 5.
TFAMSSDD diffuses on non-specific DNA significantly faster than TFAMWT and TFAMK4Q4. (A) Upper panel displays a selected frame from a fluorescence movie of alexa-555-labeled TFAMWT on dsDNA. Lower panel presents the corresponding kymograph, showing the diffusion of alexa-555-TFAMWT on dsDNA over time. (B) Histogram displaying the measured diffusion constants for TFAMWT (gray, N = 8), TFAMSSDD (red, N = 13) and TFAMK4Q4 (blue, N = 19) on dsDNA, determined from analysis of recorded kymographs for each protein variant. Data were obtained at room temperature and in a buffer of 10 mM Tris–HCl pH 7.6 with 25 mM NaCl. All errors are S.E.M.
DISCUSSION
In this study, we have explored the effects of phosphorylation and acetylation on the ability of TFAM to bind, compact and diffuse on non-specific DNA by analysing site-specific phosphoserine and acetyl-lysine mimics of TFAM using bulk (MST) and single-molecule (optical tweezers) methods. Our results are summarized in Table 1. Both approaches demonstrated that the DNA binding affinity of TFAMSSDD and TFAMK4Q4 is significantly lower than that of TFAMWT. In the case of TFAMWT, the DNA binding affinities determined here are consistent with those reported previously using the respective methods (17,22,28,31,45). We note that bulk and single-molecule studies derived different absolute binding affinity values for each species, independent of the ionic strength and buffer conditions used (Supplementary Figure S3 and Table S1). This difference is most likely attributed to the different DNA molecules employed. Single-molecule experiments used λ-DNA (∼48.5 kb), which permitted direct comparisons with previous optical tweezers studies of TFAMWT (17,25). In contrast, MST experiments employed short (∼20 bp) DNA oligonucleotides, as used for prior bulk kinetic studies of TFAMWT as well as for the crystal structures of TFAM with specific and non-specific DNA (22,27–29,31,45).
Previous work has shown that TFAM phosphorylation within HMG1 at serines 55/56, as well as the phosphoserine mimic TFAMSSDD, is associated with impaired DNA binding (35). However, those studies did not address whether HMG1 phosphorylation blocks TFAM binding to DNA, or whether it promotes TFAM release from DNA. In this study, our unique single-molecule approach enables us to distinguish between these possibilities, both in the case of TFAMSSDD and TFAMK4Q4. In this way, we reveal that the lower DNA binding affinity displayed by TFAMK4Q4 is largely explained by a reduced on-rate, while for TFAMSSDD it results from both a ∼3.5-fold lower on-rate and a ∼2-fold higher off-rate. Thus, these two PTMs induce a lower DNA binding affinity via distinct kinetic mechanisms. Phosphorylation of serine residues introduces a negative charge and adds steric bulk, which likely causes electrostatic repulsion with the phosphate backbone of DNA. Acetylation of lysine residues, meanwhile, neutralizes the positive charge of its side chains while also adding steric bulk, thereby disrupting the electrostatic interaction with the DNA backbone. Such PTMs can substantially reduce DNA binding affinity (46), and thus explain why the on-rate is reduced for both TFAMSSDD and TFAMK4Q4, relative to the wild-type protein. However, the greater electrostatic repulsion due to phosphorylation may account for the increased off-rate of TFAMSSDD, relative to that of both TFAMWT and TFAMK4Q4.
We note that our data provide evidence that the unbinding rate of TFAM from sequence non-specific DNA can sometimes deviate slightly from a mono-exponential decay (Figure 4B). This observation suggests that there are at least two intrinsic dissociation rates for TFAM–DNA disassembly. Previous work has suggested that TFAM binds to DNA as a monomer and forms dimers (31,47). Thus a possible explanation for the deviation from a mono-exponential decay is that the protein can dissociate from the DNA as either a monomer, dimer or even multimer. This might be expected given the cooperativity associated with TFAM–DNA binding shown here and previously (17). In support of this, the deviation from a mono-exponential function appears to be most apparent when the initial TFAM coverage on DNA is high (Supplementary Figure S5). The observed decay behavior could additionally reflect, in part at least, a longer timescale electrostatic binding component, as has been observed for other HMGB proteins (48).
TFAMWT has been shown to diffuse rapidly on dsDNA, and to form stable patches upon colliding (17). This likely enables TFAM to accomplish its key roles in compaction and transcription of the mitochondrial genome at the same time. Proteins can diffuse on dsDNA non-specifically by sliding parallel to the DNA axis, or by following the helical contour of the nucleic acid backbone (known as rotation-coupled diffusion). The maximum rate of rotation-coupled diffusion (due to Brownian motion) is thus intrinsically lower than for parallel diffusion. Since TFAM is thought to interact (in part at least) by intercalation into the minor groove of DNA (27,29,31), we anticipate that it exhibits rotation-coupled diffusion. Blainey et al. (49) have developed a model to predict the maximum speed for rotation-coupled diffusion, based on the size of the protein. With reference to this model, and by imaging the fluorescence from monomers of alexa-555-TFAM sliding along λ-DNA (Figure 5), we conclude that the diffusion constants for TFAMWT and TFAMK4Q4, while rapid, are both ∼6.5 and ∼4.5 times lower, respectively, than the maximum permitted for rotation-coupled diffusion. In contrast, the rate of TFAMSSDD diffusion is close to the limit for such behavior. Similar to its increase in off-rate, we relate the faster diffusion of TFAMSSDD on DNA to its weaker contact due to the presence of negative charges introduced at serine residues S55 and S56.
We envisage that enhanced sliding of phosphorylated TFAM on mtDNA provides a mechanism for fine-tuning its displacement during mtDNA replication, transcription and mt-nucleoid organization. Enhanced sliding may also facilitate promoter scanning. Here, one could imagine that phosphorylated TFAM activates transcription by de-compacting the mt-nucleoid, as well as accelerating TFAM binding at promoters. Further studies are required to determine whether phosphorylated TFAM interacts with non-phosphorylated TFAM to boost promoter scanning and transcription, and whether there are phosphatases that dephosphorylate TFAM, thereby providing another level of regulating mtDNA dynamics. It is also possible that phosphorylated TFAM or DNA-free TFAM is able to evade proteolysis (by mitochondrial Lon) by interacting with other proteins, or by additional post-translational modifications, which confer stability. This may explain recent findings showing that TFAM is stable in mice with a conditional cardiac knockout of mtRNAP, which have substantially depleted mtDNA (50).
The mechanisms by which TFAM can compact mtDNA have attracted much debate in recent years (17,22,26–32). Force-extension analysis of TFAMWT bound to dsDNA has shown that the protein decreases the DNA persistence length significantly (from ∼50 to ∼4 nm) (17). Meanwhile, application of high tensions (> 65 pN) fails to induce dissociation of the protein from the DNA (17). These observations are inconsistent with a mechanism of compaction exclusively involving the formation of loops or U-turns in the DNA (which would be expected to decrease the contour length and dissociate under high tension). Instead, the above observations have been attributed to TFAM-induced local melting of the DNA, described as the flexible-hinge mechanism and reminiscent of other non-histone architectural proteins (such as HU) (17,51,52). This does not imply that DNA looping or other mechanisms cannot additionally play a role in mtDNA compaction, particularly at lower tensions; however, it does indicate that the flexible-hinge mechanism is a fundamental part of the process. The current work shows that, importantly, the phosphoserine and acetyl-lysine mimics of TFAM do not alter this essential mechanism for DNA compaction. This is substantiated by our observations that TFAMWT, TFAMSSDD and TFAMK4Q4 all bind in an equally (positive) cooperative manner. While the phosphoserine and acetyl-lysine mimics studied here do not alter the mechanism of DNA compaction, they do change the number of TFAM proteins required to achieve a given extent of compaction. We thus propose that the lower DNA binding affinities associated with phosphorylated and acetylated TFAM may, together with Lon degradation of modified TFAM, orchestrate cycles of discrete or en masse TFAM–DNA unbinding in vivo, which can in turn tune the level of DNA compaction and reorganize the structure of the mitochondrial genome. Taken together, our findings indicate a mechanistic basis for how phosphorylation and acetylation of TFAM could be exploited in vivo to regulate its diverse and essential functions in mtDNA dynamics.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Géraldine Farge and Andreas Biebricher for helpful discussions and advice on protein labeling; Floor de Jong for assistance with preliminary experiments; I. Heller for the protein position tracking software; and H. Li and T. Liu at the Center for Advanced Proteomics Research, Rutgers—New Jersey Medical School. We are also grateful to Daniel Bogenhagen (Stony Brook University) for providing the rabbit polyclonal anti-TFAM antibody.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health (NIH) [NS046593] for the support of the Rutgers Mass Spectrometry Center for Integrative Neuroscience Research; An NIH grant [R01GM104231] (to D.T.); Nederlandse Organisatie voor Wetenschappelijk Onderzoek (NWO) through VIDI and STW-HTSM grants (to W.H.R.); New Jersey Health Foundation [PC105-13] and Dean's Biomedical Research Support Program, New Jersey Medical School (to C.K.S); A Human Frontier Science Program grant, an NWO VICI grant and a European Research Council Starting Independent Investigator grant (to G.J.L.W.). Funding for open access charge: Vrije Universiteit Amsterdam.
Conflict of interest statement. The optical tweezers and fluorescence technology used in this article are patented and licensed to LUMICKS B.V., in which G.J.L.W. has a financial interest.
REFERENCES
- 1. Iborra F.J., Kimura H., Cook P.R.. The functional organization of mitochondrial genomes in human cells. BMC Biol. 2004; 2:9–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Spelbrink J.N. Functional organization of mammalian mitochondrial DNA in nucleoids: history, recent developments, and future challenges. IUBMB Life. 2010; 62:19–32. [DOI] [PubMed] [Google Scholar]
- 3. Kukat C., Wurm C.A., Spåhr H., Falkenberg M., Larsson N.G., Jakobs S.. Super-resolution microscopy reveals that mammalian mitochondrial nucleoids have a uniform size and frequently contain a single copy of mtDNA. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:13534–13539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bogenhagen D.F., Rousseau D., Burke S.. The layered structure of human mitochondrial DNA nucleoids. J. Biol. Chem. 2008; 283:3665–3675. [DOI] [PubMed] [Google Scholar]
- 5. Wang Y., Bogenhagen D.F.. Human mitochondrial DNA nucleoids are linked to protein folding machinery and metabolic enzymes at the mitochondrial inner membrane. J. Biol. Chem. 2006; 281:25791–25802. [DOI] [PubMed] [Google Scholar]
- 6. Malarkey C.S., Churchill M.E.A.. The high mobility group box: the ultimate utility player of a cell. Trends Biochem. Sci. 2012; 37:553–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Murugesapillai D., McCauley M.J., Maher L.J. III, Williams M.C.. Single-molecule studies of high-mobility group B architectural DNA bending proteins. Biophys. Rev. 2017; 9:17–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dairaghi D.J., Shadel G.S., Clayton D.A.. Addition of a 29 residue carboxyl-terminal tail converts a simple HMG box-containing protein into a transcriptional activator. J. Mol. Biol. 1995; 249:11–28. [DOI] [PubMed] [Google Scholar]
- 9. Garstka H.L., Schmitt W.E., Schultz J., Sogl B., Silakowski B., Pérez-Martos A., Montoya J., Wiesner R.J.. Import of mitochondrial transcription factor A (TFAM) into rat liver mitochondria stimulates transcription of mitochondrial DNA. Nucleic. Acids. Res. 2003; 31:5039–5047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Asin-Cayuela J., Gustafsson C.M.. Mitochondrial transcription and its regulation in mammalian cells. Trends Biochem. Sci. 2007; 32:111–117. [DOI] [PubMed] [Google Scholar]
- 11. Shutt T.E., Lodeiro M.F., Cotney J., Cameron C.E., Shadel G.S.. Core human mitochondrial transcription apparatus is a regulated two-component system in vitro. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:12133–12138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Morozov Y.I., Parshin A.V., Agaronyan K., Cheung A.C.M., Anikin M., Cramer P., Temiakov D.. A model for transcription initiation in human mitochondria. Nucleic. Acids. Res. 2015; 43:3726–3735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hillen H.S., Morozov Y.I., Sarfallah A., Temiakov D., Cramer P.. Structural basis of mitochondrial transcription initiation. Cell. 2017; 171:1072–1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Falkenberg M., Gaspari M., Rantanen A., Trifunovic A., Larsson N.G., Gustafsson C.M.. Mitochondrial transcription factors B1 and B2 activate transcription of human mtDNA. Nat. Genet. 2002; 31:289–294. [DOI] [PubMed] [Google Scholar]
- 15. Sologub M., Litonin D., Anikin M., Mustaev A., Temiakov D.. TFB2 is a transient component of the catalytic site of the human mitochondrial RNA polymerase. Cell. 2009; 139:934–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Litonin D., Sologub M., Shi Y., Savkina M., Anikin M., Falkenberg M., Gustafsson C.M., Temiakov D.. Human mitochondrial transcription revisited: only TFAM and TFB2M are required for transcription of the mitochondrial genes in vitro. J. Biol. Chem. 2010; 285:18129–18133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Farge G., Laurens N., Broekmans O.D., van den Wildenberg S.M.J.L., Dekker L.C.M., Gaspari M., Gustafsson C.M., Peterman E.J.G., Falkenberg M., Wuite G.J.L.. Protein sliding and DNA denaturation are essential for DNA organization by human mitochondrial transcription factor A. Nat. Commun. 2012; 3:1013–1021. [DOI] [PubMed] [Google Scholar]
- 18. Traverso J.J., Manoranjan V.S., Bishop A.R., Rasmussen K.O., Voulgarakis N.K.. Allostery through protein-induced DNA bubbles. Sci. Rep. 2015; 5:9037–9042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wang Y.E., Marinov G.K., Wold B.J., Chan D.C.. Genome-wide analysis reveals coating of the mitochondrial genome by TFAM. PLoS One. 2013; 8:74513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ekstrand M.I., Falkenberg M., Rantanen A., Park C.B., Gaspari M., Hultenby K., Rustin P., Gustafsson C.M., Larsson N.G.. Mitochondrial transcription factor A regulates mtDNA copy number in mammals. Hum. Mol. Genet. 2004; 13:935–944. [DOI] [PubMed] [Google Scholar]
- 21. Kanki T., Ohgaki K., Gaspari M., Gustafsson C.M., Fukuoh A., Sasaki N., Hamasaki N., Kang D.. Architectural role of mitochondrial transcription factor A in maintenance of human mitochondrial DNA. Mol. Cell. Biol. 2004; 24:9823–9834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kaufman B.A., Durisic N., Mativetsky J.M., Costantino S., Hancock M.A., Grutter P., Shoubridge E.A.. The mitochondrial transcription factor TFAM coordinates the assembly of multiple DNA molecules into nucleoid-like structures. Mol. Biol. Cell. 2007; 18:3225–3236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kasashima K., Sumitani M., Endo H.. Human mitochondrial transcription factor A is required for the segregation of mitochondrial DNA in cultured cells. Exp. Cell Res. 2011; 317:210–220. [DOI] [PubMed] [Google Scholar]
- 24. Pohjoismäki J.L.O., Wanrooij S., Hyvärinen A.K., Goffart S., Holt I.J., Spelbrink J.N., Jacobs H.T.. Alterations to the expression level of mitochondrial transcription factor A, TFAM, modify the mode of mitochondrial DNA replication in cultured human cells. Nucleic Acids Res. 2006; 34:5815–5828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Farge G., Mehmedovic M., Baclayon M., van den Wildenberg S.M.J.L., Roos W.H., Gustafsson C.M., Wuite G.J.L., Falkenberg M.. In vitro-reconstituted nucleoids can block mitochondrial DNA replication and transcription. Cell Rep. 2014; 8:66–74. [DOI] [PubMed] [Google Scholar]
- 26. Uchida A., Murugesapillai D., Kastner M., Wang Y., Lodeiro M.F., Prabhakar S., Oliver G.V., Arnold J.J., Maher L.J. III, Williams M.C. et al. Unexpected sequences and structures of mtDNA required for efficient transcription from the first heavy-strand promoter. eLife. 2017; 6:e27283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Rubio-Cosials A., Sydow J.F., Jiménez-Menéndez N., Fernández-Millán P., Montoya J., Jacobs H.T., Coll M., Bernadó P., Solà M.. Human mitochondrial transcription factor A induces a U-turn structure in the light strand promoter. Nat. Struct. Mol. Biol. 2011; 18:1281–1289. [DOI] [PubMed] [Google Scholar]
- 28. Malarkey C.S., Bestwick M., Kuhlwilm J.E, Shadel G.S., Churchill M.E.. Transcriptional activation by mitochondrial transcription factor A involves preferential distortion of promoter DNA. Nucleic Acids Res. 2012; 40:614–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ngo H.B., Kaiser J.T., Chan D.C.. The mitochondrial transcription and packaging factor Tfam imposes a U-turn on mitochondrial DNA. Nat. Struct. Mol. Biol. 2011; 18:1290–1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kukat C., Larsson N.G.. mtDNA makes a U-turn for the mitochondrial nucleoid. Trends Cell Biol. 2013; 23:457–463. [DOI] [PubMed] [Google Scholar]
- 31. Ngo H.B., Lovely G.A., Phillips R., Chan D.C.. Distinct structural features of TFAM drive mitochondrial DNA packaging versus transcriptional activation. Nat. Commun. 2014; 5:3077–3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kukat C., Davies K.M., Wurm C.A., Spåhr H., Bonekamp N.A., Kühl I., Joos F., Polosa P.L., Park C.B., Posse V. et al. Cross-strand binding of TFAM to a single mtDNA molecule forms the mitochondrial nucleoid. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:11288–11293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Suarez J., Hu Y., Makino A., Fricovsky E., Wang H., Dillmann W.H.. Alterations in mitochondrial function and cytosolic calcium induced by hyperglycemia are restored by mitochondrial transcription factor A in cardiomyocytes. Am. J. Physiol. Cell Physiol. 2008; 295:1561–1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Dinardo M.M., Musicco C., Fracasso F., Milella F., Gadaleta M.N., Gadaleta G., Cantatorea P.. Acetylation and level of mitochondrial transcription factor A in several organs of young and old rats. Biochem. Biophys. Res. Commun. 2003; 301:187–191. [DOI] [PubMed] [Google Scholar]
- 35. Lu B., Lee J., Nie X., Li M., Morozov Y.I., Venkatesh S., Bogenhagen D.F., Temiakov D., Suzuki C.K.. Phosphorylation of human TFAM in mitochondria impairs DNA binding and promotes degradation by the AAA+ Lon protease. Mol. Cell. 2013; 49:121–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wang K.Z.Q., Zhu J., Dagda R.K., Uechi G., Cherra S.J. III, Gusdon A.M., Balasubramani M., Chu C.T.. ERK-mediated phosphorylation of TFAM downregulates mitochondrial transcription: implications for Parkinson's disease. Mitochondrion. 2014; 17:132–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wienken C.J., Baaske P., Rothbauer U., Braun D., Duhr S.. Protein-binding assays in biological liquids using microscale thermophoresis. Nat. Commun. 2010; 1:100–106. [DOI] [PubMed] [Google Scholar]
- 38. Biebricher A., Hirano S., Enzlin J.H., Wiechens N., Streicher W.W., Huttner D., Wang L.H-C., Nigg E.A., Owen-Hughes T., Ying Y. et al. PICH: a DNA translocase specially adapted for processing anaphase bridge DNA. Mol. Cell. 2013; 51:691–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Gross P., Farge G., Peterman E.J.P., Wuite G.J.L.. Combining optical tweezers, single-molecule fluorescence microscopy, and microfluidics for studies of DNA-protein interactions. Methods Enzymol. 2010; 475:427–453. [DOI] [PubMed] [Google Scholar]
- 40. Baron S., Caplanusi A., van de Ven M., Radu M., Despa S., Lambrichts I., Ameloot M., Steels P., Smets I.. Role of mitochondrial Na+ concentration, measured by CoroNa red, in the protection of metabolically inhibited MDCK cells. J. Am. Soc. Nephrol. 2005; 16:3490–3497. [DOI] [PubMed] [Google Scholar]
- 41. Azarias G., van de Ville D., Unser M., Chatton J.Y.. Spontaneous Na+ transients in individual mitochondria of intact astrocytes. Glia. 2008; 56:342–353. [DOI] [PubMed] [Google Scholar]
- 42. Broekmans O.D., King G.A., Stephens G.J., Wuite G.J.L.. DNA twist stability changes with magnesium(2+) concentration. Phys. Rev. Lett. 2016; 116:258102. [DOI] [PubMed] [Google Scholar]
- 43. McGhee J.D., von Hippel P.H.. Theoretical aspects of DNA-protein interactions: co-operative and non-co-operative binding of large ligands to a one-dimensional homogeneous lattice. J. Mol. Biol. 1974; 86:469–489. [DOI] [PubMed] [Google Scholar]
- 44. Kowalczykowski S.C., Paul L.S., Lonberg N., Newport J.W., McSwiggen J.A., von Hippel P.H.. Cooperative and noncooperative binding of protein ligands to nucleic acid lattices: experimental approaches to the determination of thermodynamic parameters. Biochemistry. 1986; 25:1226–1240. [DOI] [PubMed] [Google Scholar]
- 45. Wong T.S., Rajagopalan S., Freund S.M., Rutherford T.J., Andreeva A., Townsley F.M., Petrovich M., Fersht A.R.. Biophysical characterizations of human mitochondrial transcription factor A and its binding to tumor suppressor p53. Nucleic. Acids. Res. 2009; 37:6765–6783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Bowman G.D., Poirier M.G.. Post-translational modifications of histones that influence nucleosome dynamics. Chem. Rev. 2015; 115:2274–2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Gangelhoff T.A., Mungalachetty P.S., Nix J.C., Churchill M.E.A.. Structural analysis and DNA binding of the HMG domains of the human mitochondrial transcription factor A. Nucleic. Acids. Res. 2009; 37:3153–3164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. McCauley M.J., Rueter E.M., Rouzina I., Maher L.J. III, Williams M.C.. Single-molecule kinetics reveal microscopic mechanism by which High-Mobility Group proteins alter DNA flexibility. Nucleic. Acids. Res. 2013; 41:167–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Blainey P.C., Luo G., Kou S.C., Mangel W.F., Verdine G.L., Bagchi B., Xie X.S.. Nonspecifically bound proteins spin while diffusing along DNA. Nat. Struct. Mol. Biol. 2009; 16:1224–1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kühl I., Miranda M., Posse V., Milenkovic D., Mourier A., Siira S.J., Bonekamp N.A., Neumann U., Filipovska A., Polosa P.L. et al. POLRMT regulates the switch between replication primer formation and gene expression of mammalian mtDNA. Sci. Adv. 2016; 2:e1600963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Skoko D., Wong B., Johnson R.C., Marko J.F.. Micromechanical analysis of the binding of DNA-bending proteins HMGB1, NHP6A, and HU reveals their ability to form highly stable DNA-protein complexes. Biochemistry. 2004; 43:13867–13874. [DOI] [PubMed] [Google Scholar]
- 52. van Noort J., Verbrugge S., Goosen N., Dekker C., Dame R.T.. Dual architectural roles of HU: formation of flexible hinges and rigid filaments. Proc. Natl. Acad. Sci. U.S.A. 2004; 101:6969–6974. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.