Abstract
Many bacteria utilize simple DNA sequence repeats as a mechanism to randomly switch genes on and off. This process is called phase variation. Several phase-variable N6-adenine DNA-methyltransferases from Type III restriction-modification systems have been reported in bacterial pathogens. Random switching of DNA methyltransferases changes the global DNA methylation pattern, leading to changes in gene expression. These epigenetic regulatory systems are called phasevarions — phase-variable regulons. The extent of these phase-variable genes in the bacterial kingdom is unknown. Here, we interrogated a database of restriction-modification systems, REBASE, by searching for all simple DNA sequence repeats in mod genes that encode Type III N6-adenine DNA-methyltransferases. We report that 17.4% of Type III mod genes (662/3805) contain simple sequence repeats. Of these, only one-fifth have been previously identified. The newly discovered examples are widely distributed and include many examples in opportunistic pathogens as well as in environmental species. In many cases, multiple phasevarions exist in one genome, with examples of up to 4 independent phasevarions in some species. We found several new types of phase-variable mod genes, including the first example of a phase-variable methyltransferase in pathogenic Escherichia coli. Phasevarions are a common epigenetic regulation contingency strategy used by both pathogenic and non-pathogenic bacteria.
INTRODUCTION
Phase variation is the random, high frequency reversible switching of gene expression (1). The most common mechanism mediating phase variation of gene expression is slipped-strand mispairing of DNA that occurs in simple sequence repeats (SSRs) (1). Rates of phase variation mediated by SSRs are often several orders of magnitude greater than the base mutation rate (1). Many host-adapted bacterial pathogens contain phase-variable genes, and these often encode surface associated virulence factors that are subjected to periodic immune selection, such as iron acquisition systems (2,3), pili (4), adhesins (5,6) and lipooligosaccharide biosynthetic genes (7,8). Several bacterial pathogens also contain mod genes, encoding cytoplasmic Type III DNA methyltransferases, that exhibit phase-variable expression. In several human-adapted bacterial pathogens phase variation of these Type III DNA methyltransferases have been shown to alter the expression of multiple genes via global changes in DNA methylation (9–18). These systems are known as phasevarions (phase-variable regulon; Srikhanta et al. 2005). Phase-variable mod genes are highly conserved (>90% nucleotide sequence identity) in their 5′ and 3′ regions, but contain a highly variable central region encoding the Target Recognition Domain (TRD; also known as the DNA Recognition Domain) (19). The TRD is responsible for the sequence methylated by the Mod protein, with different TRD regions encoding different alleles of individual mod genes. Different TRDs mean different sequences are methylated, and consequently different alleles regulate different phasevarions. For example, modA, found in Haemophilus influenzae and the pathogenic Neisseria, has 21 allelic variants (9), modB from the pathogenic Neisseria has seven different alleles (18), and modH from Helicobacter pylori has 17 different alleles (14).
Phasevarion switching, controlled by on-off methyltransferase switching, differentiates the bacterial cell into two distinct phenotypic states. These states have altered virulence in animal and cell model systems of disease (20), altered expression of specific factors that are current and putative vaccine candidates (9), and altered resistance to antibiotics (9,21). The initial example of a phase-variably expressed mod gene was discovered in the first post-genomic era bioinformatics study (22) of the first genome of a free-living organism, H. influenzae KW20. In this study, all simple sequence repeats and potentially phase-variable genes in the KW20 genome were identified (Hood et al. 1996). All subsequent phase-variable mod genes were identified by examination of genome sequences for the presence of simple sequence repeats, or by identification of homologs to previously identified phase-variable methyltransferases (13,23).
Previous work studying the diversity of TRDs in bacterial pathogens has shown that horizontal transfer of TRDs drives the evolution of new methyltransferases (19), and that shuffling of TRDs within and between species is widespread (24). The diversity of TRDs found within Type III mod genes, and the high rate of horizontal gene transfer driving the evolution of new methyltransferase specificities has been well studied (24,25), but no study has yet investigated the extent of phase-variable Type III mod genes, which control phasevarions, within the bacterial domain.
Here, we present a systematic and comprehensive search for all phase-variable mod genes, by searching all possible combinations of simple sequence repeats in Type III restriction-modification systems annotated in the well-curated REBASE database of restriction-modification systems (26).
MATERIALS AND METHODS
We downloaded all 5603 mod genes with from REBASE (26) (http://rebase.neb.com/rebase/rebase.seqs.html) on 23 September 2016. After removing identical sequences, we obtained 3805 unique sequences. With a threshold of 80% nucleotide sequence identity, the genes were further divided into 2088 representative sequences using the program cd-hit (27). The list of 5603 mod genes, the 3805 unique sequences, and the subset of 2088 non-redundant representative genes (gene clusters) can be found in Supplementary Tables S1–S3, respectively. The 3805 unique sequences were then searched for simple sequence repeats by formulating all possible combinations of repeats of between one and nine repeating units, and searching each gene for these sequences. Phylogenetic analysis was carried out using the multiple sequence alignment program Muscle (28) and analyzed by RAxML (29).
Fragment length analysis of the STEC mod repeat tract
Primers were designed to anneal to conserved regions 5′ and 3′ of the CAGCGAC[n] repeat tract, with the forward primer containing a 6-carboxyfluorescein (FAM) label (STECmodF: 5′-FAM-CCAGTGAATTGAATTCATCAGAGC; STECmodR: 5′-CCGTTCCAGAACAAAGGAAATCC). PCR was carried out using genomic DNA prepared from an entire plate of the relevant STEC strain. PCR was carried out using GoTaq DNA polymerase (Promega) according to manufacturer's instructions. DNA fragment length analysis of the generated PCR products was carried out using the GeneScan system (Applied Biosystems International) by the Australian Genome Research Facility (AGRF, Brisbane, Australia).
Cloning and over-expression of the Shiga toxin-producing Escherichia coli (STEC) Type III mod
PCR products generated for cloning into the PmlI/XhoI site of pET46 cloning vector (EMD Millipore) were prepared using KOD Hot-start DNA polymerase (EMD Millipore) according to manufacturer's instructions, using STEC strain DG131/3 genomic DNA. Primers specific for the Type III locus ECSTECDG1313_5492, encoding M.Eco1313ORF5492P were designed to clone the gene containing no repeats so as to lock on the methyltransferase expression, and maintain the enterokinase cleavage site between the 6xHis-tag and the start of the gene. (forward—5′-AGTCAG CACGTGGATGATGATGATAAGACTGAATTAATTCGGGAACTGAATTCTG-3′; reverse—5′-AGTCAGCTCGAGTTACCATTGTTTTTCCGCTCCAGTGG). PmlI and Xho sites are highlighted in bold text. The sialylytransferase siaB was cloned into pET46 and expressed to serve as a non-methylating control sample as described previously (9). Over-expression of each protein was carried out using E. coli BL21 cells, which were induced by the addition of IPTG to a final concentration of 0.5 mM for 2 h at 37°C with shaking at 120 rpm.
Single-molecule, real-time (SMRT) sequencing and methylome analysis
Plasmid midi-preps from E. coli cells expressing STEC mod methyltransferase and the negative control expressing a non-methyltransferase (SiaB), were prepared using the Qiagen plasmid midi kit according to the manufacturer's instructions. SMRT sequencing and methylome analysis was carried out as previously (30,31). Briefly, DNA was sheared to an average length of approximately 5–10 kb using g-TUBEs (Covaris; Woburn, MA, USA) and SMRTbell template sequencing libraries were prepared using sheared DNA. DNA was end repaired, then ligated to hairpin adapters. Incompletely formed SMRTbell templates were degraded with a combination of Exonuclease III (New England Biolabs; Ipswich, MA, USA) and Exonuclease VII (USB; Cleveland, OH, USA). Primer was annealed and samples were sequenced on the PacBio RS II (Menlo Park, CA, USA) using standard protocols for long insert libraries. SMRT sequencing and methylome analysis was carried out by the Yale Centre for Genomic Analysis (YCGA; CT, USA).
RESULTS
In order to identify all phase-variable Type III mod genes, we searched the well curated REBASE database (26) for simple sequence repeats. All 3805 unique gene sequences were examined for repeat tracts of DNA. By searching the 3805 unique mod gene sequences found in REBASE we found 1806 genes containing at least one SSR tract. We only selected mod genes containing repeat tracts of a length that have previously been shown to lead to high rates of phase variation of the gene containing them (32). For example, mononucleotide SSR tracts of nine bases in length have been shown to phase vary at rates of 1.8–13.5 × 10−3 (33); a tetranucleotide repeat tract consisting of just three repeat units in length phase-varied at rates of between 0.5 and 2.0 × 10−6 (34). We therefore only defined a mod gene ‘phase-variable’ if the repeat tract it contained was nine bases long for mononucleotide repeat tracts (e.g. G[9]), five repeats long for dinucleotide repeats (e.g. GA[5]), and three repeat units long for repeats of three bases or longer (e.g. TCG[3], AGCC[3], etc).
By applying these strict criteria, we concluded that 662 out of 1806 of these genes are potentially phase-variable. By clustering genes with >80% identity together, we demonstrate that there are 176 unique phase-variable mod genes currently annotated in REBASE. The list of all 662 mod genes containing phase-variable SSRs, and the 176 unique phase-variable representative mod genes are presented in Supplementary Tables S4 and S5, respectively.
Phase-variable mod genes are present in a broad range of bacterial species
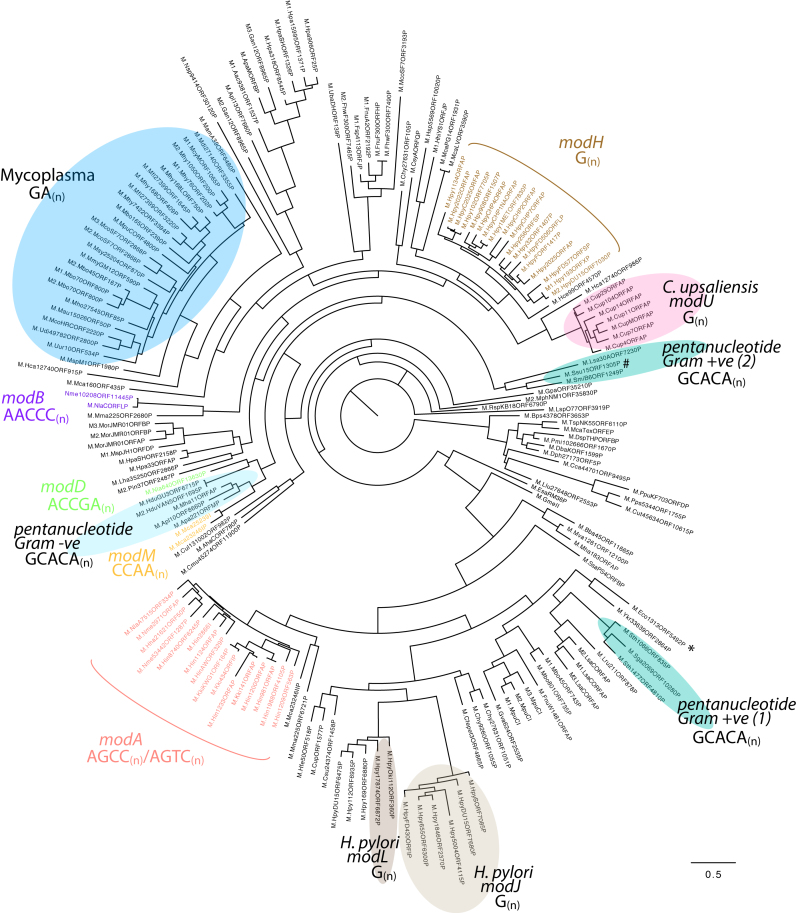
All 176 unique repeat-containing representative mod genes were aligned by using the multiple sequence alignment program Muscle (28). The resulting phylogenetic tree is shown in Figure 1. Many phase-variable mod genes have already been identified and demonstrated to control phasevarions, such as modA (9,12,15), modB (12), modD (17), modH (14) and modM (10,16). However, these known phase-variable mod genes comprise only a small proportion (39/176; 22%) of the total set of 176 representative mod genes containing repeat tracts. In addition to these well characterized mod gene groups, our phylogenetic analysis (Figure 1) reveals a highly diverse distribution of mod genes containing SSRs, in terms of the sequence of repeat tracts present, repeat tract length, and in the species that contain these phase-variable mod genes (See Supplementary Tables S4 and S5). Genes encoding potentially phase-variable mod genes are present in a diverse range of bacterial species and genera including human-adapted pathogens (groups of related mod genes have already described, e.g. modA group, found in non-typeable Haemophilus influenzae (NTHi), Neisserial spp. etc, as well as uncharacterized new groups; see below), opportunistic pathogens (e.g. Burkholderia pseudomallei [M.Bps4378ORF3653P], Fusobacterium nucleatum [M1.FnuA2ORF2192P]), commensal organisms (e.g. Gallibacterium anatis [M2.Gan12ORF8965P], Aggregatibacter actinomycetemcomitans [M1.Aac9381ORF1537P]) and environmental organisms (e.g. Clostridium thermocellum [M.RspKB18ORF6790P], Corynebacterium casei [M.Cca44701ORF9495P], and Thermosynechococcus sp. [M.TspNK55ORF6110P]). The presence of SSRs in mod genes found in environmental organisms and opportunistic pathogens is interesting as it has previously been reported that genes containing SSRs are generally restricted to small-genome, host-adapted pathogens (35).
Figure 1.
Phylogeny of the 176 representative phase-variable mod genes Sequences were aligned using Muscle (28), and phylogeny analyzed by RAxML (29). Groups highlighted with colored circles are new mod gene groups discussed in the main text, with the bacterial genera or strain, and the SSR unit, also stated. Groups with colored text represent currently described phase-variable mod genes/mod gene groups, e.g.modA, modB, etc., with the SSR unit also stated. The phase-variable mod gene from STEC strain DG131/3 is highlighted with a *; the phase-variable mod gene from S. suis strain T15 is highlighted with a # in the pentanucleotide Gram +ve (2) group.
New examples of phase-variable mod genes are present in species that contain already well-characterized phasevarions
The human adapted gastric pathogen H. pylori has previously been shown to contain the modH gene that contains 17 different alleles, and which phase-varies through changes in the length of a mononucleotide G[n] tract (14). Figure 1 shows that H. pylori contains two new phase-variable Type III mod gene groups that we propose to name modJ (mononucleotide G[n] tract; six alleles) and modL (mononucleotide G[n] tract; two alleles). The identification of these two new potentially phase-variable mod gene groups, each with multiple allelic variants, means individual strains of H. pylori can potentially contain up to three independently switching methyltransferases, each controlling a different phasevarion. This is similar to the situation in Neisseria meningitidis, which contains the modA, modB and modD genes (18), and can have major implications on vaccine development and antibiotic resistance. A phylogenetic analysis of these three mod genes, and their distribution in H. pylori strains is shown in Supplementary Figure S1.
A pentanucleotide repeat tract exists in mod genes present in a diverse group of Gram-negative and Gram-positive organisms
A pentanucleotide repeat tract GCACA[n] was identified in several unrelated mod genes present in a range of both environmental and pathogenic organisms, spread thoughout the bacterial domain. Analysis of these mod genes shows that they form three distinct clades (Figure 1; Supplementary Figure S2). Of particular note was the presence of Gram-positive organisms in this group. Although the presence of a potential phase-variable mod gene in Streptococcus thermophilus was previously noted (13), here we have identified that variation in the numbers of repeats occurs in this gene (between four and nine repeat units in strains of S. thermophilus identified), providing evidence of phase-variation. Phylogeny shows that two different alleles of the S. thermophilus mod gene are present in the sequences analyzed, and this group also contains a phase-variable mod gene from the releated species Streptococcus gallolyticus, a ruminant commensal that is linked with endocarditis and cancer in humans. We also observe phase-variable mod genes in several other Gram-positive species, including the human oral commensal Streptococcus mitis (GCACA[32]), and the human intestinal commensal Lactobacillus saerimneri (GCACA[21]). Our phylogenetic analysis shows these mod genes are also distinct genes, not different alleles of the same gene (Supplementary Figure S2). Interestingly, in our analysis of all mod genes containing a GCACA[n] repeat (Supplementary Figure S2) the S. mitis mod clusters with a the mod from L. saerimneri, and not with those from more closely related species S. thermophilus and S. gallolyticus.
Several mod genes containing a GCACA[n] repeat tract are also found in a wide variety of Gram-negative organisms, all within the Pasteurellacaea: we observe GCACA[n] repeat tract lengths of between 4 and 30 repeat units in a mod gene in Mannheimia haemolytica, a major bovine pathogen; 4 and 19 repeat units in a mod gene in Actinobacillus pleuropneumoniae, a swine respiratory pathogen; 4 and 48 repeat units in a mod gene in Haemophilus ducreyi, the cause of canker sores in humans; and a mod gene containing a GCACA[10] repeat tract in a mod gene in Avibacterium paragallinarum, a major chicken pathogen. Phylogenetic analysis (Supplementary Figure S2) shows these phase-variable mod genes are all distinct genes, but contain the GCACA[n] repeat tract, and conserved functional motifs (e.g. the DPPY catalytic and FXGXG substrate binding motifs) in the same or very similar places in each gene (Supplementary Figure S2; Figure 4). The same is true of the examples in the Gram-positive bacterial species containing a GCACA[n] repeat tract, which could imply two distinct evolutionary events – one in Gram-positive species, and one in Gram-negative species, followed by horizontal transfer and diversification. We also noted the presence of a phase-variable Type III mod gene in the Gram-positive swine pathogen Streptococcus suis (M.Ssu15ORF1305P-5) containing a CAGAG[23] repeat tract (Figure 1; Supplementary Table S1), although this is represented by just this single example. Never the less, the presence of another uncharacterized example of a mod gene containing SSRs in a Gram-positive organism indicates that this method of gene regulation is widespread in the bacterial domain.
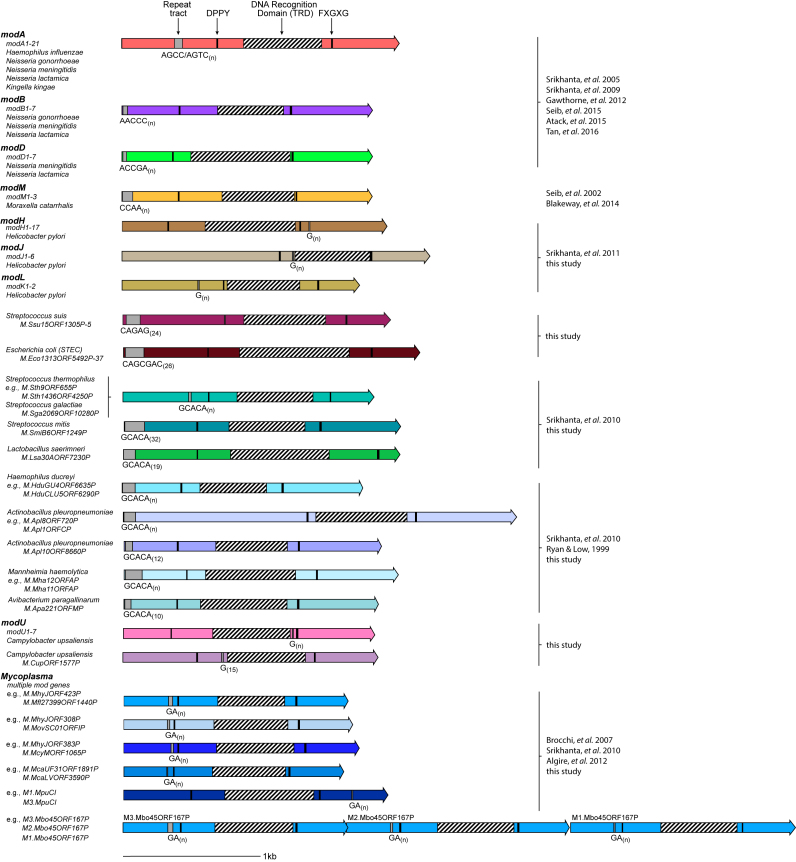
Figure 4.
Summary of a selection of representative phase-variable mod genes found in a variety of bacterial species. Individual mod genes are represented by colored arrows, with different colors indicating different, distinct mod genes/mod gene groups. The location of the repeat tract is shown by a grey box underscored with the repeating unit found in these tracts if just a single example is present. An underscore [n] indicates that multiple examples of this mod gene are present (e.g. there are 21 different alleles in the modA group, 7 alleles in the modB group), with a range of repeat tract lengths present in these examples.
A new mod gene with multiple alleles is present in Campylobacter upsaliensis
Campylobacter infection in humans can be caused by C. jejuni and C. upsaliensis (36), which are commensals of chickens and domestic animals, respectively. Our search for mod genes with SSRs identified a mononucleotide G[n] tract present in a mod gene present in several strains of C. upsaliensis. Analysis of these sequences revealed a single mod gene group, containing multiple allelic variants. We propose to name this new group modU, in line with other characterized methyltransferases that contain SSRs and exhibit phase-variable expression. Seven different alleles are present in the seqeunces surveyed (modU1–7; Supplementary Figure S3; Figures 1 and 4). Our phylogenetic analysis showed high identity level (>90%) in the conserved 5′ and 3′ regions, with a highly divergent central TRD. In previously characterized examples, a different TRD in a conserved backbone is classified as a different allele of an individual mod gene (19). The modU group of methyltransferases is closely releated to the already characterized modH group (Figure 1), found in the releated epsilon proteobacteria H. pylori. In addition to modU, our phylogenetic analysis also revealed a single example of a second phase-variable mod gene (M.CupORF1577P) that is divergent and un-related to modU. The mod gene encoding M.CupORF1577P also contains a mononucleotide G[n] tract, but it is located in a different part of the gene compared to modU, and the overall gene shows low identity (<50% nucleotide identity) to modU. M.CupORF1577P clusters closely with the newly characterized modJ and modL groups from H. pylori (Figure 1), which could imply that modU and modH have a common ancestor, and M.CupORF1577P shares a common progenitor with modJ/modL.
A large diverse group of phase-variable mod genes are present in the Mycoplasmataceae
Mycoplasmas and Ureaplasmas are small genome intracellular pathogens, responsible for a number of diseases in mammals, including pneumonia in humans (M. pneumoniae), cattle (M. bovis) and pigs (M. hyopneumoniae), pelvic inflammatory disease (M. genitalium) and urethritis (Ureaplasma) (37). The mod genes containing a dinucleotide GA[n] repeat tract have been identified in Mycoplasmas previously (13,38). They have never been shown to be phase-variably expressed, but have been discussed as potential phase-variable regulators (39). Our analysis of phase-variable mod genes from the Mycoplasmataceae shows the presence of 12 new genes, with many of the genes containing multiple allelic variants (Supplementary Figure 4). GA[n] repeat tract lengths vary from 5–25 repeat units. This group is dominated by the Mycoplasmas, with some strains containing multiple phase-variable mod genes, often located close together on the chromosome: for example M. bovis strain PG45 (ATCC 25523; accession number CP002188) (40) contains three genes all containing GA[n] repeat tracts, and all annotated as encoding separate mod genes. These three genes are co-localized on the chromosome (M1.Mbo45ORF167P = MBOVPG45_0168; M2.Mbo45ORF167P = MBOVPG45_0169; M3.Mbo45ORF167P = MBOVPG45_0170). All three of these genes contain the conserved DPPY and FXGXG Type III methyltransferase motifs required for function (41). Sequence analysis shows conserved 5′and 3′ domains, with a central, variable TRD (data not shown). Thus, these three mod genes in M. bovis are three different alleles of the same mod gene, suggesting that all will methylate a different target sequence. All contain varying numbers of GA[n] repeats, but with these SSRs located in the same part of the gene. Therefore, it is tempting to speculate that these three alleles resulted from gene duplication of a single phase-variable mod gene, then diversified through acquisition of different TRDs. This process has been shown to occur in the human pathogen non-typeable H. influenzae which contains 21 modA alleles (9,19). Some Mycoplasmas also contain multiple, separately located mod genes: for example, Mycoplasma hyopneuomiae strain J (ATCC 25934; accession number AE017243.1) contains four different GA[n] repeat tract containing mod genes (M.MhyJORF308P = MHJ_0308; M.MhyJORF383P = MHJ_0383; M.MhyJORF399P = MHJ_0399; M.MhyJORF423P = MHJ_0423). ORF399P and ORF423P appear to be different allelic variants of the same mod gene, as their 5′ and 3′ ends are highly conserved with a highly variable central TRD. The two remaining sequences, ORF308P and ORF383P are separate genes, clustering in different groups from each other and from the group containing ORF399P and ORF423P (Supplementary Figure S4). Thus, M. hyopneumoniae strain J contains four different phase-variable methyltransferases—two allelic variants of the same gene, and two additional, distinct mod genes.
A phasevariable mod gene is present in a sub-set of Shiga-toxin producing E. coli (STEC)
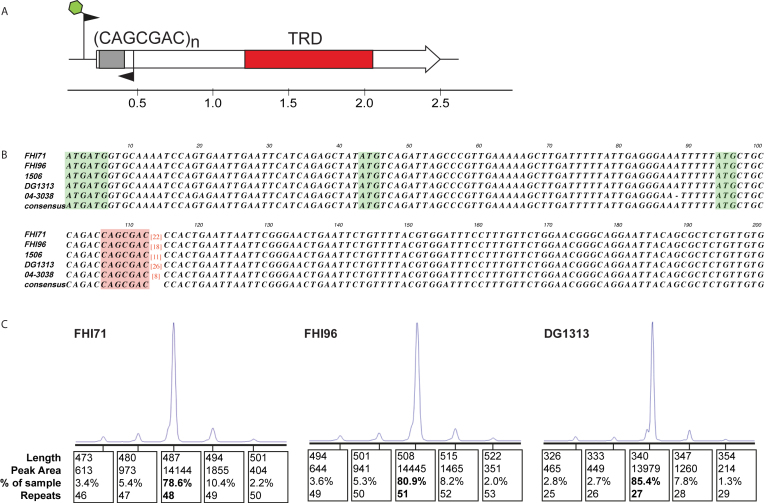
Our examination of all Type III mod genes containing SSRs revealed a single example of a phase-variable mod gene in Shiga-toxin producing E. coli (STEC) strain DG131/3. This gene, ECSTECDG1313_5492, encodes a methyltransferase (M.Eco1313ORF5492P), and contains a CAGCGAC[26] repeat in its annotated open reading frame. In order to elucidate the methyltransferase specificity of this novel phase-variable Type III mod, we cloned and over-expressed this enzyme without the CAGCGAC[26] repeat tract in order to prevent any phase-variable expression of the gene, in E. coli strain BL21, and carried out SMRT sequencing and methylome analysis using the plasmid vector as described previously (9,30). Methylome analysis demonstated that the methylated motif, 5′-GCCm6ATC, was only present when the methyltransferase M.Eco1313ORF5492P was expressed. In accordance with restriction-modification system naming conventions (26), we propose that the protein encoded by gene ECSTECDG1313_5492 be named M.Eco131I. A search of REBASE revealed this methylated motif has not currently been described in E. coli. A BLAST analysis of the NCBI whole genome shotgun (WGS) database using the ECSTECDG1313_5492 reveals this locus is present in a subset of STEC strains (Figures 1 and 2; Supplementary Table S6), with variable numbers of CAGCGAC[n] repeats present in each strain. An alignment of representative sequences from GenBank shows a common sequence containing a variable length CAGCGAC[n] tract (Figure 2B). Fragment length analysis using genomic DNA prepared from a population of three of these STEC strains—FHI71, FHI96, and DG1313—shows that the population for these strains contain individuals with variable numbers of CAGCGAC[n] repeats (Figure 2C). This approach has been used previously with phase-variable mod genes to demonstrate variable tract lengths within a bacterial population, and in every case is highly demonstrative of phase-variable expression of the encoded methyltransferase (9–12,14,17). Interestingly our results demonstrate that repeat tract length from fragment length analysis does not necessarily match that of the annotated sequence deposited in GenBank, with the annotated sequence always shorter than that demonstrated by fragment length analysis. This highlights the limitations of short-read sequencing and subsequent assembly, where long tracts of repetitive sequence appear to be ‘collapsed’ to the shortest possible length.
Figure 2.
Phase-variable mod gene present in a subset of Shiga Toxin producing Escherichia coli. (A) schematic representation of the phase-variable mod gene from STEC, with location of the CAGCGAC[n] repeat tract shown, and probable location of the central TRD (red), and location of PCR primers used for fragment length analysis of the CAGCGAC[n] repeat tract. The green hexagon on the forward primer depicts a 6-Fluoresceine (FAM) fluorescent label to allow analysis of PCR products using the GeneScan system (Applied Biosystems International); (B) alignment of representative phase-variable mod genes from five STEC strains showing variable repeat tract length contained within the conserved genomic sequence. SSR tract is highlighted in red, with actual number of CAGCGAC repeats for each strain in red. Putative start codons are highlighted in green. The start codon immediately upstream of the CAGCGAC[n] repeat tract contains a putative Shine-Dalgarno ribosome binding site immediately upstream (GAGGGAA) and (C) fragment length analysis of the mod repeat tract from three strains of STEC showing variable repeat tract lengths are present within the population of these individual strains.
Expansion of repeat units found in mod genes only occurs if this leads to variable expression of the encoded methyltransferase
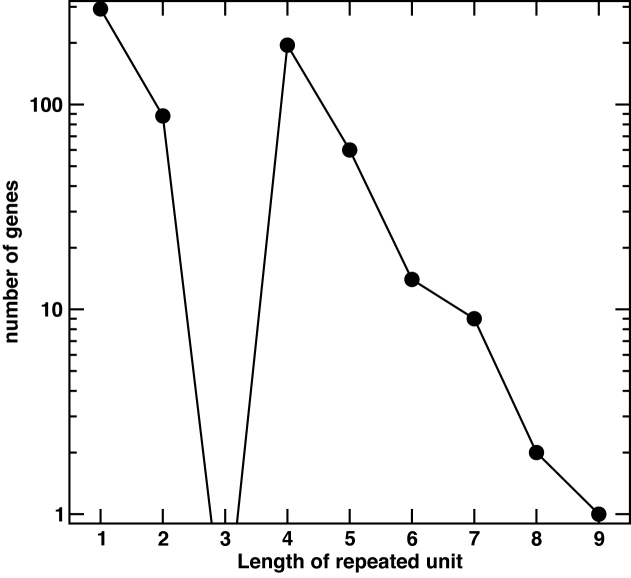
Our search of 3805 unique gene sequences showed that 662 genes contained SSRs that were composed of individual repeating units of between one and nine nucleotides within the mod open reading frame. Interestingly, we did not observe any SSR tracts in mod open reading frames of over three units of length where the repeating unit is a trimer, a hexamer is represented only 14 times, and a nonamer represented just once (Figure 3). Slipped strand mispairing leads to insertion or deletion of single repeat units during DNA replication (1), meaning a deletion or insertion of multiples of three bases would not lead to a frameshift if the repeats are in the open reading frame of the gene. As a result, loss or addition of units in these repeat tracts would not lead to phase variation of the gene (35), with less selective pressure likely exerted against expansion of tracts of this length as they do not lead to frameshifts and loss of expression (42). However, our findings demonstrate a low abundance at tri-, hexa- and nona- repeat tracts in open reading frames. These data support a hypothesis that selection and expansion of SSR tracts only occurs in those genes that would lead to phase-variable switching of mod expression; SSRs that are a multiple of 3 would not alter the reading frame and generate frame shift mutations.
Figure 3.
A comparison of the frequency of occurrence of SSR tracts containing different repeat unit lengths. All phase-variable repeat tracts from the 662 individual mod gene set (Supplementary Table S4) were analyzed for the composition of the SSR unit (one to nine nucleotide long repeating units) and this plotted against the frequency by which tracts containing those repeat units occur.
DISCUSSION
This is the first time, to our knowledge, that a systematic study has been carried out to identify mod genes that contain simple seqeunce repeats capable of mediating phase-variable expression, and thereby comprise phasevarions (phase-variable regulons). Our analysis shows that currently characterized phasevarion-controlling mod gene groups (modA, modB, modD, modH, modM) are just a small proportion (∼22%) of the now known phase-variable mod genes. Several newly identified phase-variable mod genes are present in host-adapted bacterial pathogens. These mod genes contain multiple alleles and variable repeat tract lengths, highly indicative of them controlling phasevarions (modJ and modL in H. pylori; modU in C. upsaliensis, and the large and diverse set of mod genes found in the Mycoplasmataceae). We have named these new mod gene groups in line with our previous studies (Haemophilus influenzae and Neisserial spp., modA (9); Neisserial spp., modB (12); Neisserial spp., modD (17); Helicobacter pylori, modH (14) and Moraxella catarrhalis, modM (10)) so that we can organize the information describing the distribution. The names of individual mod genes are always linked to the original genome annotation (e.g. the gene encoding the modA allelic variant modA2 in NTHi strain 723 is designated NTHI723_00580), and the naming of the encoded enzyme is determined by standard Restriction-Modification system naming conventions ((26); e.g. modA allelic variant modA2 encodes M.Hin723I). These genes are further organized into distinct mod gene groups (e.g. modA, modB, etc.) if they satisfy the criteria of being highly conserved (>90% identity) at their 5′ and 3′ regions. Within these mod gene groups, differences in the central TRD that result in an altered methylation target site, define distinct alleles within the group (e.g. ModA2/M.Hin723I methylates 5′-CCGAm6A; ModA5/M.Hin477I methylates 5′-ACm6AGC (9)). Thus, allocating individual genes into wider phasevarion related mod gene and allele classifications facilitates the study of their distribution, evolution, horizontal transfer, and relationship to bacterial virulence.
Every case where a mod gene has been identified containing varying repeat tract lengths and consisting of multiple allelic variants, this group has subsequently been shown to control phasevarions (9–12,14–16). In addition to confirming these previous findings, we also discovered phase-variable Type III mod genes present in important bacterial pathogens, including a phase-variable mod gene in Shiga-Toxin producing E. coli, which is responsible for food-poisoning that can develop into hemolytic uremic syndrome. This is the first example of a phase-variable methyltransferase identified in E. coli, and its association with only a subset of STEC isolates has intriguing implications for STEC pathobiology; a second example of a new phase-variable mod gene was observed in S. suis, a major swine pathogen and cause of zoonotic meningitis in humans. The importance of these pathogens certainly merits further investigation as to the wider presence of phasevarions in these species – in every previous example, phase-variable Type III mod genes control expression of a phasevarion, whose members include current and putative vaccine candidates and virulence factors.
As well as identifying new mod genes in a range of host-adapted bacterial pathogens, our phylogenetic analysis demonstrates that many opportunistic human and animal pathogens, vertebrate commensals, and environmental bacterial species contain phase-variable mod genes. In many of these environmental organisms and opportunistic pathogens we observe the presence of only a single, distinct phase-variable mod gene without diversification into multiple alleles. This situation could imply that there is less selective pressure to generate phenotypic diversity in these organisms (all of which have larger genomes than organisms that contain mod genes with multiple alleles) as they exist in a more predictable environment and use the conventional ‘sense and respond’ gene regulation paradigm of adaptability; i.e., these organisms contain many more two-component sensor-regulator pairs than small genome pathogens that contain phase-variable mod genes (35,43). This study also begs the question of why phasevarions exist in environmental organisms. They appear to have evolved multiple times in these organisms based on our phylogenetic analysis presented in Figure 1. However, it remains to be shown what classes of genes they regulate, and how they provide advantages to adaptation to changing environmental selective pressures that cannot be dealt with via conventional ‘sense and respond’ gene regulation strategies. Increased phenotypic diversity is an obvious advantage of phasevarions, particularly in small genome pathogens; perhaps this increased diversity provides an extra advantage in adaptation to variable environmental conditions that many of these organisms may encounter, or that cannot be sensed by conventional means, thereby requiring a contingency regulation strategy (1).
Our analysis leads us to speculate that phase-variable mod genes, and by implication phasevarions, have evolved independently at least twenty-five times. This is based on the type of repeat unit and its distinct location in each mod gene (Figure 4). We theorise that the first step towards phasevarion acquisition is the expansion of a repeat tract in a mod gene, followed by diversification into multiple allelic forms through acquisition of new TRDs. This is based on analysis of the newly identified mod groups containing multiple alleles in C. upsaliensis, the Mycoplasmataceae, and the diverse group containing a GCACA[n] repeat tract. Many of the examples of organims with a phase-variable mod gene containing a GCACA[n] repeat tract are represented by only a single allelic variant, implying that the mod gene in these organisms may have become phase-variable later in the evolutionary timeline than those mod genes represented by multiple allelic variants. For example, C. upsaliensis and the Mycoplasmataceae contain multiple allelic variants of newly identified mod genes, meaning they acquired the ability to phase-vary earlier. Previous work has demonstrated that new alleles arise by acquisition and incorporation of new sequences by horizontal gene transfer between different strains and species over time (19,24,25). Therefore the greater allelic diversity of single mod genes seen in the newly identified examples in C. upsaliensis and the Mycoplasmataceae, and in existing examples such as modA in NTHi, implies that these mod genes/gene groups have gained the ability to phase-vary earlier in the evolution of these species. Whilst previous work has focused on the evolution of methyltransferase variability (24), the current study is focused on the prevalence of SSRs within these genes, leading to phase-variable expression and therefore the presence of phasevarions in bacteria. Thus, unlike previous studies, we considered the entire mod gene as a single unit, and considered sequence similarity over the entire protein in our phylogenetic analysis, rather than separating the TRD and non-TRD regions as different entities (24). As with our previous work (19), this has led to clustering of mod genes together as a single branch/group in our phylogenetic analysis (Figure 1).
The selection and expansion of a GCACA[n] repeat unit in multiple mod genes in different locations within these genes indicates that this combination of bases may be a common progenitor sequence that is particularly prone to expansion and selection. Theoretical work using the tetra-nucleotide repeat tract AGTC[n] found in the modA1 gene of H. influenzae (44) suggests that broad conditions favor the evolution of phase-variable genes, such as the amount of time that pressures exist to produce either on or off variants, and if both states exist for long enough to provide a selective advantage (44).
Our estimate that almost one-fifth of all Type III mod genes (662/3805 = 17.4%) are phase-variable is likely to be an underestimate for a number of reasons. Our strict selection criteria for what makes a gene phase-variable likely excludes examples of phase-variable genes with short mono- and di-nucleotide repeat tracts (repeat tracts of less than nine nucleotides long for mono-nucleotide repeat tracts, and less than five repeat units long for di-nucleotide repeat tracts). For example, mon-nucleotide repeat tracts of seven and eight units in length have been shown to phase vary at rates of at least 0.65 × 10−3 in C. jejuni (33) and a repeat tract of G[7] leads to phase variation of the pptA gene of N. meningitidis at a rate of 1 × 10−2 (45). Therefore, our strict length criteria likely excludes many mod genes that contain very short (<9 nucleotides long), but never the less phase-variable, SSR tracts. Our analysis of only full-length genes will exclude examples that are phase-varied off, as these genes are not annotated as a functional methyltransferase. This could exclude as many as two-thirds of all phase-variable mod genes from our analysis. A technical issue for missing SSR tracts is that many next-generation sequencing (NGS) technologies rely on mapping short (<200 bp) reads to reference genomes, and assembling areas where SSRs are present often leads to an underestimation of the repeat tract length due to collapsing the tract down by the assembly software, or alignment to multiple places in the genome (46) as assembly software cannot distinguish between sequences (47). This can be particularly problematic in bacterial genomes as the same repeating element may be present in different genes (1). A striking example of this problem was recently highlighted: over 400Mb of repetitive sequence was missing from recent human genome assemblies (over 15% total sequence) compared to the original due to collapsing down of repetitive DNA sequences (48). Automatic trimming of repeat tract length by genome assembly software appears to be evident in our own studies of the length of the repeat tract present in the phase-variable mod gene we have described in a sub-set of Shiga Toxin producing E. coli. The characterized repeat tract length from our fragment length analysis studies is always longer than that annotated and deposited in GenBank. Consequently, any SSRs annotated from short-read sequencing technologies may not be a true representation of actual tract length found in the original bacterial population. As such, these sequences need to be confirmed by methods that can accurately discern the sequence of SSR tracts, such as Pacific Biosciences Single-Molecule, Real-Time (SMRT) long read sequencing technology (49,50). For all these reasons, our estimates to the prevalence of phase-variable mod genes are likely to be highly conservative, with many phase-variable mod genes yet to be discovered.
In summary, we have demonstrated that almost one-fifth of all Type III mod genes contain simple sequence repeats, are therefore potentially phase-variable, and consequently highly likely to control a phasevarion. A broad array of bacterial species contain phasevarions, including bacterial pathogens and environmental organisms. It appears that this form of contingency strategy is widespread throughout the entire bacterial domain. Many well characterized bacterial species contain phasevarions; this extra level of biological and genetic diversity will have a major impact on many areas of research, including the study of bacterial virulence, the development of new and novel treatments against a wide variety of important human and animal pathogens, and on vaccine development.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the Yale Centre for Genomic Analysis (YCGA; USA) for expert technical assistance in carrying out SMRT sequencing and methylome analysis. We thank Prof. James Paton, University of Adelaide, for kind provision of genomic DNA from STEC strain DG131/3. We thank Kjersti Haugum and colleagues from St. Olavs Hospital, Trondheim, Norway, for kind provision of STEC strains FHI71 and FHI96.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR online.
FUNDING
Australian National Health and Medical Research Council (NHMRC) Program [1071659]; Principal Research Fellowship [1138466 to M.P.J.]; Project Grant [1021631]; Career Development Fellowship (to K.L.S.); Project Grant [1099279 to K.L.S. and J.M.A.]; Project Grant [1121629 to Y.Z. and K.L.S.]; Australian Research Council (ARC) Discovery Grant [170104691 to M.P.J. and 180102060 to Y.Z.]; National Natural Science Foundation of China [U1611261 and 61772566 to Y.Y.]; Program for Guangdong Introducing Innovative and Entrepreneurial Teams [2016ZT06D211 to Y.Y.]. Funding for open access charge: National Health and Medical Research Council, Australia.
Conflict of interest statement. None declared.
REFERENCES
- 1. Moxon R., Bayliss C., Hood D.. Bacterial contingency loci: the role of simple sequence DNA repeats in bacterial adaptation. Ann. Rev. Genet. 2006; 40:307–333. [DOI] [PubMed] [Google Scholar]
- 2. Ren Z., Jin H., Whitby P.W., Morton D.J., Stull T.L.. Role of CCAA nucleotide repeats in regulation of hemoglobin and hemoglobin-haptoglobin binding protein genes of Haemophilus influenzae. J. Bacteriol. 1999; 181:5865–5870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Richardson A.R., Stojiljkovic I.. HmbR, a hemoglobin-binding outer membrane protein of Neisseria meningitidis, undergoes phase variation. J. Bacteriol. 1999; 181:2067–2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Blyn L.B., Braaten B.A., Low D.A.. Regulation of pap pilin phase variation by a mechanism involving differential dam methylation states. EMBO J. 1990; 9:4045–4054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Atack J.M., Winter L.E., Jurcisek J.A., Bakaletz L.O., Barenkamp S.J., Jennings M.P.. Selection and counter-selection of Hia expression reveals a key role for phase-variable expression of this adhesin in infection caused by non-typeable Haemophilus influenzae. J. Infect. Dis. 2015; 212:645–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dawid S., Barenkamp S.J., St. Geme J.W.. Variation in expression of the Haemophilus influenzae HMW adhesins: A prokaryotic system reminiscent of eukaryotes. Proc. Natl. Acad. Sci. U.S.A. 1999; 96:1077–1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fox K.L., Atack J.M., Srikhanta Y.N., Eckert A., Novotny L.A., Bakaletz L.O., Jennings M.P.. Selection for phase variation of LOS biosynthetic genes frequently occurs in progression of non-typeable Haemophilus influenzae infection from the nasopharynx to the middle ear of human patients. PLoS One. 2014; 9:e90505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Poole J., Foster E., Chaloner K., Hunt J., Jennings M.P., Bair T., Knudtson K., Christensen E., Munson R.S. Jr, Winokur P.L. et al. Analysis of nontypeable Haemophilus influenzae phase variable genes during experimental human nasopharyngeal colonization. J. Infect. Dis. 2013; 208:720–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Atack J.M., Srikhanta Y.N., Fox K.L., Jurcisek J.A., Brockman K.L., Clark T.A., Boitano M., Power P.M., Jen F.E.C., McEwan A.G. et al. A biphasic epigenetic switch controls immunoevasion, virulence and niche adaptation in non-typeable Haemophilus influenzae. Nat. Commun. 2015; 6:7828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Blakeway L.V., Power P.M., Jen F.E., Worboys S.R., Boitano M., Clark T.A., Korlach J., Bakaletz L.O., Jennings M.P., Peak I.R. et al. ModM DNA methyltransferase methylome analysis reveals a potential role for Moraxella catarrhalis phasevarions in otitis media. FASEB J. 2014; 28:5197–5207. [DOI] [PubMed] [Google Scholar]
- 11. Seib K.L., Jen F.E., Tan A., Scott A.L., Kumar R., Power P.M., Chen L.T., Wu H.J., Wang A.H., Hill D.M. et al. Specificity of the ModA11, ModA12 and ModD1 epigenetic regulator N6-adenine DNA methyltransferases of Neisseria meningitidis. Nucleic Acids Res. 2015; 43:4150–4162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Srikhanta Y.N., Dowideit S.J., Edwards J.L., Falsetta M.L., Wu H.-J., Harrison O.B., Fox K.L., Seib K.L., Maguire T.L., Wang A.H.J. et al. Phasevarions mediate random switching of gene expression in pathogenic Neisseria. PLoS Pathog. 2009; 5:e1000400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Srikhanta Y.N., Fox K.L., Jennings M.P.. The phasevarion: phase variation of type III DNA methyltransferases controls coordinated switching in multiple genes. Nat. Rev. Micro. 2010; 8:196–206. [DOI] [PubMed] [Google Scholar]
- 14. Srikhanta Y.N., Gorrell R.J., Steen J.A., Gawthorne J.A., Kwok T., Grimmond S.M., Robins-Browne R.M., Jennings M.P.. Phasevarion mediated epigenetic gene regulation in Helicobacter pylori. PLoS One. 2011; 6:e27569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Srikhanta Y.N., Maguire T.L., Stacey K.J., Grimmond S.M., Jennings M.P.. The phasevarion: A genetic system controlling coordinated, random switching of expression of multiple genes. Proc. Natl. Acad. Sci. U.S.A. 2005; 102:5547–5551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Seib K.L., Peak I.R., Jennings M.P.. Phase variable restriction-modification systems in Moraxella catarrhalis. FEMS Immunol. Med. Mic. 2002; 32:159–165. [DOI] [PubMed] [Google Scholar]
- 17. Seib K.L., Pigozzi E., Muzzi A., Gawthorne J.A., Delany I., Jennings M.P., Rappuoli R.. A novel epigenetic regulator associated with the hypervirulent Neisseria meningitidis clonal complex 41/44. FASEB J. 2011; 25:3622–3633. [DOI] [PubMed] [Google Scholar]
- 18. Tan A., Hill D.M., Harrison O.B., Srikhanta Y.N., Jennings M.P., Maiden M.C., Seib K.L.. Distribution of the type III DNA methyltransferases modA, modB and modD among Neisseria meningitidis genotypes: implications for gene regulation and virulence. Sci. Rep. 2016; 6:21015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gawthorne J.A., Beatson S.A., Srikhanta Y.N., Fox K.L., Jennings M.P.. Origin of the diversity in DNA recognition domains in phasevarion associated modA genes of pathogenic Neisseria and Haemophilus influenzae. PLoS One. 2012; 7:e32337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Brockman K.L., Jurcisek J.A., Atack J.M., Srikhanta Y.N., Jennings M.P., Bakaletz L.O.. ModA2 phasevarion switching sin nontypeable Haemophilus influenzae increases the severity of experimental otitis media. J. Infect. Dis. 2016; 214:817–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jen F.E., Seib K.L., Jennings M.P.. Phasevarions mediate epigenetic regulation of antimicrobial susceptibility in Neisseria meningitidis. Antimicrob. Agents Chemother. 2014; 58:4219–4221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hood D.W., Deadman M.E., Jennings M.P., Bisercic M., Fleischmann R.D., Venter J.C., Moxon E.R.. DNA repeats identify novel virulence genes in Haemophilus influenzae. Proc. Natl. Acad. Sci. U.S.A. 1996; 93:11121–11125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tan A., Atack J.M., Jennings M.P., Seib K.L.. The capricious nature of bacterial pathogens: phasevarions and vaccine development. Front. Immunol. 2016; 7:586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Furuta Y., Kobayashi I.. Movement of DNA sequence recognition domains between non-orthologous proteins. Nucleic Acids Res. 2012; 40:9218–9232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kojima K.K., Furuta Y., Yahara K., Fukuyo M., Shiwa Y., Nishiumi S., Yoshida M., Azuma T., Yoshikawa H., Kobayashi I.. Population evolution of Helicobacter pylori through diversification in DNA methylation and interstrain sequence homogenization. Mol. Biol. Evol. 2016; 33:2848–2859. [DOI] [PubMed] [Google Scholar]
- 26. Roberts R.J., Vincze T., Posfai J., Macelis D.. REBASE-a database for DNA restriction and modification: enzymes, genes and genomes. Nucleic Acids Res. 2015; 43:D298–D299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Li W., Godzik A.. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006; 22:1658–1659. [DOI] [PubMed] [Google Scholar]
- 28. Edgar R.C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004; 32:1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014; 30:1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Clark T.A., Murray I.A., Morgan R.D., Kislyuk A.O., Spittle K.E., Boitano M., Fomenkov A., Roberts R.J., Korlach J.. Characterization of DNA methyltransferase specificities using single-molecule, real-time DNA sequencing. Nucleic Acids Res. 2012; 40:e29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Murray I.A., Clark T.A., Morgan R.D., Boitano M., Anton B.P., Luong K., Fomenkov A., Turner S.W., Korlach J., Roberts R.J.. The methylomes of six bacteria. Nucleic Acids Res. 2012; 40:11450–11462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Cox E.C. Bacterial mutator genes and the control of spontaneous mutation. Annu. Rev. Genet. 1976; 10:135–156. [DOI] [PubMed] [Google Scholar]
- 33. Bayliss C.D., Bidmos F.A., Anjum A., Manchev V.T., Richards R.L., Grossier J.-P., Wooldridge K.G., Ketley J.M., Barrow P.A., Jones M.A. et al. Phase variable genes of Campylobacter jejuni exhibit high mutation rates and specific mutational patterns but mutability is not the major determinant of population structure during host colonization. Nucleic Acids Res. 2012; 40:5876–5889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Farabaugh P.J., Schmeissner U., Hofer M., Miller J.H.. Genetic studies of the lac repressor. J. Mol. Biol. 1978; 126:847–863. [DOI] [PubMed] [Google Scholar]
- 35. Mrazek J., Guo X., Shah A.. Simple sequence repeats in prokaryotic genomes. Proc. Natl. Acad. Sci. U.S.A. 2007; 104:8472–8477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Man S.M. The clinical importance of emerging Campylobacter species. Nat. Rev. Gastroenterol. Hepatol. 2011; 8:669–685. [DOI] [PubMed] [Google Scholar]
- 37. Razin S., Yogev D., Naot Y.. Molecular biology and pathogenicity of mycoplasmas. Microbiol. Mol. Biol. Rev. 1998; 62:1094–1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Algire M.A., Montague M.G., Vashee S., Lartigue C., Merryman C.. A Type III restriction–modification system in Mycoplasma mycoides subsp. capri. Open Biol. 2012; 2:120115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Brocchi M., Vasconcelos A.T.R.D, Zaha A.. Restriction-modification systems in Mycoplasma spp. Genet. Mol. Biol. 2007; 30:236–244. [Google Scholar]
- 40. Wise K.S., Calcutt M.J., Foecking M.F., Röske K., Madupu R., Methé B.A.. Complete genome sequence of Mycoplasma bovis type strain PG45 (ATCC 25523). Infect. Immun. 2011; 79:982–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sistla S., Rao D.N.. S-Adenosyl-L-methionine-dependent restriction enzymes. Crit. Rev. Biochem. Mol. Biol. 2004; 39:1–19. [DOI] [PubMed] [Google Scholar]
- 42. Power P.M., Sweetman W.A., Gallacher N.J., Woodhall M.R., Kumar G.A., Moxon E.R., Hood D.W.. Simple sequence repeats in Haemophilus influenzae. Infect. Genet. Evol. 2009; 9:216–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zhou K., Aertsen A., Michiels C.W.. The role of variable DNA tandem repeats in bacterial adaptation. FEMS Microbiol. Rev. 2014; 38:119–141. [DOI] [PubMed] [Google Scholar]
- 44. Palmer M.E., Lipsitch M., Moxon E.R., Bayliss C.D.. Broad conditions favor the evolution of phase-variable loci. mBio. 2013; 4:e00430–e00412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Jen F.E.C., Warren M.J., Schulz B.L., Power P.M., Swords W.E., Weiser J.N., Apicella M.A., Edwards J.L., Jennings M.P.. Dual pili post-translational modifications synergize to mediate meningococcal adherence to platelet activating factor receptor on human airway cells. PLoS Pathog. 2013; 9:e1003377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Reinert K., Langmead B., Weese D., Evers D.J.. Alignment of next-generation sequencing reads. Annu. Rev. Genomics Hum. Genet. 2015; 16:133–151. [DOI] [PubMed] [Google Scholar]
- 47. Treangen T.J., Salzberg S.L.. Repetitive DNA and next-generation sequencing: computational challenges and solutions. Nat. Rev. Genet. 2011; 13:36–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Alkan C., Sajjadian S., Eichler E.E.. Limitations of next-generation genome sequence assembly. Nat. Methods. 2011; 8:61–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Koren S., Harhay G.P., Smith T.P., Bono J.L., Harhay D.M., McVey S.D., Radune D., Bergman N.H., Phillippy A.M.. Reducing assembly complexity of microbial genomes with single-molecule sequencing. Genome Biol. 2013; 14:R101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Eid J., Fehr A., Gray J., Luong K., Lyle J., Otto G., Peluso P., Rank D., Baybayan P., Bettman B. et al. Real-time DNA sequencing from single polymerase molecules. Science. 2009; 323:133–138. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.