Abstract
Expansion of simple DNA repeats is responsible for numerous hereditary diseases in humans. The role of DNA replication, repair and transcription in the expansion process has been well documented. Here we analyzed, in a yeast experimental system, the role of RNA–DNA hybrids in genetic instability of long (GAA)n repeats, which cause Friedreich’s ataxia. Knocking out both yeast RNase H enzymes, which counteract the formation of RNA–DNA hybrids, increased (GAA)n repeat expansion and contraction rates when the repetitive sequence was transcribed. Unexpectedly, we observed a similar increase in repeat instability in RNase H-deficient cells when we either changed the direction of transcription-replication collisions, or flipped the repeat sequence such that the (UUC)n run occurred in the transcript. The increase in repeat expansions in RNase H-deficient strains was dependent on Rad52 and Pol32 proteins, suggesting that break-induced replication (BIR) is responsible for this effect. We conclude that expansions of (GAA)n repeats are induced by the formation of RNA–DNA hybrids that trigger BIR. Since this stimulation is independent of which strand of the repeat (homopurine or homopyrimidine) is in the RNA transcript, we hypothesize that triplex H-DNA structures stabilized by an RNA–DNA hybrid (H-loops), rather than conventional R-loops, could be responsible.
INTRODUCTION
The propensity of genomic loci to acquire mutations and/or undergo genetic rearrangements is linked to their transcriptional status. This phenomenon, known as transcription-associated mutagenesis (TAM) or recombination (TAR) (1,2), occurs despite the well-established role of nucleotide excision repair in removing and repairing damaged nucleotides in the transcriptional template strand (3). Many factors beyond the level of gene expression influence the rates of TAM and TAR at specific regions—including the formation of RNA–DNA hybrid structures known as R-loops (4). R-loops form at transcribed sequences when nascent RNA forms a stable hybrid with the transcriptional template strand—resulting in a three-stranded structure consisting of the RNA–DNA hybrid duplex and an extended loop of single-stranded, non-template DNA.
R-loops play important roles in regulating transcription initiation and termination (5). They are also necessary for priming replication in bacterial genomes (6) and class switch recombination at human immunoglobulin genes (7). Conversely, the mutagenic potential of R-loops has been linked to a number of human diseases (8). In particular, the formation of R-loops instigates length instability of DNA microsatellites—repetitive DNA sequences in which the repeating unit is ≤9 bp (9–12). Inheritance of abnormally long microsatellite alleles has been linked to over 30 hereditary neurological and developmental diseases (13). One such disease, Friedreich’s ataxia (FRDA), is caused by autosomal recessive inheritance of a very long (typically 600–900 repeats), non-coding (GAA)n trinucleotide repeat in the first intron of the human FXN gene. FRDA is a childhood-onset disorder characterized by degeneration of the dorsal columns of the spinal cord, scoliosis and hypertrophic cardiomyopathy (14). The mechanisms of (GAA)n repeat expansion may play important roles in disease pathogenesis either during intergenerational transmission of repeats or as they expand over the lifetime of patients (somatic instability) (15,16).
(GAA)n sequences are particularly prone to the formation of R-loops due to their homopurine/homopyrimidine nature, which allows for thermodynamically favorable formation of a purine RNA/pyrimidine DNA hybrid during transcription (17). Indeed, the formation of R-loops at (GAA)n repeats has been demonstrated in vitro (18,19) and linked to heterochromatin formation at the FXN locus in human cell lines (20). Increasing the rate of (GAA)n transcription leads to an increase in the frequency of repeat expansion in both yeast and human cells (21,22). While it has been posited that the formation of R-loops could contribute to TAM/TAR at (GAA)n sequences, it has not yet been proven experimentally in eukaryotes (19,23–25). (GAA)n repeats can also form H-DNA, a three stranded DNA structure which occurs at homopurine/homopyrimidine mirror repeats (26,27). In this intramolecular triplex, two Watson-Crick paired DNA strands form a stable complex with a third DNA strand via Hoogsteen or reverse Hoogsteen base pairing, while a complement of the third strand remains single stranded. H-DNA can be additionally stabilized by RNA or DNA paired with this free single strand in vitro (19,28,29).
The goal of this work was to examine the role of R-loop formation on (GAA)n repeat expansion rate using our well-established systems in yeast (21,30,31). To do so we knocked out the non-essential yeast RNH1 and RNH201 genes—which encode for RNase H1 and the catalytic subunit of RNase H2, respectively. RNase H enzymes eliminate RNA–DNA hybrids, including R-loops, genome-wide by cleaving their RNA portion (32). We found that (GAA)n repeat expansion and contraction rate increases significantly in rnh1Δ, rnh201Δ strains, but only when (GAA)n repeats are transcribed. We observed a similar increase in repeat expansion rate in RNase H deficient cells when we inverted our entire reporter relative to a nearby replication origin or when we inverted just the (GAA)100 repeats such that the major RNA product contained (UUC)n. We also found that an increase in (GAA)n expansions in RNase H null yeast was dependent on the function of RAD52 and POL32 genes, suggesting a role for break-induced replication (BIR). Altogether, our results imply that (GAA)n repeat expansions are induced by the formation of RNA–DNA hybrids, likely RNA-stabilized H-DNA (H-loops).
MATERIALS AND METHODS
Yeast strains
All yeast strains used in this study were derived from the parent strain CH1585 (MATa, leu2-Δ1, trp1-Δ63, ura3–52) and are listed in Supplementary Table S1. Knockouts were made using gene replacement and confirmed via polymerase chain reaction (PCR) using internal and external primers against the targeted gene (Supplementary Table S2). Strains containing the galactose-inducible, PGAL-UR-GAA100-A3 and UASGAL-GAA100-PGAL-CAN1 cassettes have been described previously (21). To construct the PGAL-UR-GAA100-A3 head-on orientation cassette we amplified the PGAL-UR-GAA100-A3 portion of plasmid pYes3-G4G1C1-Fori-T269-GAA100 (21) using PCR primers with SgrDI and BglII 5′-handles (Supplementary Table S2). This PCR insert was digested and cloned back into the pYes3-G4G1C1-Fori-T269-GAA100 plasmid such that the PGAL-UR-GAA100-A3 region was now inverted with respect to ARS306 but TRP1 remained downstream in its original orientation for subsequent selection of cassette-containing cells. To construct the PGAL-UR-TTC100-A3 cassette, two regions of the artificial intron in plasmid pYes3-G4G1C1-Fori-T269-GAA100 were amplified: (i) the repeat containing region with primers with CaiI and PspXI 5′-handles (ii) the Tet269 spacer region (30) with primers with CaiI and ClaI handles. These two PCR products were digested with CaiI and ligated together such that the repeat was inverted with respect to the Tet269 sequence. This ligated product was then cloned back into plasmid pYes3-G4G1C1-Fori-T269-GAA100 using PspXI and ClaI sites. To construct the no repeat control cassette, a 644 bp sequence from the coding part of the tetR gene of the pACYC184 plasmid (NEB) was amplified and cloned into the intron of the split URA3 gene on pYes3-G4G1C1-Fori-T269-GAA100.
The PGAL-UR-GAA128-A3 cassette for measuring contraction rates was constructed by first amplifying a (GAA)128 tract from a 5-fluoroorotic acid (5-FOA) resistant strain bearing the PGAL-UR-(GAA)100-A3 expansion cassette and cloning it into the pYES3- G4G1C1-Fori-T269-GAA100 plasmid. Next the artificial intron was balanced to ensure strains bearing the new construct are Ura− by adding a 339 bp sequence from the tetR gene on plasmid pACYC184 (NEB) in place of the 269 bp sequence present in the expansion cassette (see (30)). The UASGAL-GAA128-PGAL-HIS3 cassette for measuring contraction rates was constructed by first replacing theCAN1 portion of the pYes3-G4G1C1-T150-GAA100 plasmid (21) with the HIS3 gene amplified from plasmid pRS303 (33) using ApaI and SgrDI restriction sites. 128 (GAA)n repeats from a canavanine resistant yeast strain bearing the original UASGAL-GAA100-PGAL-CAN1 cassette were inserted into the new HIS3 plasmid using SphI and XhoI sites. All cassettes were transformed into yeast as linear fragments and their chromosomal integration and repeat lengths were confirmed using PCR and Sanger sequencing.
Measurement of mutation rates
Frozen 15% glycerol yeast stocks were first streaked out on plates containing ethanol and glycerol as a carbon source to select against petite colonies. Colonies from two independent strains for each genotype were inoculated in 5 ml liquid YEP + 2% Raffinose, grown overnight, then serially diluted in water and plated on solid YPD or YEP + 2% Galactose (YPGal) supplemented with uracil. After 40 h (YPD) or 60 h (YPGal) of growth at 30°C, 12 individual colonies were picked from these non-selective plates, suspended and serially diluted at 10× increments in water. For contraction assays, the original repeat length of each colony used in the assay was confirmed using PCR. Concentrated samples were plated on the appropriate selective media (Supplementary Table S3) to select for repeat expansion or contraction events and dilute samples were plated on YPD to determine overall cell count. Colonies were counted after three days for YPD plates and four days for selective media. For expansion experiments 96 5-FOA or canavanine resistant colonies were assessed for repeat expansion via PCR using primers A2 and B2 (Supplementary Table S2). Individual colonies were picked randomly from each plate for PCR analysis such that the proportion of colonies from each plate reflected the total number of colonies on that plate. In other words, more colonies were analyzed for PCR from selective plates with a large number of colonies than those with fewer colonies. Expansion rates were determined using the Ma-Sandri-Sarkar maximum likelihood estimator via the FluCalc webpage (34) with following inputs: Vtot—the volume of water colonies from non-selective plates were suspended in (in this case 200 μl), Csel—the number of observed expansions as determined by PCR on each plate, Ccom—the number of colonies observed on YPD plates, Dsel—the dilution factor of the cell suspension plated on selective media (typically 1 or 10), Dcom—the dilution factor of the cell suspension plated on YPD (typically 10 000), Vsel—the presumed volume corresponding to the proportion of colonies assessed via PCR (i.e. volume plated on selective media × (96/sum of all colonies counted on selective plates)), Vcom—the volume of cell suspension plated on YPD (typically 50 or 100 μl). Contraction rates were calculated similarly except the Csel input corresponded directly to the number of colonies counted on selective media. In order to confirm the accuracy of our calculations and provide a second method of rate calculation, all analyses were repeated with the rSalvador package using the ‘Newton.LD.plating’ and ‘confint.LD.plating’ commands with Csel input as ‘data’ and plating efficiencies calculated as Vsel/(Vtot*Dsel) (35). In order to generate rate numbers, the outputs of these commands were divided by the average of the total number of cells per colony picked for the assay. Assays were conducted such that the total number of cells per colony varied little between genotypes and conditions. Differences were considered highly significant if their 95% confidence intervals calculated via FluCalc did not overlap and are denoted with two asterisks. Differences were considered significant if their 84% confidence intervals calculated via rSalvador did not overlap and are denoted with a single asterisk (36,37).
Strand-specific RT-qPCR
Overnight liquid YPD cultures were diluted to an OD600 of 0.1 in 10 ml of YPD or YPGal and harvested at OD600 of 0.5. RNA was isolated using a hot phenol–chloroform extraction (38). A total of 10 μg of extracted nucleic acid (determined via nanodrop) was treated with Turbo DNase (ThermoFisher) according to the manufacturer’s protocol. RT reactions were carried out in 5 μl volumes using 50U SSIV RT (Invitrogen) in 1× buffer supplemented with 0.5 mM dNTPs, 5 mM dithiothreitol (DTT), 6 μg/ml actinomycin D (Sigma) (39), 1U RNAseIN and 0.5 μM of primers. The following primer sets were used for reverse transcription: sense; TAF10-R, RM203, antisense; TAF10-R, RM75. Following the RT reaction, samples were treated with 0.5U RNase H at 37°C for 20 min. Quantitative PCR (qPCR) was performed in 10 μl volumes using SYBR Select Master Mix (ThermoFisher) on a QuantStudio 6 Flex RTPCR system (Applied Biosystems) in 384-well plates with four technical replicates per sample. All experiments were performed three times. Quantification was achieved via standard curve based off of six 4× serial dilutions of genomic DNA. Target RNA levels were normalized to TAF10 sense RNA levels (40). Statistical analysis was performed via two-way ANOVA with Tukey’s multiple comparisons on ΔCrt (Crttarget-CrtTAF10) values using GraphPad Prism.
RESULTS
RNase H knockout increases the expansion rate of transcribed (GAA)n repeats
In order to determine the effect of R-loop formation on (GAA)n repeat expansion, we knocked out the RNH1 and RNH201 genes in yeast strains bearing our genetic construct for measuring repeat expansion rates. This cassette comprises a URA3 reporter gene that contains an artificial intron with 100 (GAA)n repeats (Figure 1A). Expansions of 10 or more repeats within the intron abrogate splicing of URA3 transcripts and render cells resistant to 5-FOA. For this study, the reporter was placed downstream of a modified galactose-inducible yeast GAL1 promoter (21). Repeat expansion rates are determined in strains carrying our cassette via Luria–Delbruck fluctuation tests followed by PCR analysis of the (GAA)n sequence to confirm expansion events (34).
Figure 1.
RNase H knockout increases the expansion rate of transcribed (GAA)n repeats. (A) Schematic of the GAA;CD system to select for repeat expansion events in yeast. A total of 100 (GAA)n repeats were cloned into an artificially split URA3 gene such that expansion events abrogate splicing and result in resistance to 5-FOA. ARS306: autonomously replicating sequence on Chr III. PGAL1: Galactose inducible promoter. TRP1: auxotrophic marker for selection of strains bearing the construct. ‘Up’ indicates the region amplified by primers used for RT-qPCR. (B) (GAA)100 expansion rates in strains bearing the construct in A. Error bars represent 95% confidence intervals. Cells were grown non-selectively on glucose (low) or galactose (high) to modulate transcription. ‘**’ Indicates non-overlapping 95% confidence intervals compared to WT under the same conditions. Numbers indicate fold change in expansion rate in rnh1Δ, rnh201Δ strains compared to WT.
We found that, regardless of the level of gene expression, rnh1Δ, rnh201Δ double mutants exhibited significantly increased rates of (GAA)100 expansion (Figure 1B). Knockout of RNH1 or RNH201 alone was not sufficient to increase expansion rate (Supplementary Table S4). As previously described, we saw a roughly 12-fold increase in repeat expansion rate when transcription was induced via galactose (21).
RNase H knockout does not influence the expansion rate of non-transcribed (GAA)n repeats
Given the established roles of RNase H2 in lagging strand replication (41,42) and ribonucleotide excision repair (43,44), we sought to confirm that our observed increase in expansion rate in rnh1Δ, rnh201Δ strains was the result of co-transcriptional RNA–DNA hybrid formation specifically. To this end, we knocked out RNH1 and RNH201 in strains bearing a different construct for measuring repeat expansion rates (Figure 2A). In this cassette 100 (GAA)n repeats are located between the upstream activating sequence (UAS) and the transcription start site (TSS) of a GAL1 promoter that controls transcription of a CAN1 reporter gene. When repeats expand, promoter activity diminishes dramatically, making cells resistant to the non-proteinogenic amino acid canavanine. In this cassette, repeats are transcribed at low levels on glucose and not transcribed at all on galactose (21).
Figure 2.
RNase H knockout does not affect the expansion rate of non-transcribed (GAA)n repeats. (A) Schematic of our system to select for expansion of non-transcribed repeats in yeast. A total of 100 (GAA)n repeats were cloned into the region between the UAS and TSS of a GAL1 promoter driving expression of the CAN1 gene. Repeat expansion events diminish promoter activity allowing for selection on media containing canvanine. ARS306: autonomously replicating sequence on Chr III. TRP1: auxotrophic marker for selection of strains bearing the construct. (B) (GAA)100 expansion rates in strains bearing the construct in A. Error bars represent 95% confidence intervals. Cells were grown non-selectively on glucose (low) or galactose (high) to modulate transcription. Differences between WT and rnh1Δ, rnh201Δ strains were not significant.
RNase H deletion had no effect on (GAA)100 repeat expansion rate when cells with the CAN1 construct were grown on galactose (Figure 2B). This indicates that transcription of the repeats is required for RNase H knockout to influence expansion rate. There was a slight, insignificant increase in expansion rate due to RNase H deletion for CAN1 strains grown on glucose-containing media (Figure 2B). The latter result is not surprising, since we have previously demonstrated that repeats are transcribed aberrantly under these conditions (21).
RNase H knockout increases expansion rate regardless of the relative orientation of transcription and replication
Several studies have shown that the orientation of transcription with respect to replication influences mutation rate. Specifically, when replication encounters transcription ‘head-on’ (RNA polymerase and the replication fork proceed in opposite directions) as opposed to ‘co-directionally’, it results in higher rates of mutagenesis and/or increased levels of R-loop formation (45–47). To test the role of orientation on (GAA)n expansion rate in our system, we inverted the URA3 cassette such that transcription and replication were oriented head-on (GAA;HO) (Figure 3A). In wild-type (WT) cells, we found that (GAA)100 expansion rate is significantly lower for the head-on orientation under conditions of high transcription (galactose) while it is the same under conditions of low transcription (glucose) (Figure 3B). Therefore, inducing transcription appears to have less of an effect on (GAA)100 expansion rate when transcription collides with replication head-on. When we deleted RNH1 and RNH201 in strains with the GAA;HO construct, we observed a significant increase in repeat expansion rate on both glucose and galactose. The magnitude of this effect was less when compared to the GAA;CD cassette under conditions of high transcription (2.6-fold versus 4.5-fold; Figures 1B and 3D). While RNase H knockout has a clear effect in both orientations, these data imply that co-directional collisions with RNA–DNA hybrid structures promote (GAA)n expansion to a greater extent than head-on collisions, arguing against the involvement of conventional R-loops.
Figure 3.
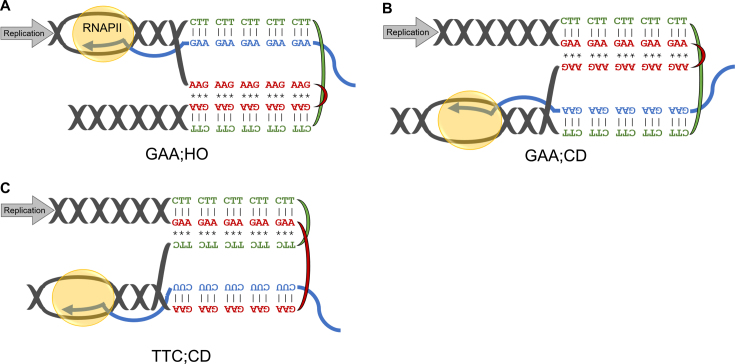
Altering the direction of transcription-replication collisions or inverting the repetitive run alone does not change the effect of RNase H deletion on repeat expansion rate. (A) Schematic of our GAA;HO construct where transcription and replication are oriented head-on. (TTC)100 serves as the lagging strand template and (GAA)100 remains on the transcriptional coding strand. (B) Repeat expansion rates in strains bearing different constructs for selection of repeat expansion events. Error bars represent 95% confidence intervals. Cells were grown non-selectively on glucose (low) or galactose (high) to modulate transcription. Numbers indicate fold increase in expansion rate due to galactose induction. (C) Schematic of our TTC;CD construct, in which only the repeats have been flipped such that (TTC)100 now serves as both the lagging strand template and the transcriptional coding strand. (D) Repeat expansion rates in WT and RNase H deficient strains bearing our different constructs for selecting for repeat expansion events. Error bars represent 95% confidence intervals. Cells were grown non-selectively on glucose (low) or galactose (high) to modulate transcription. ‘**’ Indicates non-overlapping 95% confidence intervals compared to corresponding WT strain and conditions ‘*’ Indicates non-overlapping 84% confidence intervals compared to corresponding WT strain and conditions. Numbers indicate fold change in expansion rate in rnh1Δ, rnh201Δ strains compared to WT.
RNase H knockout increases repeat expansion rate when repeats are inverted
Since inverting our URA3 construct with respect to replication simultaneously alters which repetitive strand ((TTC)100 or (GAA)100) serves as the lagging strand template, we set out to test whether inverting just the repetitive sequence with respect to replication and transcription influences repeat expansion rate. To accomplish this we flipped the repetitive sequence within our construct such that (TTC)100 served as both the lagging strand template and the transcriptional coding strand, while maintaining the co-directional orientation of replication and transcription (TTC;CD) (Figure 3C). We found that in the presence of glucose, repeat expansion rates are very similar for all three cassettes (GAA;CD, GAA;HO and TTC;CD). Transcription induction by galactose increases repeat expansion rate12.5-fold for the GAA;CD cassette, 3.9-fold for the GAA;HO cassette and 8.3-fold for the TTC;CD cassette (Figure 3B). Taken together, it appears that the nature of the repeat on the lagging strand template does not influence the expansion rate as strongly as the relative orientation of transcription and replication. Specifically, co-directional collisions lead to more expansions than head-on collisions as demonstrated by the fact that galactose induction has a greater effect on repeat expansion rate for the two CD cassettes as compared to the HO cassette (Figure 3B).
In vitro, the formation of R-loops at (GAA)n repeats was shown to depend on which strand serves as the transcriptional template (18,19). When (GAA)n repeats serve as the coding strand, homopurine (GAA)n containing transcripts form a thermodynamically favorable RNA–DNA hybrid with the (TTC)n template that stalls transcription. In contrast, when transcripts contain (UUC)n repeats, R-loops are not detected. We reasoned, therefore, that RNase H knockout would have no effect on repeat expansion rate in strains with the TTC;CD cassette (Figure 3C) if conventional R-loops were instigators of expansions. This appeared to be incorrect: rnh1Δ, rnh201Δ strains bearing the (TTC)100 cassette showed elevated rates of expansion on both glucose and galactose as compared to the WT. The magnitude of this increase was similar to the GAA;CD cassette on glucose and slightly less on galactose (Figures 1B and 3D).
(GAA)n repeats can serve as intragenic antisense promoters
Long (GAA)n repeats have previously been shown to act as bidirectional promoters when placed upstream of a promoterless gene in yeast (23). We were curious as to whether such activity could be observed at intragenic repeats such as those present in the intron of our UR-GAA-A3 construct. To address this question, we performed strand-specific RT-qPCR in the presence of actinomycin D (39) to assess the antisense promoter activity of repeats in our various constructs used for measuring expansion rates. The level of antisense transcripts for repeat-bearing URA3 cassettes relative to the antisense transcript in a no repeat control cassette was considered an appropriate measure of intragenic promoter activity. We found that repeats in both (GAA)100 (GAA;CD, GAA;HO) constructs worked as internal antisense promoters compared to the no repeat control when the activity of the upstream GAL1 promoter was induced (Figure 4A). In contrast, basal levels of antisense transcription were not significantly different between the (GAA)100 and no repeat cassettes when transcription was not induced (Figure 4A). In agreement with our earlier studies (48), sense RNA levels produced from the (TTC)100 cassette (TTC;CD) were significantly lower than those for the no repeat control on both glucose and galactose (Figure 4A). In contrast with the GAA-cassettes, we observed an intragenic promoter activity for the TTC-cassette when the strain was grown in glucose-containing media but not in galactose-containing media (Figure 4A). The reasons for these differences is unclear and beyond the scope of this study. Importantly, however, deletion of RNase H had no influence on the level of antisense transcripts for any cassette (Figure 4B), suggesting that the increased expansion rate observed in rnh1Δ, rnh201Δ strains was not a result of increased promoter activity. Overall, we see no correlation between the intrinsic (GAA)n promoter strength and repeat expansion rates in various repeat-containing cassettes.
Figure 4.
(GAA)100 repeats serve as antisense promoter sequences. (A) Strand-specific RT-qPCR results from the region upstream of the repetitive sequence (see Figure 1A) in our different constructs for measuring repeat expansion rates. Quantification was achieved using the standard curve method. Bars represent means from three biological replicates. Error bars indicate standard deviation from the mean. Sense refers to the RNA strand that contains the indicated repeats. The No repeat;CD control construct contains an intron with similar length to the repeat-bearing cassettes that does not contain a repetitive sequence. Target RNA levels were normalized to TAF10 sense RNA levels. ‘**’ Indicates P < 0.01, two-way ANOVA with Tukey’s multiple comparisons test. (B) Antisense strand-specific RT-qPCR results as displayed in A comparing WT and rnh1Δ, rnh201Δ strains.
RNA-DNA hybrid-induced (GAA)100 repeat expansion requires Rad52 and Pol32
In several experimental systems, R-loops have been shown to stimulate homologous recombination (HR) (49). This led us to predict that the stimulation of repeat expansions caused by RNA–DNA hybrids in our case might also depend on HR. To test this hypothesis we knocked out the key recombination protein Rad52 in the WT and rnh1Δ, rnh201Δ strains bearing our UR-GAA-A3 cassette in the co-directional orientation (GAA;CD) and measured repeat expansion rates. We observed a rescue of the repeat expansion phenotype caused by RNase H deletion in strains that lacked Rad52 protein, implying that HR is indeed required for the stimulation of GAA repeat expansions by RNA–DNA hybrids (Figure 5). Note that while Rad52 knockout alone leads to a small increase in the repeat expansion rate, it is not significantly different from the WT strain (Supplementary Table S4).
Figure 5.
RNA–DNA hybrid mediated repeat expansions are dependent on the function of Rad52 and Pol32 proteins. (GAA)100 expansion rates in strains bearing the GAA;CD construct. Error bars represent 95% confidence intervals. Cells were grown non-selectively on galactose (high-transcription conditions). ‘**’ Indicates non-overlapping 95% confidence intervals. ‘*’ Indicates non-overlapping 84% confidence intervals.
In order to further specify the type of recombination responsible for R-loop-induced (GAA)n expansion, we knocked out Pol32, a subunit of DNA polymerase delta required for break-induced replication (BIR) (50). Similar to rad52Δ, we observed a rescue of the elevated repeat expansion rate phenotype in the rnh1Δ, rnh201Δ, pol32Δ strain (Figure 5).
Transcription and RNA–DNA hybrids promote (GAA)n contractions by a mechanism that differs from that for expansions
Given our previous observations regarding the relationship between repeat expansion rate and gene expression levels (21,51), we were curious whether the same is true for (GAA)n repeat contractions. We, thus, modified our galactose-inducible URA3 cassette to carry 128 (GAA)n repeats in a longer artificial intron (Figure 6A). The longer length of the intron in this cassette made strains carrying the (GAA)128 repeat fully auxotrophic for uracil owing to impaired splicing. If the repeat sequence in this cassette contracts significantly, splicing is re-established, allowing for selection on media lacking uracil. We also modified our CAN1-based system such that we could measure the contraction rate of non-transcribed repetitive sequences. This was accomplished by replacing the CAN1 negative selective marker with a HIS3 positive selective marker in a cassette that contains 128 (GAA)n repeats between the UAS and TSS of the GAL1 promoter (Figure 6B). When repeats contract, it reduces the distance between the UAS and the TSS allowing for transcription of the HIS3 gene and selection on media lacking histidine.
Figure 6.
RNase H knockout increases (GAA)n contraction rate via mechanisms that differ from expansion. (A) Schematic of our construct for selecting for contraction events of transcribed (GAA)128 repeats. Our URA3 construct from Figure 1A was altered such that strains start out Ura− and become Ura+ after repeat contraction. (B) Schematic of our construct for selecting for contraction events on the non-transcribed (GAA)128 repeat. The negative selective CAN1 gene from the cassette in Figure 2A was replaced by the positive selection HIS3 marker. Strains with the (GAA)128 repeat are His−. Contraction of the repeats between the UAS and TSS leads to promoter activation and a His+ phenotype. (C) (GAA)128 contraction rates in strains bearing the construct in A. Error bars represent 95% confidence intervals. ‘**’ Indicates non-overlapping 95% confidence intervals compared to WT. ‘∧∧’ Indicates non-overlapping 95% confidence intervals compared to WT grown on glucose. (D) (GAA)128 contraction rates in strains bearing the construct in B. Error bars represent 95% confidence intervals. ‘∧∧’ Indicates non-overlapping 95% confidence intervals compared to WT grown on glucose.
Similar to repeat expansion, inducing transcription by galactose lead to a significant increase in contraction rate for both constructs (Figure 6C and D). This implies that transcriptional activity influences contraction of nearby repetitive sequences regardless of whether they serve as transcriptional templates. The magnitude of the transcription-associated increase was less than that observed for expansion (Figure 3B), and the overall rate of (GAA)128 contraction was 1–2 orders of magnitude greater than (GAA)100 expansion. This is in line with most observations of trinucleotide repeat instability, which show contractions to be the most frequent mutation type.
We then tested the effect of RNase H deletion on (GAA)n contraction rates. We found that RNase H deletion increases (GAA)128 contraction rates only for transcribed repeats, and only under conditions of low transcription (in glucose) (Figure 6C). This is one substantial difference from our expansion data, which show similar effects of RNase H knockout with both low- and high-transcription levels. Another substantial difference is that, in contrast to our expansion data, deletion of the RAD52 gene did not rescue the effect of RNase H deficiency on contraction rate (Figure 6C). Ultimately these data suggest that R-loop-induced contractions do not follow the same mechanism as expansions. Specifically, recombination due to RNA–DNA hybrid formation at (GAA)n repeats does not seem to contribute to their contraction.
DISCUSSION
The role of R-loops in trinucleotide repeat instability has been discussed in the past (reviewed in (52–54)). Most recently, it was shown that the formation of R-loops at (CAG)n repeats triggers cytosine deamination and base excision repair that ultimately leads to repeat fragility and instability in yeast (9). For (GAA)n repeats, R-loop formation has been shown to occur when they are transcribed such that the (GAA)n run is in the RNA strand, i.e. as it is in FXN gene (18,19). It was also demonstrated that increasing transcription of (GAA)n repeats in cultured human cells leads to progressive repeat expansion (22,55). Furthermore, a direct role for GAA-transcripts in promoting expansion was demonstrated in (56). In the current study, we have proven that inactivation of RNase H stimulates large-scale (GAA)n repeat instability in yeast, implying an instigative role of RNA–DNA hybrids in the process. The stimulation of (GAA)n repeat expansions in the RNase H double knockout is highly significant, yet it is much smaller than what we previously observed for mutations in the DNA replication machinery (30,57). This implies that challenges during DNA replication of (GAA)n repeats are the primary contributors to their expansion, while recombination seems to contribute to the process under special circumstances.
Our observed stimulation of (GAA)n repeat expansion by RNA–DNA hybrids appeared to depend entirely on Rad52, suggesting that some form of HR promotes repeat expansion in RNase H-deficient cells. This finding is consistent with the well-established role of R-loops in promoting recombination events (4). We also observed a rescue of expansion stimulation in POL32 knockout strains. The combined requirement of Rad52 and Pol32 strongly implies that BIR contributes to (GAA)n expansion in RNase H deletion strains.
BIR is a form of DNA double-strand break (DSB) repair that evolved to mend one-ended DSBs that arise when replication forks collapse in S-phase, or two-ended DSBs that occur in the G2 phase of the cell cycle (58). It is a highly mutagenic pathway of HR as it is prone to template switching and utilizes conservative DNA synthesis (59,60). It was previously implicated in R-loop-induced recombination in yeast (61) and we have recently discovered that BIR is primarily responsible for large-scale expansions of (CAG)n repeats in yeast (51). We hypothesize that stable RNA–DNA hybrid structures formed at (GAA)n repeats serve as a particularly potent block to DNA replication, leading to fork collapse and formation of a one-ended DSB. Subsequent repair by BIR would then cause expansions in a manner similar to (CAG)n repeats (51,58). A priori, such mechanisms of repeat instability that involve homology search and D-loop extension should be equally likely to cause expansions or contractions because they rely on ‘out-of-register’ strand invasion between repetitive sequences. If strand invasion occurs toward the downstream, 3′ end of the repetitive homologous donor sequence, you might expect a contraction. If invasion occurs all the way at the upstream 5′ end of that sequence, it should cause expansion. We found, however, that stimulation of contractions in the lack of RNase H does not depend on Rad52. This implies that HR preferentially causes repeat expansions but not contractions. This finding supports previous hypotheses that multiple template switching events, which are characteristic of BIR, might specifically favor repeat expansions (62).
What could be the nature of RNA–DNA hybrids at (GAA)n repeats that cause replication fork collapse? While we initially suspected canonical R-loops, our data argue against this idea. First, we found repeat expansion is stimulated in RNase H deficient cells regardless of whether our experimental cassette is transcribed co-directionally or head-on to replication (Figure 3D). In fact, the magnitude of this stimulation is less in the HO orientation when the cassettes are highly transcribed. These data contrast with observations that R-loops are preferably formed during head-on transcription-replication collisions (45,63,64). At the same time, our results are strikingly similar to the recently published data showing that both head-on and co-directional collision of replication with a composite RNA–DNA hybrid structure called a G-loop (G4-quadruplex DNA stabilized by an R-loop) were equally mutagenic (65). Second, and even more important, we found that inverting the (GAA)n repeat in our cassette, which placed (UUC)n runs into RNA, led to the stimulation of repeat expansions in the RNase H knockout, the magnitude of which was only slightly less than for (GAA)n RNA runs (Figure 3D). It was convincingly demonstrated that canonical R-loops are formed in vitro at (GAA)n repeats only when the (GAA)n sequence is in the RNA strand (18,19). We believe, therefore, that a complex RNA–DNA hybrid structure at (GAA)n repeats, which is distinct from a canonical R-loop, could be responsible for fork collapse and subsequent expansions in vivo.
(GAA)n repeats are known to form an intramolecular triplex structure, called H-DNA. Importantly, this repeat can adopt both H-y (Figure 7C) and H-r (Figure 7A and B) conformations in which either the pyrimidine (y) or purine (r) strand, respectively, is donated to the triplex, while its complement remains single stranded (26,27). It has been previously proposed that co-transcriptional binding of an RNA molecule to the single stranded portion of H-DNA stabilized this structure, resulting in transcription stalling (19,66). We propose that formation of RNA-stabilized H-DNA structures, which we call H-loops, during transcription of the (GAA)n repeat would stall replication resulting in a DSB and ultimately repeat instability. In the GAA;CD and GAA;HO orientations, the GAA-containing RNA would stabilize the H-r conformation of the repeat (Figure 7A and B), while in TTC;CD orientation, the UUC-containing RNA would stabilize its H-y form (Figure 7C). Our H-loop model explains why we observe stimulation of repeat expansions only for transcribed repeats, since H-loops require complementary RNA. The fact that both H-y and H-r conformations can be formed by (GAA)n repeats (26,27) makes H-loop formation independent of whether the RNA transcript is homopurine or homopyrimidine. This explains our observed lack of dependence on the nature of the major RNA product on expansion rate. Furthermore, the H-loop model explains why RNase H knockout has a greater effect on repeat instability for the GAA;CD and TTC;CD cassettes than for the GAA;HO cassette (Figures 1B and 3D). In the head-on orientation, replication encounters the RNA–DNA hybrid part of the H-loop first because the single stranded portion of H-DNA preferentially occurs 5′ to the triplex under physiological conditions (so called H-y3/H-r3 form) (Figure 7A) (67). Disruption of the RNA–DNA hybrid, which is readily accomplished by replicative helicases (68), would destabilize the triplex and dissolve the structure. In the co-directional orientation, in contrast, the replicative helicase encounters the triplex portion of the H-loop first (Figure 7B and C), which is more difficult for it to unwind (69) potentially leading to greater fork stalling and collapse.
Figure 7.
H-loop formation could occur independent of repeat orientation. (A) Potential H-loop/replication collisions for the GAA;HO cassette. When (GAA)n repeats are present in the RNA transcript, it can hybridize with the single-stranded portion of an H-r triplex leading to triplex stabilization. (B) Potential H-loop/replication collisions for the GAA;CD cassette. When (GAA)n repeats are present in the RNA transcript, it can hybridize with the single-stranded portion of an H-r triplex leading to triplex stabilization. (C) Potential H-loop/replication collisions for the TTC;CD cassette. When (UUC)n repeats are present in the RNA transcript, it can hybridize with the single-stranded portion of an H-y triplex leading to triplex stabilization. | represents Watson–Crick base pairs and ‘*’ represents Hoogsteen or reverse Hoogsteen base pairs.
The formation of stable H-loops is supported by in vitro data showing that at high negative superhelical density, which is known to exist behind transcribing RNA polymerase (70), H-DNA is strongly stabilized in the presence of an oligonucleotide complementary to the single-stranded portion of the structure (29). This stabilization persists up to pH 7 even for the H-y conformation, which requires protonation of cytosine to form stable Hoogsteen bonds (29). It is theoretically possible that the H-loop formed during transcription can ultimately resolve into other more stable structures. Grabczyk et al. (19) hypothesized that an H-loop stabilized by homopurine RNA can eventually convert into a stable R-loop (see also 71). This however cannot happen for homopyrimidine RNA because an R-loop containing pyrimidine RNA is energetically unfavorable (17). Alternatively, it has been shown that pyrimidine RNA forms very stable intermolecular triplexes with duplex DNA (17). Thus, an H-DNA stabilized by homopyrimidine RNA might convert into a long dYdR*rY intermolecular triplex. A similar conversion was, in fact, shown in the presence of single-stranded homopyrimidine DNA (72). Note, however, that this scenario is impossible for homopurine RNA, which cannot form intermolecular triplexes (73). In other words, if there were conversions between H-loops and other complex structures, the conversion would differ depending on the nature of the RNA strand. We do not think that our data are in line with the conversion of an H-loop into two separate intermolecular structures. First, an increase in the rate of repeat expansions in the RNase H double knockout is very similar regardless of whether the RNA strand is homopurine or homopyrimidine when collisions are oriented co-directionally (Figures 1B and 3D). Second, RNase H is unable to disrupt Hoogsteen interactions between RNA and DNA (74). Therefore, the elevated level of repeat expansions for the TTC;CD cassette in an RNase H knockout is hard to explain by the presence of an intermolecular dYdR*rY triplex.
In summary, we have demonstrated that RNase H knockout triggers recombination-mediated expansions of (GAA)n repeats in yeast. Altering the orientation of the repetitive sequence with respect to replication or transcription does not change the effect of RNA–DNA hybrid formation on expansion rate. These findings suggest that complex, multi-stranded RNA–DNA hybrid structures, which we call H-loops, might be instigators of HR at (GAA)n repeats. Given the fact that (GAA)n repeats have recently been shown to be replication barriers in human cell lines (24,75) and that R-loops appear to be involved in FXN silencing (20), our results contribute to the overall picture of R-loop formation in the pathobiology of FRDA.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Allen Su and Catherine Freudenreich for their helpful discussion and support throughout this project. We also thank Clara Williamson for her help in construction of mutant strains. Ryan McGinty helped pilot this study.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institute of General Medical Sciences [R01GM60987, P01GM105473]. Funding for open access charge: NIH [P01GM105473].
Conflict of interest statement. None declared.
REFERENCES
- 1. Jinks-Robertson S., Bhagwat A.S.. Transcription-associated mutagenesis. Annu. Rev. Genet. 2014; 48:341–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Gaillard H., Aguilera A.. Transcription as a threat to genome integrity. Annu. Rev. Biochem. 2016; 85:291–317. [DOI] [PubMed] [Google Scholar]
- 3. Hanawalt P.C., Spivak G.. Transcription-coupled DNA repair: two decades of progress and surprises. Nat. Rev. Mol. Cell Biol. 2008; 9:958–970. [DOI] [PubMed] [Google Scholar]
- 4. Aguilera A., Garcia-Muse T.. R loops: from transcription byproducts to threats to genome stability. Mol. Cell. 2012; 46:115–124. [DOI] [PubMed] [Google Scholar]
- 5. Ginno P.A., Lim Y.W., Lott P.L., Korf I., Chedin F.. GC skew at the 5′ and 3′ ends of human genes links R-loop formation to epigenetic regulation and transcription termination. Genome Res. 2013; 23:1590–1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Baker T.A., Kornberg A.. Transcriptional activation of initiation of replication from the E. coli chromosomal origin: an RNA-DNA hybrid near oriC. Cell. 1988; 55:113–123. [DOI] [PubMed] [Google Scholar]
- 7. Yu K., Chedin F., Hsieh C.L., Wilson T.E., Lieber M.R.. R-loops at immunoglobulin class switch regions in the chromosomes of stimulated B cells. Nat. Immunol. 2003; 4:442–451. [DOI] [PubMed] [Google Scholar]
- 8. Richard P., Manley J.L.. R loops and links to human disease. J. Mol. Biol. 2017; 429:3168–3180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Su X.A., Freudenreich C.H.. Cytosine deamination and base excision repair cause R-loop-induced CAG repeat fragility and instability in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S.A. 2017; 114:E8392–E8401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Reddy K., Schmidt M.H., Geist J.M., Thakkar N.P., Panigrahi G.B., Wang Y.H., Pearson C.E.. Processing of double-R-loops in (CAG).(CTG) and C9orf72 (GGGGCC).(GGCCCC) repeats causes instability. Nucleic Acids Res. 2014; 42:10473–10487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lin Y., Dent S.Y., Wilson J.H., Wells R.D., Napierala M.. R loops stimulate genetic instability of CTG.CAG repeats. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:692–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Nakatani R., Nakamori M., Fujimura H., Mochizuki H., Takahashi M.P.. Large expansion of CTG*CAG repeats is exacerbated by MutSbeta in human cells. Sci. Rep. 2015; 5:e11020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mirkin S.M. Expandable DNA repeats and human disease. Nature. 2007; 447:932–940. [DOI] [PubMed] [Google Scholar]
- 14. Pandolfo M. Friedreich ataxia. Handb Clin. Neurol. 2012; 103:275–294. [DOI] [PubMed] [Google Scholar]
- 15. De Biase I., Rasmussen A., Monticelli A., Al-Mahdawi S., Pook M., Cocozza S., Bidichandani S.I.. Somatic instability of the expanded GAA triplet-repeat sequence in Friedreich ataxia progresses throughout life. Genomics. 2007; 90:1–5. [DOI] [PubMed] [Google Scholar]
- 16. De Michele G., Cavalcanti F., Criscuolo C., Pianese L., Monticelli A., Filla A., Cocozza S.. Parental gender, age at birth and expansion length influence GAA repeat intergenerational instability in the X25 gene: pedigree studies and analysis of sperm from patients with Friedreich's ataxia. Hum. Mol. Genet. 1998; 7:1901–1906. [DOI] [PubMed] [Google Scholar]
- 17. Roberts R.W., Crothers D.M.. Stability and properties of double and triple helices: dramatic effects of RNA or DNA backbone composition. Science. 1992; 258:1463–1466. [DOI] [PubMed] [Google Scholar]
- 18. Reddy K., Tam M., Bowater R.P., Barber M., Tomlinson M., Nichol Edamura K., Wang Y.H., Pearson C.E.. Determinants of R-loop formation at convergent bidirectionally transcribed trinucleotide repeats. Nucleic Acids Res. 2011; 39:1749–1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Grabczyk E., Mancuso M., Sammarco M.C.. A persistent RNA.DNA hybrid formed by transcription of the Friedreich ataxia triplet repeat in live bacteria, and by T7 RNAP in vitro. Nucleic Acids Res. 2007; 35:5351–5359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Groh M., Lufino M.M., Wade-Martins R., Gromak N.. R-loops associated with triplet repeat expansions promote gene silencing in Friedreich ataxia and fragile X syndrome. PLoS Genet. 2014; 10:e1004318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Shah K.A., McGinty R.J., Egorova V.I., Mirkin S.M.. Coupling transcriptional state to large-scale repeat expansions in yeast. Cell Rep. 2014; 9:1594–1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ditch S., Sammarco M.C., Banerjee A., Grabczyk E.. Progressive GAA.TTC repeat expansion in human cell lines. PLoS Genet. 2009; 5:e1000704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhang Y., Shishkin A.A., Nishida Y., Marcinkowski-Desmond D., Saini N., Volkov K.V., Mirkin S.M., Lobachev K.S.. Genome-wide screen identifies pathways that govern GAA/TTC repeat fragility and expansions in dividing and nondividing yeast cells. Mol. Cell. 2012; 48:254–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gerhardt J., Bhalla A.D., Butler J.S., Puckett J.W., Dervan P.B., Rosenwaks Z., Napierala M.. Stalled DNA replication forks at the endogenous GAA repeats drive repeat expansion in Friedreich's ataxia cells. Cell Rep. 2016; 16:1218–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. McIvor E.I., Polak U., Napierala M.. New insights into repeat instability: role of RNA*DNA hybrids. RNA Biol. 2010; 7:551–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Potaman V.N., Oussatcheva E.A., Lyubchenko Y.L., Shlyakhtenko L.S., Bidichandani S.I., Ashizawa T., Sinden R.R.. Length-dependent structure formation in Friedreich ataxia (GAA)n*(TTC)n repeats at neutral pH. Nucleic Acids Res. 2004; 32:1224–1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Vetcher A.A., Napierala M., Iyer R.R., Chastain P.D., Griffith J.D., Wells R.D.. Sticky DNA, a long GAA.GAA.TTC triplex that is formed intramolecularly, in the sequence of intron 1 of the frataxin gene. J. Biol. Chem. 2002; 277:39217–39227. [DOI] [PubMed] [Google Scholar]
- 28. Grabczyk E., Fishman M.C.. A long purine-pyrimidine homopolymer acts as a transcriptional diode. J. Biol. Chem. 1995; 270:1791–1797. [DOI] [PubMed] [Google Scholar]
- 29. Belotserkovskii B.P., Krasilnikova M.M., Veselkov A.G., Frank-Kamenetskii M.D.. Kinetic trapping of H-DNA by oligonucleotide binding. Nucleic Acids Res. 1992; 20:1903–1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Shah K.A., Shishkin A.A., Voineagu I., Pavlov Y.I., Shcherbakova P.V., Mirkin S.M.. Role of DNA polymerases in repeat-mediated genome instability. Cell Rep. 2012; 2:1088–1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Shishkin A.A., Voineagu I., Matera R., Cherng N., Chernet B.T., Krasilnikova M.M., Narayanan V., Lobachev K.S., Mirkin S.M.. Large-scale expansions of Friedreich's ataxia GAA repeats in yeast. Mol. Cell. 2009; 35:82–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wahba L., Amon J.D., Koshland D., Vuica-Ross M.. RNase H and multiple RNA biogenesis factors cooperate to prevent RNA:DNA hybrids from generating genome instability. Mol. Cell. 2011; 44:978–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chee M.K., Haase S.B.. New and redesigned pRS plasmid shuttle vectors for genetic manipulation of Saccharomy cescerevisiae. G3. 2012; 2:515–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Radchenko E.A., McGinty R.J., Aksenova A.Y., Neil A.J., Mirkin S.M.. Quantitative analysis of the rates for repeat-mediated genome instability in a yeast experimental system. Methods Mol. Biol. 2018; 1672:421–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zheng Q. rSalvador: an R package for the fluctuation experiment. G3 (Bethesda). 2017; 7:3849–3856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zheng Q. Comparing mutation rates under the Luria-Delbruck protocol. Genetica. 2016; 144:351–359. [DOI] [PubMed] [Google Scholar]
- 37. Zheng Q. Methods for comparing mutation rates using fluctuation assay data. Mutat. Res. 2015; 777:20–22. [DOI] [PubMed] [Google Scholar]
- 38. Ares M. Isolation of total RNA from yeast cell cultures. Cold Spring Harb. Protoc. 2012; 2012:1082–1086. [DOI] [PubMed] [Google Scholar]
- 39. Perocchi F., Xu Z., Clauder-Munster S., Steinmetz L.M.. Antisense artifacts in transcriptome microarray experiments are resolved by actinomycin D. Nucleic Acids Res. 2007; 35:e128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Teste M.A., Duquenne M., Francois J.M., Parrou J.L.. Validation of reference genes for quantitative expression analysis by real-time RT-PCR in Saccharomyces cerevisiae. BMC Mol. Biol. 2009; 10:e99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Liu B., Hu J., Wang J., Kong D.. Direct visualization of RNA-DNA primer removal from Okazaki fragments provides support for Flap cleavage and exonucleolytic pathways in eukaryotic cells. J. Biol. Chem. 2017; 292:4777–4788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zheng L., Shen B.. Okazaki fragment maturation: nucleases take centre stage. J. Mol. Cell Biol. 2011; 3:23–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Rydberg B., Game J.. Excision of misincorporated ribonucleotides in DNA by RNase H (type 2) and FEN-1 in cell-free extracts. Proc. Natl. Acad. Sci. U.S.A. 2002; 99:16654–16659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Sparks J.L., Chon H., Cerritelli S.M., Kunkel T.A., Johansson E., Crouch R.J., Burgers P.M.. RNase H2-initiated ribonucleotide excision repair. Mol. Cell. 2012; 47:980–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hamperl S., Bocek M.J., Saldivar J.C., Swigut T., Cimprich K.A.. Transcription-replication conflict orientation modulates R-loop levels and activates distinct DNA damage responses. Cell. 2017; 170:774–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Sankar T.S., Wastuwidyaningtyas B.D., Dong Y., Lewis S.A., Wang J.D.. The nature of mutations induced by replication-transcription collisions. Nature. 2016; 535:178–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Prado F., Aguilera A.. Impairment of replication fork progression mediates RNA polII transcription-associated recombination. EMBO J. 2005; 24:1267–1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Krasilnikova M.M., Kireeva M.L., Petrovic V., Knijnikova N., Kashlev M., Mirkin S.M.. Effects of Friedreich's ataxia (GAA)n*(TTC)n repeats on RNA synthesis and stability. Nucleic Acids Res. 2007; 35:1075–1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Santos-Pereira J.M., Aguilera A.. R loops: new modulators of genome dynamics and function. Nat. Rev. Genet. 2015; 16:583–597. [DOI] [PubMed] [Google Scholar]
- 50. Lydeard J.R., Jain S., Yamaguchi M., Haber J.E.. Break-induced replication and telomerase-independent telomere maintenance require Pol32. Nature. 2007; 448:820–823. [DOI] [PubMed] [Google Scholar]
- 51. Kim J.C., Harris S.T., Dinter T., Shah K.A., Mirkin S.M.. The role of break-induced replication in large-scale expansions of (CAG)n/(CTG)n repeats. Nat. Struct. Mol. Biol. 2017; 24:55–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Polyzos A.A., McMurray C.T.. Close encounters: moving along bumps, breaks, and bubbles on expanded trinucleotide tracts. DNA Repair. 2017; 56:144–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Wang G., Vasquez K.M.. Effects of replication and transcription on DNA structure-related genetic instability. Genes. 2017; 8:e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Groh M., Gromak N.. Out of balance: R-loops in human disease. PLoS Genet. 2014; 10:e1004630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Soragni E., Herman D., Dent S.Y., Gottesfeld J.M., Wells R.D., Napierala M.. Long intronic GAA*TTC repeats induce epigenetic changes and reporter gene silencing in a molecular model of Friedreich ataxia. Nucleic Acids Res. 2008; 36:6056–6065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Rindler P.M., Bidichandani S.I.. Role of transcript and interplay between transcription and replication in triplet-repeat instability in mammalian cells. Nucleic Acids Res. 2011; 39:526–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Tsutakawa S., Thompson M., Arvai A., Neil A.J., Shaw S., Algasaier S., Kim J.C., Finger L., Jardine E., Gotham V. et al. Phosphate steering by Flap Endonuclease 1 promotes 5′-flap specificity and incision to prevent genome instability. Nat Commun. 2017; 8:15855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Neil A.J., Kim J.C., Mirkin S.M.. Precarious maintenance of simple DNA repeats in eukaryotes. Bioessays. 2017; 39:e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Saini N., Ramakrishnan S., Elango R., Ayyar S., Zhang Y., Deem A., Ira G., Haber J.E., Lobachev K.S., Malkova A.. Migrating bubble during break-induced replication drives conservative DNA synthesis. Nature. 2013; 502:389–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Smith C.E., Llorente B., Symington L.S.. Template switching during break-induced replication. Nature. 2007; 447:102–105. [DOI] [PubMed] [Google Scholar]
- 61. Amon J.D., Koshland D.. RNase H enables efficient repair of R-loop induced DNA damage. Elife. 2016; 5:e20533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Kramara J., Osia B., Malkova A.. Break-induced replication: an unhealthy choice for stress relief. Nat. Struct. Mol. Biol. 2017; 24:11–12. [DOI] [PubMed] [Google Scholar]
- 63. Lang K.S., Hall A.N., Merrikh C.N., Ragheb M., Tabakh H., Pollock A.J., Woodward J.J., Dreifus J.E., Merrikh H.. Replication-transcription conflicts generate R-loops that orchestrate bacterial stress survival and pathogenesis. Cell. 2017; 170:787–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Lin Y.L., Pasero P.. Transcription-replication conflicts: orientation matters. Cell. 2017; 170:603–604. [DOI] [PubMed] [Google Scholar]
- 65. Yadav P., Owiti N., Kim N.. The role of topoisomerase I in suppressing genome instability associated with a highly transcribed guanine-rich sequence is not restricted to preventing RNA:DNA hybrid accumulation. Nucleic Acids Res. 2016; 44:718–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Pandey S., Ogloblina A.M., Belotserkovskii B.P., Dolinnaya N.G., Yakubovskaya M.G., Mirkin S.M., Hanawalt P.C.. Transcription blockage by stable H-DNA analogs in vitro. Nucleic Acids Res. 2015; 43:6994–7004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Frank-Kamenetskii M.D., Mirkin S.M.. Triplex DNA structures. Annu. Rev. Biochem. 1995; 64:65–95. [DOI] [PubMed] [Google Scholar]
- 68. Shin J.H., Kelman Z.. The replicative helicases of bacteria, archaea, and eukarya can unwind RNA-DNA hybrid substrates. J. Biol. Chem. 2006; 281:26914–26921. [DOI] [PubMed] [Google Scholar]
- 69. Peleg M., Kopel V., Borowiec J.A., Manor H.. Formation of DNA triple helices inhibits DNA unwinding by the SV40 large T-antigen helicase. Nucleic Acids Res. 1995; 23:1292–1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Krasilnikov A.S., Podtelezhnikov A., Vologodskii A., Mirkin S.M.. Large-scale effects of transcriptional DNA supercoiling in vivo. J. Mol. Biol. 1999; 292:1149–1160. [DOI] [PubMed] [Google Scholar]
- 71. Belotserkovskii B.P., De Silva E., Tornaletti S., Wang G., Vasquez K.M., Hanawalt P.C.. A triplex-forming sequence from the human c-MYC promoter interferes with DNA transcription. J. Biol. Chem. 2007; 282:32433–32441. [DOI] [PubMed] [Google Scholar]
- 72. Lyamichev V.I., Mirkin S.M., Frank-Kamenetskii M.D., Cantor C.R.. A stable complex between homopyrimidine oligomers and the homologous regions of duplex DNAs. Nucleic Acids Res. 1988; 16:2165–2178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Semerad C.L., Maher L.J. 3rd. Exclusion of RNA strands from a purine motif triple helix. Nucleic Acids Res. 1994; 22:5321–5325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Fadloun A., Le Gras S., Jost B., Ziegler-Birling C., Takahashi H., Gorab E., Carninci P., Torres-Padilla M.E.. Chromatin signatures and retrotransposon profiling in mouse embryos reveal regulation of LINE-1 by RNA. Nat. Struct. Mol. Biol. 2013; 20:332–338. [DOI] [PubMed] [Google Scholar]
- 75. Stevanoni M., Palumbo E., Russo A.. The replication of frataxin gene is assured by activation of dormant origins in the presence of a GAA-repeat expansion. PLoS Genet. 2016; 12:e1006201. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.