Abstract
Arbuscular mycorrhizal fungi (AMF) are a group of soil microorganisms that establish symbioses with the vast majority of land plants. To date, generation of AMF coding information has been limited to model genera that grow well axenically; Rhizoglomus and Gigaspora. Meanwhile, data on the functional gene repertoire of most AMF families is non-existent. Here, we provide primary large-scale transcriptome data from eight poorly studied AMF species (Acaulospora morrowiae, Diversispora versiforme, Scutellospora calospora, Racocetra castanea, Paraglomus brasilianum, Ambispora leptoticha, Claroideoglomus claroideum and Funneliformis mosseae) using ultra-low input ribonucleic acid (RNA)-seq approaches. Our analyses reveals that quiescent spores of many AMF species harbour a diverse functional diversity and solidify known evolutionary relationships within the group. Our findings demonstrate that RNA-seq data obtained from low-input RNA are reliable in comparison to conventional RNA-seq experiments. Thus, our methodology can potentially be used to deepen our understanding of fungal microbial function and phylogeny using minute amounts of RNA material.
Keywords: arbuscular mycorrhizal fungi, transcriptomics, phylogenomics, orthology clustering
Introduction
Arbuscular mycorrhizal fungi (AMF) are peculiar organisms that evolved an obligate symbiotic relationship with plant roots. This mutually beneficial association occurs in more than 80% of plant species1 and is suggested to have facilitated the colonization of land by plants.2,3 In this partnership, plants provide the fungal partner with photosynthetic carbon sources and lipids in exchange for micro- and macro-elements (e.g. phosphates and nitrates) scavenged by AMF from the surrounding soil environment.4 AMF also interact with other soil microorganisms and are known drivers of ecosystem biodiversity, as they increase plant nutrient uptake and growth, along with resistance to pathogens.5,6 AMF spores and hyphae also harbour thousands of nuclei flowing in a common cytoplasm (a coenocyte) and were thought to be asexual organisms. The genomic organization of this nuclear population has long remained a puzzle,7–10 but recent data from the model species Rhizoglomus irregulare (also known as Rhizophagus irregularis and misidentified as Glomus intraradices11,12) showed the existence of conventional homokaryotic/dikaryotic patterns in this species that drive mating in sexual fungi.13,14
To date, AMF taxonomy has been based on spore morphology (presence/absence of several contrasting layers, textures and shape) and analyses of ribosomal ribonucleic acid genes (rRNA).15,16 Using the small subunit rRNA sequences (SSU), AMF were included in a monophyletic phylum called Glomeromycota,17 but recent phylogenomic data led to the erection of a new phylum called Mucoromycota, in which AMF now have their own subphyla, the Glomeromycotina.18 A total of 288 AMF species have been described, of which 60% were sequenced for at least one of ribosomal marker.19 The taxonomy of this group experience frequent reshufflings of nomenclature amendments.20,21 To alleviate this, it was proposed that AMF taxonomy should move towards a functional era22 or that phylogenomics should be used to elucidate their clade affiliations.19
Advances in sequencing technologies have geared us towards a functional and genome-based Glomeromycotina research, as evidenced by the recent publication of transcriptomes23–27 and genome drafts28,29 from two model AMF genera, namely those of R. irregulare and R. clarus, and those of sister-species G. rosea and G. margarita. Available data have provided essential information on AMF symbiosis, revealing conserved features for obligate biotrophy, effector proteins and genes involved in their molecular interactions with hosts and endosymbiotic bacteria. However, coding sequence data from hundreds of pot-cultured AMF species is virtually non-existent.
The goal of our study is to acquire primary functional and phylogenetic information from quiescent spores of poorly studied AMF. To this end, we used an approach originally designed for single cell transcriptomics.30 In principle, this methodology could generate coding sequence information from minute amounts of RNA extracted from quiescent spores cultured in pots, while simultaneously leaving-out most of the bacterial contamination.
We show that this method can lead to successful acquisition of large-scale coding data from spores of eight AMF genera (Claroideoglomus, Funneliformis, Paraglomus, Ambispora, Acaulospora, Diversispora, Dentiscutata and Scutellospora) using ultra-low amounts of RNA. This data provides a first insight into the functional diversity of species for which no coding sequence is available, and is readily usable for mapping, orthology assignment and phylogenetic analyses. Our data compares favourably against reference transcriptomes generated from in vitro cultured species, suggesting that this method could be used for comparing expression levels among different AMF life-stages or across spatial locations within cultures, including in plantae.
Materials and methods
Isolation and preparation of the biological material
In this study, quiescent spores from a total of nine AMF species were harvested, including Rhizoglomus irregulare (DAOM-234181), Funneliformis mosseae (DAOM-236685), Acaulospora morrowiae (INVAM-CR315B), Diversispora versiforme (INVAM-W475-40), Scutellospora calospora (INVAM-IL209), Racocetra castanea (BEG-1), Paraglomus brasilianum (DAOM-240472) and Ambispora leptoticha (INVAM-JA116). Spores were thoroughly identified and maintained in Agriculture and Agri-food Canada (AAC) Glomeromycotina in vivo pot culture collections. This fungal collection is commonly used by international researchers interested in all aspects of AMF biology, and are regularly checked for contamination using both morphological and molecular techniques (i.e. small subunit of the rRNA gene, SSU; H+ ATPAses).
For all species, only quiescent spores with intact morphology and lipid content were used for downstream analyses using an inverted microscope. Spores were originally collected by using a sieve cascade of descending size ranging from 500 µm to 63 µm. Lowest fractions were extracted, placed in a 50 ml Falcon tube with the addition of 50% w/w sucrose solution. After centrifugation at 5,000 g for 5 min, supernatant was collected for single spore isolation, and species identification was confirmed using morphological descriptions detailed in Supplementary Figures S1.1–S1.9, and by sequencing the SSU using DNA isolated from random spores. Claroideoglomus claroideum (DAOM-234280) was obtained from an in vitro culture on M medium31 in symbiosis with Agrobacterium-transformed Daucus carota roots. Spore surface sterilization was performed three times for each spore using successive baths of Chloramine-T 2% Tween 20 for 2 min and sterile distilled water for 1 min. Spores were incubated overnight in a solution of streptomycin 0.2 mg/ml and gentamycin 0.1 mg/ml and washed in a final bath of sterile distilled water. Sterilized spores were stored at 4°C.
RNA extraction, quantification, and cDNA synthesis
RNA was extracted using the NucleoSpin RNA XS kit (Macherey-Nagel, Germany) under a sterilized laminar flow hood. Spores were crushed in sterile nuclease-free 1.5 ml Eppendorf tubes with a micro-pestle and the RNA isolation step followed manufacturer’s recommendations. The cDNA was produced using the SMARTer Ultra Low Input RNA Kit for sequencing v4 used for single cell transcriptomic (SCT) (Takara Bio USA Inc.) following manufacturer’s recommendations. Adapters were used as template for cDNA synthesis and downstream PCR for the cDNA amplification. Two positive controls (Diluted Control RNA) provided with the kit and one negative control (no sample) were performed. The resulting cDNA was purified using the PCR-Purification kit (QIAGEN), and quantification and quality assessment was performed using a Qbit 2.0 fluorimeter with the dsDNA HS (High Sensitivity) Assay Kit.
cDNA illumina sequencing and reads assembly
The cDNA of all species was processed using the Nextera-XT DNA library preparation kit (Illumina). The resulting libraries were quality checked using a high-throughput bioanalyzer (Caliper LabChip GX instrument), and sequenced using one full lane of Illumina HiSeq 2500 instrument, using the High-Output V4 mode and paired-reads 2x125 bp (Fasteris SA, Switzerland). The generated paired-end reads were trimmed using the TrimGalore v.0.4.1 software with a Phred score cut-off of 20 and TruSeq Illumina adapter sequences were removed with CutAdapt v.1.8.3. Trimmed reads were assembled de novo using Trinity v.2.1.1.32 A genome-guided assembly was performed for R. irregulare reads obtained by SCT. In this case, paired-end reads were mapped back against the reference genome assembly and all the CDS, respectively, using the Burrows–Wheeler alignment tool (BWA v0.7.10),33 with the BWA-MEM algorithm. SAMtools v0.1.1934 was used to convert SAM files into sorted BAM. The assembly results and statistics are available in supporting information (Supplementary Table S1). Raw reads are deposited in GenBank (accession number SRX2583204-SRX2583220). Assemblies and annotations are available at https://github.com/zygolife/AMF_Phylogenomics (14 November 2017, date last accessed).
Functional annotation pipeline
SCT data and reference transcriptomes of R. irregulare and G. rosea23,25 annotated using the Trinotate v.2.0.2 annotation pipeline. The pipeline makes use of a wide range of algorithms, methods and databases for functional annotation. These include blastx and blastp35 against UniRef and SwissProt databases, HMMER3,36 the Pfam database,37 SignalP38 tmHMM,39 eggnog, GO40 and KEGG.41 Non-redundant virtual transcripts (NRVTs) for each strain were obtained by filtering the predicted Trinity genes for unique occurrences using the text manipulation tool of the Galaxy webserver.42 The annotated data set were filtered by separating NRVTs into taxonomic-affiliated groups (i.e. Glomeromycotina, Other Fungi, Other Eukaryotes, Bacteria, Archaea, Plantae and Virus). The Glomeromycotina NRVTs predicted GO-terms were used as input in the Web Gene Ontology Annotation Plot (WEGO)43 to obtain a hierarchical histogram of the GO ontology related functions. All taxonomic-sorted NRVTs were fed into the TransDecoder v.3.0.1 software to predict their ORFs (>100 aa). ORFs were used as input in the webMGA webserver44 to obtain KOG classes’ functions and protein domain families.
Orthology clustering analysis
Orthologous affiliation and shared functionality of NRVTs was obtained using FastOrtho, an OrthoMCL-based program.45 Transdecoder v.3.0.1 predicted protein sequences (>100 aa) of four NRVT classes (Glomeromycotina-, Unknown-, Other Eukaryotes-, Prokaryotes-annotated) were used as inputs with an e-value cut-off of 1e−05. The output was filtered with the Galaxy webtool42 to identify orthogroups shared among AMF strains and the number of genes present in these groups. BUSCO analyses (v.2.0) were performed on the Glomeromycota-annotated NRVTs, orthologous shared Unknown- and Other Eukaryotes-annotated NRVTs using the fungi_odb9 database and the species Rhizopus oryzae (now known as R. delemar) as reference. In total, 290 core orthologues were assessed for their presence in each transcriptome, respectively.
RNA extraction for qPCR
Quantitative real-time PCR (qPCR) was used to verify the expression of random regions of the transcriptome obtained using ultra-low input RNA methods. Expression of each region was measured in three biological replicates of R. irregulare consisting of cDNA produced using RNA from approximately 250 spores. Total RNA was extracted using the Qiagen RNeasy Plant Mini Kit (Qiagen, Venio, Netherlands) and DNA contamination was removed using the RapidOut DNA Removal Kit (ThermoFisher Scientific). RNA was subjected to RT-PCR using the iScript cDNA synthesis kit (Bio-Rad Laboratories, Hercules, CA, USA) following manufacturer’s recommendation. Six regions of the AMF transcriptome were selected (6230, 8946, 1110, 9312, 910 and α-tubulin) for analysis using qPCR and corresponding primers were designed (Supplementary Table S2). Real-time PCR reactions were carried out in a CFX 96 thermal cycler (Bio-Rad Laboratories) in technical duplicates and consisted of 4.5 µl of H2O, 1 µl each of forward and reverse primers, 7.5 µL of SsoFast 2X master mix (Bio-Rad Laboratories) and 1 µl of undiluted cDNA. The qPCR protocol consisted of an initial incubation at 95°C for 30 s followed by 40 cycles of 95°C for 5 s and 59.3°C for 5 s and a final melt curve from 60 to 95°C with 0.5°C increments. The primer efficiency set was determined using a serial dilution, temperature gradient and efficiency between 90% and 110%.
Phylogenomic analysis
A phylogenomic pipeline was applied to a set of 434 conserved genes.18 The 40 species selected include the 8 newly sequenced AMF species, 2 previously published references of R. irregulare and G. rosea, 8 other Mucoromycota representatives, 8 Dikarya fungi, 7 Zoopagomycota, four Chytridiomycota, 2 Blastocladiomycota and 1 Cryptomycota (Rozella). For AMF, the following number of partial sequences were recovered as best reciprocal hit, an approach that was shown to often outperformed projects with more complex algorithms including OrthoMCL,46 to an hmmsearch reflecting the relative completeness of these transcriptomes for these query proteins: Acaulospora morrowiae, 337 genes; Ambispora leptoticha, 206 genes; Claroideoglomus_claroideum, 313 genes; Diversispora versiforme, 222 genes; Funneliformis mosseae, 426 genes; Gigaspora rosea, 385 genes; Paraglomus occultum, 334 genes; Racocetra castanea, 428; Scutellospora calospora, 427 genes; Rhizophagus irregularis, 423 genes.
The tree was rooted with the choanoflagellate Monosiga brevicolis. The pipeline, including all associated data is archived in the github repository https://github.com/zygolife/AMF_Phylogenomics (14 November 2017, date last accessed). The approach uses HMMER336 to search the predicted proteins of each strain with profile Hidden Markov Models (HMMs) constructed from a set of conserved markers denoted ‘JGI_1086’ (https://github.com/zygolife/AMF_Phylogenomics (14 November 2017, date last accessed)). A table of the best protein match for each HMM query in each proteome is recorded. A multiple sequence alignment (MSA) is constructed for each marker gene with the single best copy protein from each strain or omitted if no sequence is of sufficient similarity. These MSAs were trimmed with trimAl47 using the automated1 option and requiring all sequences to match at least 50% of residues with an overlap with at least 60% of the rest of the sequences (-resoverlap 0.50 -seqoverlap 60). The resulting individual protein alignments were concatenated into a single alignment with one sequence per species. A bootstrapped maximum likelihood phylogenetic tree was inferred from this alignment using RAxML v8.2.848 with rapid bootstrapping (-f a -o Mbre -m PROTGAMMALG -s AMF.2016_Oct_06.JGI1086.41sp.fasaln -n Standard.AMF.2016_Oct_06.JGI1086.41sp -N autoMRE –d). The resolved tree was visualized in FigTree (http://tree.bio.ed.ac.uk/software/figtree/ (14 November 2017, date last accessed)). In parallel, the CAT+Γ4 model of amino acid substitution implemented in PhyloBayes 3 was also used.49 In this case, two concurrent chains were run in parallel and were terminated manually after 1,116 cycles. Both methodologies varied only in their placement of Chytidiomycota and Blastocladiomycota as the most basal fungal phylum.
Results and discussion
Acquisition of transcriptome data from poorly studied AMF species
Initial attempts to obtain sufficient RNA from 1 to 20 single AMF spores resulted in unsuccessful cDNA amplifications. However, subsequent RNA extractions from more spores (n = 22–175, Fig. 1) resulted in successful extractions and sequencing. The size of spores appeared to play a major role in the amount of RNA extracted, with taxa in the Gigasporaceae harbouring more RNAs than relatives with smaller spores (for variation in spore shape and size see Supplementary Figure S1). The RNA extraction ranged from a mere 3 ng for P. brasilianum to a maximum of 720 ng for S. calospora. Assembly statistics vary substantially among the data sets (Supplementary Table S1), but high RNA yield did not necessarily result in better assemblies. The lowest and highest number of transcripts and gene predictions were found for the S. calospora and D. versiforme related data, respectively. These also show the highest and lowest assembly sizes and predictions, while GC% was found to be lowest for transcriptome isolated from C. claroideum spores (30%). SCT transcriptomes from R. irregulare (260 bp) and S. calospora (1,091 bp) had the smallest and largest N50, respectively.
Figure 1.
Summary table of the growth conditions, characteristics, phylogenetic affiliations, collection identifiers and SCT protocol status of all the AMF species selected in this study. The phylogeny is based on the SSU (full)-5.8S-LSU rDNA subunit and is a representation of the tree published by Kruger et al.12 The circles correspond to successful cDNA amplification according to the range expected by the SMARTer Ultra Low Input RNA Kit for Sequencing v4 (Takara Bio USA Inc.).
SCT from single spores vs conventional culture-based procedures
To determine the suitability of the SCT approach for reference-based transcriptomics, we mapped our SCT-based ‘pot-cultured’ R. irregulare reads obtained from single spores against an available reference genome from this species.28 This analysis compares the diversity of sequence reads obtained from a few quiescent spores to those previously obtained by others on the same species using much larger amounts of RNA and conditions (axenic growth, germinating spores, in Plantae, etc.).23 Our analyses showed that 86% of SCT reads from single spores map against the reference, 72% of which are properly paired with an average coverage of 42X. Moreover, 89.6% of known R. irregularis CDS28 are mapped by our SCT reads (Supplementary Table S3) with coverage ranging from 11 to 50. Read mapping considerably improved our original R. irregularis de novo assembly (from 61,050 to 36,848 predicted genes).
When the longest isoform of each predicted gene is considered, the taxonomic distribution of NRVTs from SCT and the reference data set are largely comparable (Fig. 2A). The NRVTs and Glomeromycotina-annotated genes are 23,753 vs 25,906 and 51.8 vs 55.7% for the SCT and reference based data, respectively, and proportion of prokaryotic NRVTs is very low in both cases (0.8% and 1.3%). When transcriptomes are annotated using the same methodology, both conditions show identical GO-term functions with surprisingly comparable percentages for the vast majority of categories, around the 50% parity threshold (Fig. 2B). Mapping S. calospora and R. castanea SCT reads against an assembly and CDS from a similar Gigasporacea taxon (kindly provided by Christophe Roux) leads to similar findings. Overall, these analyses indicate that the SCT method does not introduce strong biases in functional annotation and read mapping, and could be applicable to transcriptomic analyses of other AMF stages, tissues etc.
Figure 2.
(A) Taxonomic distribution comparison of the annotated NRVTs between the R. irregulare reference transcriptome28 and the genome-guided assembly of the in vivo R. irregulare transcriptome obtained from 150 spores. Regions corresponding to the percentage of glomeromycotina with GO-terms, glomeromycotina, other fungi, other eukaryotes, prokaryotes and unknown affiliation annotated NRVTs, respectively are shown. The numbers located at the end of the histogram bars represents the total number of NRVTs. (B). Histogram representing the % of GO-terms hierarchical functions (cellular component, molecular functions and biological process) associated to the glomeromycotina with GO-terms annotated-NRVTs. The comparison is made between the reference R. irregulare transcriptome23 and the genome-guided assembly of the in vivo R. irregulare transcriptome obtained by SCT from 150 spores.
Taxonomical and functional analysis of SCT-based transcriptomes
Using identical pipelines and procedures, we compared the NRVTs taxonomic distribution and functional annotations of all SCT data with a reference transcriptome from G. rosea.25 To avoid the analysis of potential contaminants, all functional analyses were performed exclusively on Glomeromycota orthologues shared among samples analysed in this study, and no evidence of cross-contamination among our samples was found by inspecting 100 random orthologue alignments. These analyses represent a first snapshot of the coding diversity of an AMF life-stage (the spore) typically seen as dormant, from eight species for which no coding sequence is currently available. However, this data does not represent an analysis of species-specific transcript due to lack of intra-sample replication. Reference G. rosea data used for comparisons comes from a variety of conditions (in vitro, in vivo, spores, extra-radical mycelium, etc.). The G. margarita transcriptome24 was not analysed here to avoid redundancy—i.e. both taxa represent sister-species and data sets harbour essentially the same number of NRVTs; ≈ 80,00025).
Overall taxonomic distributions of SCT transcriptomes showed remarkable similarities with that of G. rosea; particularly for the best assembly (S. calospora, R. castanea, C. claroideum, F. mossea) (Fig. 3). When only Glomeromycotina-annotated NRVTs are considered, the predicted proteins can be classified into 25 KOG functional classes. In support of recent genome analyses, the most represented KOG classes for all transcriptomes were the signal transduction mechanisms and post-translational modifications, with most represented molecular functions being binding and catalytic activity. The majority of biological processes related to GO-terms were for the cellular and metabolic process (Supplementary Tables S4 and S5). Unsurprisingly, most NRVTs have no known function and/or taxonomic affiliation. Prokaryotic NRVTs are abundant in four species (Dve, Pbr, Ale and Amo) (40.3, 18.9, 24 and 26.4%, respectively) (Fig. 3, Supplementary Table S6), and many of these show high sequence similarity with known AMF or fungal prokaryotic endosymbionts,24,50,51 especially to Glomeribacter gigasporarum. Most putative bacterial transcripts are involved in translation structure and biogenesis, as well as amino-acid and energy metabolism, Supplementary Table S7).
Figure 3.
Taxonomic distribution and comparison of the annotated NRVTs between all AMF SCT transcriptomes and the G. rosea reference transcriptome.25 Regions corresponding to the percentage of Glomeromycotina with GO-terms, glomeromycotina, other fungi, other eukaryotes, prokaryotes and unknown affiliation annotated NRVTs, respectively are shown. The numbers located at the end of the histogram bars represents the total number of NRVTs.
Orthology analysis and SCT completeness
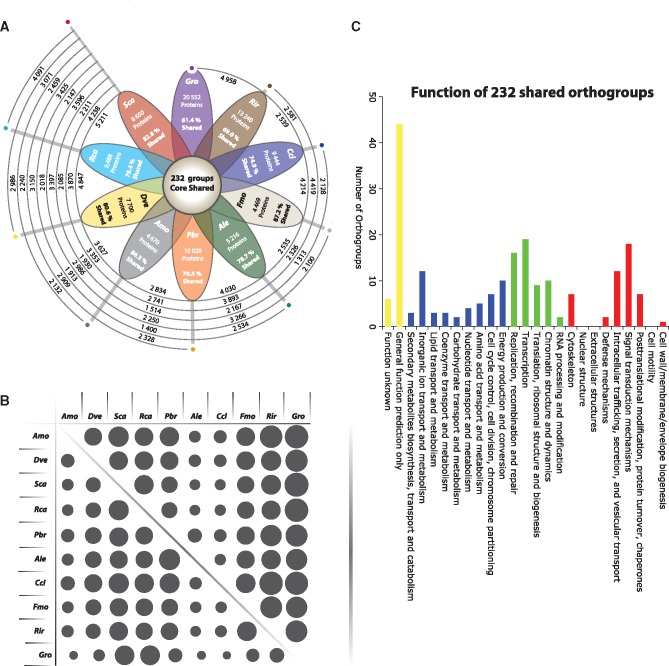
To identify conservation across data set, an orthology clustering analysis was performed on the Glomeromycotina-, Unknown- and Eukaryote-annotated NRVTs that clustered within orthogroups (Fig. 4, Supplementary Figure S2). The Glomeromycotina-annotated subgroups are surprisingly conserved among transcriptomes. Specifically, a total of 232 orthogroups were found to be shared by all transcriptomes, and the percentage of shared transcripts among SCT data sets ranged from 61.4% to 87.2%. The function of these shared AMF genes broadly reflects that of the Glomeromycotina-annotated NRVT data set. Orthogroup conservation also shows evidence of correlation with phylogenetic relationships (Supplementary Table S8). The presence of 6 orthologues involved in different biological categories was further validated using RT-qPCRs procedures using RNA extracted from R. irregularis spores (Supplementary Figure S3). Interestingly, conservation across data sets extends to NRVTs with unknown functions and Eukaryotic homologues, although none of these were found to be shared by all 10 data sets (Supplementary Figure S2 and Table S8).
Figure 4.
Gene orthology affiliations where each petal represents a single species. The total number of proteins included in the orthology analysis and the percentage of total shared protein is shown within the flower petals, along with the species identifier (Gro: Gigaspora rosea Reference; Rir: Rhizoglomus irregulare Reference; Ccl: Claroideoglomus claroideum; Fmo: Funneliformis mosseae; Ale: Ambispora leptoticha; Pbr: Paraglomus brasilianum; Amo: Acaulospora morrowiae; Dve: Diversispora versiforme; Rca: Racocetra castanea; Sca: Scutellospora calospora). The flower button shows the number of orthogroups shared among all species. The extruding spirals show the number of orthogroups shared between the individual species, where the coloured dots above the petals represent the starting point to follow the relationships, with the corresponding values always below the guiding line (towards the flower). (A) The flower shows the orthologous relationships for the Glomeromycotina-annotated proteins (>100 aa) in a clockwise manner. (B) Orthologous protein shared % matrix—linked to the orthology flowers above them. Each dot varies in size proportionally to the percentage of shared proteins between individual species in a reciprocal fashion, depending on which side of the oblique lines, the value is read. (C). Distribution of high level functional annotations from glomeromycota-shared orthogroups. Bars represent orthogroups involved in ‘Cellular Processes and Signaling’, ‘Information Storage and Processing’, ‘Metabolism’ and those that are ‘Poorly Characterized’.
BUSCO benchmark analyses, based on 290 core Eukaryotic fungal genes used by the program to infer genome completeness,52 shows strong correlation between the N50 and the performance on the benchmark genome (Rhizopus oryzae, Fig. 5). Reference transcriptomes lack between 13.1% and 22.1% of genes, compared with 21.4–77.6% for the SCT samples. Generally, reference RNA-seq data are more complete, and less fragmented, with the exception of data from S. castanea. Each transcriptome contained between 126 and 1444 (Amo and Ale, respectively) unique glomeromycotan transcripts. Their AMF origin indicates that homologues of these genes are also present in other members of the phylum. The 30 most represented data set-specific families include homologues of known AMF genes (tyrosine kinases, extracellular proteins SEL-1, proteins containing BTB/POZ and Kelch domains, TRAP230 subunit hormone receptors, Supplementary Table S9). All these homologues are also found in publicly available AMF genome data. Importantly, our data also confirms the absence of most homologues of Glomeromycotina core genes (MGCGs; a term recently coined by Tang et al. 201625; Supplementary Table S10).
Figure 5.
Transcriptome representativeness comparison among the AMF SCT data sets and the reference R. irregulare and G. rosea RNA-seq, performed using a BUSCO benchmark. In total, 290 core fungal genes were search across these data sets and evaluated for completeness, single-copy (S), duplication (D), fragmentation (F) and absence (M). The histogram representations shows the percentage of the genes (and number indicated above the bars) belonging in these categories, for each species.
Reconstruction of the AMF phylogeny using new transcriptome data
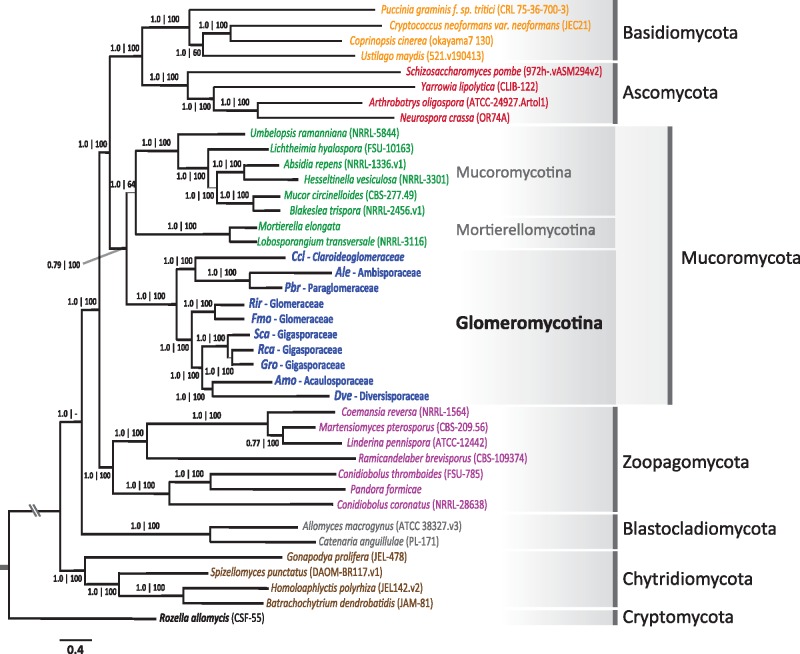
Aligning 434 putative single-copy genes (139,586 sites) found in all SCT data and distant fungal relatives in the Zoopagomycota, Blastocladiomycota, Chytridiomycota, Cryptomycota resulted in a highly supported fungal phylogeny that is consistent with recently published results based on large phylogenomic data set (Fig. 618) Monophyly of all recently proposed phyla is supported by both Maximum Likelihood and Bayesian methodologies. The Zoopagomycota is sister to the Dikarya and Mucoromycota clade. Within Mucoromyota, the Glomeromycotina is monophyletic and sister to the Mucoromycotina and Mortierellomycotina clade, though there is no strong support for their specific branching.
Figure 6.
Phylogenetic placement of the eight AMF families within Fungi. Bayesian phylogenetic tree of selected fungal representatives of the Kingdom Fungi is based on the concatenated alignment of 434 conserved orthologous proteins and more than 120 K trimmed amino acid positions. Numbers below or above branches correspond to Bayesian inference value obtained with PhyloBayes (two concurrent chains were run for 1,116 cycles using the CAT model) and bootstrap support values obtained using Maximum likelihood, respectively. The corresponding fungal phylum of each main clades are identified on the right side of the figure and is highlighted by the gray shaded area.
Within the Glomeromycotina, most known phylogenetic relationships based morphology and SSU trees are strongly supported. Specifically, our analyses confirms the basal position of the Ambisporaceae and Paraglomeraceae in the AMF tree, groups together members of the Gigasporaceae (Scutellospora, Gigaspora, Racocentra) and Glomeraceae (Rhizophagus and Funneliformis), and supports the evolutionary relationship between Acaulosporacea and Diversisporaceae.12 The only conflict with past phylogenies is represented by C. claroideum, which clusters with A. leptothica and P. brasiliensis with strong support.
SCT exposes the RNA diversity of overlooked AMF spores and new phylogenetic relationships within the glomeromycotina
Our approach based on low-input RNA produced transcriptomes with high conservation in sequence and function across the AMF phylogeny, and revealed a diverse functional diversity expressed in life-stages that are supposedly dormant. We confirm that MGCGs orthologues involved in invertase activity (SUC2 homolog) are likely absent in this phylum. The absence of the invertase orthologues in 10 transcriptomes comforts the ‘Glomeromycotina core symbiotic concept’, which proposes that the host plant is responsible for sucrose production and hydrolysis.
Spores of all species express many copies of BTB/POZ and vprBP homolog domain, which are known facilitators of the ubiquitin-protein ligase complexes.53 ‘Kelch domains’, which form general protein–protein interaction modules, are also very abundant in all transcriptomes.54 Spores also express many alpha amylase related proteins in all data sets, as well as several glycoside hydrolase, lipases and proteases. These possibly confer AMF the molecular toolkit to enzymatically breakdown complex carbohydrates (polysaccharides), multiple protein and lipid substrates, either from the environment, their host or cell energy reserves.55,56 Our study also revealed thousands of ‘unknown’ glomeromycotan coding sequences shared among all transcriptomes. Their abundance and high conservation across data sets highlights the importance of this ‘uncategorized’ realm of genes for the AMF biology.
AMF spores also share a number of transcripts encoded by putative AMF symbionts, including G. gigasporarum and mollicutes-related endosymbionts (MREs). Their genomes harbour genes linked to their host metabolism, in the intricate molecular dialogue between AMF and their intra-cellular inhabitants; particularly the expression of AMF genes involved in growth, development and transport in its fungal host.24,51,57 Our data shows that endosymbionts are transcriptionally active in AMF spores, expressing a variety of functions, including translation, amino-acid metabolism or energy production.
The multigene fungal phylogeny we report also show that acquisition of larger coding data set could result in the reorganization of certain AMF families. This is particularly true for the Claroideoglomeraceae, Ambisporaceae and Paraglomeraceae, which group together with full support in our analyses but not in rRNA based phylogenies.11 It will be interesting to see if the addition of new taxa to this multi-gene alignment—e.g. Geosiphon, Archeospora, Pacispora—will reshuffle this novel evolutionary affiliation, as the genus Claroideoglomus has been typically associated with the Glomeraceae, though with much lower support. Our low-input RNA method could also represent a stepping stone for producing future solid and durable AMF classifications,19 and could help selecting suitable protein-coding markers that are representative of the global phylogenetic signal. Such data is crucial to enter a new era of AMF classification based on functionality, as proposed by Gamper et al.22
Ultra-low input RNA methods as a tool to deepen our understanding of AMF biology and evolution
Here, we showed that the methodologies based on ultra-low input RNA represents reliable avenues to obtain large AMF coding sequence data and important functional insights from minute amounts of RNA extracted from spores of recalcitrant AMF species. These results are a solid foundation to build future knowledge on other species in the group, and deepen our understanding of this ecologically important group. The data we collected also revealed that spores from many under sampled AMF taxa express a wide panel of transcripts. Some of these could be ‘ready-made’ for germination and/or plant colonization, although it is still unclear whether all spores express these transcripts at significant levels for biological function.
From an evolutionary perspective, the wealth of data we provide uncovered phylogenetic relationships that were previously shadowed by a focus on single rRNA gene sequences. Within this context, this transcriptomics data opens the door to phylogenetic classifications that develops markers representative of the global classification and suited for functional ecological studies. In the future, the methods we used could prove useful to assess tissue-specific functionalities, and may offer further insights into the plant–fungus dialogue based on RNA-seq data. Indeed, we have shown that sequencing reads obtained from few spores results, in many cases, in information comparable to that obtained from in vitro cultures. If the trend holds, our method could be applied to study, for instance, how transcription changes along different portions of the mycelium or in plantae. Ultimately, such advances will be essential to better understand how AMF benefit the health of plants or terrestrial ecosystems; processes that are linked to crucial societal challenges such as climate change, sustainable management, pest invasion and food security.
Supplementary Material
Acknowledgements
We thank anonymous reviewers for their helpful comments on a previous version of the manuscript, and Timea Marton for assistance with RNA extraction procedures.
Funding
This work was supported by the Discovery program from the Natural Sciences and Engineering Research Council of Canada (NSERC-Discovery) and an Early Researcher Award from the Ontario Ministry of Research and Innovation (ER13-09-190) to N.C. This work was partially supported by US NSF project grant ZyGoLife DEB-1441715 to J.E.S. and DEB-1441677 to T.J. Phylogenomic analyses were performed on the high performance computing resources at the Institute for Integrative Genome Biology at University of California-Riverside supported by NSF DBI-1429826 and NIH S10-OD016290. Many of the available genomes were generated under the 1,000 Fungal Genomes project at the Joint Genome Institute. Any opinions, findings, conclusions, or recommendations expressed in this publication are those of the author(s) and do not necessarily reflect the view of the National Science Foundation. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Accession numbers
The raw sequencing reads have been deposited in GenBank under these accession number SRX2583204-SRX2583220. The assemblies and annotations are available through https://github.com/zygolife/AMF_Phylogenomics/tree/master/SCT_ALL_DATA (14 November 2017, date last accessed).
Conflict of interest
None declared.
Supplementary data
Supplementary data are available at DNARES online.
References
- 1. Smith S., Read D.. 2008, Mycorrhizal Symbiosis, 3rd edition AP (Academic Press), Cambridge, Massachusetts. [Google Scholar]
- 2. Bonfante P., Genre A.. 2010, Mechanisms underlying beneficial plant–fungus interactions in mycorrhizal symbiosis, Nat. Commun., 1, 1–11. [DOI] [PubMed] [Google Scholar]
- 3. Humphreys C.P., Franks P.J., Rees M., Bidartondo M.I., Leake J.R., Beerling D.J.. 2010, Mutualistic mycorrhiza-like symbiosis in the most ancient group of land plants, Nat. Commun., 1, 103. [DOI] [PubMed] [Google Scholar]
- 4. Parniske M. 2008, Arbuscular mycorrhiza: the mother of plant root endosymbioses, Nat. Rev. Microbiol., 6, 763–75. [DOI] [PubMed] [Google Scholar]
- 5. van der Heijden M.G.A., Klironomos J.N., Ursic M., et al. 1998, Mycorrhizal fungal diversity determines plant biodiversity, ecosystem variability and productivity, Nature, 396, 69–72. [Google Scholar]
- 6. Berruti A., Lumini E., Balestrini R., Bianciotto V.. 2015, Arbuscular mycorrhizal fungi as natural biofertilizers: let’s benefit from past successes, Front. Microbiol., 6, 1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Young J. P. 2015, Genome diversity in arbuscular mycorrhizal fungi, Curr. Opin. Plant Biol., 26, 113–9. [DOI] [PubMed] [Google Scholar]
- 8. Kuhn G., Hijri M., Sanders I.R.. 2001, Evidence for the evolution of multiple genomes in arbuscular mycorrhizal fungi, Nature, 414, 745–8. [DOI] [PubMed] [Google Scholar]
- 9. Hijri M., Sanders I.R.. 2005, Low gene copy number shows that arbuscular mycorrhizal fungi inherit genetically different nuclei, Nature, 433, 160–3. [DOI] [PubMed] [Google Scholar]
- 10. Pawlowska T.E., Taylor J.W.. 2004, Organization of genetic variation in individuals of arbuscular mycorrhizal fungi, Nature, 427, 733–7. [DOI] [PubMed] [Google Scholar]
- 11. Schussler A., Walker C.. 2010, The Glomeromycota: a species list with new families. http//www.amf-phylogeny.com (14 November 2017, date last accessed).
- 12. Kruger M., Kruger C., Walker C., Stockinger H., Schussler A.. 2012, Phylogenetic reference data for systematics and phylotaxonomy of arbuscular mycorrhizal fungi from phylum to species level, New Phytol., 193, 970–84. [DOI] [PubMed] [Google Scholar]
- 13. Corradi N., Brachmann A.. Fungal Mating in the Most Widespread Plant Symbionts? Trends Plant Sci., 64, 175–83. [DOI] [PubMed] [Google Scholar]
- 14. Ropars J., Toro K.S., Noel J., et al. 2016, Evidence for the sexual origin of heterokaryosis in arbuscular mycorrhizal fungi, Nat. Microbiol., 1, 16033.. [DOI] [PubMed] [Google Scholar]
- 15. Krüger M., Stockinger H., Krüger C., Schüßler A.. 2009, DNA-based species level detection of Glomeromycota: one PCR primer set for all arbuscular mycorrhizal fungi, New Phytol., 183, 212–23. [DOI] [PubMed] [Google Scholar]
- 16. Öpik M., Davison J., Moora M., Zobel M.. 2013, DNA-based detection and identification of Glomeromycota: the virtual taxonomy of environmental sequences, Botany, 92, 135–47. [Google Scholar]
- 17. Schussler A., Schwarzott D., Walker C.. 2001, A new fungal phylum, the Glomeromycota: phylogeny and evolution, Mycol. Res., 105, 1413–21. [Google Scholar]
- 18. Spatafora J.W., Chang Y., Benny G.L., et al. 2016, A phylum-level phylogenetic classification of zygomycete fungi based on genome-scale data, Mycologia, 108, 1028–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Öpik M., Davison J.. 2016, Uniting species- and community-oriented approaches to understand arbuscular mycorrhizal fungal diversity, Fungal Ecol., 24, 106–13. [Google Scholar]
- 20. Oehl F., Sieverding E., Palenzuela J., Ineichen K., Alves da Silva G.. 2011, Advances in Glomeromycota taxonomy and classification, IMA Fungus, 2, 191–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Redecker D., Schussler A., Stockinger H., Sturmer S.L., Morton J.B., Walker C.. 2013, An evidence-based consensus for the classification of arbuscular mycorrhizal fungi (Glomeromycota), Mycorrhiza, 23, 515–31. [DOI] [PubMed] [Google Scholar]
- 22. Gamper H.A., van der Heijden M.G.A., Kowalchuk G.A.. 2010, Molecular trait indicators: moving beyond phylogeny in arbuscular mycorrhizal ecology, New Phytol., 185, 67–82. [DOI] [PubMed] [Google Scholar]
- 23. Tisserant E., Kohler A., Dozolme-Seddas P., et al. 2012, The transcriptome of the arbuscular mycorrhizal fungus Glomus intraradices (DAOM 197198) reveals functional tradeoffs in an obligate symbiont, New Phytol., 193, 755–69. [DOI] [PubMed] [Google Scholar]
- 24. Salvioli A., Ghignone S., Novero M., et al. 2016, Symbiosis with an endobacterium increases the fitness of a mycorrhizal fungus, raising its bioenergetic potential, ISME J., 10, 130–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tang N., San Clemente H., Roy S., Becard G., Zhao B., Roux C.. 2016, A survey of the gene repertoire of gigaspora rosea unravels conserved features among glomeromycota for obligate biotrophy, Front. Microbiol., 7, 233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kikuchi Y., Hijikata N., Yokoyama K., et al. 2014, Polyphosphate accumulation is driven by transcriptome alterations that lead to near-synchronous and near-equivalent uptake of inorganic cations in an arbuscular mycorrhizal fungus, New Phytol., 204, 638–49. [DOI] [PubMed] [Google Scholar]
- 27. Sędzielewska Toro K., Brachmann A.. 2016, The effector candidate repertoire of the arbuscular mycorrhizal fungus Rhizophagus clarus, BMC Genomics, 17, 101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Tisserant E., Malbreil M., Kuo A., et al. 2013, Genome of an arbuscular mycorrhizal fungus provides insight into the oldest plant symbiosis, Proc. Natl. Acad. Sci. U S A, 110, 20117–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lin K., Limpens E., Zhang Z., et al. 2014, Single nucleus genome sequencing reveals high similarity among nuclei of an endomycorrhizal fungus, PLoS Genet, 10, e1004078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Shapiro E., Biezuner T., Linnarsson S.. 2013, Single-cell sequencing-based technologies will revolutionize whole-organism science, Nat. Rev. Genet., 14, 618–30. [DOI] [PubMed] [Google Scholar]
- 31. Toussaint J.P., St-Arnaud M., Charest C.. 2004, Nitrogen transfer and assimilation between the arbuscular mycorrhizal fungus Glomus intraradices Schenck & Smith and Ri T-DNA roots of Daucus carota L. in an in vitro compartmented system, Can. J. Microbiol., 50, 251–60. [DOI] [PubMed] [Google Scholar]
- 32. Grabherr M.G., Haas B.J., Yassour M., et al. 2011, Full-length transcriptome assembly from RNA-Seq data without a reference genome, Nat. Biotechnol., 29, 644–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Li H., Durbin R.. 2009, Fast and accurate short read alignment with Burrows-Wheeler transform, Bioinformatics, 25, 1754–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Li H., Handsaker B., Wysoker A., et al. 2009, The sequence alignment/map format and SAMtools, Bioinformatics, 25, 2078–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D. J.. 1990, Basic local alignment search tool, J. Mol. Biol., 215, 403–10. [DOI] [PubMed] [Google Scholar]
- 36. Finn R.D., Clements J., Eddy S.R.. 2011, HMMER web server: interactive sequence similarity searching, Nucleic Acids Res., 39, W29–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Finn R.D., Tate J., Mistry J., et al. 2008, The Pfam protein families database, Nucleic Acids Res., 36, D281–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Petersen T.N., Brunak S., von Heijne G., Nielsen H.. 2011, SignalP 4.0: discriminating signal peptides from transmembrane regions, Nat. Methods, 8, 785–6. [DOI] [PubMed] [Google Scholar]
- 39. Krogh A., Larsson B., von Heijne G., Sonnhammer E.L.L.. 2001, Predicting transmembrane protein topology with a hidden markov model: application to complete genomes1, J. Mol. Biol., 305, 567–80. [DOI] [PubMed] [Google Scholar]
- 40. Ashburner M., Ball C.A., Blake J.A., et al. 2000, Gene ontology: tool for the unification of biology, Nat. Genet., 25, 25–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ogata H., Goto S., Sato K., Fujibuchi W., Bono H., Kanehisa M.. 1999, KEGG: Kyoto encyclopedia of genes and genomes, Nucleic Acids Res., 27, 29–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Afgan E., Baker D., van den Beek M., et al. 2016, The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2016 update, Nucleic Acids Res., 44, W3–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ye J., Fang L., Zheng H., et al. 2006, WEGO: a web tool for plotting GO annotations, Nucleic Acids Res., 34, W293–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wu S., Zhu Z., Fu L., Niu B., Li W.. 2011, WebMGA: a customizable web server for fast metagenomic sequence analysis, BMC Genomics, 12, 444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Li L., Stoeckert C.J., Roos D.S.. 2003, OrthoMCL: identification of ortholog groups for eukaryotic genomes, Genome Res., 13, 2178–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Altenhoff A.M., Dessimoz C., Eisen J.A.. 2009, Phylogenetic and functional assessment of orthologs inference projects and methods, PLoS Comput. Biol., 5, e1000262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Capella-Gutierrez S., Marcet-Houben M., Gabaldon T.. 2012, Phylogenomics supports microsporidia as the earliest diverging clade of sequenced fungi, BMC Biol., 10, 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Stamatakis A. 2006, The RAxML 7.0.4 manual, Bioinformatics, 22, 2688–90. [DOI] [PubMed] [Google Scholar]
- 49. Lartillot N., Lepage T., Blanquart S.. 2009, PhyloBayes 3: a Bayesian software package for phylogenetic reconstruction and molecular dating, Bioinformatics, 25, 2286–8. [DOI] [PubMed] [Google Scholar]
- 50. Castillo D.M., Pawlowska T.E.. 2010, Molecular evolution in bacterial endosymbionts of fungi, Mol. Biol. Evol., 27, 622–36. [DOI] [PubMed] [Google Scholar]
- 51. Desirò A., Salvioli A., Bonfante P.. 2016, Investigating the endobacteria which thrive in arbuscular mycorrhizal fungi In: Martin F., Uroz S., (eds.), Microbial Environmental Genomics (MEG). Springer New York, New York, NY, pp. 29–53. [DOI] [PubMed] [Google Scholar]
- 52. Simão F.A., Waterhouse R.M., Ioannidis P., Kriventseva E.V., Zdobnov E.M.. 2015, BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs, Bioinformatics, 31, 3210–2. [DOI] [PubMed] [Google Scholar]
- 53. Weber H., Hellmann H.. 2009, Arabidopsis thaliana BTB/POZ-MATH proteins interact with members of the ERF/AP2 transcription factor family, FEBS J., 276, 6624–35. [DOI] [PubMed] [Google Scholar]
- 54. Sharma M., Pandey G. K.. 2016, Expansion and function of repeat domain proteins during stress and development in plants, Front. Plant Sci., 6, 1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Singh A. K., Mukhopadhyay M.. 2012, Overview of fungal lipase: a review, Appl. Biochem. Biotechnol., 166, 486–520. [DOI] [PubMed] [Google Scholar]
- 56. Murphy C., Powlowski J., Wu M., Butler G., Tsang A.. 2011, Curation of characterized glycoside hydrolases of fungal origin, Database, 2011, bar020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Bonfante P., Desiro A.. 2017, Who lives in a fungus[quest] the diversity, origins and functions of fungal endobacteria living in Mucoromycota, ISME J., 11, 1727–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.