Abstract
MicroRNAs often occur in families whose members share an identical 5′ terminal ‘seed’ sequence. The seed is a major determinant of miRNA activity, and family members are thought to act redundantly on target mRNAs with perfect seed matches, i.e. sequences complementary to the seed. However, recently sequences outside the seed were reported to promote silencing by individual miRNA family members. Here, we examine this concept and the importance of miRNA specificity for the robustness of developmental gene control. Using the let-7 miRNA family in Caenorhabditis elegans, we find that seed match imperfections can increase specificity by requiring extensive pairing outside the miRNA seed region for efficient silencing and that such specificity is needed for faithful worm development. In addition, for some target site architectures, elevated miRNA levels can compensate for a lack of complementarity outside the seed. Thus, some target sites require higher miRNA concentration for silencing than others, contrasting with a traditional binary distinction between functional and non-functional sites. We conclude that changing miRNA concentrations can alter cellular miRNA target repertoires. This diversifies possible biological outcomes of miRNA-mediated gene regulation and stresses the importance of target validation under physiological conditions to understand miRNA functions in vivo.
INTRODUCTION
MicroRNAs (miRNAs) are small RNAs of about 22 nucleotides that silence target messenger RNAs by binding to partially complementary sequences in their 3′ untranslated regions (3′UTRs). miRNAs are loaded onto an Argonaute (Ago) protein to form the core of the miRNA-induced silencing complex (miRISC), which induces decay or translational repression of the targets (1). Conceptually, miRNAs can be separated into two parts: the ‘seed’, comprising nucleotides two through eight, and the ‘seed-distal’ 3′ end (Figure 1A). The seed sequence has emerged as the main determinant for target identification (2). Usually, functional miRNA targets contain ‘seed matches’, heptamers that base pair with perfect Watson-Crick complementarity to the miRNA seed. These were found to be necessary and sufficient for silencing in studies using ectopic miRNA expression (3–5). Structural and biochemical analyses of miRISC have provided an explanation for these results: the seed of a miRNA bound by Ago exists in a pre-arranged conformation, thus reducing the entropic cost of binding and favoring duplex formation with a target (6–8).
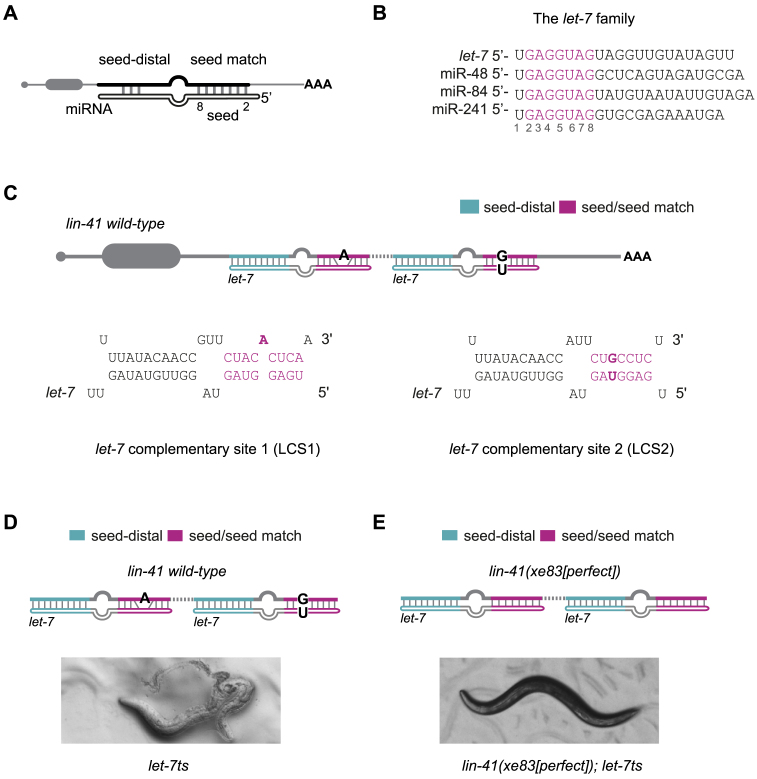
Figure 1.
let-7 becomes dispensable for viability when the lin-41 3′UTR contains perfect seed match sites. (A) Schematic drawing of a miRNA/target duplex with seed (nucleotides 2–8)/seed match and limited seed-distal pairing indicated. Top mRNA, bottom miRNA. (B) The let-7 family with the seed sequence (nucleotides 2–8) highlighted in magenta. (C) The two let-7 complementary sites (LCS1 and LCS2) in the lin-41 3′UTR of C. elegans. Each site contains an imperfect seed match (a bulged A and a G: U wobble, respectively, in bold) to the let-7 family and an extensive seed-distal pairing to let-7 only. The sites are separated by 27 nt of intervening sequence (dashed line). (D, E) Representative images of animals carrying the let-7ts mutation and (D) wild-type lin-41 or (E) the lin-41(xe83[perfect]) allele with perfect seed match to the let-7 family and unchanged seed-distal region. Animals were grown at 25°C. let-7ts: let-7(n2853) X, temperature sensitive lesion. miRNA site legend: magenta = seed/seed match; cyan = let-7 seed-distal binding.
miRNAs frequently occur in families that share the seed sequence but differ in the seed-distal part. Given the reliance of target silencing on seed matches, it is assumed that miRNA family members can function redundantly, and most computational approaches that predict miRNA targets make predictions for miRNA families rather than for individual miRNAs (2). Consequently, it was hypothesized that in order to attain specificity among family members, miRNAs require imperfect seed matches. In this scenario, an imperfect seed match impairs binding and activity of most family members, but extensive seed-distal base pairing would enable a specific family member to compensate for the unfavorable seed binding (3).
However, high-throughput biochemical capture of Ago-bound miRNA/target duplexes revealed numerous instances of interactions that frequently extended beyond the seed, to involve the seed-distal parts of the miRNA (9–12). In cell culture and in vivo assays, some of the targets that could base pair through their seed-distal parts were silenced preferentially by specific family members (9,12). Because such specificity also occurred for target sites with perfect seed matches, these findings argued that seed match imperfections might not be a requirement for miRNA family member specificity.
By contrast, specificity of miRNA silencing through seed mismatches would explain why members of the let-7 family of Caenorhabditis elegans have partially non-redundant functions. Indeed, among four members with overlapping expression patterns (13), let-7, miR-48, miR-84 and miR-241 (Figure 1B), only let-7 is essential for viability (14,15). let-7 ensures proper development of C. elegans by repressing one crucial target, lin-41 (16,17), whose 3′UTR contains two functional let-7 binding sites (let-7complementary sites, LCSs) (18). Both LCSs contain imperfect seed-matches, which yield a bulged-out nucleotide and a G:U wobble base-pair respectively (Figure 1C). Moreover, both LCSs exhibit extensive complementarity to the seed-distal sequence of let-7 but to none of its sisters. Here, we test if this miRNA site architecture ensures specific silencing by let-7 and explore miRNA site architectures as a mechanism for the selectivity of different family members towards distinct targets.
We show that extensive seed-distal pairing favors miRNA silencing by an individual miRNA family member even when the seed match is perfect, but that an imperfect seed match greatly enhances this family member specificity. Thus, we find that perturbing let-7-specific regulation of lin-41, by introducing a perfect seed match, impairs normal C. elegans development through allowing the let-7 family sisters miR-48, miR-241 and miR-84 to prematurely silence lin-41. Moreover, specificity of targets with perfect or nearly perfect seed matches can be overcome through elevated levels of a miRNA that is incapable of seed-distal pairing. Hence, although sequence-instructed, specificity is not fully hard-wired and can be altered by changes in miRNA expression levels.
Our observations are consistent with a model where let-7 family miRNAs act as rheostats (19), such that the interplay of target site architecture and miRNA abundance determine the extent of target silencing. This flexible targeting mechanism expands the regulatory potential of miRNA families and indicates that miRNA activity may differ on bona fide targets at a given miRNA concentration. Conversely, alterations in miRNA concentrations may then change the miRNA target repertoire, expanding the range of possible biological outcomes, and revealing a need for target validation under physiological conditions to understand miRNA function in vivo.
MATERIALS AND METHODS
Worm handling and strains
Worms were grown using standard methods at 25°C. The transgenic unc-54 + miRNA sites reporter strains were obtained by single-copy integration into the ttTi5605 locus on chromosome II (20). Injected plasmids were cloned using the MultiSite Gateway Technology (Thermo Fisher Scientific) and the destination vector pCFJ150 (21) or Gibson assembly (22). All strains are listed in Supplementary Table S1.
unc-54 + miRNA sites reporters
All unc-54 + miRNA sites reporters were constructed using the MultiSite Gateway Technology (Thermo Fisher Scientific) and the destination vector pCFJ150 (21) or Gibson assembly (22). First, the pGB0 vector was obtained via site-directed mutagenesis (23) of the pDONR P2R-P3_p37 vector to insert the AscI restriction site. Then, the pGB01 plasmid was obtained via LR reaction (Gateway LR Clonase II Enzyme mix, Thermo Fisher Scientific; 11791020) of the three entry vectors pdpy-30 x pGFP::H2B x pGB0 and the pCFJ150 backbone.
All the plasmids listed in Supplementary Table S2 were obtained via Gibson assembly of the digested pGB01 plasmid and gBlocks® Gene Fragments (Integrated DNA Technologies) listed below. All plasmids were verified by sequencing. Transgenic worms were obtained by single-copy integration into the ttTi5605 locus on chromosome II, following the published protocol for injection with low DNA concentration (20). We optimized our previous mCherry reference transgene (16) by replacing the artificial 3′UTR with an endogenous unc-54 3′ UTR, to achieve more physiologic and brighter expression. The resulting Pdpy-30::mCherry::H2B::unc-54 transgene was integrated on chromosome I to yield strain HW1454.
Genome editing
Mutations in the endogenous lin-41 3′UTR sequence were obtained by CRISPR-Cas9 to generate the lin-41(xe83[perfect]), lin-41(xe76[ap427_W-C]), and lin-41(xe99[48-ized]) alleles. Wild-type worms were injected as described in (24) with a mix containing 50 ng/μl pIK155, 100 ng/μl of each pGB48 and plin-41sgRNA, 20 ng/ μl repair oligo (see Supplementary Table S4), dpy-10 co-crispr mix containing 100 ng/ml pIK208 (Addgene plasmid #65630) and 20 ng/ml AF-ZF-827 oligo PAGE purified (IDT). Single F1 roller progeny of injected wild-type worms were picked to individual plates and the F2 progeny screened for the mutated allele using PCR assays and sequencing (Supplementary Table S3). The alleles were outcrossed three times to the wild-type strain.
let-7 over-expression
A let-7(++) strain (HW 1909 [xeSi287, V]) was obtained by injection of the plasmid pGB26, obtained via Gibson assembly of the PCR amplified minimal rescue fragment from (15) and the pIK37 plasmid. Transgenic worms were obtained by single-copy integration into the oxTi365 locus on chromosome V (universal MosSCI strain #EG8082 (25).
Reporter quantification
For confocal assays, worms were grown at 25°C. Let-7ts worms were maintained at 15°C and adults were transferred to 25°C for 48 h before imaging. Z-stacks of 0.313 μm μm thickness were acquired in green, red and transmitted light channels at 40× magnification on a Zeiss LSM700 confocal microscope coupled to Zeiss Zen 2010 software equipped with a multi-position tile scan macro. The z-stacks were stitched together and compiled into a single image using scripts in Matlab and Fiji (26).
For data analysis, late L4 worms were selected based on visual inspection of gonad length and vulva morphology (27). Ten to fourteen vulva cells were selected in the ‘cell counter’ macro in Fiji. Images around these seed points were de-noised using a Richardson-Lucy algorithm and segmented using an Otsu global threshold. Remaining holes were filled using a morphological filter. Signal intensity in the green channel was divided by the red signal intensity for each cell; relative signal intensities were then averaged for each worm. 10–12 vulva cells in 5–10 worms per genotype were quantified, mean signal intensity and SD were calculated and graphed using GraphPad Prism software.
Confocal analysis of LIN-29 precocious accumulation
Synchronized arrested L1 larvae of animals carrying endogenously tagged LIN-29, lin-29(xe61[lin-29::gfp::3xflag]) (28), in wild-type or lin-41(xe83[perfect]) background, were plated on food and incubated at 25°C on 2% NGM agar plates with Escherichia coli OP50 bacteria and imaged at the L3 stage (20–22 h after plating). Images were acquired in green and transmitted light channels (with Differential Interference Contrast, DIC) with 40×/1.3 oil immersion objective on a Zeiss LSM700 confocal microscope coupled to Zeiss Zen 2010 software. Further image processing was performed with Fiji (26).
RESULTS
Perfect seed matches in the lin-41 3′ UTR make let-7 miRNA dispensable for animal viability
Specific regulation of lin-41 by let-7 and not by its sisters was previously speculated (3) to derive from the imperfect seed-matches in the two let-7 miRNA Complementary Sites (LCS1 and LCS2) in the lin-41 3′UTR (Figure 1C (18,29)). When bound by let-7 family miRNAs, the seed match sequences of LCS1 and LCS2 generate an A-bulge and a G:U wobble pair. Both sites contain seed-distal complementarity to let-7, but not to its sisters. However, Broughton and colleagues recently identified a target site in the 3′UTR of dot-1.1 that appeared specific to the let-7 family member miR-48 in the absence of seed match imperfections (9). Given this unexpected finding, we tested the possibility that seed mismatches in LCS1 and LCS2 were similarly dispensable for specific recognition by let-7. To this end, we generated a lin-41 allele, lin-41(xe83[perfect]), which differs from the wild-type allele in two nucleotides: We eliminated the A bulge in LCS1 and converted the G:U wobble pair of LCS2 into a standard Watson-Crick base pair.
Strikingly, these two nucleotide changes rescued the larval lethality caused by loss of let-7, both in the let-7(mn112) null mutant strain and the let-7(n2853) temperature-sensitive strain (henceforth let-7ts), which recapitulates the let-7 null phenotype at the restrictive temperature, 25°C ((15), Figure 1D and E). Thus, ≥98% (N = 3, each with n ≥ 200 animals) of lin-41(xe83[perfect]); let-7ts double mutant animals survived into adulthood, as did 100% (N = 2, n ≥ 98 animals) of lin-41(xe83[perfect]); let-7(mn112) double mutant animals, of which 6% subsequently died as adults. These findings suggest that seed mismatches are required to restrict silencing of lin-41 to let-7, because other let-7 family members confer silencing in their absence.
A perfect seed match allows redundant activity of the let-7 sisters
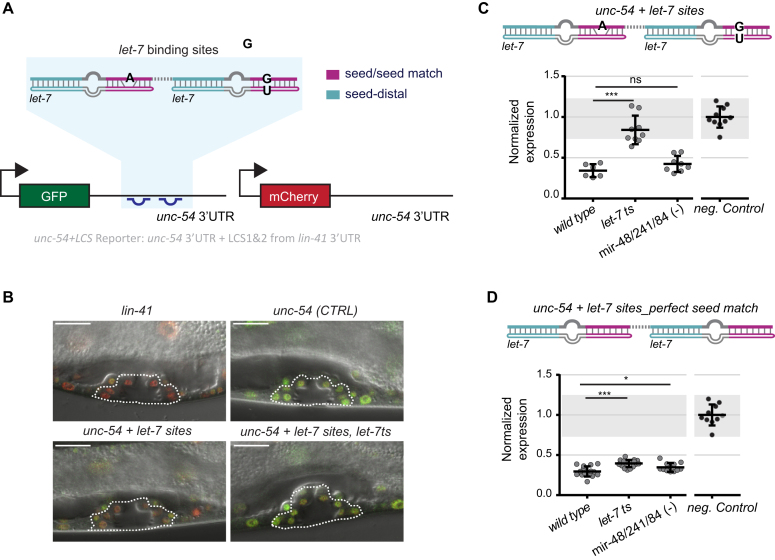
To confirm that the perfect seed matches of the lin-41(xe83[perfect]) allele allow redundant binding of the let-7 family, we monitored the activity of the four miRNAs through a GFP reporter modified from (16) (Materials and Methods). In our assay, each animal contains a red mCherry reporter, which is used as reference during image analysis, and a GFP reporter, which is the miRNA activity sensor (Figure 2A). Both reporters are driven by the ubiquitous and constitutively active dpy-30 promoter and contain the unc-54 3′UTR, generally thought to be devoid of regulatory elements. Finally, each reporter is integrated by Mos1-mediated single copy integration into a distinct genomic location (20).
Figure 2.
Redundant activity of the let-7 family in the presence of a perfect seed match. (A) Schematic of the reporters used to monitor miRNA activity in vivo. The depicted GFP transgene unc-54 + let-7 sites reporter contains 111 nucleotides of the lin-41 3′UTR (shaded in blue), which harbor the two let-7 binding sites and the 27 nt-long intervening sequence, grafted into the heterologous, unregulated unc-54 3′UTR. Worms also contain a red mCherry reporter for normalization. Transcription of the single-copy integrated reporters from the ubiquitously active dpy-30 promoter is constitutive. miRNA site legend: magenta = seed/seed match; cyan = let-7 seed-distal binding. (B) Representative confocal images of the vulvae of animals carrying the red mCherry reporter (for normalization) and GFP reporters with the indicated 3′UTRs. These are ‘lin-41 3′UTR full-length’, ‘unc-54’ (CTRL, unregulated) and ‘unc-54 + let-7 sites’ in wild-type and ‘unc-54 + let-7 sites’ in the let-7ts background. Images are merged GFP, mCherry and DIC channels. Red color indicates a greater, and green color a lesser degree of reporter repression. Dashed lines outline the vulvae of the animals, which confirm appropriate late Larval stage 4 (L4). Scale bars 15 μm. (C, D) Quantification of (C) ‘unc-54 + let-7 sites’ reporter, (D) ‘unc-54 + let-7 sites_perfect seed match’ reporter. Each dot represents the average of the GFP signal intensity, obtained by confocal imaging, divided by the mCherry intensity for a single animal per condition. 10–12 vulva cells were quantified per worm. Mean values are normalized to the average value of the GFP/mCherry ratio of the negative control unc-54 3′UTR reporter, which is not silenced. Horizontal line and error bars indicate mean values per condition ± SD. *P < 0.05 and ***P < 0.001, two-tailed unpaired t-test. For reference, data obtained for the unc-54, Neg.Control reporter are replotted in panel D; gray shading is bounded by the min-max values of this control.
To monitor let-7 activity, we generated the reporter ‘unc-54 + let-7 sites’ in which only a stretch of 111 nucleotides of the lin-41 3′UTR, comprising LCS1 and LCS2, was transplanted into the unc-54 3′UTR (Figure 2A). Silencing of this minimal target reporter by let-7 was comparable to that of a reporter containing the full-lenght lin-41 3′UTR (Figure 2B, C and Supplementary Figure S1A), confirming functionality. We focused our analysis on the vulva because lin-41 repression by let-7 in this organ is required and likely sufficient to prevent vulval rupturing (16).
As expected, the ‘unc-54 + let-7 sites’ reporter was expressed in young L1 or L2 animals (Supplementary Figure S1B), when the let-7 family levels are low (30). Moreover, it was robustly silenced in older, L4-stage larvae, when let-7 family levels are high (Figure 2B and C). Finally, it was de-silenced in let-7ts animals, but not in animals lacking the three let-7 sisters ([mir-48/mir-241(ndf51)V, mir-84(n4037)X], henceforth mir-48/241/84(–)) (Figure 2C). Therefore, the stretch of 111 nucleotides suffices for efficient and specific let-7-dependent silencing.
Next, we generated an ‘unc-54 + let-7 sites_perfect seed match’ reporter, modified to contain LCSs with perfect seed matches, as in the endogenous lin-41(xe83[perfect]) mutation (Figure 1E). Like the ‘unc-54 + let-7 sites’ reporter, the new reporter was expressed in young L1 or L2 animals (Supplementary Figure S1B), but robustly silenced in L4-stage larvae (Figure 2D). However, unlike the ‘unc-54 + let-7 sites’ reporter, the new reporter was only marginally de-repressed in L4-stage larvae lacking let-7 (let-7ts) or the three let-7 sisters (mir-48/241/84(–)) (Figure 2D).
A seed-distal match establishes specificity to one miRNA in the presence of an imperfect seed match
Taken together, the genetic interaction and the reporter assay data presented thus far validate the hypothesis that the seed mismatches in the let-7 complementarity sites of lin-41 are necessary for specific regulation of lin-41 by let-7, to the apparent exclusion of the other family members. However, this conclusion appears at odds with the results of biochemical miRNA–mRNA duplex identification, which indicate preferential target binding by individual family members even in the presence of perfect seed matches (9,12). Thus, to challenge our finding, we sought to reprogram the LCSs to another let-7 family member, miR-48, and test the effect of seed match imperfections. We chose miR-48 because its expression levels and spatial expression patterns appear very similar to those of let-7 (13,14,31).
Because structural data suggest that base pairing between nucleotides 13–16 of the miRNA and a target may be favored (8), we started out by generating a reporter with seed-distal base pairing to only these nucleotides. However, this reporter failed to be silenced even in wild-type conditions, i.e. with both let-7 and miR-48 present (Figure 3A and Supplementary Figure S2A). Hence, it appears that more extensive seed distal complementarity is required for functionality of targets with a sub-optimal seed match. Indeed, an ‘unc-54 + miR-48 sites’ reporter that emulated the LCS architecture by carrying a central bulge in the seed sequence and an extensive seed distal match to miR-48 (Supplementary Figure S2B), was silenced in L4 stage animals. Moreover, and in agreement with our predictions, the ‘unc-54 + miR-48 sites’ reporter was repressed at the L4 stage in both the presence and absence of let-7 miRNA, but became de-repressed when miR-48 was absent (Figure 3B).
Figure 3.
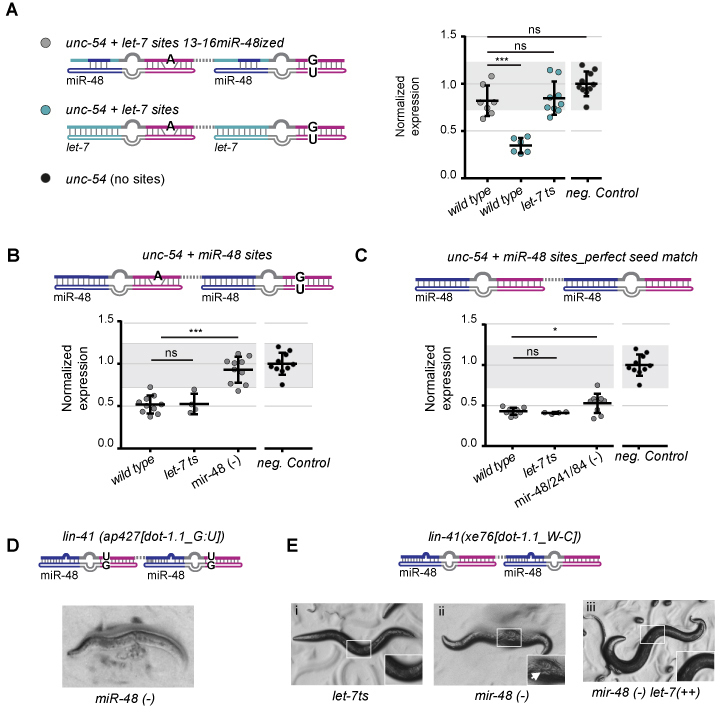
Imperfect seed matches and extensive 3′ pairing confer target specificity. (A–C) Reporter quantification as in Figure 2, from which the negative control data (black dots) are also replotted for reference; gray shading is bounded by the min-max values of the negative control. (A) The ‘unc-54 + let-7 sites 13–16miR-48-ized’ reporter contains let-7 complementary sites modified to pair miR-48 at position 13–16 but not other seed-distal nucleotides (gray dots). Results from the unmodified ‘unc-54 + let-7 sites’ reporter in wild-type and let-7ts mutant background are from Figure 2C and included for reference (cyan dots). (B) The ‘unc-54 + miR-48 sites’ reporter combines extensive seed-distal complementarity to miR-48 with seed match imperfections whereas (C) the ‘unc-54 + miR-48 sites_perfect seed match’ reporter contains extensive seed-distal complementarity to miR-48 and perfect seed matches. Horizontal line and error bars indicate mean values per condition ± SD. *P < 0.05 and ***P < 0.001, two-tailed unpaired t-test. (D) Animals carrying the lin-41(ap427[dot-1.1_G: U] (9)) allele die in the absence of miR-48. (E) Survival of strain lin-41(xe76[dot-1.1_W-C] upon manipulation of let-7 and miR-48 activity. In this strain, a U at position 8 in the two target sites of the lin-41(ap427[dot-1.1_G:U]) allele has been converted to a C, to permit Watson-Crick instead of G:U wobble base-pairing with the let-7 family seed sequence (Supplementary Figure S2C and D). This allele was crossed into a (i) let-7ts, (ii) mir-48(–) or (iii) mir-48(–) let-7(++) background, where let-7(++) denotes let-7 overexpression from a single copy integrated transgene. Insets magnify the central part of the animal body to reveal egg retention (arrow), i.e. and egg-laying defective (Egl) phenotype. let-7ts: let-7(n2853) X, temperature-sensitive lesion, grown at the restrictive temperature 25°C; mir-48(–): mir-48(n4097) V; mir-48/241/84(–): mir-48/mir-241(ndf51) V, mir-84(n4037) X.
Consistent with our results for the let-7 reporters, the specificity of the ‘unc-54 + miR-48 sites’ reporter was largely lost when we modified it to contain perfect seed matches: the resulting ‘unc-54 + miR-48 sites_perfect seed match’ reporter continued to be silenced extensively in both let7ts and mir-48/241/84(–) animals (Figure 3C). However, silencing appeared marginally impaired in the absence of the let-7 sisters (Figure 3C), mirroring an analogous result for the ‘unc-54 + let-7sites_perfect seed match’ reporter in let-7ts animals (Figure 2D). We conclude that the imperfect seed match and the extensive 3′ pairing are both important determinants for the robust target specificity of the lin-41 sites.
A G:U wobble base-pair in a peripheral seed match location promotes miRNA specificity
The duplexes formed between let-7 and lin-41 contain a bulge between nucleotides 4–5 in LCS1 and a G:U wobble base-pair at position 6 in LCS2 (Figure 1C). We wondered if such centrally located ‘imperfections’ were required for specificity. We turned to the miRNA binding site in the dot-1.1 3′UTR, which had been shown to be specific to miR-48 (9). Broughton et al. found that substitution of the let-7 complementary sites in the endogenous lin-41 3′UTR by two copies of the dot-1.1 site rendered animals insensitive to loss of let-7 (9), but made them depend on the presence of miR-48. This finding was attributed to the fact that the site features an extensive seed-distal match to miR-48 (Figure 3D and Supplementary Figure S2C). However, we noticed that the let-7 family/dot-1.1 predicted duplexes exhibited not only perfect Watson–Crick pairing from nucleotides 2–7, but also a G:U wobble pair at position 8 (Supplementary Figure S2C). Although hexameric seed match sites, with complementarity to nucleotides 2–7, are considered canonical and functional (2), genome-wide studies also suggested that they are less functional than heptameric sites that match nucleotides 2–8 (6,32,33). Since G:U wobble base pairs elsewhere in seed-seed match duplexes appear detrimental to silencing (3,4,34–36), we wondered if this ‘peripheral G:U’ in seed match position 8 might affect silencing and specificity.
To test this hypothesis, we modified the endogenous target sites in lin-41 to those of dot-1.1, but with the G:U wobbles at positions 8 converted to Watson-Crick G:C pairs, yielding allele lin-41(xe76[dot-1.1_W-C]) (Supplementary Figure S2D). We then compared the reliance of this and the lin-41(ap427[dot-1.1_G:U]) strain, which carried the unmodified G:U-wobble-containing dot-1.1 sites, on let-7 and miR-48 for survival. Whereas both strains were insensitive to loss of let-7 (Figure 3E(i) and (9)), lin-41(ap427[dot-1.1_G:U]) but not lin-41(xe76[dot-1.1_W-C]) required miR-48 for survival into adulthood (Figure 3D and E(ii)). We conclude that the G:U wobble at position eight repels binding by all let-7 family members such that only miR-48 can exert repression by compensating through extensive complementarity of its 3′ seed-distal sequence. Collectively, our data thus reveal that bulges or wobbles in different positions of a seed match can serve to avoid redundancy of the let-7 family and confer strong target specificity.
miRNA abundance affects silencing in vivo
Although our experiments provided strong evidence that seed mismatches are required for robust specificity among let-7 family members, we consistently observed evidence of residual specificity even for targets that contained a perfect seed match. In target reporters containing perfect seed matches, we observed modest but reproducible de-silencing specifically when the family member with seed-distal match was lost (Figures 2D and 3C), and phenotype (Figure 3E(ii)). In fact, although lin-41(xe76[dot-1.1_W-C]); mir-48(–) animals survived into adulthood, they exhibited an egg-laying (Egl) defect (Figure 3E (ii), 93%, n = 132), i.e. a partial vulval dysfunction that is consistent with incomplete repression of lin-41 (16).
We wondered if this partial specificity could be overridden by increased levels of another miRNA family member. Since we were unable to overexpress mir-48, we tested this possibility by overexpressing let-7. Mos1-mediated single copy integration (25) of a genomic fragment, known to rescue let-7 lethality (15), to a locus on chromosome V that is ∼5 cM apart from mir-48, yielded a ∼2-fold increase in expression levels (data not shown). Consistent with our hypothesis, lin-41(xe76[dot-1.1_W-C]) animals that over-expressed let-7 were no longer Egl in the absence of miR-48 (Figure 3E(iii), compare to E(ii)). We conclude that, in vivo, increased miRNA levels can override the specificity imparted by seed-distal pairing.
Seed match imperfections maintain specificity upon miRNA overexpression
Since the modest preferential silencing imposed by the seed-distal pairing to miR-48 could be overcome by increasing the levels of let-7 in the presence of a perfect seed match (Figure 3E (ii) and (iii)), we wondered about the effect of let-7 over-expression on sites with more extensive target specificity. Hence, we examined two reporters specific to miR-48 that harbored imperfect seed matches: the previous ‘unc-54 + miR-48 sites’ (Figures 3B and 4A) and the new ‘unc-54 + dot-1.1 sites’ reporter, obtained by inserting two copies of the binding sites from the dot-1.1 3′UTR (Figure 4B). Consistent with the in vivo data ((9) and Figure 3E), silencing of both reporters was dependent on miR-48 but not let-7 (Figures 3B, 4A, B and Supplementary Figure S2E). However, the response of the two reporters differed when we overexpressed let-7 in the absence of miR-48. The ‘unc-54 + miR-48 sites’ reporter, with central seed mismatches, was insensitive to a doubling of let-7 expression (Figure 4A). By contrast, silencing of the ‘unc-54 + dot-1.1 sites’ reporter, with peripheral seed mismatches, was restored to almost wild-type level in the same conditions (Figure 4B). This suggests that for miR-48 targets with extensive seed-distal pairing, sensitivity to let-7 levels depends on seed match quality.
Figure 4.
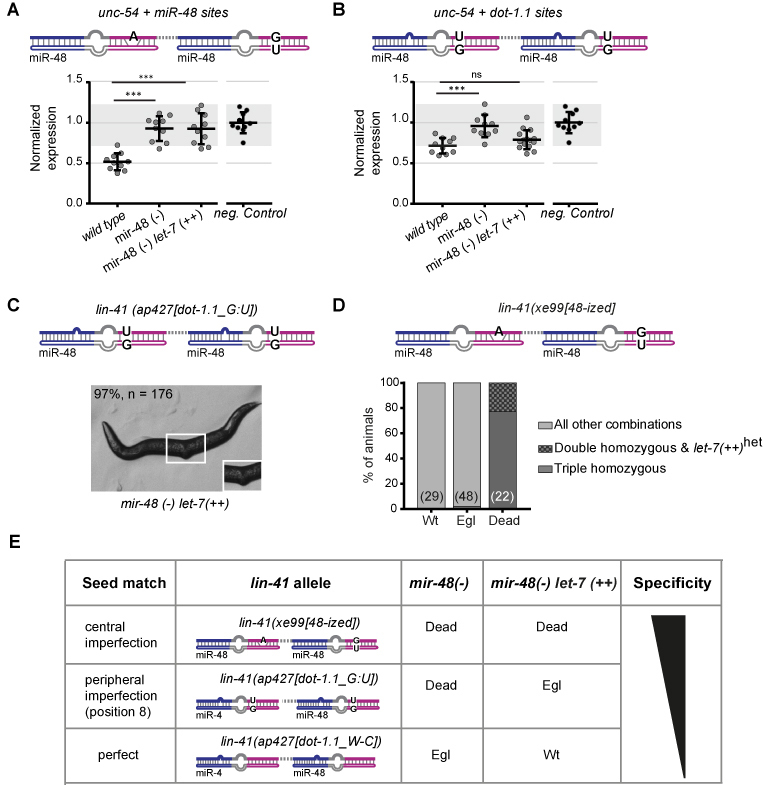
Robust miRNA specificity relies on imperfect seed matches. (A, B) Reporter quantification as in Figure 2, from which the negative control data are also replotted for reference. (A) ‘unc-54 + miR-48 sites’ reporter and (B) ‘unc-54 + dot-1.1 sites’ reporter are assayed in worms of the indicated genotypes. Horizontal line and error bars indicate mean values per condition ± SD, *P < 0.05 and ***P < 0.001, two-tailed unpaired t-test. (C) Representative image of a viable lin-41(ap427[dot-1.1_G:U]), mir-48(-) let-7(++) animal. (D) Progeny (n = 99) derived from a cross of lin-41(xe99[48-ized]) with mir-48(–) let-7(++) animals were categorized by phenotype and genotyped to determine the viability of lin-41(xe99[48-ized]); mir-48(-) let-7(++) ‘triple homozygous’ mutant animals. (E) Summary of the effect that different site architectures and miRNA abundance have on silencing lin-41 alleles ‘recoded’ towards miR-48. mir-48(–): mir-48(n4097)V; unc-54(CTRL): wild-type unc-54 3′UTR; let-7(++): let-7 over-expression allele (MosSCI, V).
To confirm this result on a functional level, we tested whether let-7 overexpression could suppress the dependence on miR-48 of animals carrying lin-41 alleles analogous to those in the miR-48-specific reporters, namely the lin-41(ap427[dot-1.1_G:U]) allele and the newly generated lin-41(xe99[48-ized]) allele (Figure 4C and D, respectively). As predicted by the reporter assay, overexpression of let-7 rendered lin-41(ap427[dot-1.1_G:U]); mir-48(–) double mutant animals viable, although Egl (Figure 4C). By contrast, we were unable to obtain viable animals of the lin-41(xe99[48-ized])I; mir-48(–) let-7 (++)V genotype (Figure 4D). Instead, we readily observed dead animals, which had burst through the vulva. Genotyping revealed that such animals were homozygous for the three alleles of interest, lin-41(xe99[48-ized]), mir-48(–), and let-7(++) (Figure 4D). [Note that mir-48(–) and let-7(++) are closely linked loci on chromosome V, explaining why we did not find dead animals that were lin-41(xe99[48-ized]); mir-48(–) double mutant but lacked the let-7 over-expression transgene.] In contrast, randomly selected wild-type animals were never doubly homozygous for lin-41(xe99[48-ized]) and mir-48(–), irrespective of let-7 transgene status, and only one Egl animal was found to be lin-41(xe99[48-ized]); mir-48(–) let-7(++) mutant. Hence, although an increase in let-7 levels can overcome the specificity to miR-48 imposed by seed-distal matches in combination with a perfect seed (Figure 3E) or in the presence of peripheral seed mismatches (Figure 4C), it cannot do so with a central seed bulge or wobble (Figure 4D), at least within the physiological ranges of the expression levels that we tested.
We conclude that specificity arises through seed-distal pairing of a miRNA, but that it is enhanced in extent and robustness by appropriate seed match architecture (Figure 4E).
Loss of miRNA specificity impairs robust development
Our results suggest that sites with central seed match imperfections, such as LCS1 and LCS2 in the lin-41 3′UTR, are extremely specific to one miRNA, even when a paralogue is highly expressed. We suspected that such robust specificity would be physiologically relevant in the case of lin-41. This is because the let-7 sisters are all expressed prior to let-7, in the L2 stage (30). Given their overlapping spatial expression patterns, lack of mechanisms to prevent let-7 sisters’ action on lin-41 might cause inappropriately early repression of lin-41, as speculated previously (2,3). Consistent with this notion, we found that the ‘unc-54 + let-7 sites_perfect seed match’ reporter was precociously repressed during the L3 stage, whereas the ‘unc-54 + let-7 sites’ reporter was still expressed at the same stage (Figure 5A).
Figure 5.
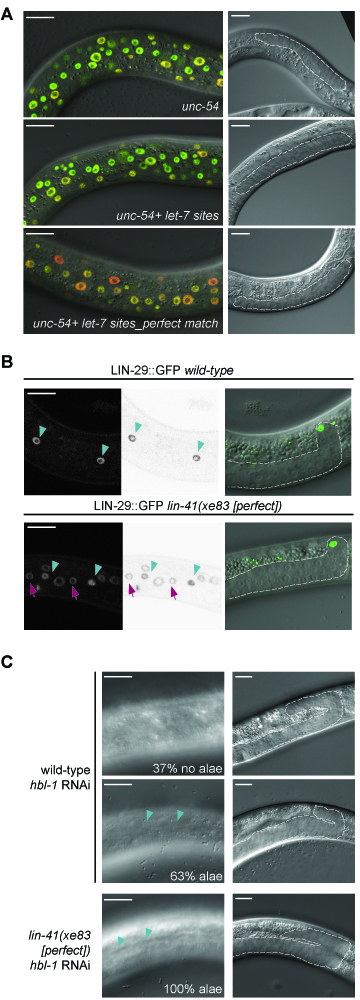
Developmental robustness requires an imperfect let-7 seed match in lin-41. (A) Representative confocal images of skin cells of animals carrying an unc-54 3′UTR reporter (top), an ‘unc-54 + let-7 sites’ (center), or an ‘unc-54 + let-7 sites_perfect seed match’ reporter (bottom). At the L3 stage, levels of miR-48 but not let-7 are already high (39). Scale bars 15 μm. (B) Microscopy images of the skin of late L3 worms expressing endogenously tagged LIN-29::GFP (xe61) (28) in wild-type and lin-41(xe83[perfect]) background. Cyan arrowheads point to LIN-29 signal in seam cells, magenta arrows to LIN-29 accumulation in hyp7 cells. Images in the middle are inverted to increase clarity. Worms are staged according to the position of the distal tip cell (green) and gonad length. Scale bars 15μm. (C) Representative images of wild-type (n = 27) or lin-41(xe83[perfect]) (n = 36) animals treated with hbl-1 RNAi. Percentages of animals with the indicated alae status at the L3/L4 transition are indicated. Gonads are outlined to confirm appropriate staging. The strains used, SX346 and HW2144, additionally contain the mjIs15 and wIs51 transgenes. Scale bars 15μm.
To test whether this precocious repression of lin-41 had physiological consequences, we examined the accumulation of LIN-29A, a target of LIN-41. In wild-type animals, LIN-41 translationally represses LIN-29A until the L4 stage, when repression is released following let-7 accumulation and consequent LIN-41 downregulation (28). Premature loss of LIN-41 activity causes inappropriately early activation of LIN-29A and thereby precocious execution of the so-called larval-to-adult transition, which includes fusion of hypodermal seam cells into a syncytium and secretion of an adult cuticular structure termed alae (17). We observed LIN-29A levels through use of a lin-29(xe61[lin-29::gfp::3xflag]) strain, in which the endogenous lin-29 locus has been edited to produce GFP-tagged LIN-29A and B isoforms, and in which loss of lin-41 activity yields a specific upregulation of only LIN-29A (28). At mid-L3 larval stage, wild-type animals have LIN-29::GFP signal only in their seam cells (Figure 5B). By contrast, animals carrying the lin-41(xe83[perfect]) allele show additional GFP expression in the major hypodermal syncytium, hyp7, at the same developmental stage (Figure 5B). Therefore, precocious downregulation of lin-41(xe83[perfect]) is responsible for premature LIN-29 translation and accumulation in the hypodermis, as described for other lin-41 loss-of-function alleles (17).
The lin-41(xe83[perfect]) animals looked superficially wild-type, but the premature upregulation of LIN-29 was sufficient to promote precocious larval-to-adult transition in a sensitized background. Specifically, the transcription factor HBL-1 inhibits larval-to-adult transition, possibly in parallel to LIN-41 (37,38), and its RNAi-mediated depletion causes partially penetrant and partially expressive precocious alae formation (Figure 5C). This phenotype was enhanced when we depleted HBL-1 in lin-41(xe83[perfect]) mutant animals, resulting in fully penetrant precocious secretion of alae (although weak or patched in some cases) (Figure 5C). We conclude that loss of specificity of repression by let-7 alone in the lin-41(xe83[perfect]) background impairs the robustness of temporal patterning through premature LIN-29 accumulation.
DISCUSSION
It has been an open question to what extent and by which mechanisms miRNA family members can function non-redundantly despite a shared seed sequence. Previously, it was proposed that redundancy was the rule (2). Rare occasions of non-redundant function were hypothesized to require targets with both an imperfect seed match and extensive seed-distal pairing to only one specific family member (3). According to this view, the seed match imperfection impairs silencing by all family members but extensive seed-distal pairing can compensate to facilitate silencing by an individual miRNA. However, this hypothesis has remained untested, and recent observations have challenged it by providing evidence that non-redundant target binding appears wide-spread and that seed-distal pairing may suffice to achieve specificity (9,12).
Our systematic study through gene editing and fluorescent reporter analysis with cell-type resolution resolves the discrepant views on specificity-promoting features for the let-7 family: We demonstrate that extensive seed-distal pairing to a specific family member suffices to generate a weak but consistent preference for silencing by this family member. However, more robust discrimination requires an imperfect seed match and depends on the quality of such imperfections: a central bulge or G:U wobble base pair, as in the lin-41 3′UTR, confers the strongest specificity, while a peripheral G:U wobble base pair, as in the dot-1.1 3′UTR, gives an intermediate level. The physiological importance of extensive, seed-mismatch-dependent specificity is evident from the decreased developmental robustness that results when perfect let-7 seed matches permit promiscuous silencing of lin-41 by the whole let-7 family.
Perfect seed matches can still be compatible with selective targeting by individual miRNAs, but the effect depends on miRNA abundance: A moderate increase in let-7 levels (∼2-fold) could overcome the specificity of a binding site that was silenced by miR-48 and had a perfect seed match. However, it only partially did so when the seed match contained a peripheral G:U wobble, and it was insufficient to override sequence-determined specificity when a site contained a central seed match imperfection. This suggests that in vivo, miRNA binding site architecture, particularly seed match quality, and miRNAs abundance act together to determine miRNA activity towards individual targets (Figure 6).
Figure 6.
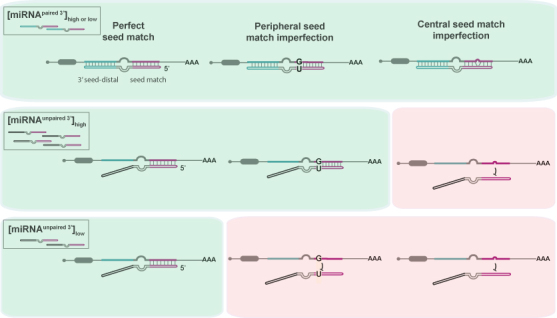
miRNA abundance and architecture of the target site determine mRNA silencing. (top) Extensive complementarity (paired 3′) between a miRNA and a target site allows for efficient and specific silencing, independently of the miRNA level and the presence of imperfections in the seed match. (middle) Abundant miRNAs can silence targets carrying a perfect seed match or a nearly-perfect seed match (e.g. a peripheral G:U wobble), even in the absence of complementarity to the sequence outside the seed. A ‘central mismatch’ repels poorly complementary miRNAs. (bottom) Lowly abundant and poorly complementary miRNAs can silence targets carrying a perfect seed match, but not the ones carrying a seed match imperfection (e.g. peripheral G:U or central bulge). Green shading: functional site; pink shading: nonfunctional site. Magenta: seed/seed match
The finding that miRNA activity is determined at the level of individual targets has implications beyond the issue of miRNA family member specificity. It contrasts with a view where a miRNA is globally either ‘on’ or ‘off’ in a cell, silencing all of its targets at sufficiently high concentrations and none at low ones. Variable, target site-dependent activity was already entertained in the early days of the miRNA field when miRNAs were likened to rheostats, whose activity is adjusted by two features, namely the extent of target site complementarity to the miRNA and miRNA abundance (19). A lack of explicit experimental testing of such context-dependent function (4) and the rising popularity of the ‘seed-match only’ model caused this hypothesis to fade from view. We propose that it is time to revisit the idea of miRNAs functioning as rheostats and subject it to further testing.
We note that target validation experiments that rely, as often done, on ectopic miRNA expression appear to make the implicit assumption that miRNAs are uniformly active on their targets. However, if the goal of target validation is to provide insights into pathway biology, physiology and/or pathology, our results and those of others (34) strongly suggest that it must be conducted in a relevant physiological context, avoiding ectopic expression or overexpression of miRNAs. Ideally, validation will also involve functional studies such as those offered by direct manipulation of individual miRNA/target interaction through genome editing. We predict that such efforts will reveal a more nuanced picture of dynamic, context-dependent miRNA target repertoires, and thereby improve our understanding of the diversity of biological outcomes that miRNA-mediated gene regulation can achieve in vivo.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Kathrin Kunzer and Lan Xu for their help with C. elegans strain generation. We are grateful to Matyas Ecsedi for initial observations on target specificity of let-7 family members. We thank Florian Aeschimann for reagents and helpful discussions and Iskra Katic for worm injections and reagents. We thank Laurent Gelman and Steven Bourke for help with confocal imaging; Roland Nitschke (Life Imaging Center, University of Freiburg, Germany) and Carl Zeiss (Jena, Germany) for sharing the macro for Multiple Position/Tile Imaging acquisitions; Raphael Thierry, Jan Eglinger and Moritz Kirschmann (University of Zurich) for help with image analysis; and Amy Pasquinelli for C. elegans strains. Some strains were provided by the Caenorhabditis Genetics Center (CGC), which is funded by the National Institutes of Health Office of Research Infrastructure Programs (P40 OD010440). We thank Matyas Ecsedi, Sarah Carl, Benjamin Towbin, Iskra Katic and Witold Filipowicz for a critical reading of the manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
NCCR RNA & Disease funded by the Swiss National Science Foundation; Novartis Research Foundation through the FMI (to H.G.); Boehringer Ingelheim Fonds PhD Fellowship (to G.B.). Funding for open access charge: Internal Funds.
Conflict of interest statement. None declared.
REFERENCES
- 1. Krol J., Loedige I., Filipowicz W.. The widespread regulation of microRNA biogenesis, function and decay. Nat. Rev. Genet. 2010; 11:597–610. [DOI] [PubMed] [Google Scholar]
- 2. Bartel D.P. MicroRNAs: target recognition and regulatory functions. Cell. 2009; 136:215–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Brennecke J., Stark A., Russell R.B., Cohen S.M.. Principles of microRNA-target recognition. PLoS Biol. 2005; 3:e85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Doench J.G., Sharp P.A.. Specificity of microRNA target selection in translational repression. Genes Dev. 2004; 18:504–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lai E.C. Micro RNAs are complementary to 3′ UTR sequence motifs that mediate negative post-transcriptional regulation. Nat. Genet. 2002; 30:363–364. [DOI] [PubMed] [Google Scholar]
- 6. Chandradoss S.D., Schirle N.T., Szczepaniak M., MacRae I.J., Joo C.. A Dynamic Search Process Underlies MicroRNA Targeting. Cell. 2015; 162:96–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Parker J.S., Parizotto E.A., Wang M., Roe S.M., Barford D.. Enhancement of the seed-target recognition step in RNA silencing by a PIWI/MID domain protein. Mol. Cell. 2009; 33:204–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Schirle N.T., Sheu-Gruttadauria J., MacRae I.J.. Structural basis for microRNA targeting. Science. 2014; 346:608–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Broughton J.P., Lovci M.T., Huang J.L., Yeo G.W., Pasquinelli A.E.. Pairing beyond the seed supports microRNA targeting specificity. Mol. Cell. 2016; 64:320–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Grosswendt S., Filipchyk A., Manzano M., Klironomos F., Schilling M., Herzog M., Gottwein E., Rajewsky N.. Unambiguous identification of miRNA:target site interactions by different types of ligation reactions. Mol. Cell. 2014; 54:1042–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Helwak A., Kudla G., Dudnakova T., Tollervey D.. Mapping the human miRNA interactome by CLASH reveals frequent noncanonical binding. Cell. 2013; 153:654–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Moore M.J., Scheel T.K., Luna J.M., Park C.Y., Fak J.J., Nishiuchi E., Rice C.M., Darnell R.B.. miRNA-target chimeras reveal miRNA 3′-end pairing as a major determinant of Argonaute target specificity. Nat. Commun. 2015; 6:8864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Roush S., Slack F.J.. The let-7 family of microRNAs. Trends Cell Biol. 2008; 18:505–516. [DOI] [PubMed] [Google Scholar]
- 14. Abbott A.L., Alvarez-Saavedra E., Miska E.A., Lau N.C., Bartel D.P., Horvitz H.R., Ambros V.. The let-7 MicroRNA family members mir-48, mir-84, and mir-241 function together to regulate developmental timing in Caenorhabditis elegans. Dev. Cell. 2005; 9:403–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Reinhart B.J., Slack F.J., Basson M., Pasquinelli A.E., Bettinger J.C., Rougvie A.E., Horvitz H.R., Ruvkun G.. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000; 403:901–906. [DOI] [PubMed] [Google Scholar]
- 16. Ecsedi M., Rausch M., Großhans H.. The let-7 microRNA directs vulval development through a single target. Dev. Cell. 2015; 32:335–344. [DOI] [PubMed] [Google Scholar]
- 17. Slack F.J., Basson M., Liu Z., Ambros V., Horvitz H.R., Ruvkun G.. The lin-41 RBCC gene acts in the C. elegans heterochronic pathway between the let-7 regulatory RNA and the LIN-29 transcription factor. Mol. Cell. 2000; 5:659–669. [DOI] [PubMed] [Google Scholar]
- 18. Vella M.C., Choi E.Y., Lin S.Y., Reinert K., Slack F.J.. The C. elegans microRNA let-7 binds to imperfect let-7 complementary sites from the lin-41 3′UTR. Genes Dev. 2004; 18:132–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bartel D.P., Chen C.Z.. Micromanagers of gene expression: the potentially widespread influence of metazoan microRNAs. Nat. Rev. Genet. 2004; 5:396–400. [DOI] [PubMed] [Google Scholar]
- 20. Frokjaer-Jensen C., Davis M.W., Ailion M., Jorgensen E.M.. Improved Mos1-mediated transgenesis in C. elegans. Nat. Methods. 2012; 9:117–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Frokjaer-Jensen C., Davis M.W., Hopkins C.E., Newman B.J., Thummel J.M., Olesen S.P., Grunnet M., Jorgensen E.M.. Single-copy insertion of transgenes in Caenorhabditis elegans. Nat. Genet. 2008; 40:1375–1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gibson D.G., Young L., Chuang R.Y., Venter J.C., Hutchison C.A. 3rd, Smith H.O.. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods. 2009; 6:343–345. [DOI] [PubMed] [Google Scholar]
- 23. Zheng L., Baumann U., Reymond J.L.. An efficient one-step site-directed and site-saturation mutagenesis protocol. Nucleic Acids Res. 2004; 32:e115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Katic I., Xu L., Ciosk R.. CRISPR/Cas9 genome editing in Caenorhabditis elegans: evaluation of templates for homology-mediated repair and knock-ins by homology-independent DNA repair. G3 (Bethesda). 2015; 5:1649–1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Frokjaer-Jensen C., Davis M.W., Sarov M., Taylor J., Flibotte S., LaBella M., Pozniakovsky A., Moerman D.G., Jorgensen E.M.. Random and targeted transgene insertion in Caenorhabditis elegans using a modified Mos1 transposon. Nat. Methods. 2014; 11:529–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B. et al. . Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012; 9:676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mok D.Z., Sternberg P.W., Inoue T.. Morphologically defined sub-stages of C. elegans vulval development in the fourth larval stage. BMC Dev. Biol. 2015; 15:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Aeschimann F., Kumari P., Bartake H., Gaidatzis D., Xu L., Ciosk R., Großhans H.. LIN41 post-transcriptionally silences mRNAs by two distinct and position-dependent mechanisms. Mol. Cell. 2017; 65:476–489. [DOI] [PubMed] [Google Scholar]
- 29. Vella M.C., Reinert K., Slack F.J.. Architecture of a validated microRNA::target interaction. Chem. Biol. 2004; 11:1619–1623. [DOI] [PubMed] [Google Scholar]
- 30. Vadla B., Kemper K., Alaimo J., Heine C., Moss E.G.. lin-28 controls the succession of cell fate choices via two distinct activities. PLoS Genet. 2012; 8:e1002588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Martinez N.J., Ow M.C., Reece-Hoyes J.S., Barrasa M.I., Ambros V.R., Walhout A.J.. Genome-scale spatiotemporal analysis of Caenorhabditis elegans microRNA promoter activity. Genome Res. 2008; 18:2005–2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Baek D., Villen J., Shin C., Camargo F.D., Gygi S.P., Bartel D.P.. The impact of microRNAs on protein output. Nature. 2008; 455:64–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Grimson A., Farh K.K., Johnston W.K., Garrett-Engele P., Lim L.P., Bartel D.P.. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol. Cell. 2007; 27:91–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Didiano D., Hobert O.. Perfect seed pairing is not a generally reliable predictor for miRNA-target interactions. Nat. Struct. Mol. Biol. 2006; 13:849–851. [DOI] [PubMed] [Google Scholar]
- 35. Lai E.C., Tam B., Rubin G.M.. Pervasive regulation of Drosophila Notch target genes by GY-box-, Brd-box-, and K-box-class microRNAs. Genes Dev. 2005; 19:1067–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wolter J.M., Le H.H., Linse A., Godlove V.A., Nguyen T.D., Kotagama K., Lynch A., Rawls A., Mangone M.. Evolutionary patterns of metazoan microRNAs reveal targeting principles in the let-7 and miR-10 families. Genome Res. 2017; 27:53–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Abrahante J.E., Daul A.L., Li M., Volk M.L., Tennessen J.M., Miller E.A., Rougvie A.E.. The Caenorhabditis elegans hunchback-like gene lin-57/hbl-1 controls developmental time and is regulated by microRNAs. Dev. Cell. 2003; 4:625–637. [DOI] [PubMed] [Google Scholar]
- 38. Lin S.Y., Johnson S.M., Abraham M., Vella M.C., Pasquinelli A., Gamberi C., Gottlieb E., Slack F.J.. The C elegans hunchback homolog, hbl-1, controls temporal patterning and is a probable microRNA target. Dev. Cell. 2003; 4:639–650. [DOI] [PubMed] [Google Scholar]
- 39. Esquela-Kerscher A., Johnson S.M., Bai L., Saito K., Partridge J., Reinert K.L., Slack F.J.. Post-embryonic expression of C. elegans microRNAs belonging to the lin-4 and let-7 families in the hypodermis and the reproductive system. Dev. Dyn. 2005; 234:868–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.