Abstract
Cajal bodies (CBs) are nuclear non-membrane bound organelles where small nuclear ribonucleoprotein particles (snRNPs) undergo their final maturation and quality control before they are released to the nucleoplasm. However, the molecular mechanism how immature snRNPs are targeted and retained in CBs has yet to be described. Here, we microinjected and expressed various snRNA deletion mutants as well as chimeric 7SK, Alu or bacterial SRP non-coding RNAs and provide evidence that Sm and SMN binding sites are necessary and sufficient for CB localization of snRNAs. We further show that Sm proteins, and specifically their GR-rich domains, are important for accumulating snRNPs in CBs. Accordingly, core snRNPs containing the Sm proteins, but not naked snRNAs, restore the formation of CBs after their depletion. Finally, we show that immature but not fully assembled snRNPs are able to induce CB formation and that microinjection of an excess of U2 snRNP-specific proteins, which promotes U2 snRNP maturation, chases U2 snRNA from CBs. We propose that the accessibility of the Sm ring represents the molecular basis for the quality control of the final maturation of snRNPs and the sequestration of immature particles in CBs.
INTRODUCTION
Spliceosomal U-rich small nuclear RNAs (snRNAs) belong to a group of small non-coding, non-polyadenylated RNAs. Five major spliceosomal snRNAs have been described. Three of these snRNAs - U2, U5 and U6 are essential components of the active spliceosome and form its catalytic RNA core. The U1 snRNA recruits the spliceosome to the 5′ exon–intron boundary and the U4 snRNA acts as a chaperone that brings the U6 snRNA to the spliceosome in a splicing inactive conformation until the spliceosome is catalytically activated. However, snRNAs do not enter the splicing reaction as naked RNAs but associate with several proteins to form complexes called small nuclear ribonucleoprotein particles (snRNPs). All spliceosomal snRNAs, except U6 and U6atac, contain a canonical uridine-rich sequence called the Sm site, which serves as a binding platform for the Sm proteins B/B′, D1, D2, D3, E, F and G which form a heptameric ring. Furthermore, snRNAs associate with a set of proteins that are specific for each snRNA. Interest in snRNP biogenesis has increased since it was shown that defects in the snRNP maturation pathway correlate with human diseases like spinal muscular atrophy or retinitis pigmentosa (1–4).
The biogenesis of an snRNP starts in the cell nucleus, where snRNAs are transcribed by RNA polymerase II. The only exceptions are the U6 and U6atac snRNAs, which are transcribed by RNA polymerase III, associate with a heptameric ring of the Like-Sm (LSm) proteins 2–8, and permanently reside in the nucleus. Soon after transcription and the initial 3′ end cleavage, snRNAs transcribed by RNA polymerase II are transported to the cytoplasm, where they associate with the SMN complex (5,6). The SMN complex recognizes, via Gemin 5, specific motifs consisting of the 5′ monomethylguanosine cap, the Sm site and the SMN binding site that is located in the stem loops found in the vicinity of the Sm site (7–10). The SMN complex also associates with Sm proteins, and in cooperation with the PRMT5 complex, assembles the Sm ring around the Sm site (11–13). After Sm ring assembly, the snRNA is finally trimmed at the 3′ end while the monomethylguanosine cap at the 5′ end is hypermethylated (14,15). Both the Sm ring and the trimethylguanosine cap are important for snRNA transport back to the nucleus and the SMN complex has been suggested to assist during the nuclear translocation of the core snRNPs (16–18).
Immature snRNPs imported into the nucleus first accumulate in nuclear structures called Cajal bodies (CBs) (19,20), but the signal that targets new snRNPs to the CB has not been identified. CBs have been proposed to serve as a compartment for final U2, U4/U6 di-snRNP and U4/U6•U5 tri-snRNP assembly and recycling (21–26) and snRNA posttranscriptional modifications ((27,28), reviewed in (29)). Recently, it has been shown that the R2TP complex is involved in the biogenesis of U4 and U5 snRNPs (30–32). Once the snRNPs are fully matured, they are released from the CB to the nucleoplasm in order to function in splicing. The final maturation of the tri-snRNP involves annealing of U4 and U6 snRNAs, addition of U4/U6-specific proteins and final interaction with U5 snRNP. The U2 snRNP is assembled in a stepwise process initiated by the interaction of the SNRPA1 (U2A') and SNRPB2 (U2B″) proteins with stem loop IV of the U2 snRNA. The heptameric SF3b protein complex subsequently binds to form 15S U2 snRNPs, followed by the trimeric SF3a complex to generate the functional 17S form of U2 snRNP (reviewed in (29)). We recently showed that snRNAs that fail to associate with snRNP-specific proteins or are unable to form U4/U6 or U4/U6•U5 particles are sequestered in CBs (33). Similarly, U2 snRNPs that fail to interact with the SF3a protein complex, and thereby complete their assembly, accumulate in CBs (34). These findings indicate a quality control mechanism that detects partially-assembled snRNPs. However, the molecular basis for the discrimination between mature and immature snRNPs has not yet been elucidated.
To determine snRNA sequences important for CB targeting and retention, we generated several snRNA variants. Mutated snRNAs were either fluorescently labeled and microinjected or transiently expressed in human cells. We also prepared chimeric non-coding RNAs to confirm sequences essential for CB targeting. We further depleted Sm proteins or expressed Sm deletion mutants to identify protein motifs that are important for CB localization of snRNPs. Finally, we microinjected snRNP assembly intermediates to determine factors important for sequestration of partially-assembled snRNPs in CBs.
MATERIALS AND METHODS
Cell culture
HeLa cells were cultured in Dulbecco's modified Eagle's medium (DMEM) containing 4.5 g glucose/l (Sigma) supplemented with 10% fetal bovine serum (FBS), 1% penicillin and streptomycin (Gibco).
Plasmids
The U4-MS2 RNA construct, where U4 is under the control of endogenous U4 promoter elements and a single MS2 loop was inserted into U4 stem loop II, was obtained from Edouard Bertrand (IGMM, CNRS, Montpellier). The U2 snRNA-MS2 construct, which includes the promoter sequence (563 nt upstream of U2 transcription start site), was amplified from HeLa genomic DNA using specific primers (Supplementary Table S1) and cloned into the pcDNA3 vector without a CMV promoter using EcoRI/BamHI restriction sites. The U2 snRNA sequence is identical to the transcript ID ENST00000616345. The MS2 loop was inserted into the stem loop IIb by site-directed mutagenesis using the primers listed in Supplementary Table S1.
The deletion constructs of U2 and U4 (U2 ΔSLI+IIa,b-MS2 and U4 Δ1–64-MS2) were created by site-directed mutagenesis using specific primers (Supplementary Table S1). All plasmids were co-transfected with A1-MS2-YFP (a kind gift of Yaron Shav-Tal, Bar-Ilan University, Ramat-Gan, Israel) using Lipofectamine3000 (Invitrogen) according to the manufacturer's protocol. Transfected HeLa cells were cultured for 24 h at 37°C.
SmB-YFP and SmD1-GFP plasmids (19) were provided by A. Lamond (University of Dundee, United Kingdom). SmD3-GFP plasmid was prepared from the total RNA of HeLa cells by RT followed by PCR using primers listed in Supplementary Table S1 and cloned into the GFP-N1 vector (Clontech) using EcoRI/BamHI restriction sites. Deletion constructs (SmBΔCtail, SmD3ΔCtail, SmD1Δ1/4GR, SmD1Δ1/2GR and SmD1ΔGR) were created by PCR using primers listed in Supplementary Table S1. D3Ala-GFP, BAla-GFP and D1Ala-GFP constructs were created by site-directed mutagenesis using specific primers (Supplementary Table S1) and verified by DNA sequencing.
Antibodies
For indirect immunostaining we used anti-coilin (5P10) antibody, kindly provided by M. Carmo-Fonseca (Institute of Molecular Medicine, Lisboa). Secondary anti-mouse antibodies conjugated with Alexa-647 (Invitrogen) were used. For immunoprecipitation we used anti-goat GFP antibodies obtained from David Drechsel (MPI-CBG Dresden, Germany). For Western blotting we used the following antibodies: mouse anti-GFP (Santa Cruz), mouse anti-U2B′ (Progen), rabbit anti-SmD1 (Abcam), rabbit anti-SmG (Abcam), rabbit anti-SF3a60/SF3A3 (Abcam), mouse anti-SF3b49 (Abcam), rabbit anti-PRPF31 (Abcam), mouse anti-GAPDH (Abcam), rabbit anti-β actin (Abcam), mouse anti-SMN (Sigma), and mouse anti-tubulin, kindly provided by P. Draber (Institute of Molecular Genetics, CAS). The anti-Sm antibody (Y12) was produced from a hybridoma cell line (a gift from Karla Neugebauer, Yale University, New Haven, USA) at the Antibody facility (Institute of Molecular Genetics, CAS). Secondary goat anti-mouse or anti-rabbit antibodies conjugated with horseradish peroxidase (Jackson ImmunoResearch Laboratories) were used.
RNAi
The siRNAs used in this study were SmB/B′, SmD1, SmG, TGS1 and SMN (20nM, Invitrogen, Supplementary Table S1). siRNAs were transfected with Oligofectamine (Invitrogen) according to the manufacturer's protocol. Cells were microinjected 48 h (siRNAs SmB/B′, SmG), 60 h (siRNA SMN, TGS1) or 72 h (siRNA SmD1) after transfection. The “Negative Control No. 5 siRNA" from Invitrogen was used as a negative control.
In vitro transcription
All DNA templates for in vitro transcription were prepared by PCR using Phusion polymerase (Biolab). The primers utilized are listed in Supplementary Table S1. All forward primers contained the T7 promoter sequence (Supplementary Table S1). ΔSm site and U2withU1Sm mutants were created by site-directed mutagenesis using ΔSm and U2withU1Sm primers (see Supplementary Table S1). Plasmids containing full-length U1, U2, U4 and U5 snRNAs (a gift from Karla Neugebauer, Yale U., New Haven, USA) were used as templates. 7SK RNA and Alu cytoplasmic RNA were cloned from total HeLa RNA isolated by TRIZOL reagent (Invitrogen) according to the manufacturer's protocol, and cDNA was synthesized using specific reverse primers and SuperScriptIII (Invitrogen). SRP RNA was isolated from total Escherichia coli as previously described (35) and cDNA was synthesized using the specific reverse primer (Supplementary Table S1) and SuperScriptIII (Invitrogen). WT RNAs and Sm+SMN mutants (7SK, Alu and SRP) were prepared by PCR using Phusion polymerase (Biolab). Fluorescently labelled RNAs were prepared as described previously (36) by in vitro transcription using MegashortscriptIII kit (Invitrogen) containing UTP-Alexa 488 (Invitrogen) and trimethylated cap analog (m32,2,7G(5′)ppp(5′)G (Jena Bioscience)) or monomethylated cap analog (m7G(5′)ppp(5′)G (Invitrogen)). After synthesis, RNA was isolated by phenol/chloroform extraction, precipitated and dissolved in nuclease-free water. RNA was diluted in solution containing dextran-TRITC 70-kDa (Sigma-Aldrich) to a final concentration 200 ng/μl.
Prediction of snRNA secondary structure
The secondary structure of all U2 snRNA mutants was analyzed by mathematical modeling (Supplementary Figure S1). Structure analysis was carried out using the Vienna RNA package (37). Minimum free energy RNA secondary structures were generated by both constrained and unconstrained prediction. For the first, RNAfold was used. The latter was accomplished using constrained RNAfold (RNAfold-C). Structures were plotted using RNAplot.
Microinjection
HeLa cells were grown on glass coverslips for 24 h and RNA was microinjected using InjectMan coupled with FemtoJet (Eppendorf) as described previously (36). Cells were then rinsed twice in PBS and fixed for 20 min at room temperature in 4% PFA/PIPES (freshly prepared).
Indirect immunofluorescence and image acquisition
HeLa cells grown on coverslips were fixed, labeled and images acquired using the the DeltaVision microscopic system (Applied Precision) coupled to an Olympus IX70 as described previously (25). Stacks of 20 z-sections with 200 nm z steps were collected per sample and subjected to mathematical deconvolution using SoftWorx software. Maximal projections of deconvolved pictures were generated by SoftWorx and are presented. For high-content microscopy, samples were scanned as described previously (33). Mean and SEM of three biological experiments were calculated and plotted. Statistical significance was analyzed by the Student's t-test.
snRNP precipitation
Immunoprecipitation was performed as previously described (38) using goat anti-GFP antibodies. RNA was extracted using phenol/chloroform, resolved on a polyacrylamide gel containing 7 M urea and silver stained. The immunoprecipitated proteins were resuspended in 30 μl of 2× sample buffer (0.25 M Tris–HCl pH 6.8, 20% glycerol, 4% SDS, 2% β-mercaptoethanol, 0.02% bromphenol blue), resolved on a 12% polyacrylamide gel and detected by western blotting.
In vitro reconstitution of snRNP cores
snRNP reconstitution were carried out as described in (39–41). Typically, 15 pmol of in vitro transcribed snRNA were assembled with 20 μg of native snRNP proteins in 30 μl of reconstitution buffer 20 mM HEPES–KOH pH 7.9, 50 mM NaCl, 5 mM MgCl2. Reconstituted snRNPs were microinjected without further purification.
Preparation of U2 snRNPs, SF3a and SF3b
Human SF3a and SF3b complexes were affinity-purified from HeLa nuclear extract (42) in G buffer (20 mM HEPES, pH 7.9, 1.5 mM MgCl2, 10% (w/v) glycerol, 0.5 mM DTE, 0.5 mM PMSF) containing 250 mM NaCl, that was first passed over an anti-m3G immunoaffinity column. The NaCl concentration was increased to 600 mM to ensure the complete dissociation of SF3a and SF3b from U2 snRNPs, and the extract was applied to affinity columns with covalently bound anti-peptide antibodies against human SF3B1 (amino acids 99–113) or SF3A2 (amino acids 444–458). Bound complexes were eluted with an excess of the cognate peptide in G buffer containing 600 mM NaCl, further purified by gel filtration on a Superose 6 column (SF3b) or on a Superdex 200 column (SF3a), and then concentrated by ultracentrifugation. To isolate 12S U2 snRNPs, a mixture of anti-m3G affinity purified spliceosomal snRNPs were first separated by 10–30% (v/v) glycerol gradient centrifugation in G buffer containing 150 mM NaCl. The 12S peak (containing both 12S U1 and U2 snRNPs) was subsequently applied to a HiTrap Heparin HP column (GE Healthcare) and the bound snRNPs were eluted with a linear salt gradient (50–1000 mM NaCl in buffer containing 20 mM Tris, pH 7.9, 1.5 mM MgCl2, 1 mM DTT). The eluted 12S U2 snRNPs were subsequently concentrated by ultracentifugation. 15S U2 snRNPs were generated by combining equal molar amounts of purified 12S U2 and SF3b, whereas 17S U2 snRNPs were generated by combining equal molar amounts of purified 12S U2, SF3b and SF3a, and then incubating for 1h on ice. Gradient centrifugation confirmed that the vast majority of the 12S U2 snRNPs were converted to 15S or 17S complexes, under these conditions (data not shown).
RESULTS
Sm and SMN binding sites are necessary for targeting microinjected U2 snRNA to Cajal bodies
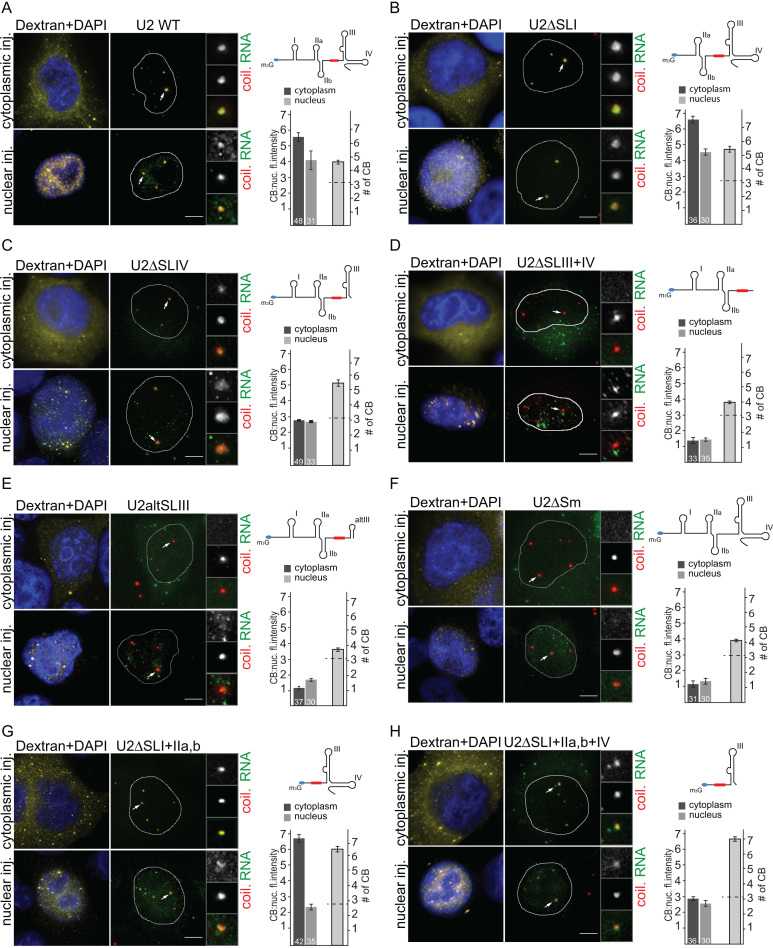
To determine snRNA sequences that are necessary for targeting snRNAs to CBs, we utilized the U2 snRNA as a model RNA molecule. We prepared several deletion mutants of U2 snRNA and transcribed snRNAs in vitro in the presence of UTP-Alexa488 and a trimethylated 5′ cap analog. In vitro transcribed snRNAs were microinjected into HeLa cells together with TRITC-labeled dextran-70 kDa that does not cross the nuclear membrane and serves as a marker of nuclear or cytoplasmic injection. We microinjected snRNA into either the nucleus or the cytoplasm in order to determine whether different sequences are required for CB targeting depending on whether the snRNA is imported from the cytoplasm or is directly delivered to the nucleus. Cells were incubated for 60 min following injection, fixed and coilin, a marker of CBs, was detected by indirect immunofluorescence. Wild-type (WT) U2 snRNA accumulated in CBs after both cytoplasmic and nuclear injection (Figure 1A). Nuclear localization of cytoplasmatically injected RNAs indicated that WT U2 snRNAs acquired the Sm-ring and were imported into the nucleus.
Figure 1.
Sm and SMN binding sites are necessary to target the microinjected U2 snRNA to Cajal bodies. (A–H) In vitro transcribed WT U2 snRNA or deletion mutants thereof were microinjected into the cytoplasm or into the nucleus of HeLa cells. U2 snRNA was labeled with UTP-Alexa-488 (green), coilin, a marker of CBs, was immunolabeled by Alexa-647 (red). Dextran-TRITC 70 kDa (yellow) was used to monitor nuclear or cytoplasmic injection, DNA was stained by DAPI (blue). Small red box in U2 snRNA scheme represents the Sm site. The intensity of the RNA signal in CBs versus the nucleoplasm was determined for each CB, and the average and SEM are shown in graphs next to the micrographs (number of microinjected cells is indicated in the bar). Number of CBs in microinjected cells was counted and plotted. Dotted line indicates number of CBs in control non-injected cells. The scale bar represents 10 μm.
Subsequently we assayed the importance of different snRNA domains for CB targeting. Predicted secondary structure models of individual deletion mutants are shown in Supplementary Figure S1. We first deleted stem loop I (U2ΔSLI), which is important for binding of the SF3A3 (SF3a60) protein from the SF3a complex (43). We observed that there was a strong accumulation of U2ΔSLI in CBs (Figure 1B). Deletion of the stem loop IV (U2ΔSLIV), which contains part of the U2 SMN binding motif and interacts with the U2-specific dimer, SNRPA1 (U2A')/ SNRPB2 (U2B″) (44,45), decreased CB localization in comparison to WT U2 (Figure 1C). Next, we removed both stem loops III and IV at the 3′ end of U2 (U2ΔSLIII+IV), which together form the binding platform for the SMN complex (46). The deletion of both stem loops completely inhibited CB accumulation (Figure 1D). The U2ΔSLIII+IV RNA remained in the cytoplasm after cytoplasmic injection, indicating that the Sm ring was not formed and that snRNA without the Sm ring was not able to cross the nuclear membrane. In native snRNAs, the Sm site is found between two stem loops, which is a spatial organization that is missing in the U2ΔSLIII+IV RNA. Therefore, we replaced stem loops III and IV with a shortened stem loop III (U2altSLIII) that lacked 18 central nucleotides, 120–137, which were previously shown to bind the SMN complex (47). After cytoplasmic injection, the U2altSLIII RNA was partially retained in the cytoplasm and CB localization was significantly reduced (Figure 1E). Following nuclear injection, U2altSLIII snRNAs did not accumulate in CBs but remained in the nucleus. In some cases after nuclear injection we observed accumulation of snRNAs lacking SMN or Sm sites (see below) in nuclear dots that did not co-localize with either coilin, Gemin2 or PML (Figures 1 and 2 and data not shown). These data suggest that the SMN complex is directly or indirectly, via Sm-ring assembly, required for CB localization of the U2 snRNA.
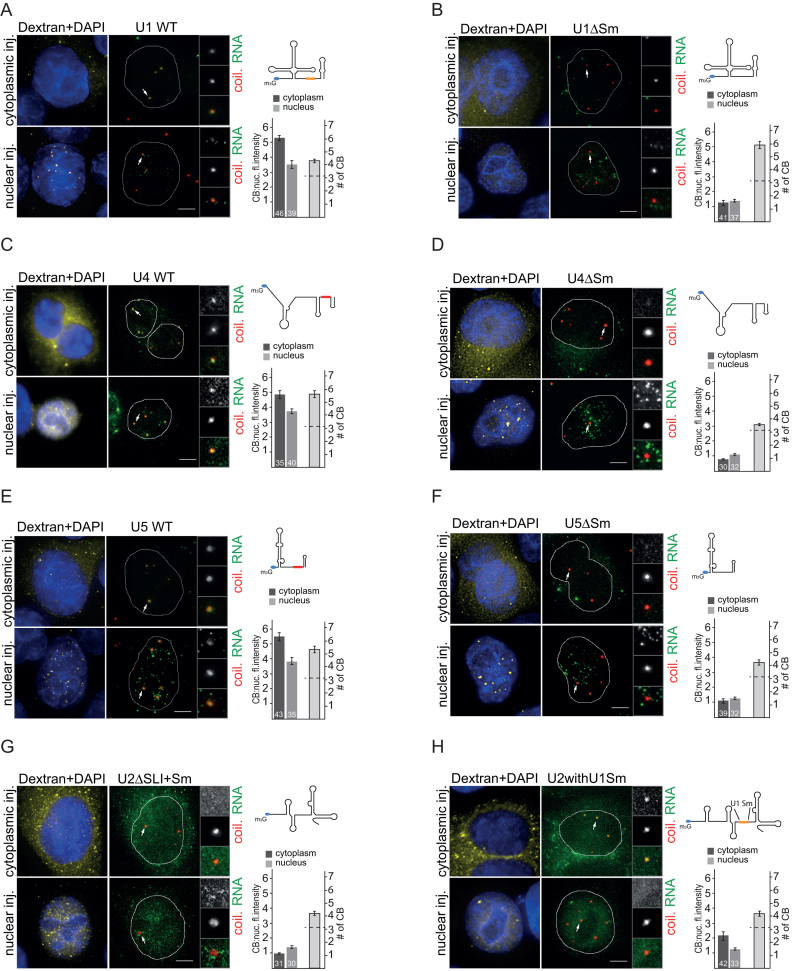
Figure 2.
The Sm site is necessary for Cajal body targeting of U1, U4 and U5 snRNAs. (A–F) WT or ΔSm U1, U4 and U5 snRNAs were transcribed in vitro and microinjected into the cytoplasm or the nucleus of HeLa cells (green). CBs are marked by coilin immunolabeling (red). Legend is the same as in Figure 1. Small red boxes in snRNA schemes represent the canonical Sm site, the orange box depicts the non-canonical U1 Sm site. (G, H) U2ΔSLI+Sm and U2 snRNA with U1-like Sm site were injected into the nucleus and the cytoplasm of Hela cells. Legend is the same as in Figure 1.
To test whether the sequence binding the Sm ring is important for CB targeting, we prepared U2 snRNA lacking the Sm site (U2ΔSm). Consistent with it lacking an Sm ring, the U2ΔSm RNA injected into the cytoplasm was not imported into the nucleus (Figure 1F). The U2ΔSm RNA injected into the nucleus mimicked the behavior of RNAs that lacked the SMN binding site (U2ΔSLIII+IV and U2altSLIII); that is, the deletion of the Sm site completely abolished CB localization. These data collectively demonstrate that Sm and SMN binding sites are both necessary for U2 snRNA targeting to CBs.
To test whether the Sm and SMN binding sites are sufficient for CB targeting, we deleted the first 94 nt of U2 snRNA that contain stem loops I and IIa,b, keeping only the Sm and SMN sites (U2ΔSLI+IIa,b). This RNA was localized to CBs similar to WT U2 snRNA after cytoplasmic injection (Figure 1G). CB localization was reduced when U2ΔSLI+IIa,b RNA was injected into the nucleus but it still accumulated in CBs at levels two times higher than the U2ΔSm or U2ΔSLIII+IV RNAs. The stem loop IV binds SNRPB2 and SNRPA1, which might still target U2 snRNA to CBs. Therefore, we further deleted stem loop IV (U2ΔSLI+IIa,b+IV). This minimal RNA of only 55 nucleotides was localized to CBs, but CB accumulation was lower than observed with WT U2 (Figure 1H).
Finally, we set out to probe whether the 5′ cap was involved in the CB accumulation of the U2 snRNA. To test this, we prepared all U2 snRNA constructs described above with a mono-methylguanosine at the 5′ end and injected them into the nucleus and the cytoplasm. As shown in Supplementary Figure S2, the mono-methylguanosine-capped U2 snRNAs were readily detectable in CBs in a manner similar to that observed with the tri-methylguanosine-capped U2 snRNAs. These data indicate that the methylation status of the U2 snRNA 5′ guanosine cap does not affect its localization to CBs. Taken together, our results suggest that Sm and SMN binding sites are dominant factors essential for U2 snRNA targeting and retention in CBs.
The Sm site is a general snRNA CB targeting sequence
Next we tested whether the Sm site is required for CB targeting of other snRNAs (U1, U4 and U5). We microinjected WT snRNAs and snRNAs without the Sm site (Figure 2). WT snRNAs accumulated in CBs in all cases (Figure 2A, C and E). snRNAs microinjected into the nucleus exhibited weaker CB localization than snRNAs microinjected into the cytoplasm. Surprisingly, microinjected U1 snRNA also accumulated in CBs, even though it was shown previously by in situ hybridization that endogenous U1 snRNA accumulates in CBs to a much lower extent than other snRNAs, and instead localizes to nuclear structures called gems (48–50). In contrast to the WT snRNAs, none of the snRNAs lacking the Sm site accumulated in CBs (Figure 2B, D and F). After cytoplasmic microinjection, ΔSm snRNAs were retained in the cytoplasm, which is consistent with the inhibition of Sm ring formation and nuclear import.
Previous work has shown that inhibition of the final U2 snRNP maturation step by the knockdown of SF3A3 (SF3a60) results in the accumulation of U2 snRNA in CBs (51). To test whether CB localization of the U2 snRNA lacking the SF3A3 binding site (U2ΔSLI) also depends on the Sm site, we prepared RNA that lacked both the SF3A3 and Sm binding sites (U2ΔSLI+Sm) and injected this RNA into the nucleus as well as the cytoplasm (Figure 2G). We did not observe any CB accumulation, which demonstrates that the Sm site is required for CB targeting of mutated snRNAs that are unable to form a mature snRNP particle.
The U1 snRNA Sm site (AUUUGUG) differs from the canonical Sm site found in U2, U4 and U5 snRNAs (AUUUUUG). To test whether the non-canonical U1 Sm site can act as a CB targeting signal in the context of other snRNAs, we replaced the U2 Sm site with the U1 Sm site (U2withU1Sm). This chimeric snRNA localized partially to CBs only after cytoplasmic microinjection. In the case of nuclear microinjection, the U2withU1Sm RNA did not localize to CBs, but was distributed throughout the nucleoplasm (Figure 2H). This finding suggests that the U1 Sm site cannot fully replace the canonical Sm site, at least when the U1 Sm site has been inserted into U2 snRNA. This result is consistent with observations that the U1-specific protein SNRP70 (U1–70K) interacts with the SMN complex and enhances Sm ring formation specifically on the U1 snRNA (50,52).
Sm and SMN sites are sufficient to target non-coding RNAs into Cajal bodies
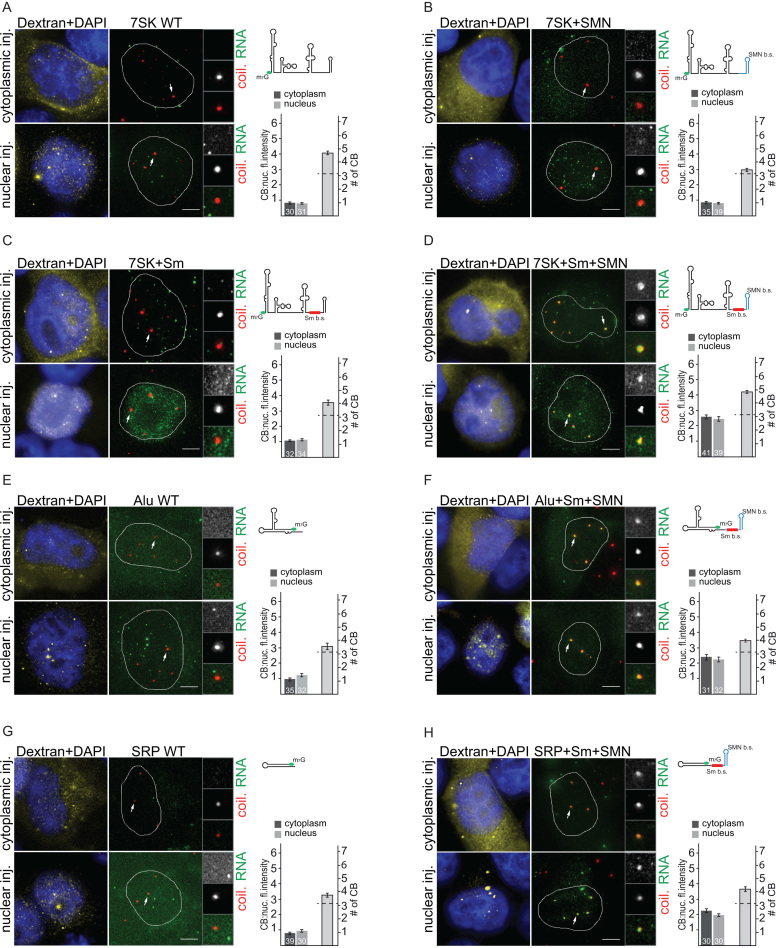
To test whether Sm and SMN sites are sufficient to target non-CB RNAs to the CB, we fused the Sm and SMN sites with several non-coding RNAs, including human 7SK and Alu RNAs, and E. coli SRP RNA. First, we utilized the 7SK RNA, which is an abundant nuclear non-coding RNA important for regulation of RNA polymerase II, that under physiological conditions does not accumulate in CBs (49,53). We prepared three different chimeric 7SK RNAs: (i) 7SK+Sm site RNA containing the consensus Sm site inserted between two stem loops at the 3′ end; (ii) 7SK+SMNsite RNA where the 3′ end stem loop was replaced with a stem loop from Herpes saimiri virus (HSUR1) RNA, which binds the SMN complex (54); and (iii) 7SK+Sm+SMNsites RNA containing both Sm and SMN sites. All 7SK RNAs were microinjected into the nucleus or the cytoplasm of HeLa cells (Figure 3A–D). We found that neither WT nor chimeric RNAs that contained either the Sm site or the SMN site accumulated in CBs (Figure 3A–C). However, the 7SK+Sm+SMNsites RNA which possessed both Sm and SMN binding sequences, efficiently localized in CBs after both nuclear and cytoplasmic injections (Figure 3D). To further confirm this finding, we attached Sm + SMN sequences to Alu RNA, a 120nt RNA component of the cytoplasmic Alu RNP, which is involved in translation regulation (55,56). While WT Alu RNA did not accumulate in CBs (Figure 3E), chimeric Alu+Sm+SMN RNA localized to CBs after nuclear and cytoplasmic injections (Figure 3F). Finally, we utilized a 114nt non-coding RNA from bacteria (E. coli SRP RNA). Microinjected WT SRP RNA did not localize to CBs (Figure 3G), but the addition of Sm+SMN sites targeted this RNA to CBs (Figure 3H). This experiment shows that SMN and Sm sites are together sufficient to target various non-coding RNAs to CBs.
Figure 3.
Sm and SMN sites are sufficient to target non-coding RNAs into Cajal bodies. (A–D) In vitro transcribed 7SK RNAs (WT or chimeras containing Sm, SMN or both sites) were microinjected into the nucleus or cytoplasm of HeLa cells. Legend is the same as in Figure 1. (E, F) WT Alu or Alu+Sm+SMN sites RNAs were microinjected into the nucleus or cytoplasm of HeLa cells. Legend is the same as in Figure 1. (G, H) WT E. coli SRP RNA or SRP+Sm+SMN sites RNA were microinjected into the nucleus or cytoplasm of HeLa cells. Legend is the same as in Figure 1.
Minimal transiently expressed snRNAs containing Sm and SMN sites accumulate in Cajal bodies
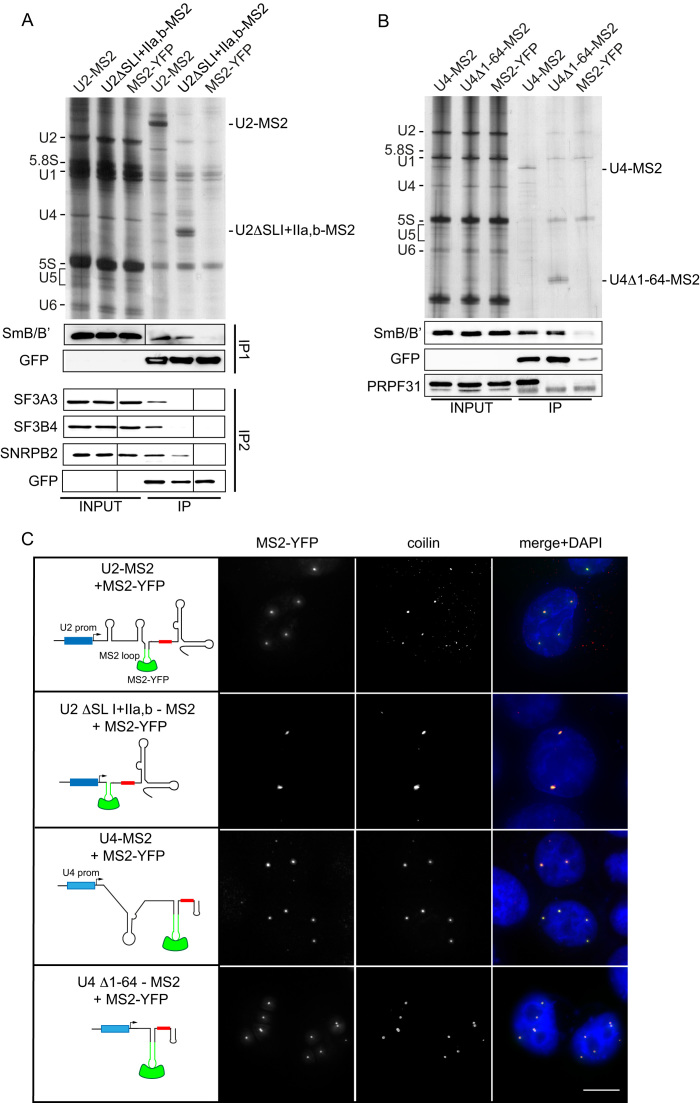
Our experiments demonstrate the importance of Sm and SMN sites for CB accumulation of microinjected RNAs. To test whether these sequences are important also for CB targeting of endogenously expressed snRNAs, we utilized a system devised by Edouard Bertrand and colleagues in which the MS2 binding site was inserted to detect transiently expressed U4 snRNAs (57). Similarly, we cloned the MS2 site into the stem loop IIb of U2 snRNA driven by the U2 promoter (U2-MS2) to distinguish the transiently expressed U2 snRNA from its endogenous counterpart. To prepare minimized U2 snRNA, we deleted the first 94 nucleotides from the U2-MS2 RNA leaving only the MS2 loop followed by the Sm site and the SMN site containing stem loops III and IV (U2ΔSLI+IIa,b-MS2). Similarly, the first 64 nucleotides were deleted from U4-MS2 creating a minimized U4 RNA (U4Δ1–64-MS2) that contains the Sm site between stem loops II and III, which together serve as the SMN binding site (58).
U2-MS2 and U4-MS2 constructs were co-transfected with MS2-YFP and after 24 h, cells were subjected to immunoprecipitation using anti-GFP antibodies, which also recognize YFP. Immunoprecipitation followed by RNA detection showed that the deletion mutants were expressed at levels similar to WT snRNAs (Figure 4A and B). Western blot analysis of proteins co-precipitating with MS2 snRNAs revealed the association of U2-MS2 with Sm proteins, SNRPB2 (U2B″), SF3A3 (SF3a60) and SF3B4 (SF3b49) (Figure 4A) and the U4-MS2 RNA with Sm proteins and PRPF31 (Figure 4B). Although only a small subset of snRNP-specific proteins was tested, these results suggest that the MS2 insertion does not block the association of snRNAs with snRNP-specific proteins. These data also confirm that U2 snRNA lacking stem loops I and II does not interact with the SF3a and SF3b complexes and thus mimics immature snRNPs. Similarly, the U4 snRNA without stem loop I does not bind the PRPF31 protein. Next, MS2-tagged snRNAs were expressed together with MS2-YFP, cells were fixed after 24 h and coilin was detected by indirect immunofluorescence (Figure 4C). Both minimized U2 and U4 snRNAs localized to CBs in a similar manner as the WT snRNAs, which was also observed in our microinjection experiments, demonstrating that Sm + SMN sites are sufficient for targeting snRNAs to CBs. It should be noted that we cannot test the role of Sm and SMN sites in CB targeting directly via their depletion because snRNA expressed without these sequences would be retained in the cytoplasm and would not reach the nucleus.
Figure 4.
Transiently-expressed, truncated snRNAs containing Sm and SMN binding sites accumulate in Cajal bodies. (A) Immunoprecipitation of U2-MS2 and the deletion mutant U2ΔSLI+IIa,b-MS2. RNAs were immunoprecipitated via MS2-YFP by anti-GFP antibodies and detected by silver staining. Co-precipitated proteins were analyzed by Western blotting. With the full-length U2-MS2 RNA we detected all tested proteins (SmB/B′, SF3A3, SNRPB2 and SF3B4), while only SmB/B′ and SNRPB2 co-precipitated with the U2ΔSLI+IIa,b-MS2 RNA. (B) Immunoprecipitation of U4-MS2 and the deletion mutant U4Δ1–64-MS2. Legend as in (A). With the full-length U4-MS2 RNA we detected both tested proteins (SmB/B′ and PRPF31), while only SmB/B′ co-precipitated with U4Δ1–64-MS2 RNA. (C) Hela cells were co-transfected with U2 or U4 snRNAs containing the MS2 loop (green stem loop) and MS2-YFP (green). Coilin was used as a marker of CBs (red). Small red box in snRNA schemes represents the canonical Sm site and blue boxes represent endogenous U2 or U4 promoters. DNA was stained by DAPI. The scale bar represents 10 μm.
Sm proteins are essential for snRNA Cajal body targeting
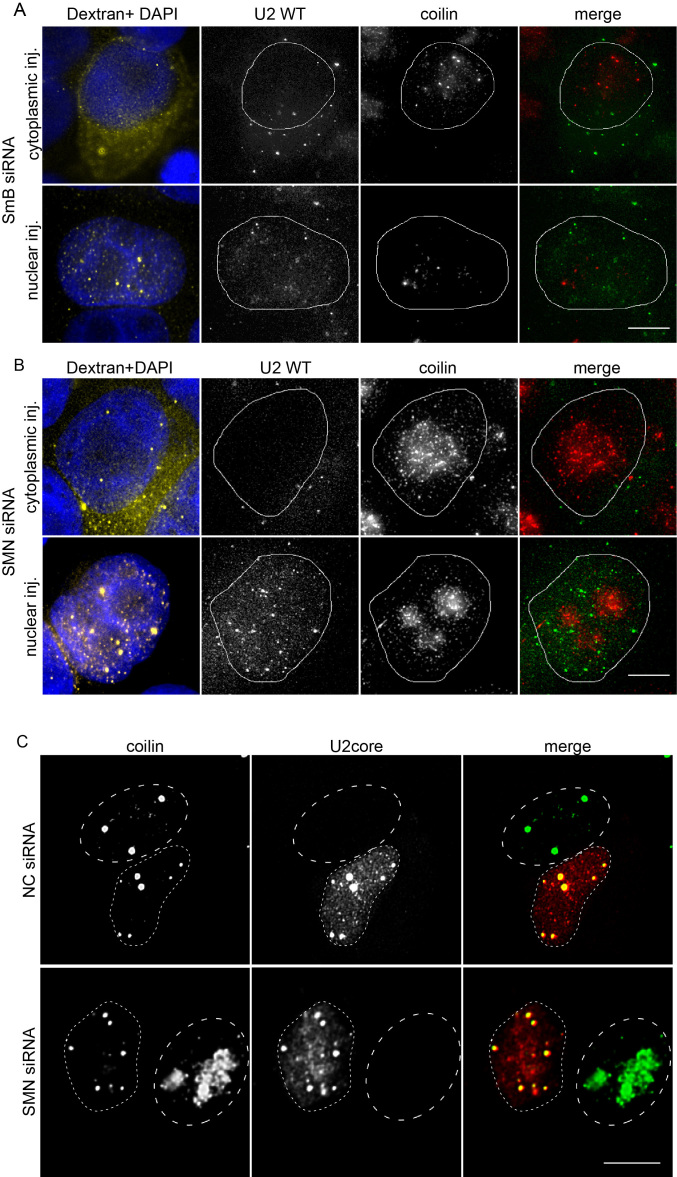
Our experiments provide evidence that SMN and Sm sites are essential for snRNA targeting into CBs. These minimal sequences were previously shown to be sufficient for SMN complex binding and Sm ring assembly (54,58). To test whether the snRNA sequence per se or the Sm ring is essential for targeting snRNAs to CBs, we depleted one of the Sm proteins, SmB/B′, by RNA interference (Supplementary Figure S3D). When WT U2 snRNA was microinjected into the cytoplasm, SmB/B′ knockdown completely inhibited its CB localization and the U2 snRNA was retained in the cytoplasm, confirming the efficiency of the knockdown and the inhibition of Sm ring assembly (Figure 5A). After nuclear microinjection, WT U2 snRNA remained in the nucleus but did not accumulate in CBs. We observed the same phenotype in cells depleted of SmD1 and SmG (Supplementary Figure S3A, B), but not in the cells treated with a negative control siRNA (Supplementary Figure S3C). It should be noted that depletion of Sm B/B′ had a negative effect on CB integrity and in a fraction of cells, coilin partially accumulated in nucleoli. However, CBs were still present in a significant number of cells treated for 48h with the anti-SmB/B′ siRNA. These experiments suggest that Sm proteins assembled around the Sm site are recognized as a CB targeting signal.
Figure 5.
SmB/B′ but not the SMN protein is essential for Cajal body targeting of snRNA. (A) Depletion of the SmB/B′ protein disrupts targeting of microinjected snRNAs into the Cajal body. SmB/B′ protein was depleted by RNAi and Alexa-488 labeled WT U2 snRNA (green) subsequently microinjected into HeLa cells. Dextran-TRITC 70 kDa was used as a marker of microinjection (yellow). DNA was stained by DAPI (blue). (B, C) SMN protein was depleted by siRNA and either Alexa-488 labeled WT U2 snRNA (B) or core U2 snRNP (C) were microinjected into HeLa cells. Depletion of the SMN protein disrupts CBs, which can be rescued by cytoplasmic microinjection of core U2 snRNP (red). Coilin was used as a marker of CB (green). (C) Dotted lines mark the nucleus of microinjected cells; dashed lines mark the nucleus of non-microinjected cells. The scale bar represents 10 μm.
Both SMN and Sm sites are recognized by the SMN complex (7–10). Therefore, we decided to test whether the SMN complex was directly required for snRNP targeting to CBs. However, the SMN complex is involved in Sm ring assembly, which we showed above as a CB targeting signal (Figure 5A). In addition, SMN complex depletion results in dissociation of CBs (59,60). To overcome these obstacles, we utilized the fact that incomplete snRNPs are able to induce formation of CBs in cells lacking these nuclear structures (33). We knocked down the SMN protein, which resulted in CB disappearance, and injected either U2 snRNA alone or in vitro assembled U2 core snRNPs (snRNA+Sm ring) (Figure 5B and C). Injection of in vitro synthesized snRNA did not restore CBs and snRNA remained in the compartment where it was injected without any specific localization (Figure 5B). Treatment with a negative control siRNA did not have any effect on U2 snRNA localization to CBs (data not shown, see also Supplementary Figure S3C). In contrast, injection of core snRNPs restored CBs and injected snRNPs accumulated in CBs (Figure 5C). These data show the essential role of Sm proteins in targeting U2 snRNA to CBs, and suggest that the SMN complex is not directly involved in snRNP CB localization.
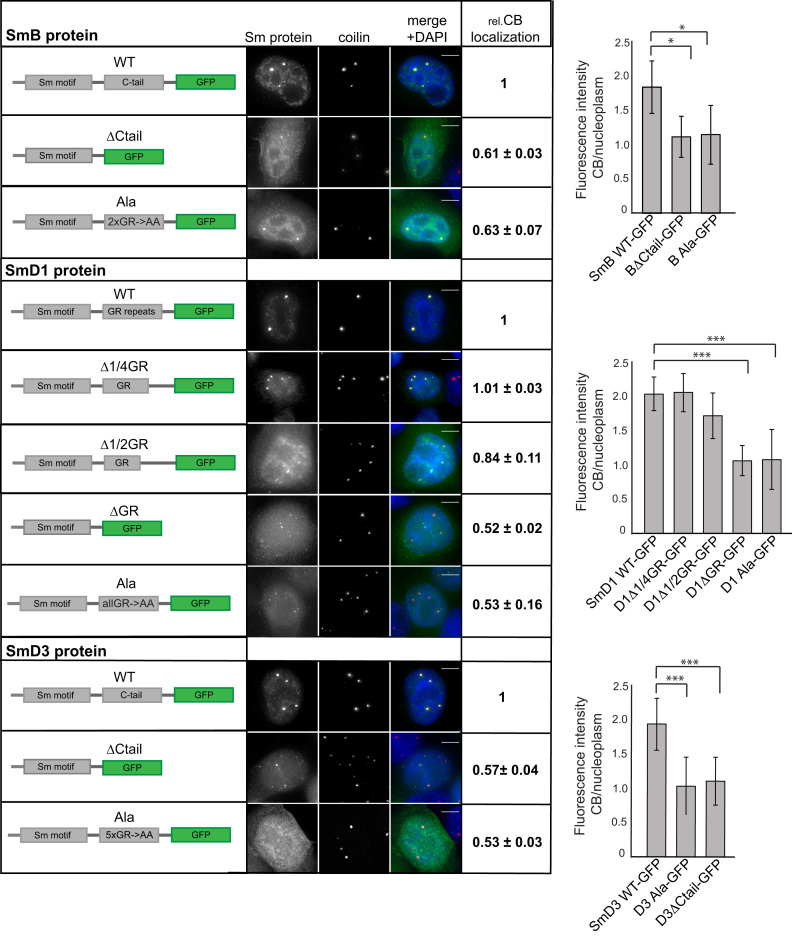
C-terminal GR-rich tails of SmB, SmD1 and SmD3 are important for Cajal body localization
To specify protein motifs that are important for CB targeting of Sm proteins, we deleted several non-Sm fold domains, namely the GR domain of SmD1 and C-terminal tails of SmD3 and SmB, which contain several GR dipeptides (Figure 6). We tagged the proteins with GFP, transiently expressed them in HeLa cells, and assayed Sm-GFP interaction with snRNAs by immunoprecipitation (Supplementary Figure S4) and CB localization by fluorescence microscopy (Figure 6). Cells were transfected with the same amount of DNA but some mutants were expressed at higher levels than others likely due to better protein and/or mRNA stability. Deletion of the C-terminal tails from SmD3 (aa 110–126) and SmB (aa 170–231) did not prevent SmD3 and SmB interactions with snRNAs (Supplementary Figure S4A and B) but significantly reduced CB localization (Figure 6). The C-terminal tails of SmD3 and SmB proteins contain GR dipeptides. To test whether GR repeats are important for CB targeting, we substituted two GR repeats in SmB (107–108 and 111–112) (BAla) that are predicted to be methylated by PRMT5 (Uniprot P14678 and (61)). In SmD3 we substituted amino acids 110–120 containing several GR repeats (GRGRGMGRGN) with a corresponding stretch of alanine residues (D3Ala). The GR substitutions resulted in the same phenotype as the C-tail deletion. The D3Ala mutant was immunoprecipitated together with snRNA, albeit to a lesser extend than WT SmD3, while its CB localization was strongly reduced (Figure 6 and Supplementary Figure S3C). Similarly, the BAla mutant was precipitated with the same amount of snRNA as the wild-type SmB (Supplementary Figure S4C) but accumulated less efficiently in CBs (Figure 6). The third Sm protein in the Sm ring containing GR repeats is SmD1. Therefore, we deleted the GR domain from SmD1 (aa 97–119) as well, which led to a reduced interaction with snRNAs (Supplementary Figure S4D) and less CB localization (Figure 6). Next, we removed a half (aa 107–119) (D1Δ1/2GR) or a quarter (aa 113 -119) (D1Δ1/4GR) of the GR domain. Partial deletion of the GR repeats did not block association with snRNAs (Supplementary Figure S4D) and only a partial reduction in CB localization was observed with SmD1Δ1/2GR (Figure 6). Finally, we mutated the last C-terminal 23 amino acids (aa 97–119) of SmD1, which mostly consists of GR repeats, into alanines (D1Ala). Substitution of GR with AA did not inhibit the association of the SmD1Ala mutant with snRNAs (Supplementary Figure S4D), but its CB targeting was significantly reduced (Figure 6). These data demonstrate that GR repeats in the C-terminal domains of SmB, D1 and D3 are important for CB targeting of Sm proteins and likely the whole snRNP.
Figure 6.
C-terminal tails of SmB, D1 and D3 are important for Cajal body localization. Cells were transfected with plasmids expressing SmB, D1 or D3 protein variants tagged with GFP. Coilin was used as a marker of CBs (red). DNA was stained with DAPI. The scale bar represents 10 μm. Intensity of GFP signal in CBs vs. the nucleoplasm was determined by high-content microscopy. Values normalized to the WT proteins are shown in the table next to the micrographs and non-normalized values are shown in graphs. The average of three experiments and SEM are shown. Statistical significance was assayed by the two tailed t-test and data with a P value < 0.1 are marked by * and P < 0.001 by ***.
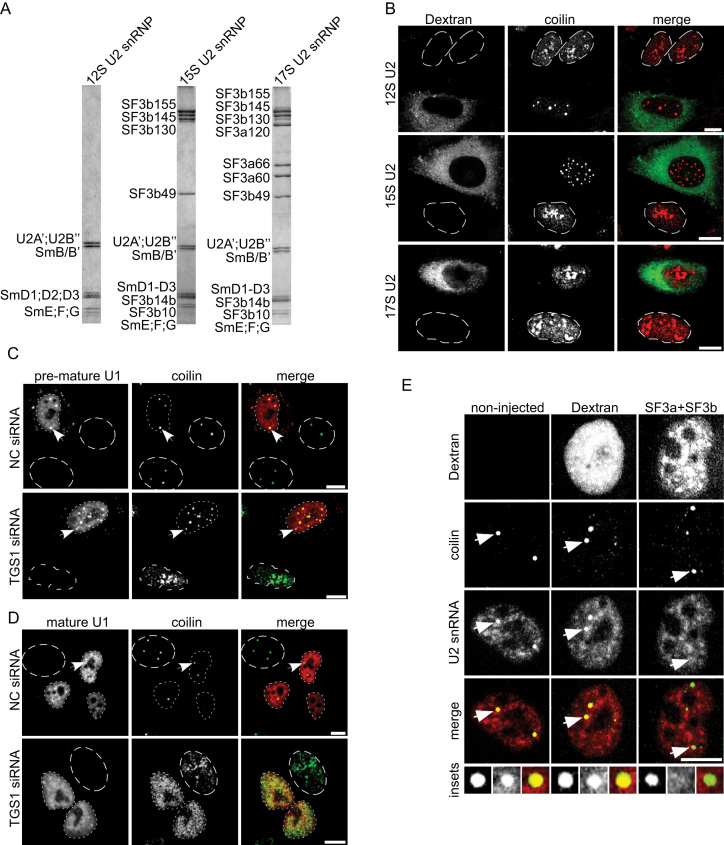
Partially-assembled but not mature U1 and U2 snRNP are targeted to CBs
Our data show that the Sm ring is essential for targeting snRNPs to CBs and that core snRNPs are able to induce CB formation even in cells where the biogenesis of snRNP was inhibited. To test whether mature snRNPs are able to restore CB formation, we induced the dissociation of CBs by siRNA-mediated knockdown of the TGS1 protein. TGS1 is the methyltranferase that hypermethylates the 5′ cap of snRNAs and its depletion results in the disintegration of canonical CBs and redistribution of coilin into multiple small foci scattered throughout the nucleoplasm and also coilin accumulation in nucleoli (59,62) (see also Figure 7). To obtain fully- and partially-assembled U2 snRNPs, we purified endogenous 12S U2 snRNP and reconstituted in vitro partially-assembled 15S and mature 17S U2 snRNP particles containing additionally either the SF3b complex (15S) or SF3a and SF3b complexes (17S). Analysis of their protein composition by SDS PAGE after gradient centrifugation confirmed that the reconstituted particles corresponded to 15S and 17S U2 snRNPs respectively (Figure 7A). Next, the 12S, 15S and 17S U2 snRNPs were microinjected into TGS1-depleted cells along with FITC-labelled Dextran. Similar to core U2 snRNPs (Figure 5C), microinjection of the pre-mature 12S and 15S U2 snRNPs resulted in the reformation of coilin positive nuclear foci (Figure 7B). Characterization of these coilin positive nuclear foci revealed the presence of canonical CB residents like the snoRNP-specific proteins fibrillarin and NOP58, and the U4/U6 recycling factor SART3 (Supplementary Figure S5) indicating that these structures correspond to bona fide CBs. In sharp contrast, we did not observe any coilin positive foci upon 17S U2 snRNP microinjection, and like its distribution in the non-microinjected cells, coilin remained dispersed in the nucleoplasm and enriched in the nucleolus. Similar results were obtained with U1 snRNPs; microinjection of an in vitro-reconstituted core U1 snRNP (U1 snRNA+Sm proteins) resulted in the formation of CBs in TGS1 knockdown cells (Figure 7C), while microinjection of an endogenous, purified mature U1 snRNP did not (Figure 7D). Taken together these data show that only partially-assembled snRNPs can induce CB formation and localize to CBs.
Figure 7.
Partially-assembled snRNP particles induce formation of CBs. (A) Purified 12S U2 snRNP and in vitro reconstituted 15S and 17S U2 snRNPs were analyzed by SDS-PAGE and proteins were visualized by Coomassie staining. (B) TGS1 was knocked down by siRNA and cells were microinjected into the cytoplasm with native 12S U2 snRNP (top panel), in vitro reconstituted 15S U2 snRNP (middle panel) or mature 17S U2 snRNP (bottom panel). FITC-Dextran served as microinjection marker (green) and coilin was visualized by immunostaining (red). (C, D) TGS1 was knocked down by siRNA and cells were microinjected into the cytoplasm with digoxygenin-labeled in vitro-reconstituted U1 snRNP (C) or a native Cyan3-labelled mature U1 snRNP (D) and examined by immunofluorescence 2 h post microinjection using the anti-coilin antibody (C, D) and anti-digoxygenin antibodies (C). Dotted lines mark the nucleus of microinjected cells; dashed lines mark the nucleus of non-microinjected cells. (E) HeLa cells were microinjected in the nucleus with FITC-Dextran (middle panel) or FITC-Dextran together with an equimolar mix of purified SF3a + SF3b complexes (right panel). Left panel shows control non-injected cell. The U2 snRNA was stained by FISH (red) and coilin by immunostaining (green). Insets show a magnified picture of CBs indicated by arrows. Scale bars: 10 μm.
U2 snRNP is released from CBs upon association with its specific proteins
Our data strongly suggest that only pre-mature U2 snRNP, not assembled with both SF3a and b, can accumulate in CBs. We thus assayed whether U2 snRNPs, detected in CBs under physiological conditions, can be chased out of CBs by an excess of SF3a/b complexes. We microinjected ∼1 × 106 copies of an equimolar mixture of SF3a+SF3b complexes into HeLa cell nuclei and assayed for the presence of the U2 snRNA in CBs by fluorescence in-situ hybridization (Figure 7E). In non-microinjected cells, as well as in cells microinjected with Dextran alone, the overall U2 snRNA distribution was not affected and was readily detected in CBs. Strikingly, microinjection of SF3a + SF3b resulted in a complete loss of the U2 snRNA signal in the CBs in over 90% of the cells examined (n = 42). These findings suggest that the excess of injected SF3a+b complexes dissociates endogenous U2 snRNPs from CBs likely by promoting U2 snRNP maturation. These findings are consistent with the idea that U2 snRNPs that accumulate in CBs are pre-mature particles lacking SF3a and/or SF3b proteins and that addition of SF3a/b complexes releases them from CBs.
DISCUSSION
SnRNP biogenesis starts in the cell nucleus by snRNA transcription, continues in the cytoplasm where the snRNA acquires a ring of seven Sm proteins, and then this core snRNP returns to the nucleus where the assembly of the snRNP is finalized by addition of snRNP-specific proteins, which is a prerequisite for its subsequent participation in splicing. Following nuclear import, core snRNPs first appear in CBs where the final steps of their maturation occur (reviewed in (29)). While the Sm ring has been established as an essential nuclear import signal (63,64), it has not been clear what targets core snRNPs to CBs. In addition, inhibition of the final snRNP maturation step leads to sequestration of partially-assembled particles in CBs (33). However, the molecular mechanism that discriminates between partially- and fully-assembled snRNPs is unknown. Here, we provide several lines of evidence that the Sm ring is an essential factor important for CB targeting and retention. These include our observations that (i) Sm and SMN sites in snRNAs, which are essential for Sm ring assembly, are both necessary and sufficient for CB targeting (Figures 1–4), (ii) depletion of several Sm proteins prevents accumulation of snRNAs in CBs (Figure 5 and Supplementary Figure S2) and (iii) core snRNPs (snRNA + Sm proteins) but not naked snRNAs are targeted to CBs in the absence of the SMN protein (Figure 5).
These data are in agreement with microinjection experiments showing that minimal U1 snRNPs partially accumulate in CBs (41) and a recent finding in zebrafish that U7 snRNP with a canonical Sm ring is localized to CBs (65). We further demonstrate that GR repeats in C-terminal domains of the SmB, D1 and D3 proteins are important for the accumulation of these proteins in CBs. Interestingly, despite the integration of the mutated Sm proteins into an otherwise normal Sm ring that contains other GR repeat-containing Sm proteins, their localization in CBs is reduced. A similar observation was described recently, where replacement of LSm10 and LSm11 with SmD1 and SmD2 in the U7-specific Sm ring retargeted the U7 snRNP to CBs (65). WT U7 snRNP contains two out of three of the GR repeat-containing Sm proteins (SmD3 and SmB), but under physiological conditions U7 snRNP is not localized in CBs. This and our data suggest that the GR domains of different Sm proteins together form the CB localization signal.
The minimal sequence containing Sm and SMN binding sites has been previously shown to bind the SMN complex (54,58). The SMN complex was proposed to facilitate nuclear import of newly assembled core snRNPs and target them via interaction with coilin to CBs (17,66–68). This suggests that SMN could be directly involved in CB localization of core snRNPs. Here we show that the depletion of Sm proteins blocks CB localization of WT U2 snRNA, which contains SMN binding sites, suggesting that the SMN complex alone is not sufficient to target snRNAs to CBs (Figure 5). Moreover, core snRNPs containing an Sm ring efficiently accumulate in CBs even when the SMN protein is depleted (Figure 5). These results show that the SMN complex is not directly involved in CB localization and that the Sm ring is sufficient to target snRNAs to CBs.
Previous studies have shown a direct interaction between coilin and Sm proteins (68,69). Coilin interacts with Sm proteins via the Sm fold, however the interaction is substantially enhanced by the C-terminal tails of Sm proteins (68). The C-terminus of coilin contains a Tudor domain (70), which in other proteins interacts with methylated arginines (71). It is tempting to speculate that the coilin Tudor domain binds dimethylated arginines in GR repeats found in the C-termini of SmB/B′, D1 and D3. However, it should be noted that the isolated coilin Tudor domain did not exhibit any dimethylated arginine binding activity in vitro (70). Thus, it is likely that other factors modulate association of core snRNPs with CBs. In this respect, coilin has been shown to directly bind snRNA (72,73), which might provide an additional signal for CB localization of snRNAs. However, the molecular details remain to be established.
It has been previously shown that immature and partially-assembled snRNPs accumulate in CBs (20,21,31,33,51). Based on our results that the Sm ring serves as an essential CB targeting signal, we propose a model where snRNPs bearing an exposed Sm ring are retained in CBs. This model is supported by the following findings: (i) immature U1 and U2 snRNPs induce formation of CBs in cells lacking these nuclear structures while mature particles fail to generate CBs (Figure 7); (ii) microinjection of an excess of SF3a/SF3b factors diminishes the accumulation of U2 snRNAs in CBs (Figure 7E) and (iii) the CB accumulation of U2ΔSLI RNA, which does not interact with the SF3a complex and mimics pre-mature snRNPs, requires the presence of an Sm site (Figure 2). We hypothesize that interactions of CB factors with the unprotected Sm ring and specifically with the GR repeats of Sm proteins provide the molecular basis for the cellular mechanism controlling the final steps of snRNP assembly. Core snRNPs are sequestered in CBs until specific proteins (e.g. the SF3a trimeric complex in the case of U2) are bound or the composite U4/U6•U5 tri-snRNP is formed. The binding of snRNP-specific proteins weakens the interaction between Sm proteins and CB factors, allowing the mature snRNP to leave the CB. A high-resolution structure of the 17S U2 snRNP is not currently available, but the SF3a and b complexes are known to interact with the Sm core (24,74) and their association with the U2 snRNP, by masking the Sm core, may result in the release of the mature U2 snRNP from CBs. In addition, the high resolution structure of the tri-snRNP shows that the U4 and U5 Sm rings are positioned on the periphery of the tri-snRNP, with the side of the Sm ring that exposes the C-terminal tails in close proximity to snRNP specific proteins (75,76). In line with this, the N-terminal part of the U1 specific protein U1–70K has been shown to wrap around the Sm core complex upon binding to the U1 snRNP (77,78). This illustrates well how association of specific proteins may destabilize the association of snRNPs with CBs by shielding the Sm core.
Taken together, our data reveal a new role for Sm proteins in targeting snRNPs to CBs. Of particular importance are the GR repeats present at the C-terminus of several Sm proteins, which appear to function as CB targeting and retention signals. In addition, we provide evidence that partially-assembled snRNPs induce CB formation, while the fully assembled particles do not. Based on these data, we have proposed a model that the interaction of CB factors with the exposed Sm ring, enables cells to discriminate between mature and immature snRNPs, and sequester partially-assembled snRNPs in CBs until their assembly is completed.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Karla Neugebauer, Edouard Bertrand, David Drechsel, Angus Lamond, Yaron Shav-Tal, Pavel Draber and Maria Carmo-Fonseca for providing us with reagents and Gabi Heyne for excellent technical assistance. We are grateful to members of the Stanek lab for critical reading.
Author Contribution: A.R. designed and performed most of the experiments (Figures 1–6) and participated in the writing of the manuscript; C.W. isolated 12S, 15S and 17S U2 snRNPs, C.G. prepared and microinjected core, pre-mature and mature snRNP particles (Figures 5 and 7); K.K. prepared minimal U4snRNA-MS2 and performed immunoprecipitation with U4-MS2, U2-MS2 constructs and Sm mutants and participated in the writing of the manuscript; J.P. carried out the snRNA structure prediction and D.S., R.L. and C.G. designed and evaluated experiments and wrote the manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Czech Academy of Sciences [RVO68378050]; Czech Science Foundation [15-00790S]; Czech Ministry of Education, Youth and Sports [LH14033]; Sonderforschungsbereich SFB 860; Light Microscopy Core Facility, IMG CAS, Prague, Czech Republic [Czech-Bioimaging - LM2015062]; ‘Centre of Model Organisms’ OPPK [CZ.2.16/3.1.00/21547]; ‘Biomodels for health’ [LO1419]. Funding for open access charge: Czech Science Foundation [15-00790S].
Conflict of interest statement. None declared.
REFERENCES
- 1. Pellizzoni L., Charroux B., Dreyfuss G.. SMN mutants of spinal muscular atrophy patients are defective in binding to snRNP proteins. Proc. Natl. Acad. Sci. U.S.A. 1999; 96:11167–11172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Eggert C., Chari A., Laggerbauer B., Fischer U.. Spinal muscular atrophy: the RNP connection. Trends Mol. Med. 2006; 12:113–121. [DOI] [PubMed] [Google Scholar]
- 3. Ruzickova S., Stanek D.. Mutations in spliceosomal proteins and retina degeneration. RNA Biol. 2017; 14:544–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Matera A.G., Shpargel K.B.. Pumping RNA: nuclear bodybuilding along the RNP pipeline. Curr. Opin. Cell Biol. 2006; 18:317–324. [DOI] [PubMed] [Google Scholar]
- 5. Gruss O.J., Meduri R., Schilling M., Fischer U.. UsnRNP biogenesis: mechanisms and regulation. Chromosoma. 2017; 126:577–593. [DOI] [PubMed] [Google Scholar]
- 6. Massenet S., Pellizzoni L., Paushkin S., Mattaj I.W., Dreyfuss G.. The SMN complex is associated with snRNPs throughout their cytoplasmic assembly pathway. Mol. Cell. Biol. 2002; 22:6533–6541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Jin W., Wang Y., Liu C.P., Yang N., Jin M., Cong Y., Wang M., Xu R.M.. Structural basis for snRNA recognition by the double-WD40 repeat domain of Gemin5. Genes Dev. 2016; 30:2391–2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Xu C., Ishikawa H., Izumikawa K., Li L., He H., Nobe Y., Yamauchi Y., Shahjee H.M., Wu X.H., Yu Y.T. et al. Structural insights into Gemin5-guided selection of pre-snRNAs for snRNP assembly. Genes Dev. 2016; 30:2376–2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Golembe T.J., Yong J., Dreyfuss G.. Specific sequence features, recognized by the SMN complex, identify snRNAs and determine their fate as snRNPs. Mol. Cell. Biol. 2005; 25:10989–11004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Yong J., Kasim M., Bachorik J.L., Wan L., Dreyfuss G.. Gemin5 delivers snRNA precursors to the SMN complex for snRNP biogenesis. Mol. Cell. 2010; 38:551–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chari A., Golas M.M., Klingenhager M., Neuenkirchen N., Sander B., Englbrecht C., Sickmann A., Stark H., Fischer U.. An assembly chaperone collaborates with the SMN complex to generate spliceosomal SnRNPs. Cell. 2008; 135:497–509. [DOI] [PubMed] [Google Scholar]
- 12. Grimm C., Chari A., Pelz J.P., Kuper J., Kisker C., Diederichs K., Stark H., Schindelin H., Fischer U.. Structural basis of assembly chaperone-mediated snRNP formation. Mol. Cell. 2013; 49:692–703. [DOI] [PubMed] [Google Scholar]
- 13. Fischer U., Englbrecht C., Chari A.. Biogenesis of spliceosomal small nuclear ribonucleoproteins. Wiley Interdiscipl. Rev. RNA. 2011; 2:718–731. [DOI] [PubMed] [Google Scholar]
- 14. Mattaj I.W. Cap trimethylation of U snRNA is cytoplasmic and dependent on U snRNP protein binding. Cell. 1986; 46:905–911. [DOI] [PubMed] [Google Scholar]
- 15. Matera A.G., Wang Z.. A day in the life of the spliceosome. Nat. Rev. Mol. Cell Biol. 2014; 15:108–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hamm J., Darzynkiewicz E., Tahara S.M., Mattaj I.W.. The trimethylguanosine cap structure of U1 snRNA is a component of a bipartite nuclear targeting signal. Cell. 1990; 62:569–577. [DOI] [PubMed] [Google Scholar]
- 17. Narayanan U., Achsel T., Lührmann R., Matera A.G.. Coupled in vitro import of U snRNPs and SMN, the spinal muscular atrophy protein. Mol. Cell. 2004; 16:223–234. [DOI] [PubMed] [Google Scholar]
- 18. Raimer A.C., Gray K.M., Matera A.G.. SMN - A chaperone for nuclear RNP social occasions?. RNA Biol. 2016; 14:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sleeman J.E., Lamond A.I.. Newly assembled snRNPs associate with coiled bodies before speckles, suggesting a nuclear snRNP maturation pathway. Curr. Biol. 1999; 9:1065–1074. [DOI] [PubMed] [Google Scholar]
- 20. Ospina J.K., Gonsalvez G.B., Bednenko J., Darzynkiewicz E., Gerace L., Matera A.G.. Cross-talk between snurportin1 subdomains. Mol. Biol. Cell. 2005; 16:4660–4671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Schaffert N., Hossbach M., Heintzmann R., Achsel T., Luhrmann R.. RNAi knockdown of hPrp31 leads to an accumulation of U4/U6 di-snRNPs in Cajal bodies. EMBO J. 2004; 23:3000–3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Stanek D., Neugebauer K.M.. Detection of snRNP assembly intermediates in Cajal bodies by fluorescence resonance energy transfer. J. Cell Biol. 2004; 166:1015–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Stanek D., Rader S.D., Klingauf M., Neugebauer K.M.. Targeting of U4/U6 small nuclear RNP assembly factor SART3/p110 to Cajal bodies. J. Cell Biol. 2003; 160:505–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Nesic D., Kramer A.. Domains in human splicing factors SF3a60 and SF3a66 required for binding to SF3a120, assembly of the 17S U2 snRNP, and prespliceosome formation. Mol. Cell. Biol. 2001; 21:6406–6417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Novotny I., Blazikova M., Stanek D., Herman P., Malinsky J.. In vivo kinetics of U4/U6.U5 tri-snRNP formation in Cajal bodies. Mol. Biol. Cell. 2011; 22:513–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Will C.L., Urlaub H., Achsel T., Gentzel M., Wilm M., Luhrmann R.. Characterization of novel SF3b and 17S U2 snRNP proteins, including a human Prp5p homologue and an SF3b DEAD-box protein. EMBO J. 2002; 21:4978–4988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Darzacq X., Jady B.E., Verheggen C., Kiss A.M., Bertrand E., Kiss T.. Cajal body-speci c small nuclear RNAs: a novel class of 2 0 -O-methylation and pseudouridylation guide RNAs. EMBO J. 2002; 21:2746–2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Jady B.E., Darzacq X., Tucker K.E., Matera A.G., Bertrand E., Kiss T.. Modification of Sm small nuclear RNAs occurs in the nucleoplasmic Cajal body following import from the cytoplasm. EMBO J. 2003; 22:1878–1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Stanek D. Cajal bodies and snRNPs - friends with benefits. RNA Biol. 2017; 14:671–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Malinova A., Cvackova Z., Mateju D., Horejsi Z., Abeza C., Vandermoere F., Bertrand E., Stanek D., Verheggen C.. Assembly of the U5 snRNP component PRPF8 is controlled by the HSP90/R2TP chaperones. J. Cell Biol. 2017; 216:1579–1596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bizarro J., Dodre M., Huttin A., Charpentier B., Schlotter F., Branlant C., Verheggen C., Massenet S., Bertrand E.. NUFIP and the HSP90/R2TP chaperone bind the SMN complex and facilitate assembly of U4-specific proteins. Nucleic Acids Res. 2015; 43:8973–8989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Cloutier P., Poitras C., Durand M., Hekmat O., Fiola-Masson E., Bouchard A., Faubert D., Chabot B., Coulombe B.. R2TP/Prefoldin-like component RUVBL1/RUVBL2 directly interacts with ZNHIT2 to regulate assembly of U5 small nuclear ribonucleoprotein. Nat. Commun. 2017; 8:15615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Novotny I., Malinova A., Stejskalova E., Mateju D., Klimesova K., Roithova A., Sveda M., Knejzlik Z., Stanek D.. SART3-dependent accumulation of incomplete spliceosomal snRNPs in Cajal bodies. Cell Rep. 2015; 10:429–440. [DOI] [PubMed] [Google Scholar]
- 34. Nesic D., Tanackovic G., Kramer A.. A role for Cajal bodies in the final steps of U2 snRNP biogenesis. J. Cell Sci. 2004; 117:4423–4433. [DOI] [PubMed] [Google Scholar]
- 35. Hnilicova J., Jirat Matejckova J., Sikova M., Pospisil J., Halada P., Panek J., Krasny L.. Ms1, a novel sRNA interacting with the RNA polymerase core in mycobacteria. Nucleic Acids Res. 2014; 42:11763–11776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Klingauf M., Stanek D., Neugebauer K.M.. Enhancement of U4/U6 small nuclear ribonucleoprotein particle association in Cajal bodies predicted by mathematical modeling. Mol. Biol. Cell. 2006; 17:4972–4981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lorenz R., Bernhart S.H., Honer Zu Siederdissen C., Tafer H., Flamm C., Stadler P.F., Hofacker I.L.. ViennaRNA Package 2.0. Algorithms Mol. Biol. 2011; 6:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Huranova M., Hnilicova J., Fleischer B., Cvackova Z., Stanek D.. A mutation linked to retinitis pigmentosa in HPRP31 causes protein instability and impairs its interactions with spliceosomal snRNPs. Hum. Mol. Genet. 2009; 18:2014–2023. [DOI] [PubMed] [Google Scholar]
- 39. Sumpter V., Kahrs A., Fischer U., Kornstadt U., Luhrmann R.. Invitro reconstitution of U1 and U2 Snrnps from isolated proteins and Snrna. Mol. Biol. Rep. 1992; 16:229–240. [DOI] [PubMed] [Google Scholar]
- 40. Segault V., Will C.L., Sproat B.S., Luhrmann R.. In vitro reconstitution of mammalian U2 and U5 snRNPs active in splicing: Sm proteins are functionally interchangeable and are essential for the formation of functional U2 and U5 snRNPs. EMBO J. 1995; 14:4010–4021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Malatesta M., Fakan S., Fischer U.. The Sm core domain mediates targeting of U1 snRNP to subnuclear compartments involved in transcription and splicing. Exp. Cell Res. 1999; 249:189–198. [DOI] [PubMed] [Google Scholar]
- 42. Dignam J.D., Lebovitz R.M., Roeder R.G.. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983; 11:1475–1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Brosi R., Groning K., Behrens S.E., Luhrmann R., Kramer A.. Interaction of mammalian splicing factor SF3a with U2 snRNP and relation of its 60-kD subunit to yeast PRP9. Science. 1993; 262:102–105. [DOI] [PubMed] [Google Scholar]
- 44. Boelens W., Scherly D., Beijer R.P., Jansen E.J., Dathan N.A., Mattaj I.W., van Venrooij W.J.. A weak interaction between the U2A' protein and U2 snRNA helps to stabilize their complex with the U2B" protein. Nucleic Acids Res. 1991; 19:455–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Williams S.G., Hall K.B.. Human U2B″ protein binding to snRNA stemloops. Biophys. Chem. 2011; 159:82–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Yong J., Golembe T.J., Battle D.J., Pellizzoni L., Dreyfuss G.. snRNAs contain specific SMN-binding domains that are essential for snRNP assembly. Mol. Cell. Biol. 2004; 24:2747–2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Battle D.J., Kasim M., Yong J., Lotti F., Lau C.K., Mouaikel J., Zhang Z., Han K., Wan L., Dreyfuss G.. The SMN complex: an assembly machine for RNPs. Cold Spring Harb. Symp. Quant. Biol. 2006; 71:313–320. [DOI] [PubMed] [Google Scholar]
- 48. Carmo-Fonseca M., Tollervey D., Pepperkok R., Barabino S.M., Merdes A., Brunner C., Zamore P.D., Green M.R., Hurt E., Lamond A.I.. Mammalian nuclei contain foci which are highly enriched in components of the pre-mRNA splicing machinery. EMBO J. 1991; 10:195–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Matera A.G., Ward D.C.. Nucleoplasmic organization of small nuclear ribonucleoproteins in cultured human cells. J. Cell Biol. 1993; 121:715–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Stejskalova E., Stanek D.. The splicing factor U1-70K interacts with the SMN complex and is required for nuclear gem integrity. J. Cell Sci. 2014; 127:3909–3915. [DOI] [PubMed] [Google Scholar]
- 51. Tanackovic G., Kramer A.. Human Splicing Factor SF3a, but Not SF1, Is Essential for Pre-mRNA Splicing In Vivo. Mol. Biol. Cell. 2005; 16:1366–1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. So B.R., Wan L., Zhang Z., Li P., Babiash E., Duan J., Younis I., Dreyfuss G.. A U1 snRNP-specific assembly pathway reveals the SMN complex as a versatile hub for RNP exchange. Nat. Struct. Mol. Biol. 2016; 23:225–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Peterlin B.M., Brogie J.E., Price D.H.. 7SK snRNA: a noncoding RNA that plays a major role in regulating eukaryotic transcription. Wiley Interdiscipl. Rev. RNA. 2012; 3:92–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Golembe T.J., Yong J., Battle D.J., Feng W., Wan L., Dreyfuss G.. Lymphotropic Herpesvirus saimiri uses the SMN complex to assemble Sm cores on its small RNAs. Mol. Cell. Biol. 2005; 25:602–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Maraia R.J., Driscoll C.T., Bilyeu T., Hsu K., Darlington G.J.. Multiple dispersed loci produce small cytoplasmic Alu RNA. Mol. Cell. Biol. 1993; 13:4233–4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Ivanova E., Berger A., Scherrer A., Alkalaeva E., Strub K.. Alu RNA regulates the cellular pool of active ribosomes by targeted delivery of SRP9/14 to 40S subunits. Nucleic Acids Res. 2015; 43:2874–2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Hallais M., Pontvianne F., Andersen P.R., Clerici M., Lener D., Benbahouche Nel H., Gostan T., Vandermoere F., Robert M.C., Cusack S. et al. CBC-ARS2 stimulates 3′-end maturation of multiple RNA families and favors cap-proximal processing. Nat. Struct. Mol. Biol. 2013; 20:1358–1366. [DOI] [PubMed] [Google Scholar]
- 58. Yong J., Wan L., Dreyfuss G.. Why do cells need an assembly machine for RNA-protein complexes?. Trends Cell Biol. 2004; 14:226–232. [DOI] [PubMed] [Google Scholar]
- 59. Lemm I., Girard C., Kuhn A.N., Watkins N.J., Schneider M., Bordonne R., Luhrmann R.. Ongoing U snRNP biogenesis is required for the integrity of Cajal bodies. Mol. Biol. Cell. 2006; 17:3221–3231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Shpargel K.B., Matera A.G.. Gemin proteins are required for efficient assembly of Sm-class ribonucleoproteins. Proc. Natl. Acad. Sci. U.S.A. 2005; 102:17372–17377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Friesen W.J., Paushkin S., Wyce A., Massenet S., Pesiridis G.S., Van Duyne G., Rappsilber J., Mann M., Dreyfuss G.. The methylosome, a 20S complex containing JBP1 and pICln, produces dimethylarginine-modified Sm proteins. Mol. Cell. Biol. 2001; 21:8289–8300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Mouaikel J., Verheggen C., Bertrand E., Tazi J., Bordonne R.. Hypermethylation of the cap structure of both yeast snRNAs and snoRNAs requires a conserved methyltransferase that is localized to the nucleolus. Mol. Cell. 2002; 9:891–901. [DOI] [PubMed] [Google Scholar]
- 63. Fischer U., Sumpter V., Sekine M., Satoh T., Luhrmann R.. Nucleo-cytoplasmic transport of U snRNPs: definition of a nuclear location signal in the Sm core domain that binds a transport receptor independently of the m3G cap. EMBO J. 1993; 12:573–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Girard C., Mouaikel J., Neel H., Bertrand E., Bordonne R.. Nuclear localization properties of a conserved protuberance in the Sm core complex. Exp. Cell Res. 2004; 299:199–208. [DOI] [PubMed] [Google Scholar]
- 65. Heyn P., Salmonowicz H., Rodenfels J., Neugebauer K.M.. Activation of transcription enforces the formation of distinct nuclear bodies in zebrafish embryos. RNA Biol. 2017; 14:752–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Narayanan U., Ospina J.K., Frey M.R., Hebert M.D., Matera A.G.. SMN, the Spinal Muscular Atrophy Protein, Forms a Pre-Import Snrnp Complex with Snurportin1 and Importin beta. Hum. Mol. Genet. 2002; 11:1785–1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Hebert M.D., Szymczyk P.W., Shpargel K.B., Matera A.G.. Coilin forms the bridge between Cajal bodies and SMN, the spinal muscular atrophy protein. Genes Dev. 2001; 15:2720–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Xu H., Pillai R.S., Azzouz T.N., Shpargel K.B., Kambach C., Hebert M.D., Schumperli D., Matera A.G.. The C-terminal domain of coilin interacts with Sm proteins and U snRNPs. Chromosoma. 2005; 114:155–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Toyota C.G., Davis M.D., Cosman A.M., Hebert M.D.. Coilin phosphorylation mediates interaction with SMN and SmB′. Chromosoma. 2010; 119:205–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Shanbhag R., Kurabi A., Kwan J.J., Donaldson L.W.. Solution structure of the carboxy-terminal Tudor domain from human Coilin. FEBS Lett. 2010; 584:4351–4356. [DOI] [PubMed] [Google Scholar]
- 71. Pek J.W., Anand A., Kai T.. Tudor domain proteins in development. Development. 2012; 139:2255–2266. [DOI] [PubMed] [Google Scholar]
- 72. Machyna M., Kehr S., Straube K., Kappei D., Buchholz F., Butter F., Ule J., Hertel J., Stadler P., Neugebauer K.M.. Global identification of coilin binding partners reveals hundreds of small non-coding RNAs that traffic through Cajal bodies. Mol. Cell. 2014; 56:389–399. [DOI] [PubMed] [Google Scholar]
- 73. Broome H.J., Hebert M.D.. Coilin displays differential affinity for specific RNAs in vivo and is linked to telomerase RNA biogenesis. J. Mol. Biol. 2013; 425:713–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Kramer A., Gruter P., Groning K., Kastner B.. Combined biochemical and electron microscopic analyses reveal the architecture of the mammalian U2 snRNP. J. Cell Biol. 1999; 145:1355–1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Nguyen T.H.D., Galej W.P., Bai X.-C., Oubridge C., Newman A.J., Scheres S.H.W., Nagai K.. Cryo-EM structure of the yeast U4/U6.U5 tri-snRNP at 3.7 A resolution. Nature. 2016; 530:298–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Agafonov D.E., Kastner B., Dybkov O., Hofele R.V., Liu W.T., Urlaub H., Luhrmann R., Stark H.. Molecular architecture of the human U4/U6.U5 tri-snRNP. Science. 2016; 351:1416–1420. [DOI] [PubMed] [Google Scholar]
- 77. Pomeranz Krummel D.A., Oubridge C., Leung A.K., Li J., Nagai K.. Crystal structure of human spliceosomal U1 snRNP at 5.5 A resolution. Nature. 2009; 458:475–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Weber G., Trowitzsch S., Kastner B., Luhrmann R., Wahl M.C.. Functional organization of the Sm core in the crystal structure of human U1 snRNP. EMBO J. 2010; 29:4172–4184. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.