Abstract
Completion of eukaryal genomes can be difficult task with the highly repetitive sequences along the chromosomes and short read lengths of second-generation sequencing. Saccharomyces cerevisiae strain CEN.PK113-7D, widely used as a model organism and a cell factory, was selected for this study to demonstrate the superior capability of very long sequence reads for de novo genome assembly. We generated long reads using two common third-generation sequencing technologies (Oxford Nanopore Technology (ONT) and Pacific Biosciences (PacBio)) and used short reads obtained using Illumina sequencing for error correction. Assembly of the reads derived from all three technologies resulted in complete sequences for all 16 yeast chromosomes, as well as the mitochondrial chromosome, in one step. Further, we identified three types of DNA methylation (5mC, 4mC and 6mA). Comparison between the reference strain S288C and strain CEN.PK113-7D identified chromosomal rearrangements against a background of similar gene content between the two strains. We identified full-length transcripts through ONT direct RNA sequencing technology. This allows for the identification of transcriptional landscapes, including untranslated regions (UTRs) (5′ UTR and 3′ UTR) as well as differential gene expression quantification. About 91% of the predicted transcripts could be consistently detected across biological replicates grown either on glucose or ethanol. Direct RNA sequencing identified many polyadenylated non-coding RNAs, rRNAs, telomere-RNA, long non-coding RNA and antisense RNA. This work demonstrates a strategy to obtain complete genome sequences and transcriptional landscapes that can be applied to other eukaryal organisms.
INTRODUCTION
The genome of the most well studied eukaryotic model organism, Saccharomyces cerevisiae strain S288c, was sequenced and released in 1996; it was the first complete, high quality genome sequence of an eukaryal organism (1). Since then, the development of DNA sequencing technologies has yielded scientific breakthroughs that enable us to obtain and analyze genomic DNA sequences at a faster, more economical pace (2). As of August 2017, the NCBI genome database lists >500 Saccharomyces genomes and 4600 eukaryal sequenced genomes. However, <1% (35 genomes) of these are classified as ‘complete genomes’, which harbor contiguous chromosomal sequence(s) without gaps (definition by NCBI); this includes 1 animal (Caenorhabditis elegans), 29 fungi and 5 protists. All of these have relatively small genome sizes, most <100 Mb. Thus, >99% of the eukaryal genomes are drafts, and virtually all of the larger genomes are incomplete. There are many limitations of draft genomes, which can lead to misinterpretations (e.g. see (3)). Complete genome status should be the objective for a genome-sequencing project, even though draft genomes can provide most of the coding sequences that are sufficient to gain functional insights about an organism. Once a genome is obtained, transcriptional analysis can be performed to improve gene annotation and identify dynamic signatures of gene expression. Traditional RNA-Seq has been widely employed in several studies as a powerful tool for transcriptomics (4).
The small percentage of complete genome sequences in both eukaryotes (as mentioned above) and prokaryotes (5) strongly indicates the difficulties and high cost of properly locating and ordering DNA segments obtained from assemblers as well as resolving ambiguities or discrepancies among reads for a completely assembled genome sequence (3). One major limitation lies in reads generated from DNA sequencing technologies. First-generation sequencing technology (6) can generate moderately long and accurate reads, but is a slow and expensive method for obtaining the high sequencing depth required for genome assemblers. Second-generation sequencing technologies (7) (such as Illumina, 454 and Ion-torrent) can generate massive amounts of reads with high accuracy, although the reads are too short to allow for the de novo assembly of complete genomes (8), resulting in pieces of DNA rather than chromosomal-sized contiguous sequences. Nevertheless, DNA spanning technologies from BioNanoGenomics, 10X Genomics, and Dovetail cHiCago sequencing company can produce long pieces of DNA sequence (with a mean span length of 30–250 kb depending on the technology) from short reads. Third-generation sequencing technologies can generate very long reads at the single-molecule level, though the error rate is high. Pacific Biosciences (PacBio) has developed Single Molecule Real Time (SMART) technology that offers two sequencing strategies—continuous long read (CLR) and circular consensus long read (CCS). The error rate of raw reads derived from CLR approach is around 13% (7,9). The high level of error can be reduced to ≤1% (7) with the CCS approach (multiple passing), which involves sequencing shorter DNA pieces, typically lower than 25 kb, several times. Oxford Nanopore Technologies (ONT) has developed a portable sequencing device called MinION that is able to perform single-molecule DNA sequencing (10) and, recently, cDNA sequencing (11). The DNA sequencing by ONT also offers two chemistries—1D and 1D2 for the latest version of flow cell R9.4/R9.5. The raw reads generated by 1D chemistry have a sequencing error rate similar to PacBio CLR, with possible read lengths of more than 300 kb (10). By using 1D2 chemistry, the mean error rate can be improved to <4%, although the throughput will be reduced by half when compared to the 1D chemistry. In addition, both PacBio and ONT can directly detect DNA methylation (12–14), providing additional valuable information for epigenetics.
The S. cerevisiae strain CEN.PK113-7D, the offspring of parental strains ENY.WA-1A and MC996A, is used extensively in academic and industrial research, especially in metabolic engineering and systems biology, due to a combination of ease of genetic manipulation and a fast growth rate (15). Based on systems biology analysis by Canelas et al. (16), the phenotypic differences between CEN.PK113-7D and S288C are mainly observed in protein metabolism and ergosterol biosynthesis. Having a high quality complete genome for this strain is important for a detailed mechanistic understanding at the systems biology level. Otero et al. (17) first performed whole-genome sequencing of the CEN.PK113-7D strain using short reads (35 bp) with 18X coverage to identify single nucleotide variations (SNVs) compared to the S288c strain. Some of these SNVs were related to metabolic differences between the two strains. Later, Nijkamp et al. (18) performed a de novo assembly of the CEN.PK113-7D strain genome, with sequences from a GS FLX+ system, 454 Life Sciences (average read length of 350 bp) and Illumina short reads (2 × 50 bp). The result was a draft genome sequence, with 565 contiguous DNA sequences (contigs) instead of the contiguous 16 chromosomes. Recently, third-generation sequencing was used to sequence the genomes of S. cerevisiae strain S288C and other isolates (19–21). The promising results from the de novo assembly of ONT+Illumina and PacBio+Illumina gave a high degree of sequence scaffold continuity—>99% accuracy when compared to the reference genome sequence of S288C. However, the sequence of all 16 chromosomes was still not complete (19).
In this study, we performed whole-genome sequencing of the CEN.PK113-7D strain with a combination of three sequencing technologies: PacBio, ONT and Illumina (22) to obtain a complete genome sequence by de novo assembly. We also performed a genome comparison between the S288C and CEN.PK113-7D strains. Further, we identified the transcriptional landscapes of the CEN.PK113-7D strain under diauxic growth conditions, using ONT direct RNA sequencing technology.
MATERIALS AND METHODS
Genomic DNA extraction and cell cultivation
Saccharomyces cerevisiae CEN.PK113-7D (MATa MAL2-8c SUC2, obtained from Dr Peter Kötter, Frankfurt, Germany) was cultivated overnight in 15 ml of yeast extract peptone dextrose (YPD) medium (10 g/l yeast extract, 20 g/l of peptone and 20 g/l of glucose). We used the Blood & Cell Culture DNA Mini Kit (Qiagen, Hilden, Germany) to extract genomic DNA from 3 ml of the overnight yeast culture (∼5 × 108 cells). The protocol recommended by the manufacturer was modified for yeast cells in the following steps: (i) the lyticase digestion was extended to 1 h, (2) the spheroplasts were centrifuged at 2000 × g for 5 min, (iii) proteinase K digestion was performed at 60°C for 2.5 h, (iv) RNase A was added after the proteinase K digestion, (v) RNase A incubation was performed overnight at 37°C and the final elution volume was reduced to 1 ml of buffer QF and (vi) after the precipitation with isopropanol, the DNA was spooled by inverting the tube, recovered with a pipette tip, washed in 1 ml of cold ethanol 70%, dried at room temperature for 10–20 min and dissolved in 0.1× TE buffer (1 mM Tris, 0.1 mM EDTA, pH 8.0).
RNA extraction and cell cultivation
We extracted RNA from S. cerevisiae CEN.PK113-7D cultivated in 50 ml of defined media as previously described (23) with 20 g/l of glucose, 7.5 g/l of (NH4)2SO4, 14.4 g/l of KH2PO4 and the pH adjusted to 6.5 with NaOH. We sampled the culture on two different time points: mid-exponential growth on glucose (∼4.3 × 107 cells/ml) and oxidative growth on the ethanol/fermentation products (∼2.6 × 108 cells/ml). At each sampling point, we quickly transferred 15 ml of the sample into a 50 ml conical tube half-filled with ice pellets, centrifuged it at 2000 × g for 5 min, snap froze it on liquid nitrogen and stored it at -80°C. We used the RNeasy Mini Kit (Qiagen, Germany) to extract RNA from 3 to 4 × 108 frozen cells with the protocol recommended by the manufacturer. The cells were disrupted in 2-ml tubes filled with 500 mg of acid-washed glass beads (425–600 μm particle size of Lysing Matrix C, MP Biomedicals) using a FastPrep-24 Instrument (MP Biomedicals, California, USA) at 6.0 m/s for 40 s. We used the total RNA obtained from the three biological replicates for direct RNA sequencing following manufacturer recommendation on the starting amount of poly-A RNA of 500 ng.
Library preparation and genomic DNA sequencing by PacBio
We produced one PacBio library using the SMRTbell™ Template Prep Kit version 1.0 according to manufacturer's instructions. In brief, we sheared 10 μg of genomic DNA per library into 20 kb fragments using the Megaruptor system, followed by an exo VII treatment, DNA damage repair, and end-repair before ligation of hairpin adaptors to generate a SMRTbell™ library for circular consensus sequencing. We then subjected the library to exo treatment and PB AMPure bead wash procedures for clean-up before it was size-selected with the BluePippin system with a cut-off value of 9000 bp. We used one SMRTcell™ to sequence the DNA library on the PacBio Sequel instrument using the Sequel 2.0 polymerase and 600 min of movie time. The high quality PacBio reads are deposited in an SRA database under BioProject:PRJNA398797, SRP116559.
Library preparation and genomic DNA sequencing by ONT
We performed genomic sequencing using the Rapid Sequencing Kit for genomic DNA SQK-RAD002 (ONT, USA), The protocol of the library preparation is provided in Supplementary link. The DNA library was eluted and loaded onto a flow cell for sequencing. We accomplished the flow cell loading in three steps: (i) draw back a small volume to remove any bubbles, (ii) prime the flow cell and (iii) add 75 μl of sample to the flow cell via the sample port in a dropwise fashion. We sequenced the genomic DNA on a single R9.5/FLO-MIN107 flow cell on a MinION Mk1B for 48 h. We further base-called the signal files (.fast5) using Albacore version 1.2.6 (ONT, USA). The high quality DNA reads from ONT are deposited in an SRA database under accession number BioProject: PRJNA398797, SRP116559.
Library preparation and direct RNA sequencing by ONT
We performed direct RNA sequencing using the Direct RNA Sequencing protocol for the MinION with the SQK-RNA001 kit (ONT, USA), which recommends 500 ng of poly-A RNA for input. We purified poly-A RNA from total RNA of either glucose condition or ethanol conditions. In all, we used about 222–550 ng of poly-A RNA purified from glucose and ethanol conditions as the input for library preparation, in adherence to the kit protocol. The protocol of the library preparation is provided in Supplementary link. We then loaded the library onto a flow cell (the same way of the DNA sequencing described previously) and sequenced the polyadenylated RNA on a single R9.5/FLO-MIN107 flow cell on a MinION Mk1B for 48 h. For base calling, we used the local-based software Albacore version 2.1.0. The high quality RNA reads from ONT are deposited in an SRA database under accession number BioProject:PRJNA398797, SRP116559.
Bioinformatics and statistical analysis
De novo assembly and polishing
We first filtered the raw reads from both ONT sequencings using a mean quality score cutoff of 9 in the Albacore software (version 1.2.6) to obtain ONT high quality ONT reads. We obtained high quality PacBio reads from SMART link software using the default setting. Only reads longer than 500 bases were kept as high-quality reads and used for further analyses. The high quality reads from both ONT and PacBio were identified in their overlap using GraphMap software version 0.52 (24).
For de novo assembly of the reads, we used Canu software version 1.5 (25) (at default parameters) with three strategies: (i) use ONT reads alone, (ii) use PacBio reads alone and (iii) use both ONT and PacBio reads. We will call the contigs obtained from the genome assemblies ONT_assembly, PacBio_assembly and OP_assembly, respectively. We used Pilon software version 1.22 (26) to further polish the assembled contigs with Illumina reads of our previous published data (22) to obtain a high quality genome.
Genome comparison, computational annotation, and methylation analysis
With the complete S288C genome and annotation information (version R64) from the Saccharomyces Genome Database (SGD), we used MUMMER software version 3 for global genome comparisons of the assemblies with the S. cerevisiae strain S288C genome (27). We selected the best assembly contigs result (OP_assembly) to perform genome annotations. We annotated the open reading frames (ORFs) of coding sequences (CDSs) and RNA non-coding sequences on the CEN.PK113-7D genome by the similarity search using the ORF sequences of the S288C query against the CEN.PK113-7D genome sequence using Blat software version 36 (28). In addition, we employed ab initio CDS calling using AUGUSTUS software version 2.5.5 (29) to identify possible new CDSs in the CEN.PK113-7D genome that were probably not present in S288C. For the local genome comparisons, we used LAST software version 1.04.00 (30) to identify synteny, inversion, and translocation events between S288C and CEN.PK113-7D chromosomes. Further, we called the possible DNA methylations at the signal level of DNA sequencing using Nanopolish (default parameters) to identify 5mC methylation (14) for ONT reads. For PacBio reads, we used blasr (31) and employed kinetic tools from SMRT link software version 4.0 to identify 4mC and 6mA methylations, using a cut-off of P-value of 0.001 and >30 reads coverage. We took the results derived from the genome's features and comparisons and summarized and plotted them for global visualization using Circos software version 0.69–4 (32).
Transcriptional landscape analysis
We first filtered the raw reads obtained from direct RNA sequencing with Albacore (version 1.2.6) using a quality score cutoff of 8 to obtain high quality reads. Then we employed GraphMap software version 0.5.2 (24) to align the high quality reads on the CEN.PK113-7D complete genome to identify transcriptional landscapes. We used two strategies—direct chromosome alignment and transcript model guided alignment—to map the direct RNA sequence reads. We quantified the gene expression levels based on the transcript model guided alignments by counting the number of mapped reads with respect to the transcript location using bedtools software version 2.26 (33). We performed the differential gene expression analysis in ethanol versus glucose conditions with the negative binomial statistic approach on the DESeq2 package (34). The P-value of each individual transcript was corrected for multiple testing using the Benjamini–Hochburg method to generate adjusted P-values. We used the PIANO package (36) to perform the gene set analysis of Gene Ontology (GO), which is the control vocabularies describe gene function, and relationships that are organized in a hierarchical structure (35). We selected the GO terms that have adjusted enrichment P-value less than 10e-6 and plotted a heatmap. In addition, we re-analyzed the Illumina RNA-Seq data from our previous study (22) by only mapping the reads on the CEN.PK113-7D complete genome using Stamy aligner version 1.0.31 with the default parameters (37). To compare the dynamic range of direct RNA sequencing (this study) with traditional RNA-Seq (Illumina RNA-Seq data from our previous study (22)), we calculated the mean coverage depth based on the mapped reads for each transcript for both datasets (see detail of calculation in the Supplementary text). We used the distribution of the mean coverage depth of each biological replicate for the dynamic range comparison. To identify UTR regions, we developed a Python script in-house for mapping the 5′ and 3′ ends of gene boundary detected by direct RNA sequencing by searching for a sharp reduction in signals at both ends of mapped reads. The regions between gene boundaries and ORFs can be defined as 5′ and 3′ UTRs at 5′ and 3′ ends of a given transcript, respectively.
The details of all bioinformatics commands used and the Python script are provided in Supplementary text.
RESULTS
Third-generation sequencing long reads
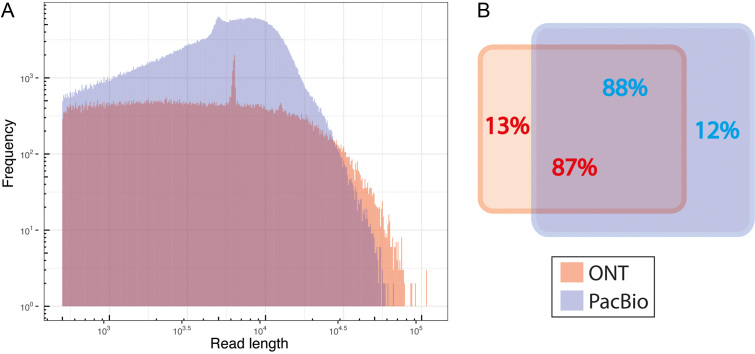
We generated high quality sequencing reads with third-generation sequencing using both ONT and PacBio. We obtained about 130 000 reads from ONT MinION, corresponding to 830 million bases (Mb) of data, with an N50 (the shortest sequence length at 50% of sequenced bases) of 12 500 bases; this corresponds to a 69-fold genome coverage for the yeast genome. Using the CCS chemistry of PacBio, we generated a higher number of reads (∼739 000), with 4 900 Mbp of data with an N50 of 8700 bases. Although the PacBio had shorter average read length, the larger number of reads resulted in a 408-fold coverage, about six times greater than obtained with the ONT sequencing. The details of the third-generation sequencing reads are provided in the supplementary Table TS1. The distribution of the read lengths (Figure 1A) shows that ONT generated longer reads than PacBio. We investigated the overlap of reads between ONT and PacBio and found a high level of overlap (Figure 1B), even though the number of reads generated from PacBio were 5.6 times more than ONT. Surprisingly, about 13% and 12% of the reads were specific (non-overlapping) for ONT and PacBio sequencing, respectively. The non-overlapping reads may reflect differences in sample preparation; the PacBio library preparation has a DNA size selection procedure, whereas ONT does not have any size selection.
Figure 1.
Summary of DNA sequencing reads from ONT and PacBio. (A) Histogram plot showing the distribution of read length of high quality of DNA sequencing reads. ( B) The read overlap plot between ONT and PacBio. The red and blue colors represent the DNA sequencing reads from ONT and PacBio, respectively.
De novo genome assembly
We performed de novo assembly using the Canu software (25), with three strategies: (i) use ONT reads only, (ii) use PacBio reads only and (iii) use both ONT and PacBio reads. For each strategy, we polished the assembly (base correction) using short reads (Illumina) and the Pilon software (26). The resulting de novo assembly for all three methods produced full-length, contiguous DNA sequences for nearly all of the chromosomes and the mitochondria genome, with a length comparable to the S288C chromosomes (see the assemblies statistic in supplementary Table TS2). Notably, in all three cases, the assembled 2-micron plasmid is much longer than the known length of around 6.3 kb, as shown in Table 1. Interestingly, the ONT_assembly (obtained from strategy 1 has the best results, in terms of the correct number of known chromosomes (18 contigs = 16 chromosomes plus mitochondria plus 2-micron plasmid). The PacBio_assembly (obtained from strategy 2) has 19 contigs, caused by a broken mitochondrial chromosome. The OP_assembly (obtained from the strategy 3), has the highest number of contigs (21 contigs), with three additional pieces of telomere DNA and two additional pieces associated with the 2-micron plasmid, which had a similar size when ONT or PacBio reads were used alone. Unexpectedly, a contig derived from OP_assembly joined ChrVII with ChrXIII at their telomeric regions, as shown in Supplementary Figure S1. We investigated the mapped reads from ONT, PacBio, and Illumina on the joined chromosome region and found a clear breakpoint, then separated ChrVII and ChrXIII.
Table 1. The summary of de novo assembly results of S. cerevisiae strain CEN.PK113-7D obtained from the three strategies comparing it to the genome of S. cerevisiae strain S288C.
| Feature | CEN.PK113-7D | S288C | ||
|---|---|---|---|---|
| ONT_assembly (69X) | PacBio_assembly (408X) | OP_assembly(477X) | SGD | |
| chrI | 224 821 | 241 274 | 235 019 | 230 218 |
| chrII | 806 426 | 820 406 | 827 088 | 813 184 |
| chrIII | 319 119 | 369 115 | 367 275 | 316 620 |
| chrIV | 1 504 163 | 1 518 811 | 1 518 534 | 1 531 933 |
| chrV | 577 655 | 593 818 | 579 053 | 576 874 |
| chrVI | 272 158 | 278 189 | 286 399 | 270 161 |
| chrVII | 1 123 142 | 1 138 579 | 113 7891 (**205 1454) | 1 090 940 |
| chrVIII | 560 935 | 577 405 | 577 241 | 562 643 |
| chrIX | 441 593 | 461 772 | 452 809 | 439 888 |
| chrX | 764 537 | 759 892 | 777 694 | 745 751 |
| chrXI | 679 352 | 690 875 | 680 699 | 666 816 |
| chrXII | 1 117 833 | 1 078 292 | 1 114 766 | 1 078 177 |
| chrXIII | 912 255 | 913 070 | 913563 (**2051454) | 924 431 |
| chrXIV | 776 471 | 801 157 | 778 019 | 784 333 |
| chrXV | 1 091 066 | 1 101 379 | 1 105 746 | 1 091 291 |
| chrXVI | 948 593 | 965 730 | 979 195 | 948 066 |
| chrmt | 86 132 | *53 964 | 86 343 | 85 779 |
| *27 063 | ||||
| Total length (main+mt) | 12 206 251 | 12 462 516 | 12 417 334 | 12 157 105 |
| 2-micron | 147 349 | 71 725 | 144 927 | 6 318 |
| 2-micron | – | – | 61 136 | – |
| telomere | – | – | 76 064 | – |
| telomere | – | – | 49 485 | – |
| telomere | – | – | 45615 | – |
| # contigs intotal | 18 | 19 | 21 | 18 |
| # Ab initio CDS | 5 624 | 5 531 | 5 554 | 5 465 |
| Avg identity*** | 99.3 | 99.6 | 99.6 | NA |
*The length of the two broken contigs of mitochondrial chromosome.
**The length of the missed assembly contig before manually broken.
***Average identity of the assembly contig comparing with Illumina read before the polishing step.
NA = not applicable.
The 2-micron plasmid sequence obtained from all of our assemblies (ONT, PacBio and OP) is longer than the reported length of the 2-micron plasmid for strain S288C. We further investigated the long 2-micron contigs and found that the Canu assembler (25) had difficulty discriminating the extra depth from the multi-copy 2-micron plasmids (see Supplementary Figure S2).
The complete genomes (all chromosomes) from the three assembly strategies were very similar, with a DNA identity of 99.95% and a similar number of CDS ORFs by the ab initio method AUGUSTUS (29). We decided to use OP_assembly to represent the whole genome of S. cerevisiae strain CEN.PK113-7D for further analysis and comparison because we believe that combining the reads will give the highest sequencing depth, leading to a high confidence genome sequencing. Moreover, the OP_assembly has the highest average identity (see Table 1) when compared to Illumina reads if the broken mitochondria chromosome found using PacBio_assembly is not considered. We further evaluated the assembly completeness by identifying telomeric repeats, which we found on both ends of all of the main chromosomes as illustrated in Figure 2. The S. cerevisiae strain CEN.PK113-7D complete genome has been deposited in Genbank (accession numbers CP022966–CP022982).
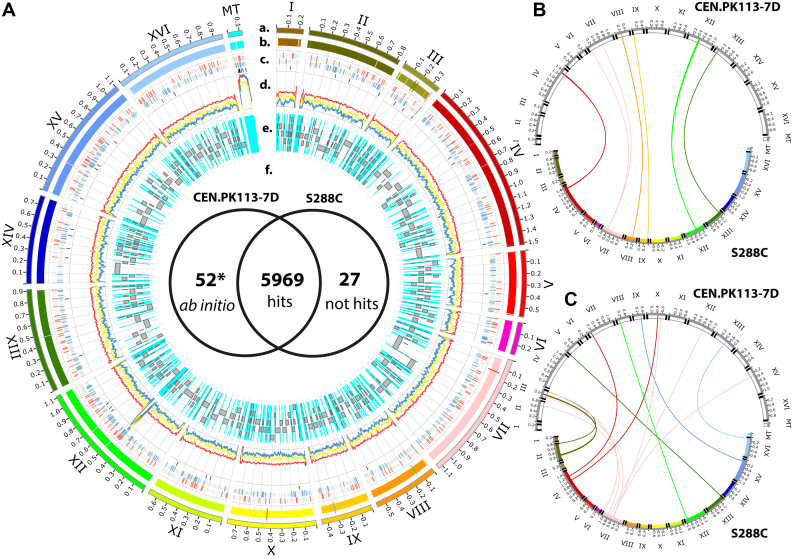
Figure 2.
The complete CEN.PK113-7D genome obtained from de novo assembly and its comparisons. (A) A Circos plot shows the genome comparisons. Lane a) The CEN.PK113-7D chromosomes (I-XVI) and the mitochondrial chromosome (mt) are plotted in different colors. Lane (b) S288C chromosomes and the crossed vertical lines represent the confidence chromosome rearrangement regions (>1 kb) between the two stains. Lane (c) The crossed vertical line plot shows the location on the chromosome of DNA methylation sites 4mC, 6mA and 5mC illustrated from the outer ring to the inner ring, respectively. The methylation sites that do not locate on the upstream region of ORFs are plotted in gray. The red and blue crossed vertical lines represent the methylation sites located on the upstream region of ORFs of 4mC and 6mA, respectively. Lane (d) DNA sequencing depth coverage plots, yellow, red and blue represent the data obtained from Illumina, PacBio and ONT reads, respectively. Lane (e) Gray bars represent the location of the assembled contigs obtained from Nijkamp et al. The cyan color represents the missing regions (gaps) that cannot be captured by the short reads assembly from Nijkamp et al. Lane (f) The Venn diagram compares the hits of S288C CDS ORFs hit on the CEN.PK113-7D genome. The star indicates the additional ORFs from ab initio gene calling obtained with AUGUSTUS software. On the right-hand side, the circos plot shows the results obtained from chromosomal rearrangement analysis between CEN.PK113-7D and S288C for synteny in panel (B) and translocation in panel (C). The chromosomes of CEN.PK113-7D are plotted in white on the top. The chromosomes of S288C are plotted in different colors on the bottom. The telomere regions were marked on the end of each chromosome in black. A close-up of the inversion is provided in Supplementary Figure S5.
Comparative genomics between the S288C and CEN.PK113-7D
The CEN.PK113-7D genome was first compared to the S288C genome by MUMMER (27), yielding a global average DNA identity of 99.5%, (see the dot plot in Supplementary Figure S3). The number of identified SNVs are 24 071; this number is comparable with previous reports (18,22). Interestingly, the number of identified insertion-deletions (INDELs) detected is 13 732, which is around four times higher than reported from experiments using short reads (18,22), possibly due to homopolymer problems commonly observed when using PacBio and ONT.
The complete CEN.PK113-7D genome is shown in Figure 2A.a. The read coverage from the ONT, PacBio, and Illumina, as illustrated in Figure 2A.d, reveals the unusual high sequencing depth, linked with high DNA copy numbers for the mitochondrial chromosome, and also in the middle of chromosome XII, which contains a cluster of repeated rRNA genes. In S288C, rRNA genes are also found in the long repeat region of 9.1 kb on Chromosome XII. To ensure that the assembly results are valid, we investigated the read alignment over the region. We found some long reads that span over the long repeat region, as seen in Supplementary Figure S4. The larger mitochondrial DNA content has been previously reported to show an increase in cell growth and nuclear DNA replication (38), reflecting the mid-log phase sampling point.
DNA methylation plays important roles in various cellular regulation pathways and is also known to be responsible for epigenetic modification, which is associated with human diseases (39). Methylation in the upstream region of coding sequences can slow down the transcription process. S. cerevisiae has been used as an expression host to study higher eukaryote 5-methylcytosine (5mC), because the yeast is thought to contain no 5mC, as reported by Hattman et al. (40) and Capuono et al. (41). Using third-generation DNA sequencing technologies, the 4-methylcytosine (4mC) and 6-methyladenine (6mA) can be captured on the PacBio reads (12) and 5mC can be captured on ONT reads (14). Results of DNA methylation analysis are illustrated in Figure 2A.c. The Nanopolish software (14) identified only 40 5mCs, compared to thousands of methylation sites for 4mC and 6mA; none of the 5mCs are located in the upstream region of ORFs for the CEN.PK113-7D genome; this is consistent with other results obtained experimentally by LC–MS/MS methods (41). SMART link software identified 6946 4mC and 4688 6mA with 359 sites and 297 sites located in the upstream region of ORFs, defined as 200 bp before the start codon and corresponding to the typical length of the yeast core promoter, as reported by Lubliner et al. (42).
All assembled contigs from the previous study of Nijkamp et al. (18) can be almost perfectly mapped to our assembled genome with a 99.8% DNA identity, as illustrated in Figure 2A.e, indicating the comprehensive quality of our genome. The result from the assembly based on short reads (in Figure 2A.e) shows the difficulty in mapping the terminal regions of the chromosome, close to telomeres. Moreover, the assembly based on short reads missed the mitochondrial chromosome and the middle region of chromosome XII, where we found the unusual sequencing depth in our study.
Due to the high percentage DNA identity of the two yeast genomes (CEN.PK113-7D and S288C), which are the same species, we used the annotated protein-encoding ORFs (5,996 ORFs) of the strain S288C genome to directly query the CEN.PK113-7D genome using the Blat software (28), and identified 5,969 ORFs that hit as illustrated in Figure 2A.f. The hits resulted in 6,173 loci (annotated as CDS ORFs) on the CEN.PK113-7D chromosomes, indicating some genes had been duplicated. We found that 23 ORFs were absent in the CEN.PK113-7D genome (see supplementary Table TS3). This is less than previously reported by Nijkamp et al. (18), indicating problems from unknown gaps that possibly derived from collapsed tandem repeats in the assembly based on short reads (see supplementary Table TS3). Eighteen of the absent ORFs are in the set of previously reported missing genes; only five of the absent ORFs are uniquely identified in this study, possibly due to the different versions of S288C genome annotation used in the two studies. To look for possible additional ORFs in CEN.PK113-7D, we employed ab initio gene calling (29), and yielded 52 ORFs that have high similarity to known proteins in the Uniprot database, indicating a high confidence for these additional ORFs (see supplementary Table TS3). Furthermore, all 417 genes of non-translated RNAs (e.g. tRNA, rRNA, snRNA, snoRNA) of S288C hits on the CEN.PK113-7D genome by direct sequence queries resulted in identification of 412 loci in the CEN.PK113-7D genome.
We used LAST software (30) for a detailed chromosome comparison between the CEN.PK113-7D and S288C genomes and identified a total of 555 regions of chromosomal rearrangements. Considering only the regions >1 kb, there are 35 regions identified as synteny, translocation, or inversion of the chromosomes illustrated in Figure 2A.b (see supplementary Table TS4). We further examined the 32 regions that contain ORFs and found 12 synteny regions on chromosomes IV, VIII, IX, and XII as well as two-inversion regions on chromosome VII, as illustrated in Figure 2B (see Supplementary Figure S5). The two largest synteny regions are 50 kb on chromosome IV with 28 ORFs and 13.5 kb on chromosome IX with 7 ORFs. The two two-inversion regions carry three retrotransposon-related ORFs. We also found 19 chromosome translocations with 35 ORFs on 9 chromosomes, as illustrated in Figure 2C. Interestingly, chromosome VII of S288C translocates into many chromosomes of CEN.PK113-7D (see supplementary Table TS4).
CEN.PK113-7D transcriptional landscape and quantification
We explored the transcriptional landscape using direct RNA sequencing over two metabolic stages of diauxic growth: respiro-fermentative growth on glucose and oxidative growth on ethanol. Averaged across four biological replicates, we obtained ∼530,000 high quality reads with N50 of 1150 bases, corresponding to ∼509 MB (59X of total transcripts length) for growth on glucose and ∼623 000 high quality reads with N50 of 1263 bases, corresponding to ∼623 MB (72X of total transcripts length) for growth on ethanol (see detail in supplementary Table TS5). We then evaluated the error rate of the aligned direct RNA sequence reads based on the reference genome sequence following Quick et al. (43) and found that, on average, the direct RNA sequencing read has 88% identity and 12% error (see detail in supplementary table TS6).
As shown in Figure 3A, the distribution of high quality direct RNA sequencing reads obtained from both growth conditions have similar shapes, indicating a transcriptome signature of CEN.PK113-7D that can be captured by sequencing. Moreover, the distribution of direct RNA sequencing reads agrees with the distribution of transcript lengths obtained from gene annotations. The direct RNA sequencing reads were further aligned to the CEN.PK113-7D genome, and the level of expression of individual transcripts, with respect to the gene calling and annotation, were determined by simply counting the number of mapped reads on the individual transcripts. We found that ∼91% of the predicted transcripts (5433 from 5994) can be consistently detected across the four biological replicates of growth on either glucose or ethanol. Under the same criterion, out of the 492 non-translated transcripts, almost all of them (398 or 81%) did not pass the criterion (see supplementary Table TS7). The absence of non-translated transcripts is likely due to the experimental method of extracting transcripts, which was based only on the presence of a poly(A) tail by the poly(A) selection strategy. This would exclude polymerase III transcripts. We further explored the mapped direct RNA sequence reads to the 479 known spliced genes in the genome and found that 80 spliced genes (17%) were not expressed at all in any of the growth conditions used in the experiments. We found only 10 spliced genes (2%) that had direct RNA sequence reads covering less than 95% of total exon length. The rest (389 spliced genes) had direct RNA sequence reads mapped covering their exons (see supplementary Table TS8).
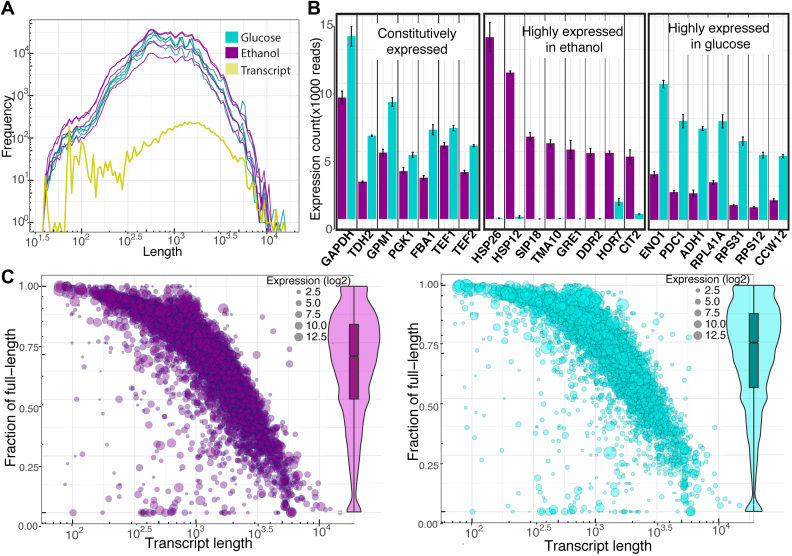
Figure 3.
Summary of the direct RNA sequencing data. (A) The histogram plot shows the distribution of read length of high quality reads obtained from yeast cell growth ethanol (magenta) and glucose (cyan), respectively, with the distribution of expected transcript lengths derived from the ORFs annotation. (B) Bar plots of the detected highly expressed transcripts are presented as an average normalized count with standard error over four biological replicates for each growth condition. The constitutively expressed, highly expressed in ethanol growth and highly expressed in glucose growth are illustrated in the left middle and right box, respectively. (C) The bubble scatter plots show the relationship between the fraction of detected full-length transcripts by the direct RNA sequencing with the transcript length and the level transcript expression. The violin-boxplots on the right show the overall distribution of the fraction of detected full-length transcripts.
Only a few transcripts are highly expressed. There are only 22 transcripts with >5000 direct RNA sequencing reads mapped for either growth condition, as illustrated in Figure 3B. As expected, the well-known glyceraldehyde-3-phosphate dehydrogenase (GAPDH) is one of the most abundant mRNAs. It is the most abundant transcript during growth on glucose, and the third-most abundant during growth on ethanol. Besides this, TDH2, which is the homolog of GAPDH, is also highly expressed under both growth conditions. The three key transcripts of enzymes for glycolytic pathways, the 3-phosphoglycerate kinase (PGK1), glycerate phosphomutase (GPM1) and fructose-1,6-bisphosphatase aldolase (FBA1) are highly expressed under both growth conditions. In addition, transcripts encoding the translation elongation factor TEF1 and the paralog TEF2 were found to be constitutively high expressed. The constitutively high expression of PGK1 and TEF1,2 are in agreement with a study by Partow et al., who reported high performance of the promoters of these transcripts in a yeast expression vector (44). The three transcripts of enolase (ENO1), the major form of pyruvate decarboxylase (PDC1) that is key for alcoholic fermentation, and alcohol dehydrogenase (ADH1) involving in ethanol production, were specifically highly expressed during growth on glucose, as expected, clearly reflecting the respiro-fermentative metabolism. Faster growth of cells on glucose than on ethanol resulted in overexpression of three ribosome-related transcripts (RPL41A, RPS31, RPS12) and a transcript coding cell wall mannoprotein (CCW12). On the other hand, highly expressed transcripts encoding heat shock proteins (HSP26, HSP12), oxidative stress protection, and overexpression of many stress related transcripts (SIP18, TMA10, GRE1, DDR2, HOR7) were specifically observed in growth on ethanol, reflecting oxidative stress. Moreover, CIT2 encoding citrate synthase (peroxisomal isozyme) was overexpressed during growth on ethanol, indicating that the glyoxylate shunt is active.
The ONT technology enables very long sequencing reads, a capability we explored in detection of full-length transcripts from the obtained direct RNA sequencing data. The direct RNA sequence reads that have 95% covered of the total transcript length were considered the full-length transcript reads and were used to calculate the fraction of full-length transcript detected. As seen in the violin-boxplots in Figure 3C, most of the detected transcripts have around 70% full length, with a small influence by the growth condition (see supplementary Figure S6 for violin-boxplot of detected full-length transcripts for individual sample). As expected, the fraction of detected full-length transcripts declined with increasing transcript length but independent of expression level, as illustrated in the bubble plots of Figure 3C (see supplementary Figure S7 for the plot of detected full-length transcripts versus expression level of transcript in detail). It is interesting that the direct RNA sequencing can detect full-length transcripts over 5kb. The heterogeneity of individual transcript lengths may reflect information about RNA turnover.
An important goal of transcriptome analysis is differential gene expression identification. We first evaluated the intrinsic variability of transcriptome data using principle component analysis and found clear separation (90% of variance capture by PC1) between the two growth conditions as illustrated in Figure 4A. A simple count of the number of direct RNA sequence reads mapped to individual transcripts can be used as a proxy for quantification of gene expression that is a very similar approach to the traditional short read RNA-Seq. Therefore, we employed the DESeq2 method (34) to estimate the transcript-level statistic of transcriptional changes between growth on ethanol as summarized in the MA plot and violin-boxplot of adjusted P-values illustrated in Figure 4B and C. We further evaluated the biological sense of differential gene expression results using gene-set enrichment analysis (36), as illustrated in Figure 4D. The identified enrichment GO terms show reasonable explanations, in terms of known physiology of the classic diauxic growth pattern in yeast. For example, the GO terms related to transcription and translation processes were up-regulated in growth on glucose, which is in agreement with the higher growth rate on glucose than on ethanol. It is known that after glucose depletion, ethanol, which is a fermentative product, will be utilized through oxidative metabolism; this is in agreement with the up-regulated GO terms related to TCA cycle, glyoxylate shunt, and mitochondria electron transport. Lack of nutrients and accumulation of toxic metabolites in the growth on ethanol products were revealed by the up-regulated GO terms, responses to stress, catabolic processes, and beta-oxidation.
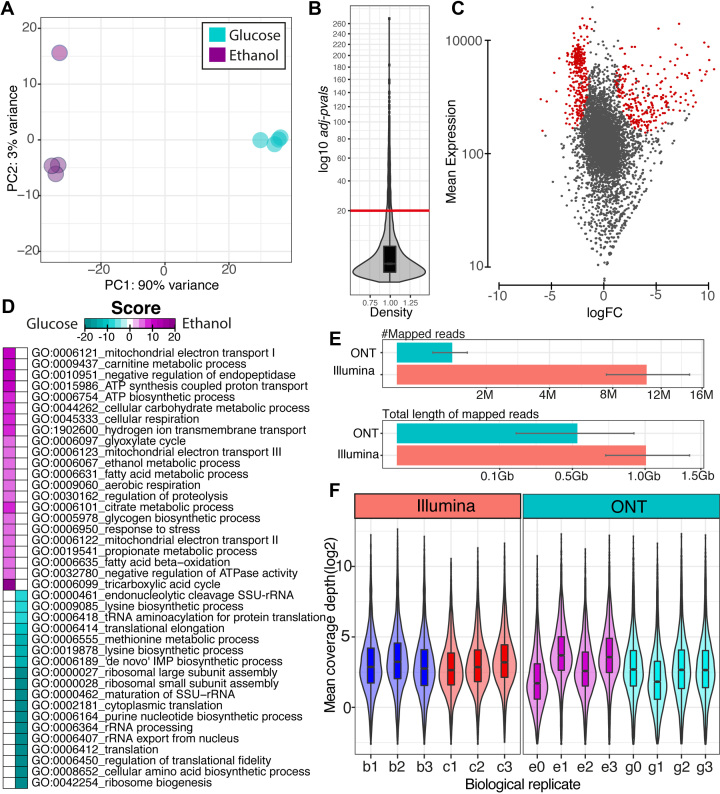
Figure 4.
The summary results of transcript quantification and differential gene expression analysis. (A) Principle component analysis plot of individual sample (circle) of yeast cell growth ethanol (magenta) and glucose (cyan color). (B) Violin-boxplot (square root transformed y-axis) shows the distribution of statistical adjusted P-values calculated using the DEseq2 method. The red line represents the yeast cell growth ethanol (magenta) and glucose (cyan color) adjusted P-value cut-off of 1e–20. (C) MA plot obtained from DEseq2 package. The red dots represent the transcripts that had adjusted P-values lower than the cut-off. The logFC represents the expression ratio of ethanol growth over glucose growth. (D) Heatmap illustration of the directional enrichment score of gene-set enrichment analysis of gene ontology using the PIANO package. Magenta represents the up-regulated scores on ethanol growth and cyan represents the up-regulated scores on glucose growth. (E) Bar plots show the comparison of DNA sequencing library size between ONT and Illumina datasets in terms of number of reads and amount in gigabases. The average values are presented with standard error over quadruplicate for each growth condition. (F) Violin-boxplots show the comparison of dynamic range in library size (Gb) corrected read count (log2) of the Illumina and ONT datasets across biological replicates (e = ethanol growth, g = glucose growth, b = batch growth, c = chemostat growth).
We further compared the dynamic range of transcript detection between direct RNA sequencing using ONT and traditional RNA-Seq (using Illumina technology obtained from our previous study (22)). Based on the read mapping results as shown in Figure 4E, the number of mapped reads obtained from the Illumina dataset is about ten times higher than the ONT dataset due to the different read lengths (200 bp for Illumina, compared to >1000 bp for ONT). Therefore, the total length of mapped reads (that is, the total number of bp sequenced) was used instead to fairly compare the sequencing depth. We found that the ONT dataset has about half amount of the Illumina dataset, corresponding to about 64X and 118X of transcripts length, respectively (see Supplementary Table TS9 for more details). The distribution of the library size-corrected mean coverage depth across the transcripts for each biological replicate of Illumina and ONT dataset is illustrated in Figure 4F to compare dynamic ranges (see supplementary Figure S8 for the same data without library size correction). Both datasets have similar dynamic ranges across the different biological replicates, except e0 and g1, which have much lower sequencing depths than the other replicates (see Supplementary Table TS9). The dynamic range comparable to the lower half of the sequencing depth of direct RNA sequencing data might be reflective of the different methodologies. For RNA-Seq, the RNA is first converted to cDNA, then amplified, sequenced, and mapped back to the transcript. In contrast, for direct RNA sequencing, the RNA is sequenced directly.
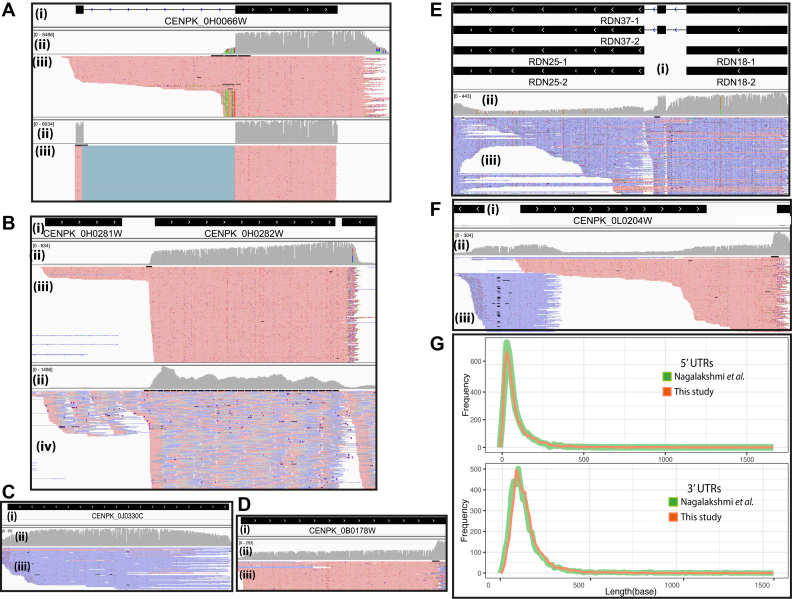
We examined regions of chromosomes to get an idea of the local transcriptional landscape structures. Figure 5A illustrates the simultaneous detection of mature and premature mRNA for the CENPK_0H0066W (RPL27A, Ribosomal 60S subunit protein L27A) locus. Figure 5A shows results for mapping the reads using GraphMap software (24) with non-guided mapping (Figure 5A, upper panel), compared to the transcript model guided mapping (Figure 5A, lower panel), which results in a very clean mapping signal for the exons. In another example (Figure 5B), we found an unexpected region that shows evidence of a polymerase II missed termination on the first ORF, which continues to transcribe until the termination of the second gene. These two genes are located in the region of 492 500 to 494 500 on Chromosome VIII. The two ORFs, CENPK_0H0281W (PTH1, Peptidyl-Trna Hydrolase) and CENPK_0H0282W (ERG9, Farnesyl-diphosphate farnesyl transferase), are illustrated in Figure 5B. We then compared the Polymerase II missed termination on the locus between direct RNA sequence reads (upper panel) with ‘traditional’ RNA-Seq results from short reads (lower panel). The long reads give a clear signal in support of read-through from the first transcript. In contrast, the short reads that are aligned in the region between the two ORFs are not firm covered, resulting in a lower-confidence signal possibly leading to a missed identification. We further explored the region and found that CENPK_0H0281W has no polyadenylation signal sequence (see Supplementary Figure S9); thus, it is likely that properly terminated full length transcripts of this gene would not be enriched in the poly(T) purification step. This reveals uncommon transcriptional regulation of the second gene (homolog to ERG9), which is a key gene in the sterol biosynthesis pathway in yeast. Moreover, the coverage plots clearly show that direct RNA sequence reads provide a homogeneously distributed signal over any ORF transcript. This even distribution is not seen in traditional RNA-Seq results. The ‘bumpy’ grey distribution seen in Figure 5B lower panel, compared to the relatively smooth grey band in the upper panel means the short reads have a less homogeneous distribution, possibly due to uneven amplification that needs further investigation at a similar sequencing depth.
Figure 5.
Transcriptional landscape structure examples illustrated by the snapshots of mapped reads using the IGV software. For panels A–F, i) shows the transcript structure(s), ii) indicates depth coverage, iii) details the mapped long reads of direct RNA sequence data. Red and blue represent the forward and reverse direction of mapped reads, respectively. iv) Shows details of mapped short reads of RNA-Seq data. (A) The different read alignment strategy shows that the non-guided exon alignment, illustrated in the upper panel, visually detects pre-mature transcripts that miss in guided exon alignment, illustrated in the lower panel. (B) The presence of dual transcribed of THI1 and ERG9. (C) The evidence of telomere RNA and (D) The evidence of polyadenylated long non-coding RNA. (E) The evidence of polyadenylated ribosomal RNA. (F) The presence of antisense transcript. (G) The length distribution of 5′UTRs (upper panel) and 3′UTRs (lower panel) of this study compared with Nagalakshmi et al. represented in red and green, respectively.
Surprisingly, we found some high-confidence non-coding exon ORFs that have direct RNA sequence reads mapped (Figure 5C, D, and E). The locus CENPK_0L0245C, with homologs to 35S rRNA (RDN37-1,2), predicted to be processed to 25S and 18S rRNA genes, has direct RNA sequence reads mapped indicating polyadenylation of the transcripts from this locus, and these rRNA genes might be transcribed by polymerase II rather than polymerase III. These findings are consistent with the discovery of polyadenylation on yeast rRNA by Kukai and coworkers (45) that used specific primers to probe polyadenylated rRNA. Interestingly, we found a signal of direct rRNA sequence reads in the region around the locus CENPK_0L0245C on both ethanol and glucose conditions (see Supplementary Figure S10). This region of chromosome XII has an unusually high DNA sequencing coverage depth, as shown in Figure 2A.d, and contains many copies of rRNA genes, based on predicted gene annotations. We found the polyadenylated rRNA transcript of telomerase RNA gene (TCL1), required for telomere replication at the locus CENPK_0B0178W, that is consistent with the study of Chapon et al. (46). In addition, we found polyadenylation of a long non-coding RNA transcript, which is the key regulation of the molecule (47), on the locus CENPK_0J0330C homologs to LTR1 involved in mating-type control of gametogenesis. We also found a polyadenylated antisense transcript from the 5′ region of CENPL_0L0204W homolog to regulation of RNA polymerase I (RRN5), an encoding transcription factor member of upstream activation factor (UAF) family (Figure 5F).
It is well-known that UTRs can impact mRNA processing, gene expression, and protein synthesis. We identified most of the 5′ UTRs and 3′ UTRs of CEN.PK113-7D using our direct RNA sequencing data, and compared it to those found in strain S288C, as reported by Nagalakshmi et al. (48). As shown in the histograms in Figure 5G, the identified 5′ and 3′ UTRs of the both studies are in agreement.
DISCUSSION
DNA library prep and read length is important for assembly
De novo assembly of eukaryotic genomes, which have large genome sizes and many repetitive regions, has made it almost impossible to obtain complete contiguous sequences of chromosomes with current short-read-based technology. The short read lengths obtained from second-generation sequencing make the problem mathematically hard in spite of high coverage, even though several algorithms have been developed to help overcome this problem (49). In our study, we used third-generation sequencing methods (with both ONT and PacBio) that yield very long reads, leading to successful de novo assembly of complete sequences of all 16 main yeast chromosomes in one step. Interestingly, even though the sequencing depth coverage of ONT reads was 6-fold lower than for the PacBio reads, the ONT_assembly yielded contiguous chromosome sequences for all 16 chromosomes. The PacBio_assembly had a small problem assembling the mitochondria chromosome, which could be the result of excessive DNA sequencing depth in combination with the short length of the mitochondrial chromosome. We could possibly improve this by adjusting the coverage depth when performing the assembly. The slightly better results obtained with ONT assembly compared to PacBio_assembly could be due to the longer N50 of the reads, which would provide longer pieces of DNA to anchor contigs The shorter read length distribution of PacBio reflects a different way of preparing the starting DNA for sequencing library preparation. Based on the PacBio sequencing protocol, we sheared the DNA before we made the sequencing library. This optimizes the CCS chemistry, as performance drops for DNA fragments longer than 25 kb. In contrast, we did not shear DNA for ONT sequencing library preparation. It is likely that shearing of the DNA is not completely random, and assembly across some breakage ‘hot spots’ could be difficult. Further, it is reasonable to assume that having very long reads is an important factor to obtain the complete sequence of eukaryal organisms through de novo assembly.
Repetitive regions and high copy numbers of small pieces of DNA in the genome cause difficulty with de novo assembly
Large eukaryal genomes typically contain several highly repetitive regions; these represent one of the biggest technical challenges when performing de novo assembly on short reads (50). The yeast S. cerevisiae, which is the subject of this study, has few repetitive regions (compared to plant or animal genomes, for example), except for telomere regions. These repetitive regions caused assembly errors in joining the ends of chromosomes VII and XIII when the sequencing depth was increased by combining the ONT and PacBio reads. In addition, the OP_assembly resulted in an additional three contigs from telomeres that cannot be joined to any chromosome. This kind of problem will be amplified in larger eukaryal genomes, such as human genomes, which contain repetitive sequences in more than half of the genome (51). Obtaining very long reads that can cover repetitive regions will, however, reduce assembly difficulties.
S. cerevisiae has many copies of the 2-micron plasmid, with a size of around 6.3 kb, which is shorter than the N50 length of ONT and PacBio reads. Interestingly, the most abundant reads’ lengths, observed as a spike in the histogram in Figure 1A, are similar to the size of the 2-micron plasmids. We further found that more than 7,899 ONT reads and >1400 PacBio reads cover the full-length of the 2-micron plasmid; however, few of the PacBio reads that match the 2-micron plasmid are full-length (see the distribution in Supplementary Figure S11). The high abundance of 2-micron plasmid reads might confuse the assembler into connecting them into longer multi-copy plasmid chimeric assemblies (see Supplementary Figure S2). Therefore, natural plasmids, which are commonly found in fungi and some plants (52), need to be carefully annotated during genome assembly.
Genome annotation: the next important step
Genome annotation is the next important step after genome sequences are achieved. Annotation is a challenging and time-consuming task that requires manual curation from experts and the research community to obtain high-quality results (53). Even with the most studied eukaryote, S. cerevisiae, high quality gene annotation requires curation. Therefore, we have provided the CEN.PK113-7D genome browser for the yeast community to curate and validate the current annotation. The browser includes processed information of the complete genome sequence, automated gene annotation, DNA methylation sites, direct RNA sequence alignments, and 5′ and 3′ UTR location prediction using the JBrowse software (54). The CEN.PK113-7D genome browser is freely available at http://genomebrowser.uams.edu/cenpk1137/.
Direct RNA sequencing enables single molecule quantification
Traditional RNA-Seq by short reads requires the conversion of transcripts to complementary DNA (cDNA) and amplification before measurement through second-generation sequencing. These two procedures introduce artifacts and biases as seen in the non-uniform signal of coverage plots in Figure 5B (ii), lower panel; however, direct RNA sequencing provides a solution—single molecule detection (Figure 5B (ii), upper panel).
The dynamic total RNA and mRNA concentrations in the cell at different cellular states directly impact analysis of the transcriptome, and have been simplified under the assumption that cells produce similar levels of RNA per cell as well as using similar amounts of total RNA as the beginning step without internal spike control, leading to erroneous interpretations, as reported by Loven et al. (55). Therefore, using a known amount of mRNA as starting material without amplification through direct RNA sequencing gave an accurate transcript quantification on differential gene expression analysis using negative binomial statistic and functional enrichment analysis. Furthermore, our differential analysis results suggested that there is no need to develop a new statistical analysis pipeline to analyze direct RNA sequence data, as existing tools can be employed effectively.
The error-prone long reads obtained from direct RNA sequencing are the main limitation in studying RNA editing and modifications. However, the long reads are good for transcript abundance detection whether or not a reference genome is available. As reported in this study, however, around 30% of detected transcripts are not full-length, which may possibly impact transcript quantification if there is not a reference genome available. The full-length transcript detection capability of direct RNA sequencing allows us to (i) accurately identify the structure and boundary of the transcript, (ii) detect unexpected transcriptional events and (iii) capture transcript heterogeneities and dynamics, which are important phenomena in elucidating transcriptional regulations. The standard direct RNA sequence relies on the enrichment of poly-A transcripts; thus, only polyadenylated transcripts can be detected. This means that probing the eukaryotic transcripts derived from polymerase I and III, as well as prokaryotic transcripts, is not covered in the current protocol.
CONCLUSION
Here we show that combining long reads of third-generation sequencing technology with matured bioinformatics analysis allows for full assembly of an eukaryal genome. We demonstrated that the superior scaffolding of long reads, obtained from careful extraction of high molecular weight DNA that minimizes shearing, enables the de novo assembly of a high quality, complete eukaryotic genome sequence. These results imply the transition from a ‘draft genome era’ to the ‘complete genome era’, allowing for a solid foundation for comparative genomics. Nevertheless, there is the major boundary of financial feasibility or sequencing cost, as the cost per bp sequenced of third-generation sequencing technologies is still more expensive than second-generation sequencing. Transcriptional landscape identification by direct RNA sequencing enables accurate determination of the encoded mRNA location, differential gene expression quantification, and structure identification of polyadenylated transcripts, free from the bias of DNA amplification. We believe that Direct RNA sequencing will become a versatile tool for transcriptome analysis in the ‘complete genome era’ of the future. It should be noted that the results presented here are for a relatively small, well defined organism (yeast). However, in dealing with larger genomes from animals and plants, a combination of higher sequencing depths and longer reads may be needed to overcome their bigger genome size, higher complexity, and higher ploidy for genome assembly. Similarly, transcriptome analysis through direct RNA sequencing for higher eukaryotes will require more sequencing depth than for S. cerevisiae, which has a low number of spliced genes, to be able to quantify transcriptional isoforms.
AVAILABILITY
The data have deposited in an SRA database under BioProject: PRJNA398797, SRP116559. The CEN.PK113-7D genome browser is freely available at http://genomebrowser.uams.edu/cenpk1137/.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to acknowledge support of the National Genomics Infrastructure (NGI)/Uppsala Genome Center and UPPMAX for providing assistance in massive parallel sequencing and computational infrastructure. Work performed at NGI/Uppsala Genome Center was funded by RFI/VR and Science for Life Laboratory, Sweden. The Swedish Research Council (VR-2013-4504) is acknowledged for partial financial support of IN. The authors would like to thank the reviewers—especially Nicolas Delhomme, PhD, Researcher at SLU and Manager of the UPSC Bioinformatics Platform—for their valuable comments, which helped improve the manuscript. This manuscript was edited by the Office of Grants and Scientific Publications at the University of Arkansas for Medical Sciences.
Author Contributions: I.N. and J.N. designed and conceived the genome sequencing project. P.J., I.N. performed computational analysis. T.W. performed MinION sequencing for DNA and direct RNA sequencing as well as genome sequence annotation and submission. R.P. performed cell cultivation, DNA extraction and RNA extraction and coordinated with the sequencing facility for PacBio sequencing. P.P. assisted with computational analysis. D.W.U. participated in design and supervised the study. I.N., P.J. and T.W. wrote the first version of the manuscript. All authors have read and approved the final version.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR online.
FUNDING
Arkansas Research Alliance; Helen Adams & Arkansas Research Alliance Professor & Chair; (NIH/NIGMS) [1P20GM121293]; Knut and Alice Wallenberg Foundation; Novo Nordisk Foundation. Funding for open access charge: UAMS startup fund.
Conflict of interest statement. None declared.
REFERENCES
- 1. Goffeau A., Barrell B.G., Bussey H., Davis R.W., Dujon B., Feldmann H., Galibert F., Hoheisel J.D., Jacq C., Johnston M. et al. Life with 6000 genes. Science. 1996; 274:546–567. [DOI] [PubMed] [Google Scholar]
- 2. Heather J.M., Chain B.. The sequence of sequencers: the history of sequencing DNA. Genomics. 2016; 107:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Mardis E., McPherson J., Martienssen R., Wilson R.K., McCombie W.R.. What is finished, and why does it matter. Genome Res. 2002; 12:669–671. [DOI] [PubMed] [Google Scholar]
- 4. Wang Z., Gerstein M., Snyder M.. RNA-Seq: a revolutionary tool for transcriptomics. Nat. Rev. Genet. 2009; 10:57–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Land M., Hauser L., Jun S.R., Nookaew I., Leuze M.R., Ahn T.H., Karpinets T., Lund O., Kora G., Wassenaar T. et al. Insights from 20 years of bacterial genome sequencing. Funct. Integr. Genomics. 2015; 15:141–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Smith L.M., Sanders J.Z., Kaiser R.J., Hughes P., Dodd C., Connell C.R., Heiner C., Kent S.B., Hood L.E.. Fluorescence detection in automated DNA sequence analysis. Nature. 1986; 321:674–679. [DOI] [PubMed] [Google Scholar]
- 7. Goodwin S., McPherson J.D., McCombie W.R.. Coming of age: ten years of next-generation sequencing technologies. Nat. Rev. Genet. 2016; 17:333–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Baker M. De novo genome assembly: what every biologist should know. Nat. Methods. 2012; 9:333–337. [Google Scholar]
- 9. Rhoads A., Au K.F.. PacBio sequencing and its applications. Genomics Proteomics Bioinformatics. 2015; 13:278–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Jain M., Olsen H.E., Paten B., Akeson M.. The Oxford Nanopore MinION: delivery of nanopore sequencing to the genomics community. Genome Biol. 2016; 17:239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Byrne A., Beaudin A.E., Olsen H.E., Jain M., Cole C., Palmer T., DuBois R.M., Forsberg E.C., Akeson M., Vollmers C.. Nanopore long-read RNAseq reveals widespread transcriptional variation among the surface receptors of individual B cells. Nat. Commun. 2017; 8:16027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Flusberg B.A., Webster D.R., Lee J.H., Travers K.J., Olivares E.C., Clark T.A., Korlach J., Turner S.W.. Direct detection of DNA methylation during single-molecule, real-time sequencing. Nat. Methods. 2010; 7:461–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Rand A.C., Jain M., Eizenga J.M., Musselman-Brown A., Olsen H.E., Akeson M., Paten B.. Mapping DNA methylation with high-throughput nanopore sequencing. Nat. Methods. 2017; 14:411–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Simpson J.T., Workman R.E., Zuzarte P.C., David M., Dursi L.J., Timp W.. Detecting DNA cytosine methylation using nanopore sequencing. Nat. Methods. 2017; 14:407–410. [DOI] [PubMed] [Google Scholar]
- 15. van Dijken J.P., Bauer J., Brambilla L., Duboc P., Francois J.M., Gancedo C., Giuseppin M.L., Heijnen J.J., Hoare M., Lange H.C. et al. An interlaboratory comparison of physiological and genetic properties of four Saccharomyces cerevisiae strains. Enzyme Microb. Technol. 2000; 26:706–714. [DOI] [PubMed] [Google Scholar]
- 16. Canelas A.B., Harrison N., Fazio A., Zhang J., Pitkanen J.P., van den Brink J., Bakker B.M., Bogner L., Bouwman J., Castrillo J.I. et al. Integrated multilaboratory systems biology reveals differences in protein metabolism between two reference yeast strains. Nat. Commun. 2010; 1:145. [DOI] [PubMed] [Google Scholar]
- 17. Otero J.M., Vongsangnak W., Asadollahi M.A., Olivares-Hernandes R., Maury J., Farinelli L., Barlocher L., Osteras M., Schalk M., Clark A. et al. Whole genome sequencing of Saccharomyces cerevisiae: from genotype to phenotype for improved metabolic engineering applications. BMC Genomics. 2010; 11:723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Nijkamp J.F., van den Broek M., Datema E., de Kok S., Bosman L., Luttik M.A., Daran-Lapujade P., Vongsangnak W., Nielsen J., Heijne W.H. et al. De novo sequencing, assembly and analysis of the genome of the laboratory strain Saccharomyces cerevisiae CEN.PK113-7D, a model for modern industrial biotechnology. Microb. Cell Fact. 2012; 11:36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Giordano F., Aigrain L., Quail M.A., Coupland P., Bonfield J.K., Davies R.M., Tischler G., Jackson D.K., Keane T.M., Li J. et al. De novo yeast genome assemblies from MinION, PacBio and MiSeq platforms. Sci. Rep. 2017; 7:3935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Istace B., Friedrich A., d’Agata L., Faye S., Payen E., Beluche O., Caradec C., Davidas S., Cruaud C., Liti G. et al. de novo assembly and population genomic survey of natural yeast isolates with the Oxford Nanopore MinION sequencer. Gigascience. 2017; 6:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Goodwin S., Gurtowski J., Ethe-Sayers S., Deshpande P., Schatz M.C., McCombie W.R.. Oxford Nanopore sequencing, hybrid error correction, and de novo assembly of a eukaryotic genome. Genome Res. 2015; 25:1750–1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Nookaew I., Papini M., Pornputtapong N., Scalcinati G., Fagerberg L., Uhlen M., Nielsen J.. A comprehensive comparison of RNA-Seq-based transcriptome analysis from reads to differential gene expression and cross-comparison with microarrays: a case study in Saccharomyces cerevisiae. Nucleic Acids Res. 2012; 40:10084–10097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Verduyn C., Postma E., Scheffers W.A., Van Dijken J.P.. Effect of benzoic acid on metabolic fluxes in yeasts: a continuous-culture study on the regulation of respiration and alcoholic fermentation. Yeast. 1992; 8:501–517. [DOI] [PubMed] [Google Scholar]
- 24. Sovic I., Sikic M., Wilm A., Fenlon S.N., Chen S., Nagarajan N.. Fast and sensitive mapping of nanopore sequencing reads with GraphMap. Nat. Commun. 2016; 7:11307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Koren S., Walenz B.P., Berlin K., Miller J.R., Bergman N.H., Phillippy A.M.. Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 2017; 27:722–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Walker B.J., Abeel T., Shea T., Priest M., Abouelliel A., Sakthikumar S., Cuomo C.A., Zeng Q., Wortman J., Young S.K. et al. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One. 2014; 9:e112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kurtz S., Phillippy A., Delcher A.L., Smoot M., Shumway M., Antonescu C., Salzberg S.L.. Versatile and open software for comparing large genomes. Genome Biol. 2004; 5:R12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kent W.J. BLAT–the BLAST-like alignment tool. Genome Res. 2002; 12:656–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Stanke M., Schoffmann O., Morgenstern B., Waack S.. Gene prediction in eukaryotes with a generalized hidden Markov model that uses hints from external sources. BMC Bioinformatics. 2006; 7:62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kielbasa S.M., Wan R., Sato K., Horton P., Frith M.C.. Adaptive seeds tame genomic sequence comparison. Genome Res. 2011; 21:487–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Chaisson M.J., Tesler G.. Mapping single molecule sequencing reads using basic local alignment with successive refinement (BLASR): application and theory. BMC Bioinformatics. 2012; 13:238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Krzywinski M., Schein J., Birol I., Connors J., Gascoyne R., Horsman D., Jones S.J., Marra M.A.. Circos: an information aesthetic for comparative genomics. Genome Res. 2009; 19:1639–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Quinlan A.R., Hall I.M.. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010; 26:841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Love M.I., Huber W., Anders S.. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014; 15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T. et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000; 25:25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Varemo L., Nielsen J., Nookaew I.. Enriching the gene set analysis of genome-wide data by incorporating directionality of gene expression and combining statistical hypotheses and methods. Nucleic Acids Res. 2013; 41:4378–4391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lunter G., Goodson M.. Stampy: a statistical algorithm for sensitive and fast mapping of Illumina sequence reads. Genome Res. 2011; 21:936–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Blank H.M., Li C., Mueller J.E., Bogomolnaya L.M., Bryk M., Polymenis M.. An increase in mitochondrial DNA promotes nuclear DNA replication in yeast. PLoS Genet. 2008; 4:e1000047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Robertson K.D. DNA methylation and human disease. Nat. Rev. Genet. 2005; 6:597–610. [DOI] [PubMed] [Google Scholar]
- 40. Hattman S., Kenny C., Berger L., Pratt K.. Comparative study of DNA methylation in three unicellular eucaryotes. J. Bacteriol. 1978; 135:1156–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Capuano F., Mulleder M., Kok R., Blom H.J., Ralser M.. Cytosine DNA methylation is found in Drosophila melanogaster but absent in Saccharomyces cerevisiae, Schizosaccharomyces pombe, and other yeast species. Anal. Chem. 2014; 86:3697–3702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lubliner S., Regev I., Lotan-Pompan M., Edelheit S., Weinberger A., Segal E.. Core promoter sequence in yeast is a major determinant of expression level. Genome Res. 2015; 25:1008–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Quick J., Quinlan A.R., Loman N.J.. A reference bacterial genome dataset generated on the MinION portable single-molecule nanopore sequencer. GigaScience. 2014; 3:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Partow S., Siewers V., Bjorn S., Nielsen J., Maury J.. Characterization of different promoters for designing a new expression vector in Saccharomyces cerevisiae. Yeast. 2010; 27:955–964. [DOI] [PubMed] [Google Scholar]
- 45. Kuai L., Fang F., Butler J.S., Sherman F.. Polyadenylation of rRNA in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S.A. 2004; 101:8581–8586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Chapon C., Cech T.R., Zaug A.J.. Polyadenylation of telomerase RNA in budding yeast. RNA. 1997; 3:1337–1351. [PMC free article] [PubMed] [Google Scholar]
- 47. Beaulieu Y.B., Kleinman C.L., Landry-Voyer A.M., Majewski J., Bachand F.. Polyadenylation-dependent control of long non-coding RNA expression by the poly(A)-binding protein nuclear 1. PLoS Genet. 2012; 8:e1003078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Nagalakshmi U., Wang Z., Waern K., Shou C., Raha D., Gerstein M., Snyder M.. The transcriptional landscape of the yeast genome defined by RNA sequencing. Science. 2008; 320:1344–1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Henson J., Tischler G., Ning Z.. Next-generation sequencing and large genome assemblies. Pharmacogenomics. 2012; 13:901–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Treangen T.J., Salzberg S.L.. Repetitive DNA and next-generation sequencing: computational challenges and solutions. Nat. Rev. Genet. 2011; 13:36–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. de Koning A.P., Gu W., Castoe T.A., Batzer M.A., Pollock D.D.. Repetitive elements may comprise over two-thirds of the human genome. PLoS Genet. 2011; 7:e1002384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Griffiths A.J. Natural plasmids of filamentous fungi. Microbiol Rev. 1995; 59:673–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Yandell M., Ence D.. A beginner's guide to eukaryotic genome annotation. Nat. Rev. Genet. 2012; 13:329–342. [DOI] [PubMed] [Google Scholar]
- 54. Buels R., Yao E., Diesh C.M., Hayes R.D., Munoz-Torres M., Helt G., Goodstein D.M., Elsik C.G., Lewis S.E., Stein L. et al. JBrowse: a dynamic web platform for genome visualization and analysis. Genome Biol. 2016; 17:66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Loven J., Orlando D.A., Sigova A.A., Lin C.Y., Rahl P.B., Burge C.B., Levens D.L., Lee T.I., Young R.A.. Revisiting global gene expression analysis. Cell. 2012; 151:476–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.