Abstract
Phi29 (Φ29) DNA polymerase is an enzyme commonly used in DNA amplification methods such as rolling circle amplification (RCA) and multiple strand displacement amplification (MDA), as well as in DNA sequencing methods such as single molecule real time (SMRT) sequencing. Here, we report the ability of phi29 DNA polymerase to amplify RNA-containing circular substrates during RCA. We found that circular substrates with single RNA substitutions are amplified at a similar amplification rate as non-chimeric DNA substrates, and that consecutive RNA pyrimidines were generally preferred over purines. We observed RCA suppression with higher number of ribonucleotide substitutions, which was partially restored by interspacing RNA bases with DNA. We show that supplementing manganese ions as cofactor supports replication of RNAs during RCA. Sequencing of the RCA products demonstrated accurate base incorporation at the RNA base with both Mn2+ and Mg2+ as cofactors during replication, proving reverse transcriptase activity of the phi29 DNA polymerase. In summary, the ability of phi29 DNA polymerase to accept RNA-containing substrates broadens the spectrum of applications for phi29 DNA polymerase-mediated RCA. These include amplification of chimeric circular probes, such as padlock probes and molecular inversion probes.
INTRODUCTION
Most DNA polymerases used in molecular biology methods are repair enzymes. The reason for this is that they are single protein enzymes, unlike replication enzymes that consists of large assemblies of proteins. Phi29 DNA polymerase is an exception. Composed of a single subunit, this DNA replication enzyme has proven invaluable in many molecular methods (1). Compared to the repair enzymes, it has an exceptionally high replication processivity and nucleotide incorporation rate (2) that in combination with a half-life of 11 h (3), can generate hundreds of kilobases long DNA products without dissociation from the substrate (2,3). The 3′→ 5′ proofreading activity of the phi29 DNA polymerase provides a high replication fidelity and also promotes its processivity (4). Finally, it is equipped with efficient strand displacement activity (2). These qualities are utilized in MDA to generate fairly unbiased whole genome amplification of high molecular weight input DNA (5). In the SMRT sequencing technology, the enzyme is used in a single-molecule sequencing reaction where acceptance of labelled nucleotides by individual phi29 DNA polymerases is observed while replicating a circular DNA substrate (6). Finally, RCA is an isothermal amplification method using short circular DNA molecules (7–9). RCA using phi29 DNA polymerase has found numerous applications, ranging from nucleic acid detection methods, to oligonucleotide synthesis (10), and library preparation for next generation sequencing technology (11–16). Here, DNA circles are generated in various ways using different substrates. Genomic DNA fragments can be circularized with the use of selector probes (11,12) to generate targeted libraries for NGS and mRNA-derived cDNA molecules can be circularized by Circ-ligase (16) yielding sequencing substrates in situ. Alternatively, nucleic acid detection methods based on RCA can use target specific circularizable DNA probes, so called padlock probes (3,17), or molecular inversion probes (18,19). Padlock probes are 80–100 nucleotide long linear DNA oligonucleotides that form nicked or gapped circles when bound to target DNA or RNA (17). Upon ligase-mediated circularization, probes can be amplified by RCA. The reaction can be initiated by supplying a circle-complementary primer, or by the target DNA or RNA itself (3,20–22).
Some DNA polymerases, like Tth (23) have been shown to display reverse transcriptase (RT) activities at elevated Mn2+ concentration. Such activity permits one-step cDNA synthesis and PCR, reducing risk for contamination and experiment time (24). However, comparing to traditional two-step system, one-step PCR is less sensitive and suffers from lower incorporation fidelity (25). Escherichia coli wild-type DNA polymerase I can also engage in RT under selected conditions (26,27). However, it is mostly used in DNA nick translation and second strand cDNA synthesis. Engineered enzymes—for example KB17 DNA polymerase producing RNA/DNA chimeric libraries decoded by mass spectrometry (28)—can be used for certain specific applications. Other polymerases like Bst DNA polymerase I Large Fragment are equipped with intrinsic strand displacement and RT activity (29) permitting one-step isothermal RNA sensing (30). Low processivity Bst polymerase can be used in large DNA amounts synthesis during hyperbranched RCA (hRCA) (9,31), although as not a replication enzyme, it cannot be used for efficient single-substrate RCA. Phi29 DNA polymerase is a preferred enzyme for isothermal amplification of circular substrates but its RT activity has not been properly examined. Tang et al. have studied the inhibition of RCA when individual DNA and RNA analogues are introduced into circular DNA templates (32). They found that single 2′-O-methylated RNA analogues inhibit the RCA reaction while single 2′-fluoro-RNA and LNA substitutions do not have a significant effect on the RCA efficiency (32). However, the RNA dependent DNA synthesis ability of phi29 DNA polymerase has not been studied. In a separate study, we have used RNA nucleotide-containing padlock probes for increased ligation efficiency on RNA templates (Krzywkowski et al. manuscript in preparation). Contradictory to the opinion in literature that phi29 DNA polymerase cannot amplify RNA (33), we found that the enzyme readily accepts RNA containing circular substrates. This observation motivated us to characterize the ability of phi29 DNA polymerase to replicate RNA containing circular templates. We hypothesized that phi29 DNA polymerase either ignores the RNA base, generating a ‘deletion’ in the RCA product, randomly incorporates nucleotides at the RNA template positon, or reverse transcribes the RNA base to a faithful DNA copy. In the present work, we demonstrate limited reverse transcriptase activity of phi29 DNA polymerase. We analyse the ability of the phi29 DNA polymerase to reverse transcribe different number of RNA bases, organized in various sequence patterns. Moreover, by sequencing the RCA products, we compare the velocity and incorporation fidelity of RCA using RNA-containing circular substrates in the presence of Mg2+ and Mn2+ cofactors, providing evidence for limited, yet highly accurate, reverse transcription activity of the phi29 DNA polymerase.
MATERIALS AND METHODS
Oligonucleotides used in the experiment
All oligonucleotides used in the present work were purchased from IDT (Integrated DNA Technologies, Inc., Coralville, IA, USA). Padlock probes were purchased as 4 nmol 5′-phosphorylated RNA or DNA Ultramers®. Decorator DNA oligonucleotides were HPLC purified, with 5′ conjugated fluorophore (Supplementary Table S1). Padlock probes contained RNA substitutions on the terminal 3′-OH(rN) or in the backbone (Supplementary Tables S1 and S2). Ligation reactions were performed on synthetic KRAS mRNA template. Padlock probes were designed such that upon hybridization to RNA, probes undergo circularisation forming a nick between terminal arms. For convenient size assessment of rolling circle products (RCP), a reporter sequence was embedded in the sequence linking the probe arms (backbone). Complementary decorators were used for RCP staining by hybridising to the reporter sequence.
Real-time RCA and data analysis
To assess the effect of RNA bases on RCA performance by phi29 polymerase, 20 nM of padlock probes were mixed with 10 nM of RNA template supplemented with 0.4 U/μl RNase Inhibitor (DNA Gdansk), 0.25 U/μl of PBCV-1 DNA ligase (SplintR, M0375S, NEB) in the respective buffer in a final volume of 15 μl. The reactions were incubated at 37°C for 30 min and heat inactivated at 65°C for 3 min. Following the ligation, 2 μl ligation volume (circles) was added to 18 μl RCA reaction mix containing 0.1 U/μl phi29 DNA polymerase (Monserate Biotechnology Group) 1 × phi29 reaction buffer (Thermo Fisher), 125 μM dNTP (DNA Gdansk), 0.2 μg/μl BSA (NEB) and 1 × SYBR Gold (S11194, Invitrogen) to a final circles’ concentration of 1 nM. To ensure simultaneous initiation of RCA in all samples, circles were put in the tube lids and spun down using table-top centrifuge into the pre-dispensed master mix. RCA initiated immediately and SYBR Gold incorporation was monitored using a Mx3005P qPCR System (Agilent Genomics) at 37°C for 60 min, followed by phi29 DNA polymerase inactivation at 65°C for 2 min.
Real-time amplification data (fluorescence build-up during 60 min RCA) for duplicated reactions were exported and processed in Microsoft® Excel 15.41 and Prism 7. Amplification curves were presented either as raw average from duplicated samples (e.g. Figure 2C) or curves for each duplicated experiment were first smoothed (each data point and two nearest neighboring points were averaged in a sliding window fashion), linear regression function was calculated where RCA velocity was linear and maximal and slope of regression was used as final velocity approximation (for example, Figure 3B).
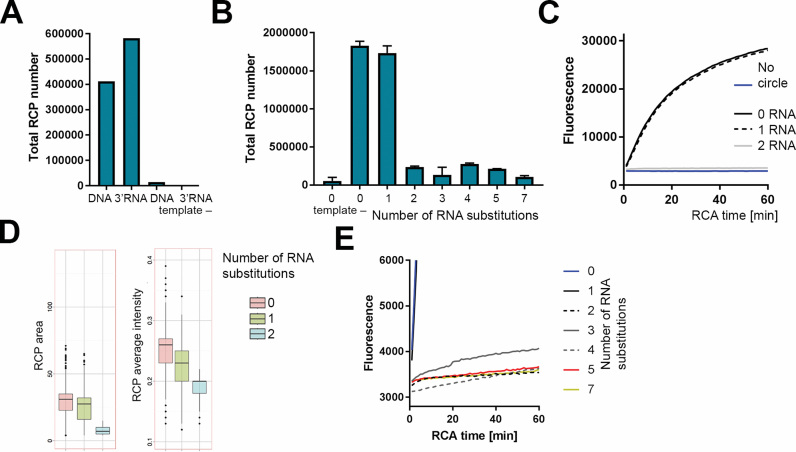
Figure 2.
Effect of RNA substitutions in circular templates on rolling circle amplification with phi29 DNA polymerase. (A) Total amount of RCA products (y-axis) generated for padlock probes with/without a terminal 3′ RNA and in the absence of synthetic RNA ligation template (template -). (B) Circles with 0–7 RNA substitutions in the backbone were amplified and digitally counted. The y-axis shows the number of rolling circle products (RCPs); error bars ± S.D.; n = 2. The same RCA reactions with chimeric circles were also monitored in real-time by measuring SYBR Gold incorporation on qPCR instrument (C and E). (C) RCA reaction curves of circles with 0, 1 and 2 RNA substitutions. (D) RCPs from C were imaged on microscope slides and size and intensity of individual RCPs were quantified. Black line, median; upper whisker, highest value that is within 1.5 the interquartile range of the hinge; lower whisker, lowest value within 1.5 the interquartile range of the hinge. (E) Real-time data of the same RCA reactions as in B with 0–7 RNA substitutes are displayed. Representative samples are presented from a duplicated experiment. To highlight the initial stages of RCA and to show the difference between the samples with low RCA efficiency, fluorescence intensity readout between 3000 and 6000 is presented.
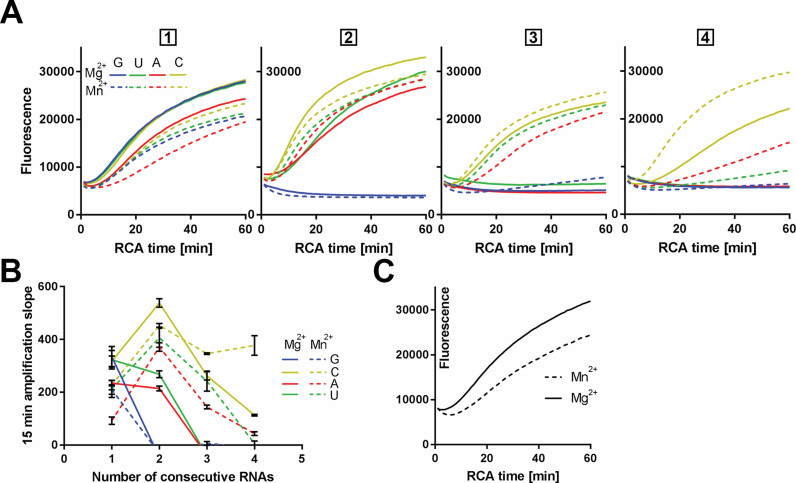
Figure 3.
Phi29 DNA polymerase exhibits higher RCA rate with circles containing pyrimidine RNA substitutions. (A) Real-time RCA curves of circles containing 1, 2, 3 or 4 consecutive RNA substations of rG, rU, rA, rC RNA bases are displayed (number of consecutive substitutions is indicated above plots). Rate of RCA was monitored by measuring fluorescence build-up (y-axis) resulted from SYBR Gold incorporation into RCPs. Averaged fluorescence intensity for each RCA time point was calculated from a duplicated experiment. RCA was conducted in the presence of Mg2+ and Mn2+ (solid and dashed lines respectively). (B) Linear, early stage RCA velocity (y-axis) is presented for PLPs from (A) in the presence of Mg2+ (solid lines) and Mn2+ (dashed lines). (C) RCA for the control PLP (non-chimeric DNA circle, with Mg2+ (solid) and Mn2+ (dashed line) are displayed.
To study the influence of Mn2+ and Mg2+ on phi29 DNA polymerase RNA-dependent RCA velocity and fidelity, a divalent cation-free 10 × amplification buffer was used in RCA (500 mM Tris–HCl, 100 mM (NH4)2SO4, 1% Tween20), supplied with 1 mM Mn2+ or 10 mM Mg2+.
Finally, to investigate whether RCA efficiency of circles with single rU RNA base, as well as di-, tri- and four-nucleotide long rUs can be stimulated by addition of reverse transcriptase, various RNaseH(–) reverse transcriptases were added to the RCA reaction mixture.
Morphological assessment of RCP size and intensity
To measure RCA product size and fluorescence intensity, the RCA products from the real-time RCA reaction were diluted to a final concentration of 20 pM, labelled with decorator probes at a final concentration of 5 nM in standard hybridization conditions (20). 10 μl of the fluorescently labeled RCA products were spotted onto Superfrost glass slides (Thermo Fisher), spread out by a 20 × 20 mm coverslip (Menzel Gläser) and left to bind electrostatically to the positively charged surface during a 15 min incubation. Coverslips were removed, slides were briefly washed in PBS, mounted in mounting medium and imaged on a Zeiss Axioplan fluorescent microscope with 20× magnification in the Cy3 channel. Images were exported as original black–white (BW) pictures and processed with Cell Profiler software (34). Briefly, each image was pre-processed using automated top-hat filtering. Objects were identified using manually adjusted thresholding and separated based on observed fluorescence intensity. The average fluorescence intensity and object size was recorded, exported as a csv file and processed in R! Studio.
Sequencing of RCA products generated from RNA-containing circles
Monomerization
In order to sequence the incorporated bases within the RCA products that correspond to the RNA bases within the circular templates, the RCA products were first monomerized by restriction digestion. First, RCA products from the real-time RCA measurements, as described above, were diluted in PBS-Tween 0.05% to a concentration of 100 pM. Next, RCA products were digested with AluI restriction enzyme in a reaction mixture containing 1 × phi29 DNA polymerase buffer, 0.2 μg/μl BSA, 100 nM restriction oligonucleotide (Supplementary Table S2), 120 mU/μl AluI (NEB) and RCA products at a final concentration of 10 pM during 10 min incubation at 37°C. Subsequently, the enzyme was heat inactivated at 65°C for 2 min. After complete digestion of the 10 pM RCA products, the RCA monomer concentration is approximately 10 nM (1 h RCA of an 80 base circle yields ∼1000× amplification). The RCA monomers were diluted to 100 pM in PBS–Tween 0.05%.
Sequencing library preparation of RCA monomers
The RCA monomers were first tagged with Illumina adapter sequences during a PCR reaction, containing 1 × Taq DNA polymerase buffer (NEB), 1.5 mM MgCl2 (NEB), 250 μM dNTP, 1 × SYBR Gold, 25 mU/μl Taq DNA polymerase (NEB), 0.5 μM forward primer PE1 (Supplementary Table S2), 0.5 μM reverse primer PE2 (Supplementary Table S2) and final concentration of 10 pM RCA monomers. The PCR was initiated with 5 min denaturation at 95°C and cycled between 95°C for 15 s, 55°C for 30 s and 70°C for 20 s for 20 cycles. The reaction was monitored using qPCR instrument and it was stopped before the amplification reached saturation. After the first PCR ('Extension (PCR)' step in the Figure 4A), 1 μl of the PCR products were transferred into index PCR mixture containing 1 × Phusion HF Buffer (Thermo Scientific), 200 μM dNTP (Thermo Scientific), 1% DMSO, 250 nM index PCR primers (Supplementary Table S3), each sample was labelled with unique combination of one of seven different forward and one of four different reverse index primers and programmed for an initial 2 min at 95°C, and 2 cycles of 95°C for 15 s, 60°C for 1 min and 72°C for 1 min, and an extra cycle of 72°C for 3 min. The indexed PCR products after 200-fold dilution served as templates in a new PCR mixture containing 1 × Phusion HF Buffer (Thermo Scientific), 200 μM dNTP, 1% DMSO, 500 nM P5 and P7 primers. The PCR was programed at 2 min at 95°C, and 15 cycles of 95°C for 15 s, 60°C for 30 s and 72°C for 30 s. The PCR products were pooled, purified using QIAquick PCR Purification Kit and sequenced by NSQ® 500 High Output Kit v2 (75 CYS) in NextSeq® 550 system (Illumina). The reads containing correct primer sequences at the expected positions (Figure 4) were analysed in Python and sequence logos created in Weblogo 3 (http://weblogo.threeplusone.com/create.cgi).
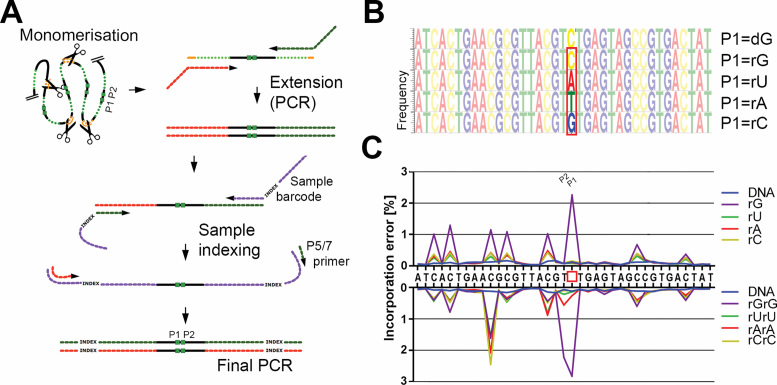
Figure 4.
DNA sequencing-based analysis of rolling circle products reveals reverse transcription activity of phi29 DNA polymerase. (A) After RCA, short DNA oligonucleotides were hybridized to an AluI restriction site in the RCA products and RCPs were digested with AluI restriction enzyme, resulting in RCA monomers. Following digestion, monomers were PCR-amplified using primers containing Ilumina adapter sequences. PCR products were extended using IIlumina indexed primers. Finally, sequencing library was prepared using indexed primers-specific P5/7 PCR primers. The region of interest containing RNA substitutions in the original padlock probe sequence is indicated with green boxes. (B) Logos showing sequencing frequencies for each position within RCA monomers generated from the control DNA circle (P1 = dG), and circles containing single rG, rU, rA and rC substitutions at the RNA position (P1). Positions P1 and P2 are indicated and position P1 was additionally highlighted with the red box. (C) Incorporation of incorrect nucleotides for every position in the sequenced monomers from (B). Error rates, calculated as Incorporation error [%] = 1 – number of reads with expected nucleotide/total number of reads, is presented for padlock probes with single- (upper plot) and double-RNA substitutions (lower plots). P1 position for the first RNA substitution is indicated with the box.
RESULTS
Phi29 DNA polymerase accepts chimeric circles as rolling circle amplification (RCA) templates
In a separate study, we have explored RNA templated padlock probe end-joining with PBCV-1 DNA ligase and T4 RNA ligase 2 (Krzywkowski et al. manuscript in preparation). We have found increased ligation efficiency for 3′ RNA containing padlock probes and observed that circularized chimeras are good substrates for RCA. To explore the mode though which the phi29 DNA polymerase replicates chimeric substrates, we circularized a variety of RNA/DNA chimeric padlock probes—containing 1–7 RNA substitutions in a DNA probe backbone—and used the circularized probes as templates for RCA (Figure 1).
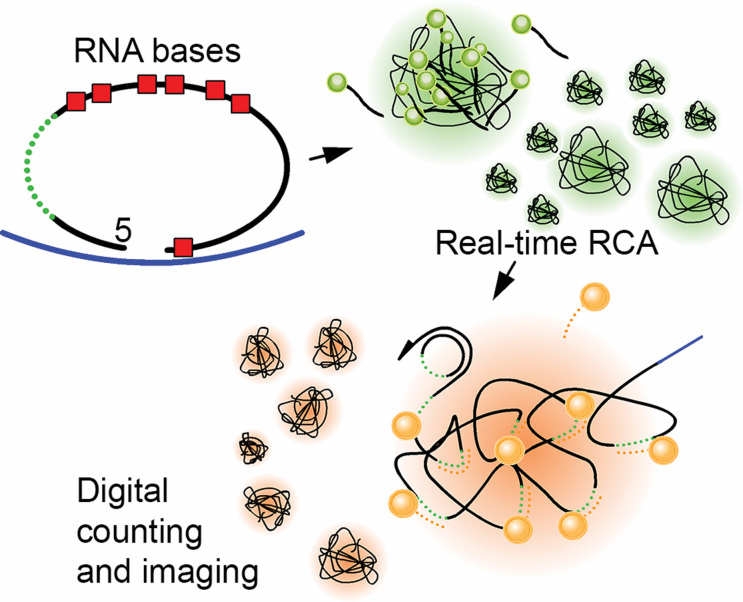
Figure 1.
Experimental procedure. Padlock probes, with various RNA substitutions (red boxes) at the 3′ terminus or in the probe backbone were circularised on synthetic RNA templates (blue) by ligation and used as circular substrates in rolling circle amplification (RCA). During RCA, replication was monitored in real-time by SYBR Gold incorporation into rolling circle products (RCPs) (green). For counting and size quantification, RCPs were further labelled with Cy3- decorator probes (orange, dotted) by hybridization to the reporter sequence (green, dotted). RCPs were then imaged and digitally quantified.
The RCA reaction was monitored in real-time by SYBR Gold incorporation. Additionally, the RCA products were digitally counted as described in (20,35), and their size and intensity assessed. We observed that PBCV-1 readily joined both pure DNA probe and chimeric 3′-(rN)/5′-(N) probes, and that phi29 DNA polymerase accepted both pure DNA and RNA-containing circles as templates for RCA (Figure 2A). When no template was added during the ligation step, probes were not ligated and no amplification was detected (Figure 2A).
Next, we aimed to investigate RCA efficiency of chimeric DNA/RNA circles without the bias of the ligation reaction. For this purpose, we added RNA substitutions in the circle backbone. Consequently, all probes had the same target-complementary arm sequences (contributing to forming the ligation substrate), and were ligated on the same RNA target. We then investigated how an increasing number of RNA substitutions affects the RCA reaction efficiency by quantitating RCA products (Figure 2B) after completion of the amplification reaction. As an independent way of measuring RCA rate, we also monitored the amplification reaction in real-time (Figure 2C).
We observed no effect on RCA efficiency when a single rG was substituted in the circle backbone (Figure 2B and C). A strong inhibition of RCA was observed when circles were substituted with two consecutive rG nucleotides (Figure 2B and C). For circles with more than these 2 RNA substitutions, no amplification was detected (Figure 2B and E). The average size and intensity of RCPs imaged using epifluorescence microscopy decreased for circles with 2 RNA substitutes (Figure 2D).
We have additionally investigated whether RCA of RNA-rich circles can be recovered by supplying a dedicated reverse transcriptase during the RCA reaction. Three different M-MuLV RT variants in different concentrations were examined (Supplementary Figure S1). However, under the reaction conditions used, RCA activity did not recover in the presence of the reverse transcriptase (Supplementary Figure S1).
Phi29 DNA polymerase preferentially replicates RNA pyrimidines during RCA
In the previous experiment, DNA bases in the circles’ backbone were substituted with RNA bases. Strong amplification inhibition was observed for circles with rGrG and rGrGrG substitutions. In order to study if there is some sequence dependency in RCA efficiency of RNA containing circles, we monitored RCA rate in real-time using circles with single rU/rA/rC/rG substitutions, as well as, di-, tri- and tetra-nucleotide long homo-nucleotide stretches (Supplementary Table S2). We observed efficient RCA for all single RNA substitutions (Figure 3A, solid lines). For the dinucleotide RNA substitutions, we observed the highest RCA rate for rCrC circles followed by rUrU circles, and rArA, while rGrG circles were not amplified. For tri- and tetranucleotide RNA circles, only rC circles generated detectable RCA (Figure 3A, solid lines in third and fourth panel). For rC RNA substations, the RCA rate decreased by ∼50% for each additional rC (Figure 3B, solid yellow line).
Manganese ions increase RNA-dependent RCA activity of phi29 DNA polymerase
As certain DNA-dependent DNA polymerases are able to reverse transcribe RNA in the presence of Mn2+ (23), we compared phi29 DNA polymerase RCA rates with Mg2+ and Mn2+. Using Mn2+ as cofactor, phi29 DNA polymerase in addition to rC also efficiently amplified single, di- and tri-nucleotide rU and rA stretches (Figure 3B, dashed lines). Interestingly, amplification rates of rCrC-, rUrU- and rArA- circles were higher when compared to single RNA substituted circles, which was also true for rCrC with Mg2+ as cofactor (Figure 3B). To investigate whether RCA can be recovered if multiple RNA bases are mixed with DNA-bases, multiple chimeric constructs were amplified with Mg2+ and Mn2+ (Supplementary Figure S2). According to our observations, phi29 DNA polymerase was able to engage in efficient Mn2+-dependent RCA when RNA bases were interspaced with DNA (Supplementary Figure S2). Interestingly, circular chimeric substrate with as many as 8 RNA bases was well amplified when substitutions were organised in a uniformly dispersed pattern (Supplementary Figure S2).
Sequencing of rolling circle products demonstrates ability of phi29 DNA polymerase to reverse transcribe RNA
Since the phi29 polymerase amplification rate was inversely proportional to the number of RNA bases in the substrate, we hypothesized that the enzyme may ignore RNA positions during RCA, introducing single, or double nucleotide deletions in amplified product. To validate this hypothesis, circles containing single and double homopolymeric RNA substitutions (Supplementary Table S2) were amplified and the RCA rate was monitored in real-time. Following amplification, sequencing libraries were prepared from the different RCA products, and then sequenced using Illumina NextSeq® 550 system (Figure 4A). Full length sequencing reads were extracted from the dataset and the proportion of extracted/total reads was calculated for all samples (Supplementary Table S4). Sequencing reads were aligned and base frequency was calculated for each position in the padlock probe backbone (Figure 4B).
According to our observations, phi29 DNA polymerase incorporated the expected DNA nucleotides in the RCA products, where the templates contained a RNA base. For circles with no RNA substitutions, >99% of sequenced monomers showed correctly incorporated base at P1 padlock probe region highlighted in the Figure 4B (0.125%, error for position P1 for DNA padlock probe). When P1 position was substituted with rA, rC, rG or rU, the average error was 0.111%, 0.153%, 2.259% and 0.084% respectively (Figure 4C, upper panel, red, yellow, violet and green lines). While the rA, rC, and rU were copied with same accuracy as DNA (as measured by sequencing), rG stands out with higher replication error. Interestingly, this higher incorporation error was observed not only for the P1 position, but for all guanosine bases in the padlock probe backbone (visible as high error rate peaks in the upper panel of Figure 4C) and higher thymine frequencies for the rG padlock probe logo graph in the Figure 4B). When both P1 and P2 positions were substituted with rArA, rCrC, rGrG and rUrU, error rate for the P1/P2 site was 0.269%/0.561%, 0.107%/0.109%, 2.827%/2.231% and 0.144%/0.220% respectively (Figure 4C, lower panel). Similarly to circles with a single rG substitution in P1, all dinucleotide RNA substrates demonstrated a higher error rate for non-RNA guanosine across probe backbone sequence. All sequencing alignment plots are presented in the Supplementary Figure S3. To investigate if more efficient Mn2+-dependent RCA RT is counterbalanced with lower amplification fidelity—observed for other DNA polymerases (25)—another sequencing experiment was conducted. Interestingly, similar incorporation error rates were observed for RNA and non-RNA positions in all padlock probes tested when Mn2+ were used as cofactor during RCA (Supplementary Figure S4).
DISCUSSION
In this work, we demonstrated limited reverse transcription activity of phi29 DNA polymerase. We show that single RNA substitutes in circular templates have no impact on RCA efficiency. We have found, however, that amplification was suppressed when longer stretches of RNA bases were present in the circular template sequence. In order to characterize this novel activity of phi29 DNA polymerase, we amplified circular templates containing either one, two, three or four consecutive RNA bases rA, rG, rC or rU with phi29 DNA polymerase and monitored the RCA rate in real-time. Moreover, we tested various combinations of different RNA bases and interspacing RNA bases with DNA bases. Our data demonstrated a preference for circular substrates containing pyrimidine RNA bases, since circles with four consecutive pyrimidine cytosine bases could still be efficiently amplified. Interestingly, interspacing 3, and even eight RNA substitutions with DNA bases led to a partial recovery of the RCA efficiency, indicating that RCA of RNA containing circles is restricted to single RNA base substitutions or very short stretches of consecutive RNA bases. Because certain DNA polymerases have the ability to engage in reverse transcription in the presence of Mn2+, we attempted to stimulate phi29 DNA polymerase RCA of RNA bases with Mn2+ as a cofactor. Our data demonstrated that phi29 DNA polymerase, when supplied Mn2+, can be used to replicate longer homo- and heteropolymeric RNA stretches. However, the general inability to reverse transcribe rGs (for both Mn2+ and Mg2+) limits its application to RT-RCA reactions to designed sequences. The attempt to increase RCA efficiency of circles containing longer stretches of RNA bases by addition of reverse transcriptase failed.
In order to identify the mode by which the phi29 DNA polymerase copies RNA containing circles, we sequenced the RCA products. Our data clearly illustrates that the mode by which the polymerase copies RNA containing circles, is reverse transcription, as we found the matching DNA base incorporated into the RCA products with high frequency (>99% for rA, rU and rC, ∼98% for rG). The overall incorporation accuracy on RNA substitutes was not different from the accuracy on pure DNA substrates. This proves that the mode, by which phi29 DNA polymerase copies RNA containing circles, is neither ‘base-jumping/skipping’, nor random incorporation of nucleotides, but indeed reverse transcription.
This, to the best of our knowledge, previously unknown activity of phi29 DNA polymerase, to reverse transcribe RNA bases in otherwise DNA circles, opens up a range of new applications. Certain ligases, such as T4 RNA ligase II, have been shown to possess increased end-joining activity for 3′-RNA chimeric substrates (36,37). The reverse transcriptase activity of phi29 DNA polymerase opens up the possibility to utilize chimeric ligation probes, such as padlock and molecular inversion probes, as more efficient ligation substrates. The increased ligation efficiency, combined with efficient RCA of chimeric circles, may enable high sensitivity analyses of nucleic acids. In our experiments, the reverse transcription activity seemed limited to only short stretches of RNA bases, and with a preference for pyrimidine bases. However, the activity on longer stretches and purine RNA bases was improved by dispersing the RNAs in the padlock probe primary sequence, and by altering the RCA reaction conditions. Our study demonstrated competent reverse transcription of as many as eight RNA bases in the padlock probe backbone provided that Mn2+ were present in the RCA reaction. In this study we report phi29 DNA polymerase reverse transcriptase activity and its base and cofactor preference. An investigation to reach an understanding of the mechanisms involved is an interesting subject for future studies.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Dr Kamila Klamecka for valuable comments and corrections of the manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
This work was supported by The Swedish Research Council (VR), Formas Strong Research Environment [Biobridges, project no. 221-2011-1692], and the SSF project FLU-ID (project no: SBE13-0125). Funding for open access charge: Vetenskapsrådet.
Conflict of interest statement. One or more embodiments of one or more patents and patent applications filed by authors may encompass the methods and data presented in this manuscript.
REFERENCES
- 1. Berman A.J., Kamtekar S., Goodman J.L., Lázaro J.M., de Vega M., Blanco L., Salas M., Steitz T.A.. Structures of phi29 DNA polymerase complexed with substrate: the mechanism of translocation in B-family polymerases. EMBO J. 2007; 26:3494–3505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Blanco L., Bernad A., Lázaro J.M., Martín G., Garmendia C., Salas M., Bernads A., Lharo J.M., Martins G.. Highly efficient DNA synthesis by the Phage Φ29 DNA polymerase. J. Biol. Chem. 1989; 264:8935–8940. [PubMed] [Google Scholar]
- 3. Banér J., Nilsson M., Mendel-Hartvig M., Landegren U.. Signal amplification of padlock probes by rolling circle replication. Nucleic Acids Res. 1998; 26:5073–5078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Esteban J. a., Salas M., Blanco L.. Fidelity of phi29 DNA Polymerase. J. Biol. Chem. 1993; 268:2719–2726. [PubMed] [Google Scholar]
- 5. Paez J.G., Lin M., Beroukhim R., Lee J.C., Zhao X., D R.J., Gabriel S., Herman P., Sasaki H., Altshuler D. et al. Genome coverage and sequence fidelity of Φ29 polymerase-based multiple strand displacement whole genome amplification. Nucleic Acids Res. 2004; 32:e71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Korlach J., Marks P.J., Cicero R.L., Gray J.J., Murphy D.L., Roitman D.B., Pham T.T., Otto G.a, Foquet M., Turner S.W.. Selective aluminum passivation for targeted immobilization of single DNA polymerase molecules in zero-mode waveguide nanostructures. Proc. Natl. Acad. Sci. U.S.A. 2008; 105:1176–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Liu D., Daubendiek S.L., Zillman M.A., Ryan K., Kool E.T.. Rolling circle DNA synthesis: Small circular oligonucleotides as efficient templates for DNA polymerases. J. Am. Chem. Soc. 1996; 118:1587–1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Fire A., Xu S.Q.. Rolling replication of short DNA circles. Proc. Natl. Acad. Sci. U.S.A. 1995; 92:4641–4645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lizardi P.M., Huang X., Zhu Z., Bray-Ward P., Thomas D.C., Ward D.C.. Mutation detection and single-molecule counting using isothermal rolling-circle amplification. Nat. Genet. 1998; 19:225–232. [DOI] [PubMed] [Google Scholar]
- 10. Ducani C., Kaul C., Moche M., Shih W.M., Högberg B.. Enzymatic production of ‘monoclonal stoichiometric’ single-stranded DNA oligonucleotides. Nat. Methods. 2013; 10:647–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dahl F. Multiplex amplification enabled by selective circularization of large sets of genomic DNA fragments. Nucleic Acids Res. 2005; 33:e71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dahl F., Stenberg J., Fredriksson S., Welch K., Zhang M., Nilsson M., Bicknell D., Bodmer W.F., Davis R.W., Ji H.. Multigene amplification and massively parallel sequencing for cancer mutation discovery. Proc. Natl. Acad. Sci. U.S.A. 2007; 104:9387–9392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Drmanac R., Sparks A.B., Callow M.J., Halpern A.L., Burns N.L., Kermani B.G., Carnevali P., Nazarenko I., Nilsen G.B., Yeung G. et al. Human genome sequencing using unchained base reads on self-assembling DNA nanoarrays. Science. 2009; 1469:78–81. [DOI] [PubMed] [Google Scholar]
- 14. Pihlak A., Baurén G., Hersoug E., Lönnerberg P., Metsis A., Linnarsson S.. Rapid genome sequencing with short universal tiling probes. Nat. Biotechnol. 2008; 26:676–684. [DOI] [PubMed] [Google Scholar]
- 15. Ke R., Mignardi M., Pacureanu A., Svedlund J., Botling J., Wählby C., Nilsson M.. In situ sequencing for RNA analysis in preserved tissue and cells. Nat. Methods. 2013; 10:857–860. [DOI] [PubMed] [Google Scholar]
- 16. Lee J.H., Daugharthy E.R., Scheiman J., Kalhor R., Amamoto R., Peters D.T., Turczyk B.M., Marblestone A.H., Yang J.L., Ferrante T.C. et al. Highly multiplexed subcellular RNA sequencing in situ. Science. 2014; 343:1360–1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Nilsson M., Malmgren H., Samiotaki M., Kwiatkowski M., Chowdhary B.P., Landegren U.. Padlock probes: circularizing oligonucleotides for localized DNA detection. Science. 1994; 265:2085–2088. [DOI] [PubMed] [Google Scholar]
- 18. Hardenbol P., Yu F., Belmont J., MacKenzie J., Bruckner C., Brundage T., Boudreau A., Chow S., Eberle J., Erbilgin A. et al. Highly multiplexed molecular inversion probe genotyping: Over 10, 000 targeted SNPs genotyped in a single tube assay. Genome Res. 2005; 15:269–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hardenbol P., Banér J., Jain M., Nilsson M., Namsaraev E.A., Karlin-Neumann G.A., Fakhrai-Rad H., Ronaghi M., Willis T.D., Landegren U. et al. Multiplexed genotyping with sequence-tagged molecular inversion probes. Nat. Biotechnol. 2003; 21:673–678. [DOI] [PubMed] [Google Scholar]
- 20. Krzywkowski T., Nilsson M.. Fidelity of RNA templated end-joining by chlorella virus DNA ligase and a novel iLock assay with improved direct RNA detection accuracy. Nucleic Acids Res. 2017; 45:e161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Merkiene E., Gaidamaviciute E., Riauba L., Janulaitis A., Lagunavicius A.. Direct detection of RNA in vitro and in situ by target-primed RCA: The impact of E. coli RNase III on the detection efficiency of RNA sequences distanced far from the 3′-end. RNA. 2010; 16:1508–1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lagunavicius A., Merkiene E., Kiveryte Z., Savaneviciute A., Zimbaite-Ruskuliene V., Radzvilavicius T., Janulaitis A.. Novel application of Phi29 DNA polymerase: RNA detection and analysis in vitro and in situ by target RNA-primed RCA. RNA. 2009; 15:765–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Myers T.W., Gelfand D.H.. Reverse transcription and DNA amplification by a Thermus thermophilus DNA polymerase. Biochemistry. 1991; 30:7661–7666. [DOI] [PubMed] [Google Scholar]
- 24. Chiocchia G., Smith K.. Highly sensitive method to detect mRNAs in individual cells by direct RT-PCR using Tth DNA polymerase. Biotechniques. 1997; 22:312–318. [DOI] [PubMed] [Google Scholar]
- 25. Bustin S.A. Absolute quantification of mRNA using real-time reverse transcription polymerase chain reaction assays. J. Mol. Endocrinol. 2000; 25:169–193. [DOI] [PubMed] [Google Scholar]
- 26. Loeb L.A., Tartof K.D., Travaglini E.C.. Copying Natural RNAs with E. coli DNA Polymerase I. Nature. 1973; 242:66–69. [DOI] [PubMed] [Google Scholar]
- 27. Karkas J.D. Reverse transcription by Escherichia coli DNA polymerase I. Proc. Natl. Acad. Sci. U.S.A. 1973; 70:3834–3538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mauger F., Bauer K., Calloway C.D., Semhoun J., Nishimoto T., Myers T.W., Gelfand D.H., Gut I.G.. DNA sequencing by MALDI-TOF MS using alkali cleavage of RNA/DNA chimeras. Nucleic Acids Res. 2007; 35:e62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Shi C., Shen X., Niu S., Ma C.. Innate reverse transcriptase activity of DNA polymerase for isothermal RNA direct detection. J. Am. Chem. Soc. 2015; 137:13804–13806. [DOI] [PubMed] [Google Scholar]
- 30. Lee M.-S., Lin Y.-C., Lai G.-H., Lai S.-Y., Chen H.-J., Wang M.-Y.. One-step reverse-transcription loop-mediated isothermal amplification for detection of infectious bursal disease virus. Can. J. Vet. Res. 2011; 75:122–127. [PMC free article] [PubMed] [Google Scholar]
- 31. Hamidi S.V., Ghourchian H.. Colorimetric monitoring of rolling circle amplification for detection of H5N1 influenza virus using metal indicator. Biosens. Bioelectron. 2015; 72:121–126. [DOI] [PubMed] [Google Scholar]
- 32. Tang S., Wei H., Hu T., Jiang J., Chang J., Guan Y., Zhao G.. Suppression of rolling circle amplification by nucleotide analogs in circular template for three DNA polymerases. Biosci. Biotechnol. Biochem. 2016; 80:1555–1561. [DOI] [PubMed] [Google Scholar]
- 33. Berthet N., Reinhardt A.K., Leclercq I., van Ooyen S., Batéjat C., Dickinson P., Stamboliyska R., Old I.G., Kong K.a, Dacheux L. et al. Phi29 polymerase based random amplification of viral RNA as an alternative to random RT-PCR. BMC Mol. Biol. 2008; 9:77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Carpenter A.E., Jones T.R., Lamprecht M.R., Clarke C., Kang I.H., Friman O., Guertin D.A., Chang J.H., Lindquist R.A., Moffat J. et al. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 2006; 7:R100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Jarvius J., Melin J., Go J., Stenberg J., Fredriksson S., Gonzalez-rey C., Bertilsson S.. Digital quantification using amplified single-molecule detection. Nat. Methods. 2006; 3:725–727. [DOI] [PubMed] [Google Scholar]
- 36. Nandakumar J., Ho C.K., Lima C.D., Shuman S.. RNA substrate specificity and structure-guided mutational analysis of bacteriophage T4 RNA ligase 2. J. Biol. Chem. 2004; 279:31337–31347. [DOI] [PubMed] [Google Scholar]
- 37. Bullard D.R., Bowater R.P.. Direct comparison of nick-joining activity of the nucleic acid ligases from bacteriophage T4. Biochem. J. 2006; 398:135–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.