Nucleic Acids Research (2017) 45:4131–4141, https://doi.org/10.1093/nar/gkw1284
The authors have identified an error in Figure 1 and wish to replace it with the figure provided below.
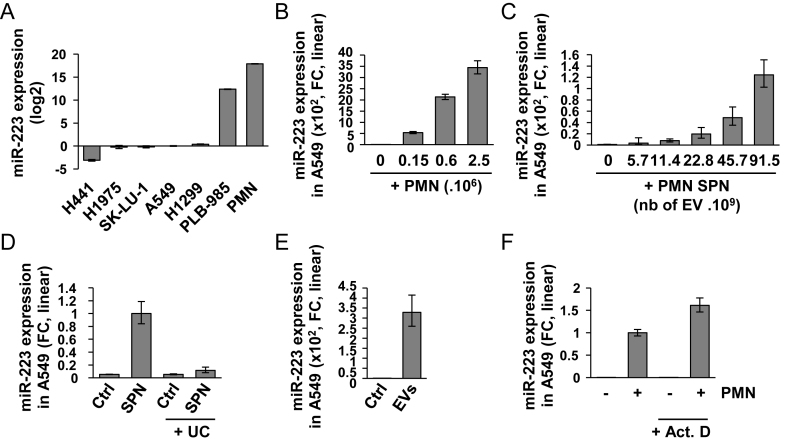
Figure 1.
Ex-miR-223-3p is engulfed into recipient cells (A) Relative quantification analysis of ex-miRNA-223-3p expression in H441, H1975, SK-LU-1, A549, H1299, PLB-985 and PMN cells. Cell lines were harvested at 70% confluency. PMN were isolated from blood as described in the ‘Materials and Methods’ section. (B) Relative quantification of the expression of ex-miR-223-3p in A549 cells after overnight co-culture with increasing numbers of PMN and extensive washes. (C) Relative quantification of the expression of ex-miR-223-3p in A549 cells following incubation with increasing amounts of SPN. (D) Relative quantification of the expression of ex-miR-223-3p in A549 cells following incubation with SPN or SPN depleted of EVs by ultra-centrifugation (UC). (E) Relative quantification of the expression of ex-miR-223-3p in A549 cells incubated overnight with EVs isolated from SPN of PLB-985 cells. (F) Relative quantification of the expression of ex-miR-223-3p in A549 cells co-cultured overnight with PMN and treated with actinomycin D (Act. D) at 10 μg/ml. Cells were extensively washed and harvested. (A–E) For all experiments, the levels of miR-223-3p were normalized using U6 snRNA. (F) Spike-in were used for normalization (see the ‘Materials and Methods’ section). (A–F) Results are representative of three biological replicates, ‘centre values’ as mean and error bars as s.d.
In the published Figure 1, the panels D, E and F were inverted. Panel D should be panel F, E should be D, and F should be E. In addition, the scales of the vertical axis in panels C, D and E have been adjusted. These corrections do not affect the results or conclusions of the article.